Abstract
Background:
The relationship between dyslipidemia and type 2 diabetes mellitus (T2DM) has been frequently reported. Lipoprotein lipase (LPL) is considered to be an effective gene in regulating lipid profile. MicroRNAs (miRNAs) are small noncoding RNAs involved in posttranscriptional regulation of gene expression. In the present study, we have evaluated rs13702 (C/T) polymorphism located in miRNA-410 binding site of LPL gene in subset of Iranian T2DM patients and their normal counterparts.
Materials and Methods:
In this case–control study, 102 T2DM patients and 98 healthy controls were worked out for rs13702 single nucleotide polymorphism genotypes. High resolution meting (HRM) analysis was used for genotyping.
Results:
C allele of rs13702 C/T polymorphism located in miRNA-410 binding site in LPL gene was detected to be significantly associated with T2DM (C allele; odds ratios (OR) = 1.729 (95% confidential intervals (CI) = 1.184–2.523); P = 0.005) also its CC genotype (OR = 3.28 (95% CI 8.68–1.24); P = 0.010) showed the same association.
Conclusion:
Correlation of rs13702 C allele with susceptibility to T2DM may be due to the higher level of LPL that leads to increased plasma fatty acids and its entry into peripheral tissues such as skeletal muscle, liver, and adipocytes causing development of insulin resistance and ultimately T2DM.
Keywords: Lipoprotein lipase, microRNA, rs13702, type 2 diabetes
Introduction
Type 2 diabetes mellitus (T2DM) includes >90% of all types of diabetes. It is one of the most common metabolic disorders in the world. The number of people with diabetes was estimated to be 366 million in 2011. It is predicted that the number of patients will reach 552 million in 2030.[1] MicroRNAs (miRNAs) are a subset of small, endogenous, noncoding RNA molecules (21–23 nucleotides in length) that play a critical role in posttranscriptional gene regulation by binding to the 3' untranslated region (UTR) of most of the mammalian genes.[2] Recently, emerging studies have shown that miRNAs play major roles in pathogenesis of T2DM.[3] Genome wide association studies (GWAS) through evaluation of single nucleotide polymorphisms (SNP) proved very helpful to discover susceptibility loci related to T2DM. Emerging evidences indicating that SNPs in miRNA genes or miRNA target sites (miRSNPs) may alter the structure and expression of the mature miRNA or affect binding affinity of miRNAs to their target mRNAs and thus influence target gene expression. Recent studies show that miRSNPs are of high impact in T2DM development.[4,5,6]
The lipoprotein lipase (LPL) gene is located at 8p22 and known as an important regulator of fat storage in the body. This function is accomplished by the removal of triglycerides (TGs) from the blood stream and transfers them into the fat cells. The main factor in development of hypertriglyceridemia is impaired clearance, presumably because of LPL dysfunction. Impaired LPL system is associated with insulin resistance and T2DM. LPL also binds to chylomicron residues which facilitate their transfer to the liver where removal of the lipoproteins by low-density lipoprotein (LDL) receptors are undertaking.[7]
Reports on LPL polymorphisms and their association with lipid profile in different populations revealed significant differences based on their ethnic and geographical origins.[8,9] However, studies on association of genetic polymorphisms of LPL and T2DM are very limited.
Evaluation of the rs13702 (C/T) polymorphism, a SNP located in the 3′ UTR of LPL gene with no record of previous studies in Iranian population has undertaken in this study. We also examined other anthropometric characteristics of cases and controls in relation to T2DM and individuals' rs13702 genotypes.
