Abstract
Next generation sequencing (NGS) technologies have revolutionized the genomics field and are becoming more commonplace for identification of human infectious diseases. However, due to the low abundance of viral nucleic acids (NAs) in relation to host, viral identification using direct NGS technologies often lacks sufficient sensitivity. Here, we describe an approach based on two complementary enrichment strategies that significantly improves the sensitivity of NGS-based virus identification. To start, we developed two sets of DNA probes to enrich virus NAs associated with respiratory diseases. The first set of probes spans the genomes, allowing for identification of known viruses and full genome sequencing, while the second set targets regions conserved among viral families or genera, providing the ability to detect both known and potentially novel members of those virus groups. Efficiency of enrichment was assessed by NGS testing reference virus and clinical samples with known infection. We show significant improvement in viral identification using enriched NGS compared to unenriched NGS. Without enrichment, we observed an average of 0.3% targeted viral reads per sample. However, after enrichment, 50%–99% of the reads per sample were the targeted viral reads for both the reference isolates and clinical specimens using both probe sets. Importantly, dramatic improvements on genome coverage were also observed following virus-specific probe enrichment. The methods described here provide improved sensitivity for virus identification by NGS, allowing for a more comprehensive analysis of disease etiology.
Numerous infectious diseases, including 25%–50% of acute lower respiratory tract illnesses, go undiagnosed (Garau and Calbo 2008). Failures in detection may result from the limited repertoire of routine diagnostics or the inability of commonly used PCR methods to pick up variants of known pathogens or unknown and previously unrecognized pathogens. Using next generation sequencing (NGS) for viral detection and discovery provides significant advantages to traditional viral diagnostic assays—full genome sequences may be obtained and targets are not limited by pre-existing knowledge of the pathogen. However, one of the greatest challenges to NGS as a routine, affordable, diagnostic tool is the relative paucity of viral nucleic acids (NAs) present in clinical samples.
In an effort to increase the proportion of viral sequences obtained by NGS, several techniques for non-sequence-specific viral enrichment have been implemented. Ultracentrifugation, sample filtration, and viral culture have been used to enrich viral particles prior to nucleic acid extraction (Breitbart and Rohwer 2005; Duhaime and Sullivan 2012). Additionally, host genomic DNA depletion by DNase treatment or ribosomal RNA depletion have been used to reduce levels of host NAs in a sample (Allander et al. 2001). Although these methods show modest levels of improvement in detection of viral target sequence reads, they do not achieve the desired sensitivity.
An alternative sequence-specific approach involves hybridization-based sequence capture of specific NAs using 80- to 120-mer DNA or RNA probes (Lovett et al. 1991; Albert et al. 2007; Hodges et al. 2007; Okou et al. 2007). Several groups have employed this method for virus-specific capture and genome enrichment prior to NGS (Depledge et al. 2011; Mate et al. 2015; Olp et al. 2015). Using the SureSelectXT Target Enrichment System, Depledge et al. (2011) enriched human herpesviruses from clinical samples, finding significant improvement in sequencing depth.Two studies have described the use of enrichment for characterization of Ebola virus and Kaposi's sarcoma-associated herpesvirus using Illumina TruSeq RNA Access and Agilent SureSelectXT Target Enrichment, respectively (Mate et al. 2015; Olp et al. 2015). Collectively, these strategies result in increased target sequence reads and improved depth of coverage.
Sequence-specific enrichment probes have also been used in the study of the human virome (Briese et al. 2015; Wylie et al. 2015). Briese and colleagues developed the Virome Capture Sequencing Platform for vertebrate viruses, where the authors designed ∼2 million viral enrichment probes to target all virus taxa where at least one virus was known to infect vertebrates (Briese et al. 2015). This method resulted in a 100- to 1000-fold increase in the number of targeted viral reads and reduction in host background. Similarly, Wylie et al. (2015) reported the ViroCap enrichment system, where a custom panel of 2.1 million genus- and virus-specific probes targeted against vertebrate viruses from 34 families allowed for viral enrichment in clinical specimen libraries. In both studies, the probe design targeting known sequences was limited in identification of highly divergent viruses.
Here, we report an approach for target-based enrichment for sensitive detection of a broad spectrum of respiratory viruses by NGS. We used two complementary panels of oligonucleotide probes targeted against representative common respiratory viruses as a proof of concept. They are (1) virus-specific probes that span the full genome of common respiratory viruses, and (2) conserved viral group probes that target against conserved regions from each of nine viral families or subfamilies that are associated with respiratory diseases. The virus-specific probes allow for full genome sequencing, which contributes to a more complete and confident identification and characterization of target viruses. The conserved viral group probes enable detection of divergent viruses with the potential to recognize novel viruses within these known viral families. The combination of these two enrichment approaches is expected to increase the number of usable reads per sample and per sequencing run, significantly improving the sensitivity and value of NGS for viral detection, discovery, and sequence characterization.
