Summary
An AT motif-dependent axis, modulated by the transcription factor Zfhx3, influences the circadian clock in mice. In particular, gain of function of Zfhx3 significantly shortens circadian rhythms and alters the transcriptional activity of an important class of neuropeptides that controls intercellular signaling in the suprachiasmatic nucleus (SCN) of the hypothalamus. The ZFHX3/AT axis revealed an important, largely cell-nonautonomous control of the circadian clock. Here, by studying the recently identified circadian mouse mutant Zfhx3Sci/+, we identify significant effects on sleep homeostasis, a phenomenon that is outside the canonical circadian clock system and that is modulated by the activity of those neuropeptides at a circuit level. We show that the Zfhx3Sci/+ mutation accelerates the circadian clock at both the hourly scale (i.e., advancing circadian rhythms) and the seconds-to-minutes scale (i.e., anticipating behavioral responses) in mice. The in vivo results are accompanied by a significant presence of sleep targets among protein-protein interactions of the Zfhx3Sci/+-dependent network.
Introduction
The intrinsic circadian clock significantly influences the performance of multiple biological processes. Research on circadian biology has predominantly investigated the classical transcriptional-translational feedback loop (TTFL), in which core clock genes are cell autonomously regulated via the E-box DNA motif. Recently, a new circadian axis was identified (Parsons et al., 2015). Using a circadian-driven forward genetics approach in mice, we discovered a point mutation in exon 9 of the zinc finger homeobox 3 (Zfhx3) gene, a transcription factor that is highly expressed in the suprachiasmatic nucleus (SCN) of the hypothalamus. The mutation, which results in a G→T transversion, causes shortening of the circadian period in short circuit (Sci) mice. This experimental model revealed an important consensus AT motif that functions as a clock-regulated transcriptional axis. Interestingly, in searching for transcriptional changes in the SCN caused by the Zfhx3Sci/+ mutation, we identified a significant functional module of closely connected genes. In particular, the Zfhx3Sci/+ mutation downregulates important neuropeptides, such as vasoactive intestinal peptide (Vip), its receptor (Vipr2), and the neuropeptide receptor prokineticin receptor 2 (Prokr2). These peptidergic systems are known to play fundamental roles in sleep-wake regulation (Hu et al., 2011; Piggins and Cutler, 2003) and in circadian cell-cell signaling (Maywood et al., 2006). The interplay between the circadian clock and sleep processes over 24 hr influences many behavioral/cognitive traits, such as timing at the scale of seconds to minutes (Lassi et al., 2012; Agostino et al., 2011). The ability to perceive and produce short-interval behavioral responses is known as “interval timing,” and it is a primary property of many cognitive functions (e.g., attention and decision making). For example, in conditioning behaviors, interval timing permits the association between a stimulus and a reward (or a punishment): in risk assessment, the association between a choice and the payoff and, in feeding opportunities, the association between two meals. Both the circadian clock (hourly scale) and interval timing (seconds-to-minutes scale) are basic mechanisms for anticipatory behaviors involved in many daily life behaviors.
Here, we studied sleep in Zfhx3Sci/+ mice and littermate controls and discovered that they exhibit significant alterations in sleep homeostasis. We also investigated the function of short-interval timing in working-for-food cognitive tasks and discovered that, as for the circadian clock, the interval timing mechanism also runs faster in Zfhx3Sci/+ mice. The exploration of gene enrichment ontologies revealed a significant presence of sleep terms among Zfhx3Sci/+ SCN targets compared with wild-type SCNs.