Materials and Methods
Study subjects
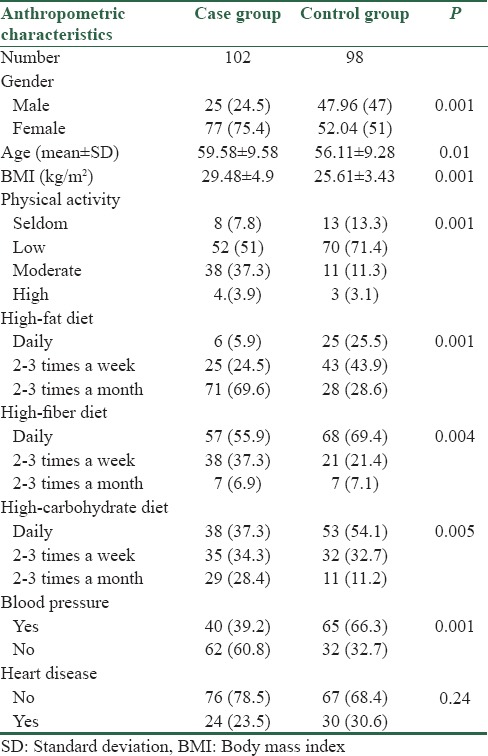
This case-control study conducted on 102 T2DM patients (25 men and 77 women; age 59.58 ± 9.58 years; fasting plasma glucose 151.44 ± 62.07 mg/dL; body mass index (BMI) 29.48 ± 4.9 kg/m2); [Table 1] and 98 healthy participants (47 men and 51 women; age 56.11 ± 9.28 years; fasting plasma glucose 90.40 ± 6.17 mg/dL; BMI 25.61 ± 3.43 kg/m2); [Table 1]. Cases and controls were randomly selected from the Isfahan endocrine and metabolism research center. T2DM patients were selected on the basis of the world health organization (WHO) criteria (fasting plasma glucose >126 mg/dL, and/or 2 h oral glucose tolerance test >200 mg/dL and essentially all were on drug therapy for T2DM). Again according to the WHO criteria control individuals were selected on the basis of items like: no history of T2DM in the first degree relatives and fasting blood glucose < 110 mg/dl. However, we precociously selected those with fasting blood sugar of < 100 mg/dl (90.40 ± 6.17 mg/dl). Informed consent was obtained from all participants in accordance with the Isfahan University of Medical Sciences Ethics Committee.
Table 1.
Anthropocentric characteristics of sample population participated in this study
Genotyping of single nucleotide polymorphisms rs13702 polymorphism
DNA was extracted from 0.5 ml EDTA anticoagulated peripheral blood using GenetBio kit (Korea). The quality and quantity of the extracted DNA was determined by using agarose gel electrophoresis and spectrophotometry at wavelengths of 260 and 280 nm. Genotype analysis of rs13072 was performed by real-time polymerase chain reaction high resolution melting method (HRM) using QuantiFast SYBR®Green kits (Qiagen, Germany) and Rotor-Gene 6000 analyzer device.
The thermal profile of the reaction applied was as follow: preincubation at 95°C for 15 min, 40 cycles of 95°C for 15 s, 60°C and 72°C both for 20 s, finally for HRM analysis, the temperature profile was increased from 75°C to 90°C at the rate of 0.2°C/s. Melting curves were normalized between the two temperature ranges compared with samples of defined genotypes as standard. To determine sample genotypes to use them in HRM analysis as standard genotypes, some samples were subjected to genotype assessment by direct sequencing.
Statistical analysis
For statistical analysis, SPSS windows software (version 20.0; SPSS, Chicago, IL, USA) was used. Hardy–Weinberg equilibrium assumption was performed to compare the observed and expected genotype frequencies using χ2 test. Logistic regression analysis was accomplished to investigate association between genotypes and T2DM and calculate specific odds ratios (ORs), 95% confidential intervals (CIs), and P values. Mean age of cases and controls, cholesterol, triglyceride, and fasting plasma glucose were compared using t-test. Alleles and genotypes comparison was performed using χ2 test. Aadjustments made for gender, age, and BMI as covariates between the study participants.
Results
Anthropometric data of the participants, including age, gender, BMI, physical activity, blood pressure, heart disease, and diet are summarized in Table 1. No deviation from Hardy–Weinberg equilibrium was detected among the cases or controls. A statistically significant difference was observed for the physical activity (P = 0.001), blood pressure (P = 0.001), diet; high-fat diet (P = 0.001), high-carbohydrate diet (P = 0.005), and high-fiber diet (P = 0.004) variables, between the controls and case group.