Results
Enrichment analysis of reference viruses with two hybridization probe sets
Both sets of enriching probes provide significant improvement, increasing both the overall rate of virus detection and the percent targeted viral reads (PTRs) per sample by NGS. We first evaluated the enrichment by virus-specific probes using representative reference viruses from viral families associated with human respiratory diseases (Supplemental Table S1). Prior to library preparation, we determined the virus load (Ct) where real-time PCR assays were available. Viral Ct values ranged from about 21 to 33, depending on the available reference virus NA stock, with most Ct values at or around 25. For each sample, we generated two parallel libraries from the same viral NA template using different sequencing indices. This allowed us to assess samples with and without enrichment on the same sequencing run.
The virus-specific probe pool improved detection of target virus for all of the reference virus NAs tested when compared to their unenriched matches, except the HBoV1 virus (Fig. 1). Overall enrichment with virus-specific probes results in a 7285-fold median increase in PTRs (increase ranges from 33- to 188,019-fold) (Supplemental Table S2). Notably, 18 of the 26 (69%) samples improved to 50%–99% PTRs following enrichment: 229E, NL63, OC43, AdV E4, HPeV6, HRV B14, HPIV1, HPIV2, HPIV3, RSV A2, RSV B1, HMPV83, all five representative influenza A viruses, and one influenza B (B/Yamagata) virus (Fig. 1; Supplemental Table S2). Three samples—AdV B11, HMPV75, and HPeV1—were enriched to 20%–45% PTRs. The overall linear genome coverage and depth of coverage was improved dramatically, as 73% of the samples generated a linear genome coverage of more than 85% (Supplemental Table S2), which provided sufficient sequence information to confirm virus identity. Enrichment of HKU1, AdV C2, influenza B/Victoria, and HBoV3 was less efficient (<10% PTRs) (Fig. 1; Supplemental Table S2), and enrichment of HBoV1 was not successful.
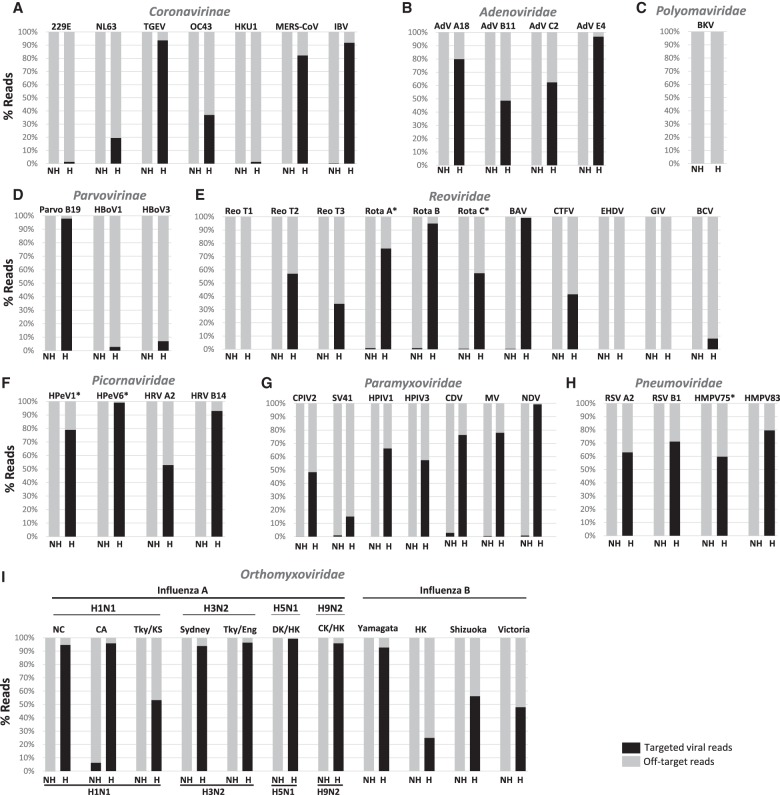
Figure 1.
Distribution of sequence reads for reference samples enriched with virus-specific probes. The frequency of reads identified by Kraken for each sample with and without enrichment (H: hybridized; NH: nonhybridized) is shown in bar graphs for each viral family/subfamily tested: (A) Coronavirinae; (B) Adenoviridae; (C) Parvovirinae; (D) Picornaviridae; (E) Paramyxoviridae; (F) Pneumoviridae; (G) Orthomyxoviridae. (*) Frequency of reads obtained from BWA-MEM read mapping. Abbreviations of virus names are listed in Supplemental Table S1.