Results
The Zfhx3Sci/+-Dependent Gene Network Includes a Significant Presence of Sleep Targets and Cell Response Signaling Pathways
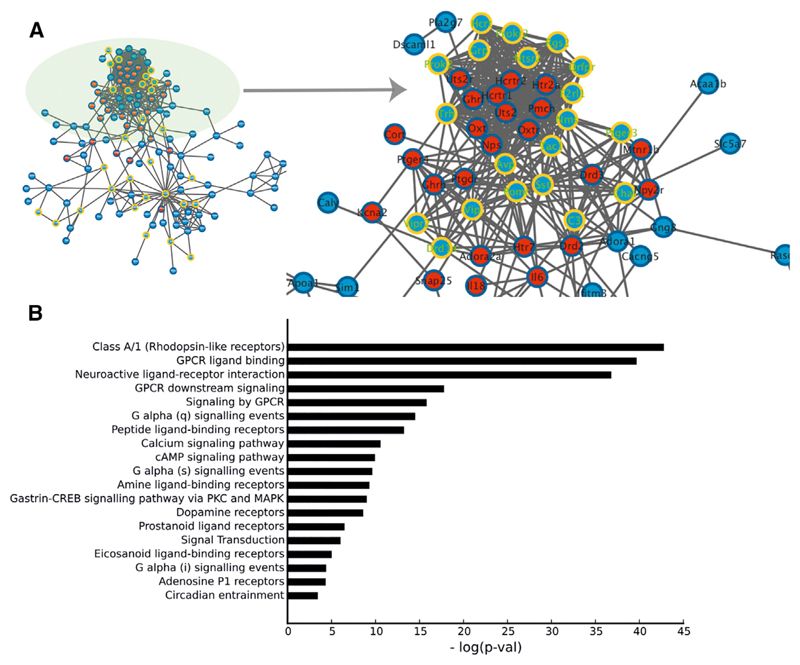
Specific analyses of the RNA sequencing data (reported in Parsons et al., 2015) in the SCN of Zfhx3Sci/+ and wild-type control mice allowed us to investigate whether the mutation affected specific pathways associated with sleep. A densely connected subnetwork (module 1; Parsons et al., 2015) displayed a significant presence of important sleep and sleep-wake neuropeptides (Figure 1A). Here, the interaction data showed that known sleep genes (red nodes in Figure 1A) were highly connected with module 1 (p < 0.001) compared with the other modules in the Zfhx3 differential expression network. We further explored regulatory processes by searching for functional involvement in up- and downstream regulatory mechanisms. In particular, we searched for classes that were enriched with Zfhx3-modulated genes. We observed an overrepresentation of proteins associated with the cellular membrane, in signal transduction and the integration of extracellular signals (Figure 1B). In particular, rhodopsin-like receptors and guanine nucleotide-binding (G) proteins were the main pathways targeted by the gene network affected by the Zfhx3Sci/+ mutation.
Figure 1. Protein-Protein Interactions.
(A) Zfhx3 and sleep genes. The blue nodes are differentially expressed Zfhx3 transcripts taken from Parsons et al. (2015). The red nodes are known mouse genes annotated with the “sleep” Gene Ontology term obtained from Amigo. The yellow ring nodes are differentially expressed transcripts containing a predicted AT motif. The circle highlights the densely connected subnetwork containing significant neuropeptides.
(B) Results from the pathway analysis are represented by significance levels, from the most significant (top bar) to the least significant (bottom bar). The bar length represents the inverse logarithm of p values. Significant differences are indicated as follows: **p value < 0.01.