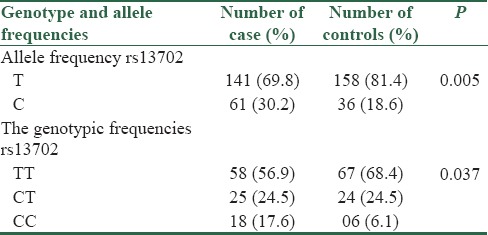
The frequencies of the CC, CT, and TT genotypes in the case group were 17.6%, 24.5%, and 56.9%, respectively, and the genotype frequencies in the control group was 6.1%, 24.5%, and 68.4%, respectively.
The distribution of rs13702 C/T polymorphism genotypes significantly differed between the controls and case group (CC vs. TT + CT) [Table 2]. The CC genotypes of rs13702 C/T polymorphism within the LPL gene compared to TT + CT genotypes indicate an association between rs13702 and T2DM among our study population (CC genotype OR [95% CI] = 3.28 P = 0.010). Furthermore, we studied the allelic frequency distribution of rs13702 C/T polymorphism among the cases and controls. The study results showed that the C allele of the rs13702 is more frequent in the case group than control group (30.2% vs. 18.6%) and also we found that C allele of the rs13702 C/T polymorphism had a potential association with an increased risk of T2DM (C allele; OR = 1.729, 95% CI = 1.184–2.523, P = 0.005). Genotypic and allelic frequency of case and control group had no significant association with lipid profile (TGs, cholesterol, high-density lipoprotein (HDL), and LDL of patients in our population.
Table 2.
Association between genotypes and allele frequency with type 2 diabetes risk
Discussion
In the present study, association between rs13702 C/T polymorphism located in 3' UTR reign of LPL gene and T2DM was investigated in a subset of Iranian population from Isfahan province located at central part of the country. CC genotype of this SNP showed significant association with the risk of T2DM in our study population. Moreover, it was understood that the CT genotype was expressively associated with protection against T2DM development. Furthermore in this study, no significant relationship was detected between participants' lipid profile and rs13702 C/T genotypes.
LPL is a protein with multiple functions that are produced and secreted by parenchymal cells such as brain, heart, fat tissues, skeletal muscles, as well as blood macrophages. LPL hydrolyzes circulating core TGs of very low-density lipoprotein and chylomicron, in fact LPL plays a key role in metabolism and transportation of blood lipids.[10] It also creates free fatty acids (FFA) for storage in adipose tissue, oxidation, and utilization in the heart and other tissues.[11]
The rs13702 C/T polymorphism is located in the miR-410 target site. The existence of allele C in this position destroys the binding site of miR-410. This destruction leads to increased expression of LPL.[12] LPL overexpression leads to increased fat metabolites, consequently FFA accumulated in several tissues like liver and muscles. Given that these tissues are major responsive organs to insulin, the accumulation of fat metabolites can lead to insulin resistance in liver and muscles.[13] Insulin resistance is considered as one of the major causes of developing Type 2 diabetes and its side effects. However, increased expression of LPL in adipose tissue will improve glucose metabolism and insulin tolerance.[14] On the other hand some evidence represent that in people with T2DM, LPL activity decreases, which ultimately leads to increased TGs and decreased levels of blood HDL.[15]
An experimental study in 2008 was indicated no significant relationship exists between rs13702 C/T polymorphism and diabetes.[16] Nevertheless, Rendel et al. postulated a hypothesis for FFA and glucose uptake by tissues as early as 1963. They stated FFA can inhibit glucose uptake by insulin stimulated cells through increasing the oxidation of fatty acids and reducing carbohydrate oxidation in myocardium and rat diaphragm.[17]
Nowadays, several mechanisms have been reported to explain how FFA creates insulin resistance:
Lipid metabolites: Investigations suggest that increased FFA will increase diacylglycerol (DAG) in skeletal muscle tissue. DAG activates two isoforms of protein kinase C (PKC), PKC β2 and δPKC. In fact, PKC is a serine-threonine kinase which causes insulin resistance by reducing tyrosine phosphorylation in insulin receptor substrate (IRS)[18,19]