The same set of 25 out of 26 reference virus sequencing libraries described above was also used to test the efficiency of enrichment with the conserved viral group probe set. In order to evaluate the enrichment for divergent viruses by the conserved viral group probe set, we included an additional 27 reference viruses of human and animal origin in these experiments (Supplemental Table S1). We were able to detect a total of 48 of 52 target viruses in the enriched samples, while the majority of viruses were difficult to confirm in the unenriched match (Fig. 2; Supplemental Table S3). The PTRs per sample is also greatly increased following enrichment. Of the 48 samples that tested positive, we found 17 with >80% PTRs, 17 with 50%–80% PTRs, and 14 with <50% PTRs following enrichment. Enrichment also resulted in an 8990-fold median increase in PTRs (range from 0 to 1,211,475-fold), with an increase across all families (Fig. 2; Supplemental Table S3). Notably, enrichment with the conserved viral group probe set resulted in detection of all 25 human respiratory viruses targeted by the virus-specific probes, as well as an additional 23 viruses that were not targeted by the virus-specific probes. Examples include Middle East respiratory syndrome coronavirus (MERS-CoV), avian infectious bronchitis virus (IBV), canine parainfluenza virus 2 (CPIV2), simian virus 41 (SV41), canine distemper virus (CDV), Colorado tick fever virus (CTFV), Bunyip Creek virus (BCV), and several avian influenza viruses (Fig. 2). Four viruses—BK polyomavirus (BKV), mammalian orthoreovirus type 1 (Reo T1), epizootic hemorrhagic disease virus (EHDV), and Great Island virus (GIV)—were not enriched.
Figure 2.
Distribution of sequence reads for reference samples enriched with conserved viral group probes. The frequency of reads identified by Kraken for each sample with and without enrichment (H: hybridized; NH: nonhybridized) is shown in bar graphs for each viral family/subfamily tested: (A) Coronavirinae; (B) Adenoviridae; (C) Polyomaviridae; (D) Parvovirinae; (E) Reoviridae; (F) Picornaviridae; (G) Paramyxoviridae; (H) Pneumoviridae; (I) Orthomyxoviridae. (*) Frequency of reads obtained from BWA-MEM read mapping. Abbreviations of virus names are listed in Supplemental Table S1.
Improvement on sensitivity and viral genome coverage
An important consideration for the viral genome capture protocol is how it affects the overall sensitivity of virus detection by NGS. To assess the relative sensitivity, we compared the highest sample Ct value with positive targeted viral reads and linear genome coverage before and after enrichment by the virus-specific probe set on viruses at various viral loads (Fig. 3A; Supplemental Table S4). Representative reference viruses selected for the Coronavirinae, Adenoviridae, Pneumoviridae, and Orthomyxoviridae families/subfamilies are OC43, AdV E4, RSV A2, and influenza A/Sydney/05/97 (H3N2), respectively. We prepared libraries from 10-fold dilutions of these reference virus samples that had been spiked into equivalent amounts of human RNA. The resulting viral Ct ranged from 22.1 to >38. As in previous experiments, we generated parallel, independently barcoded libraries using the same viral NA mixture in order to compare targeted viral reads with and without enrichment on the same sequencing run. Using these libraries, we were able to measure the detection limit following viral genome enrichment.
Figure 3.
Sensitivity of enrichment in hybridization. Samples were prepared as 10-fold serial dilutions of reference viral nucleic acids spiked into a constant amount of human RNA prior to library preparation. The frequency of reads identified by Kraken is shown in bar graphs for each sample with and without enrichment (H: hybridized; NH: nonhybridized) for (A) virus-specific probe enrichment and (B) conserved viral group probe enrichment. From the same sequencing run, the linear genome coverage is shown (C) for samples with enrichment (diagonal stripes) or without enrichment (white) with virus-specific probes. Viral Ct and (average) depth of coverage are shown below the bar graphs. Abbreviations of virus names are listed in Supplemental Table S1.
