Correction
Following the publication of this article [1], the authors noticed that Fig. 3 was missing. In that figure, one of the numbers corresponding to the Halomonas chemoreceptors was missing: namely, chemoreceptor 07070. The correct version of Fig. 3 has been included in this Correction.
Fig. 3.
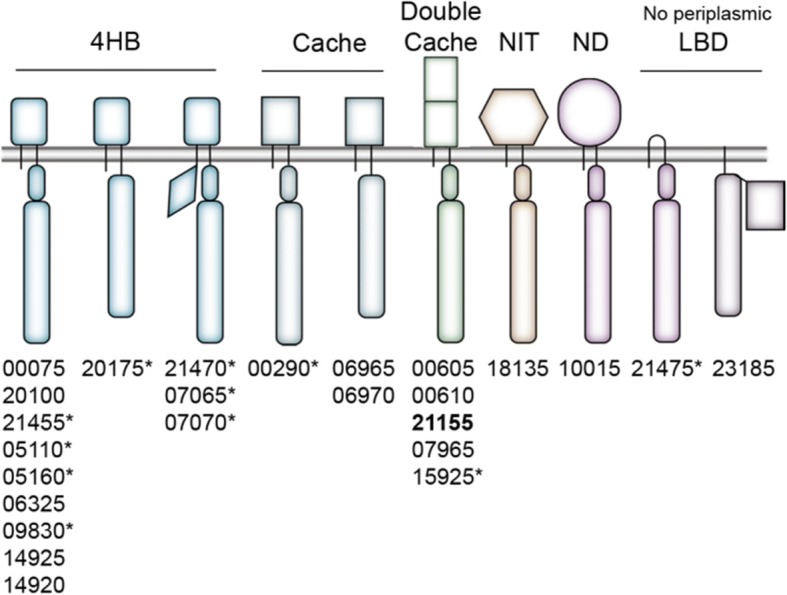
Schematic representation of chemoreceptors encoded in the genome of H. titanicae KHS3. MCPs are grouped according to the predicted structure of the periplasmic LBD: 4-helix bundle (4HB, rectangle with curved edges), Cache (rectangle), double Cache (double rectangle), nitrate-nitrite sensing fold (NIT, hexagon), not determined (ND, circle) and those with no periplasmic LBD. The gray horizontal bar represents the cytoplasmic membrane. MCP cytoplasmic subdomains are represented by a long rectangle (conserved cytoplasmic domain or signaling domain), an oval representing the HAMP domain, a diamond shape representing PAS domain. The rectangle at the C-terminus of RO22_23185 represents a Cache domain. The corresponding MCP ID numbers are listed below each kind of MCP (all ID numbers should be preceded by “RO22_” following the IMG gene annotation). Asterisks indicate the presence of C-terminal pentapeptide for interaction with CheR. All MCPs belong to the 36H family except one, which belongs to the 40H family (shown in bold)
Footnotes
The original article can be found online at 10.1186/s12864-018-4655-4
Reference
- 1.Gasperotti AF, et al. BMC Genomics. 2018;19:266. doi: 10.1186/s12864-018-4655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