Inflammation: Studies have shown that FFA can lead to the activation of NF-κB proinflammatory pathways in skeletal muscles. The activation of NF-κB pathway can increase expression of a number of proinflammatory cytokines such as tumor necrosis factor-α, IL1 β and IL6
Oxidative stress: Studies have shown that an increase in FFA can lead to the activation of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase in adipocytes. This will increase the production of reactive oxygen species. On the other hand, free oxygen radicals lead to the intensification of oxidative stress in adipose tissue by producing a series of cytokines. In addition, these cytokines are able to create insulin resistance [19]
Endoplasmic reticulum stress: Studies on the side-effects of the increase FFA in plasma indicate that the increase in FFA can induce endoplasmic reticulum (ER) stress in adipocytes,[20] liver cells [21] and pancreatic beta cells.[22] Studies have shown that ER stress activates c-Jun N-terminal kinase (JNK) pathway and subsequently causes insulin resistance.[23]
Overall, rs13702 C allele destructs the binding site of miR-410 and leads to overexpression of LPL which ultimately lead to n resistance in target tissues. However, this study determined no relationship between genotype and lipid profile, but it should be noted that lipid metabolism is very complex and affected by numerous regulatory genes. In fact, rs13702 C allele causes overexpression of LPL and slowly reduces serum triglyceride level and moves FFA to peripheral tissues. High level of FFA in peripheral tissues such as skeletal muscles and liver can lead to insulin resistance through mechanisms described above. These mechanisms can be considered as a beginning point of T2DM development and its subsequent complications.
Financial support and sponsorship
Financial support provided by Isfahan University of Medical Sciences is gratefully acknowledged
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
We would like to appreciate generous cooperation of Isfahan endocrine and metabolism research center, Isfahan University of Medical Sciences, Isfahan Iran.
References
- 1.Whiting DR, Guariguata L, Weil C, Shaw J. IDF diabetes atlas: Global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract. 2011;94:311–21. doi: 10.1016/j.diabres.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 2.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147–54. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong L, Zhu J, Han W, Jiang X, Xu M, Zhao Y, et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A clinical study. Acta Diabetol. 2011;48:61–9. doi: 10.1007/s00592-010-0226-0. [DOI] [PubMed] [Google Scholar]
- 4.Gong W, Xiao D, Ming G, Yin J, Zhou H, Liu Z, et al. Type 2 diabetes mellitus-related genetic polymorphisms in microRNAs and microRNA target sites. J Diabetes. 2014;6:279–89. doi: 10.1111/1753-0407.12143. [DOI] [PubMed] [Google Scholar]
- 5.Elek Z, Németh N, Nagy G, Németh H, Somogyi A, Hosszufalusi N, et al. Micro-RNA binding site polymorphisms in the WFS1 gene are risk factors of diabetes mellitus. PLoS One. 2015;10:e0139519. doi: 10.1371/journal.pone.0139519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao X, Ye Q, Xu K, Cheng J, Gao Y, Li Q, et al. Single-nucleotide polymorphisms inside microRNA target sites influence the susceptibility to type 2 diabetes. J Hum Genet. 2013;58:135–41. doi: 10.1038/jhg.2012.146. [DOI] [PubMed] [Google Scholar]
- 7.Olivecrona G. Role of lipoprotein lipase in lipid metabolism. Curr Opin Lipidol. 2016;27:233–41. doi: 10.1097/MOL.0000000000000297. [DOI] [PubMed] [Google Scholar]
- 8.Rodrigues R, Artieda M, Tejedor D, Martínez A, Konstantinova P, Petry H, et al. Pathogenic classification of LPL gene variants reported to be associated with LPL deficiency. J Clin Lipidol. 2016;10:394–409. doi: 10.1016/j.jacl.2015.12.015. [DOI] [PubMed] [Google Scholar]