We used a relative read count threshold (0.01%) based on an empirical overall barcode contamination rate to exclude low viral reads due to potential barcode contamination. Samples with a low number of viral reads (<0.01% of the highest number of reads obtained for any sample of the same virus in the same run) were labeled as negative (Supplemental Tables S4, S5). We determined the endpoint of detection as the highest sample Ct with positive targeted viral reads. Endpoints were assessed both prior to and after enrichment, and improvement in endpoint detection was described as the log difference between the two. For OC43, using virus-specific probes, we could detect the virus from pre-enrichment samples at Ct 24.1, while from post-enrichment samples, we detected the virus at Ct 36, a >3-log improvement in endpoint detection compared to the unenriched sample (Fig. 3A; Supplemental Table S4). With the conserved viral group probes, we observed a 2-log improvement in detection for OC43 and detected OC43 at Ct 32 post-enrichment (Fig. 3B; Supplemental Table S5). We observed 3-log improvement in detection for AdV E4 (Ct 33), RSV A2 (Ct 39), and influenza A/Sydney/05/97 (H3N2) (Ct 39) when using the virus-specific probes (Fig. 3A; Supplemental Table S4). With the conserved viral group probe enrichment, at least 3-log improvement was achieved for AdV E4 at Ct 33 and influenza A/Sydney/05/97 (H3N2) at Ct 39, and 2 logs for RSV A2 at Ct 39 (Fig. 3B; Supplemental Table S5). Overall, these results highlighted that these enrichment strategies resulted in a robust improvement in the relative endpoint sensitivity of viral detection by NGS.
We also assessed improvements on linear genome coverage for this set of viruses using the virus-specific probe set. For both enriched and unenriched samples, linear genome coverage decreased relative to the reduced viral loads (Fig. 3C; Supplemental Table S4). To show the effect that enrichment has on genome coverage, we measured the fold change in coverage between unenriched (nonhybridized) and enriched (hybridized) samples. The overall linear coverage was greatly increased for enriched samples, up to a 184-fold increase above unenriched samples (Supplemental Table S4). Additionally, we obtained near full genome coverage for OC43, RSV A2, and AdV E4 viruses at their top two viral loads tested (around Ct 30) and for influenza A Sydney/05/97 (H3N2) at its top three viral loads tested (around Ct 36). This genome sequence information allowed us to confirm the individual virus identity. For unenriched samples, we did not observe full genome coverage from any sample except influenza A Sydney/05/97, which had full genome coverage at its top one viral load tested (around Ct 26.4) (Fig. 3C; Supplemental Table S4).
Enrichment analysis of clinical samples with two hybridization probe sets
Following the validation of enrichment using reference samples, we proceeded to test enrichment of NAs from a collection of clinical samples confirmed to contain known viruses (Supplemental Table S1). We selected between one and 11 clinical samples with known viral infection from each viral family/subfamily to test the performance of our two sets of enriching probes. Some of the virus-specific probes, such as probes for HPIV4 that were not evaluated using reference viruses, were evaluated using clinical samples. Viral loads (where available) range from a Ct value of 17.4 to 32 (Supplemental Table S1). Again, for each clinical sample, we generated two parallel and separately barcoded libraries to assess samples with and without enrichment on the same sequencing run. We observed improvements in the rate of target virus detection in these clinical samples, for both sets of probes, compared to unenriched samples (Figs. 4, 5; Supplemental Tables S6, S7). Many viruses were nearly undetectable in the unenriched specimens—all samples had <1% PTRs and 10 samples had <100 targeted viral reads (Supplemental Table S6, S7).
Figure 4.
Distribution of sequence reads for clinical samples enriched with virus-specific probes. The frequency of reads identified by Kraken for each sample with and without enrichment (H:hybridized; NH: nonhybridized) is shown in bar graphs for each viral family/subfamily tested: (A) Coronavirinae; (B) Adenoviridae; (C) Parvovirinae; (D) Picornaviridae; (E) Paramyxoviridae; (F) Pneumoviridae; (G) Orthomyxoviridae. Viral Ct is shown below the bar graphs. (ND) Ct not available. Abbreviations of virus names are listed in Supplemental Table S1.
Figure 5.
Distribution of sequence reads for clinical samples enriched with conserved viral group probes. The frequency of reads identified by Kraken for each sample with and without enrichment (H: hybridized; NH: nonhybridized) is shown in bar graphs for each viral family/subfamily tested: (A) Coronavirinae; (B) Adenoviridae; (C) Parvovirinae; (D) Polyomaviridae; (E) Reoviridae; (F) Picornaviridae; (G) Paramyxoviridae; (H) Pneumoviridae; and (I) Orthomyxoviridae. Viral Ct is shown below the bar graphs. (ND) Ct not available. (*) Frequency of reads obtained from BWA-MEM read mapping. Abbreviations of virus names are listed in Supplemental Table S1.
Virus-specific probe enriched viral reads for 20 of the 22 clinical samples tested (Fig. 4; Supplemental Table S6). Collectively, virus-specific probe enrichment leads to a median 2308-fold increase in PTRs above unenriched (range of 0- to 136,310-fold) (Supplemental Table S6). Nearly all clinical samples display high levels of virus sequence enrichment following hybridization, with the exception of HRV A62 and HRV C6. Of the 22 sample libraries tested for enrichment with virus-specific probes, we identified 11 samples with >80% PTRs, eight samples with 50%–80% PTRs, one sample with <50% PTRs, and two samples with rhinoviruses A62 and C6 that failed to enrich efficiently. We also observed an overall improvement on average depth of coverage for all of the clinical samples after enrichment as well as on linear genome coverage (>50%) for all clinical samples except the HRV B14 sample that was less efficiently enriched (Supplemental Table S6). This improved genome coverage provided sequence information needed to confirm each virus identity.
In cases of coinfection with two or more viruses at different titers, there could be issues with one enriched virus saturating the available reads, obscuring detection of the other. To simulate this situation, we generated experimentally mixed samples that contain two viruses, RSVA and OC43, at differing titers. These viruses were sourced from a bank of single-infection clinical samples in order to keep the comparison in the range of relevant clinical parameters. In one mixed sample, RSVA is present with higher viral loads (Ct 28) and OC43 with lower viral loads (Ct 32), while in the other mixed sample, OC43 is present with higher viral loads (Ct 28) and RSVA with lower viral loads (Ct 32). We observed substantial enrichment of both viruses in both samples, although with the lower viral load viruses, there was an unsurprising corresponding lower viral read (Table 1). Genome coverage was not affected in the mixed samples, and 95%–100% genome coverage was obtained for both viruses in both samples (Table 1).
Table 1.
Virus-specific probe hybridization for mixed clinical samples with known viral infection
With the conserved viral group probes, we observed viral read enrichment in 30 of the 33 clinical samples tested, leading to a median increase in PTRs of 15,252-fold above unenriched (range of 21- to 554,254-fold) (Supplemental Table S7). For unenriched samples, 29 of 33 contain <1% PTRs, and 23 of 33 contain fewer than 100 total viral reads. Following enrichment with the conserved probe set, we observed 16 samples with >80% PTRs, eight samples with 50%–80% PTRs, six samples with <50% PTRs, and three samples which failed to enrich. There were dramatic improvements in detection for coronaviruses OC43, HKU1, MERS-CoV, and camel CoV HKU23; adenoviruses AdV C and AdV E; parvovirus HBoV1; rotavirus A; and all viruses tested for picornaviruses, paramyxoviruses, pneumoviruses, and influenza A and B viruses (Fig. 5; Supplemental Table S7). Enrichment was inefficient for clinical samples with known infection of coronavirus 229E and two polyomaviruses (Fig. 5).
Discussion
The utility and convenience of NGS-based sequencing for routine pathogen detection and discovery is hindered by the abundance of host and commensal NAs, which results in reduced sensitivity for virus detection by NGS. In this study, we show greatly improved NGS-based virus detection following enrichment of viral NAs using two complementary sets of probes, testing both viral reference and clinical samples. Our virus-specific probes also improve linear genome coverage with greater depth. The information gained by increased or full genome sequencing allows for detection of viral mutations or minor variants which may prove useful in viral transmission and evolution studies.
Notably, we observe that our virus-specific probes are effective when they share 90% or more nucleotide similarity with the probe sequences. This is consistent with other observations for hybridization-based capture performance (Briese et al. 2015). This high level of hybridization stringency likely contributes to the inability to enrich related viruses with insufficient sequence homology, including members of the picornavirus family. For example, HRV A62 and HRV C6 in clinical samples do not enrich (Fig. 4D)—there is only a 70% match in nucleotide identity between the genome sequences of HRV A89 (NC_001617.1), the virus strain used for probe design, and HRV A62, the virus tested. Similarly, there is only an average of 70% match in nucleotide identity between HRV C (NC_009996.1), the species used for probe design, and HRV C6, the species tested, based on the partial sequences available for HRV C6. As expected, we observed that HMPV83, which was used for the HMPV probe design, was enriched more efficiently than HMPV75 virus. In addition, viruses 229E, mammalian orthoreovirus type 1, epizootic hemorrhagic disease virus, and Great Island virus were not enriched by the conserved viral group probe set. The enrichment failure for these viruses could be due to the probe sequence issues, and thus the conserved probe set for coronaviruses and reoviruses may need to be further optimized.
The ability to obtain high-quality genome sequences is a major benefit of using this method. Linear genome coverage (percent of the genome sequenced) and the depth of sequencing at each base (number of supporting reads) are both important. In this study, we demonstrate that virus-specific probe enrichment significantly improves linear genome coverage as well as sequencing depth in viruses with Ct values in the high 20s into the 30s (Supplemental Tables S4, S6). The increased depth of coverage over a full or nearly full genome makes virus classification more accurate, as we can be more confident at every base. Even when samples with low viral loads had low linear genome coverage, the sequence islands with high read depth scattered along the genome can be used for subsequent PCRs to fill the genome gaps for virus classification and typing.