- 9.Daoud MS, Ataya FS, Fouad D, Alhazzani A, Shehata AI, Al-Jafari AA, et al. Associations of three lipoprotein lipase gene polymorphisms, lipid profiles and coronary artery disease. Biomed Rep. 2013;1:573–82. doi: 10.3892/br.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu G, Xu JN, Liu D, Ding Q, Liu MN, Chen R, et al. Regulation of plasma lipid homeostasis by hepatic lipoprotein lipase in adult mice. J Lipid Res. 2016;57:1155–61. doi: 10.1194/jlr.M065011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassing HC, Surendran RP, Mooij HL, Stroes ES, Nieuwdorp M, Dallinga-Thie GM, et al. Pathophysiology of hypertriglyceridemia. Biochim Biophys Acta. 2012;1821:826–32. doi: 10.1016/j.bbalip.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Richardson K, Nettleton JA, Rotllan N, Tanaka T, Smith CE, Lai CQ, et al. Gain-of-function lipoprotein lipase variant rs13702 modulates lipid traits through disruption of a microRNA-410 seed site. Am J Hum Genet. 2013;92:5–14. doi: 10.1016/j.ajhg.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JK, Fillmore JJ, Chen Y, Yu C, Moore IK, Pypaert M, et al. Tissue-specific overexpression of lipoprotein lipase causes tissue-specific insulin resistance. Proc Natl Acad Sci U S A. 2001;98:7522–7. doi: 10.1073/pnas.121164498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walton RG, Zhu B, Unal R, Spencer M, Sunkara M, Morris AJ, et al. Increasing adipocyte lipoprotein lipase improves glucose metabolism in high fat diet-induced obesity. J Biol Chem. 2015;290:11547–56. doi: 10.1074/jbc.M114.628487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalmár T, Seres I, Balogh Z, Káplár M, Winkler G, Paragh G, et al. Correlation between the activities of lipoprotein lipase and paraoxonase in type 2 diabetes mellitus. Diabetes Metab. 2005;31:574–80. doi: 10.1016/s1262-3636(07)70233-1. [DOI] [PubMed] [Google Scholar]
- 16.Cho YS, Go MJ, Han HR, Cha SH, Kim HT, Min H, et al. Association of lipoprotein lipase (LPL) single nucleotide polymorphisms with type 2 diabetes mellitus. Exp Mol Med. 2008;40:523–32. doi: 10.3858/emm.2008.40.5.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Randle PJ, Garland PB, Hales CN, Newsholme EA. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet. 1963;1:785–9. doi: 10.1016/s0140-6736(63)91500-9. [DOI] [PubMed] [Google Scholar]
- 18.Boden G, Lebed B, Schatz M, Homko C, Lemieux S. Effects of acute changes of plasma free fatty acids on intramyocellular fat content and insulin resistance in healthy subjects. Diabetes. 2001;50:1612–7. doi: 10.2337/diabetes.50.7.1612. [DOI] [PubMed] [Google Scholar]
- 19.Furukawa S, Fujita T, Shimabukuro M, Iwaki M, Yamada Y, Nakajima Y, et al. Increased oxidative stress in obesity and its impact on metabolic syndrome. J Clin Invest. 2004;114:1752–61. doi: 10.1172/JCI21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo W, Wong S, Xie W, Lei T, Luo Z. Palmitate modulates intracellular signaling, induces endoplasmic reticulum stress, and causes apoptosis in mouse 3T3-L1 and rat primary preadipocytes. Am J Physiol Endocrinol Metab. 2007;293:E576–86. doi: 10.1152/ajpendo.00523.2006. [DOI] [PubMed] [Google Scholar]
- 21.Wei Y, Wang D, Topczewski F, Pagliassotti MJ. Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am J Physiol Endocrinol Metab. 2006;291:E275–81. doi: 10.1152/ajpendo.00644.2005. [DOI] [PubMed] [Google Scholar]
- 22.Karaskov E, Scott C, Zhang L, Teodoro T, Ravazzola M, Volchuk A, et al. Chronic palmitate but not oleate exposure induces endoplasmic reticulum stress, which may contribute to INS-1 pancreatic beta-cell apoptosis. Endocrinology. 2006;147:3398–407. doi: 10.1210/en.2005-1494. [DOI] [PubMed] [Google Scholar]
- 23.Kharroubi I, Ladrière L, Cardozo AK, Dogusan Z, Cnop M, Eizirik DL, et al. Free fatty acids and cytokines induce pancreatic beta-cell apoptosis by different mechanisms: Role of nuclear factor-kappaB and endoplasmic reticulum stress. Endocrinology. 2004;145:5087–96. doi: 10.1210/en.2004-0478. [DOI] [PubMed] [Google Scholar]