Importantly, in this study we show that the conserved viral group probes are capable of detecting divergent and potentially unknown viruses. Their design focuses on conserved regions of the viral genome within a viral family, subfamily, or genus. Using these probes will not result in full genome sequences but will allow for enrichment of a more diverse collection of viruses, including both known and potentially novel viruses. For example, the recently discovered viruses MERS-CoV, camel coronavirus HKU23, Colorado tick fever virus, and Bunyip Creek virus were enriched using the conserved viral group probe set. Another benefit of the conserved viral group probe pool is that it requires only a modest number of oligonucleotide probes to be synthesized, compared to the millions of probes that would be required for enriching a broad range of viruses. We used 346 probes with the ability to detect a broad range of viruses, including both known and potentially novel viruses associated with human respiratory disease, significantly reducing the cost burden for pathogen discovery efforts.
The Illumina RNA Access library preparation kit used in this pilot study generates libraries only from chemically fragmented RNA. Thus, we would only expect to enrich DNA virus sequences that were being actively transcribed into RNA. Accordingly, many DNA viruses (e.g., polyomaviruses and parvoviruses) included in this study did not enrich as efficiently as RNA viruses. Adenoviruses, the parvovirus B19 reference sample (Fig. 2), and HBoV1 clinical samples (Figs. 4, 5) enriched efficiently possibly because of the presence of RNA transcript from the DNA virus genome. Efforts are under way to evaluate alternative kits that work well for both RNA and DNA viruses.
Overall, our study presents a method that can reduce background noise and improve NGS sensitivity, which ultimately reduces cost and reduces the complexity of NGS-based virus sequencing from unknown samples. The complementary pair of respiratory probe panels are able to enrich full genomes of commonly known respiratory viruses and enrich key signatures of divergent or novel viruses simultaneously, so that each sequencing run is more likely to give useful information. The use of commercially available reagents and an easy-to-follow protocol makes this method easily adaptable in a variety of laboratory settings, and it may be easily automated for a high-throughput setting. Further, we are able to enrich viruses from nine families or subfamilies associated with common respiratory infection using the smaller conserved viral group probe pool which contains just 346 probes. We observe that the addition of more probes (increasing the conserved viral group probe pool from 138 to 346 probes) does not decrease the efficiency of enrichment. The work presented here is a pilot study based on a selection of representative respiratory viruses. We will update the virus-specific probe set to include probes against additional respiratory viruses and update the probes that did not work as well as expected. This adaptability also paves the way for larger panels, including the development of a pan viral probe set for pan viral genome enrichment. Taken together, the approaches described here offer an efficient and comprehensive method to harness the power of NGS in routine laboratory and clinical virus detection and discovery. The enriched NGS coupled with the rapid diagnosis methods by real-time PCR will provide a powerful and comprehensive tool for outbreak investigation of both known and unknown infectious etiologies.
Methods
Viral samples and nucleic acid template preparation
A combination of reference virus samples and clinical specimens used in this study are described in Supplemental Table S1. Clinical specimens are respiratory specimens (nasopharyngeal swab, oropharyngeal swab, or lung tissue) with known infection and were selected from previous human respiratory etiology studies, de-identified, and nucleic acids were extracted as described below.
Between one and 11 reference viruses of human or animal origin were tested for each family/subfamily. Reference viruses that match the viruses used for virus-specific probe design (Supplemental Table S8) were selected, including coronaviruses 229E, NL63, OC43, and HKU1; AdV C2 and AdV E4; pneumoviruses RSVA, RSVB, and HMPV 83; paramyxoviruses HPIV1, HPIV2, HPIV3; bocaviruses HBoV1 and HBoV3; influenza A virus A/Chicken/HongKong (H9N2); picornaviruses HPeV1, HPeV6, and rhinovirus HRV B14. In some cases a match was unavailable; therefore, we selected the closest available viruses. For example, we used AdV B11 in place of the adenovirus B1 that the probes were based on. For influenza A virus, we selected the same serotype: A/New Caledonia/20/99 and A/California/07/2009 (pandemic) for H1N1; A/Sydney/05/97 (H3N2) and A/Avian Turkey/England for H3N2; and A/Avian Duck/Hong Kong for H5N1. For influenza B virus, we included both B/Yamagata and B/Victoria lineages, while the probes were only based on B/Lee/40.
NAs were extracted using the QIAamp MinElute Virus Spin Kit (Qiagen) according to the manufacturer's instructions. NAs eluted in nuclease-free water were aliquoted and stored at −80°C. RNA was extracted from the human lung carcinoma cell line A549 (ATCC CCL-185) using TRIzol LS (Thermo Fisher Scientific) according to the manufacturer's instructions, and stored at −80°C.
A 1:1 mixture by volume of reference viral NAs and human RNA was made to provide reference samples of known host/virus content. For these mixtures, reference virus NAs were added to A549 cellular RNA (the final concentration of the mixed RNA was ∼10 ng/µL, with RNase P Ct of around 28, and viral Ct of 23–25 unless the only available reference virus had a viral Ct higher than 25). To evaluate the relative sensitivity of enrichment, reference virus NAs were first serially diluted (1:10), then mixed with a constant amount of A549 RNA (samples were prepared using a 1:1 ratio by volume). NAs extracted from clinical specimens were either used directly (undiluted) or spiked into A549 RNA as described above.
Real-time PCR
RT-PCRs were performed using either AgPath-ID One-Step RT-PCR (Thermo Fisher Scientific) or SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase (Thermo Fisher Scientific) according to the manufacturer's recommended protocol, using primers and probes described previously (Supplemental Table S9). In some cases, we obtained the Ct of the sample from another lab when available.
Hybridization probe design
We designed two different, complementary panels of oligonucleotide probes, virus-specific probes and conserved viral group probes, for hybridization-based enrichment of common respiratory viruses as described below.
Virus-specific probes were designed to span the genome, with tiling probes targeted against a selection of common respiratory viruses (Supplemental Table S8) as previously described (Dehority et al. 2017). Prior to probe design and selection, viral sequences were masked for low-complexity sequences and common repetitive elements in the human transcriptome (e.g., transposable elements). Probes otherwise cover protein-coding sequences of the viral genomes, generally in a tiled, end-to-end design, with exceptions in cases where there was masked low-complexity or repetitive sequence. A final cocktail of 5361 80-mer oligonucleotides was generated.
Conserved viral group probes targeted against viruses associated with respiratory diseases (Supplemental Table S10) were designed using the same algorithm as described previously (Tong et al. 2008). For each taxon group, sequence alignments were generated, and conserved regions among all genomes of each viral family or genus available in GenBank were identified. Consensus and degenerate sequences were selected and single strand, 5′-end biotinylated 80- to 131-mer oligonucleotides were synthesized; oligonucleotides were normalized to 10 µM and pooled together in a cocktail. A total of 346 conserved probes for this respiratory viral panel were generated. The composition of the conserved enrichment probe cocktail includes probes for members of Adenoviridae (38), Coronavirinae (45), Orthomyxoviridae (31), Paramyxoviridae (47), Parvovirinae (38), Picornaviridae (54), Pneumoviridae (9), Polyomaviridae (36), and Reoviridae (48), with the number of oligonucleotides shown in parentheses.
Library preparation and target enrichment
Library preparation and target enrichment were performed using a TruSeq RNA Access Library Prep kit (currently, TruSeq RNA Exome kit; Illumina) which contains reagents for both library preparation and hybridization (Dehority et al. 2017). Briefly, 10–50 ng of viral NA mixture or up to 5 µL of clinical sample NAs were used for library preparation. Libraries were prepared in parallel with individual indices for sequencing with or without hybridization enrichment. Either virus-specific probes or conserved probes were used for hybridization enrichment. The manufacturer's protocol was followed with the following modifications: (1) first-strand cDNA synthesis was performed with SuperScript III Reverse Transcriptase (Thermo Fisher Scientific) rather than SuperScript II Reverse Transcriptase (Thermo Fisher Scientific); (2) between 15 and 17 cycles were used in the first PCR; (3) coding exome capture oligonucleotides were replaced with either the conserved probes or virus-specific probes; (4) for hybridization using conserved probes, 1.2 pmol probe/200 ng library (up to 4.8 pmol probe) were used, and multiplexing was varied from four to eight libraries per hybridization; (5) for hybridization using virus-specific probes, 5 µL of probe mixture were used, and multiplexing was varied from four to 12 libraries per hybridization; (6) for hybridization with both conserved and virus-specific probes, the second PCR amplification was increased from 10 cycles to 17–22 cycles.
NGS
Libraries generated from parallel hybridized and nonhybridized samples were normalized and combined at equimolar quantities to denature. The final loading concentration was 9 pM, with 1%–5% PhiX added. Samples were run on an Illumina MiSeq according to the manufacturer's instructions, yielding a median of ∼500,000 sequencing reads per sample.
Data analysis and bioinformatics pipeline
FASTQ files were analyzed by k-mer matching and read mapping using the following steps. Reads were trimmed for adapters and for quality using Cutadapt 1.8.1 (Martin 2011). Trimmed reads were then classified as human, bacterial, archaeal, viral, and PhiX using Kraken 0.10.5 (Wood and Salzberg 2014) and a database consisting of the human genome (GCF_000001405.26, 12/17/2013) and bacteria and virus genomes from RefSeq (Release 67, 9/8/2014). In parallel, trimmed reads were mapped to chosen references using Bowtie 2 2.2.9 (Langmead and Salzberg 2012) or BWA-MEM (Li and Durbin 2009). Resulting read numbers were used to generate charts as well as fold enrichment, where possible. For simplicity, targeted viral reads and total nontarget reads are depicted in results. We used a relative read count threshold (0.01%) to exclude low targeted viral reads because of potential barcode contamination. The samples with targeted viral reads <0.01% of the highest number of reads obtained for any sample of the same virus in the same MiSeq run were called negative.
Data access
The sequence data from this study have been submitted to the NCBI BioProject database (BioProject; https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA448596.
Supplementary Material
Acknowledgments
This work was made possible through support from the Advanced Molecular Detection (AMD) program at Centers for Disease Control and Prevention (CDC). The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the Centers for Disease Control and Prevention or the US Department of Health and Human Services.
Author contributions: S.T. conceived the project. B.M.O., Y.L., Y.T., J.Z., and K.Q. performed the experiments and generated the data. B.M.O., Y.L., and C.R.P. analyzed the data. B.M.O. and Y.L. wrote the manuscript. C.R.P., Y.T., K.Q., J.Z., D.L.D., S.M.G., G.P.S., and S.T. reviewed and edited the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.226316.117.
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. 2007. Direct selection of human genomic loci by microarray hybridization. Nat Methods 4: 903–905. [DOI] [PubMed] [Google Scholar]
- Allander T, Emerson SU, Engle RE, Purcell RH, Bukh J. 2001. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc Natl Acad Sci 98: 11609–11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Rohwer F. 2005. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques 39: 729–736. [DOI] [PubMed] [Google Scholar]
- Briese T, Kapoor A, Mishra N, Jain K, Kumar A, Jabado OJ, Lipkin WI. 2015. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. MBio 6: e01491-01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehority WN, Eickman MM, Schwalm KC, Gross SM, Schroth GP, Young SA, Dinwiddie DL. 2017. Complete genome sequence of a KI polyomavirus isolated from an otherwise healthy child with severe lower respiratory tract infection. J Med Virol 89: 926–930. [DOI] [PubMed] [Google Scholar]
- Depledge DP, Palser AL, Watson SJ, Lai IY, Gray ER, Grant P, Kanda RK, Leproust E, Kellam P, Breuer J. 2011. Specific capture and whole-genome sequencing of viruses from clinical samples. PLoS One 6: e27805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhaime MB, Sullivan MB. 2012. Ocean viruses: rigorously evaluating the metagenomic sample-to-sequence pipeline. Virology 434: 181–186. [DOI] [PubMed] [Google Scholar]
- Garau J, Calbo E. 2008. Community-acquired pneumonia. Lancet 371: 455. [DOI] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, et al. 2007. Genome-wide in situ exon capture for selective resequencing. Nat Genet 39: 1522–1527. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Meth 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett M, Kere J, Hinton LM. 1991. Direct selection: a method for the isolation of cDNAs encoded by large genomic regions. Proc Natl Acad Sci 88: 9628–9632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Mate SE, Kugelman JR, Nyenswah TG, Ladner JT, Wiley MR, Cordier-Lassalle T, Christie A, Schroth GP, Gross SM, Davies-Wayne GJ, et al. 2015. Molecular evidence of sexual transmission of Ebola virus. N Engl J Med 373: 2448–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. 2007. Microarray-based genomic selection for high-throughput resequencing. Nat Methods 4: 907–909. [DOI] [PubMed] [Google Scholar]
- Olp LN, Jeanniard A, Marimo C, West JT, Wood C. 2015. Whole-genome sequencing of Kaposi's sarcoma-associated herpesvirus from Zambian Kaposi's sarcoma biopsy specimens reveals unique viral diversity. J Virol 89: 12299–12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Chern SW, Li Y, Pallansch MA, Anderson LJ. 2008. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J Clin Microbiol 46: 2652–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15: R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie TN, Wylie KM, Herter BN, Storch GA. 2015. Enhanced virome sequencing using targeted sequence capture. Genome Res 25: 1910–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.