Zfhx3Sci/+ Mouse Mutants Show Defects in Sleep Homeostasis
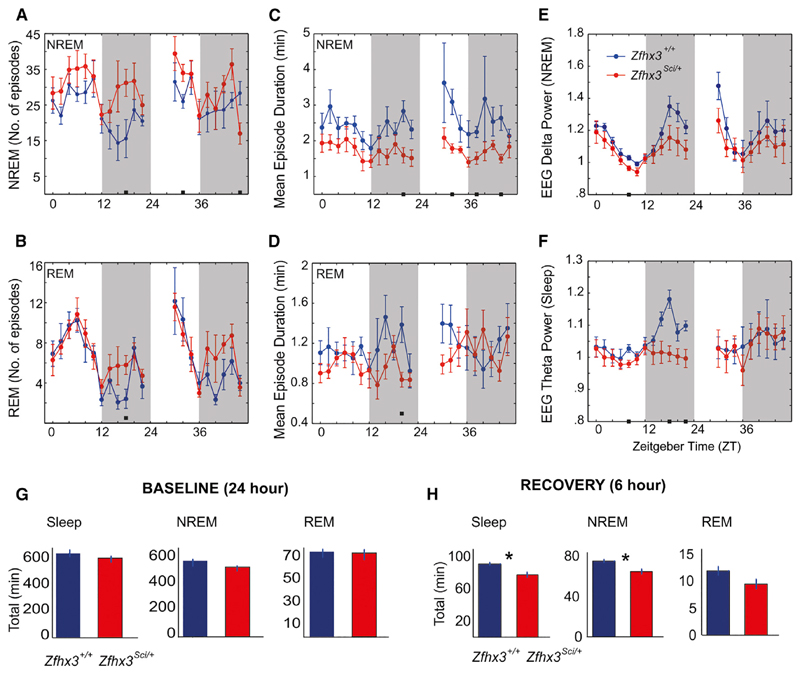
In order to probe the role of the Zfhx3 mutation in sleep-wake cycles, we studied the daily profile of wakefulness and two major states of sleep: rapid eye movement (REM) and non-REM (NREM) sleep (see Experimental Procedures). We found that Zfhx3Sci/+ mice showed significant differences in their sleep architecture compared with their littermate controls. The mutants showed a significantly greater number of overall sleep episodes during the light (p < 0.01) and dark phases (p < 0.05) of the baseline profile. In particular, by examining the distribution of NREM (Figure 2A) and REM (Figure 2B) sleep episodes, we found that genotypic differences were related to an increase in NREM sleep episodes, whereas REM sleep was unaltered between the two groups. These different sleep profiles between mutant and wild-type mice indicate that Zfhx3Sci/+ mice exhibit more fragmented sleep patterns, which was confirmed by their shorter mean sleep episodes compared with those of Zfhx3+/+ mice during the light phase of the baseline (p < 0.01) and during the recovery period (light phase: p = 0.06; dark phase: p < 0.05). These differences were precisely replicated in terms of the mean duration of NREM sleep episodes (Figure 2C), but not in REM (Figure 2D) patterns. However, fragmentation affects NREM/REM sleep quality by altering electroencephalogram (EEG) delta (Figure 2E) and theta power (Figure 2F). EEG delta power, which is the main index of sleep homeostasis, was reduced in Zfhx3Sci/+ mice during the last part of the dark phase (Figure 2E). As a consequence, we observed also a reduced sleep rebound following sleep deprivation (SD) (Table 1). Overall, Zfhx3Sci/+ mice showed a significant reduction in sleep duration and NREM sleep duration during the first 6 hr of rebound after SD, but not in baseline (Figures 2G and 2H; Table 1).
Figure 2. Sleep Homeostasis Is Altered in Zfhx3Sci/+ Compared with Zfhx3+/+ Mice.
Each graph shows the time course of sleep parameters in 1-hr bins, across 48 hr. The first 24 hr represent the baseline of the experiment, followed by 6 hr of sleep deprivation and 18 hr of recovery.
(A and B) The number of episodes of (A) NREM and (B) REM sleep are plotted in the first column.
(C and D) In the second column, the graphs display the mean duration of the episodes of (C) NREM and (D) REM sleep.
(E and F) The overall delta (E) and theta (F) power in NREM and REM sleep epochs are respectively shown.
(G and H) The histograms represent total sleep, NREM, and REM sleep duration in Zfhx3Sci/+ and Zfhx3+/+ mice during baseline (G) conditions and during the first 6 hr of recovery (H) after sleep deprivation.
Graphs are presented as the mean ± SEM. The white and black stripes in each graph indicate light and dark phases, respectively. Filled small black squares within graphs indicate statistically significant differences between Zfhx3sci/+ mice and their littermate controls. Significant differences are indicated as follows: *p < 0.05.
Table 1. Sleep Architecture Comparison between Genotypes.
| NREM (min) | |||
| Baseline | Post-SD | Difference | |
| +/+ | 180 ± 10 | 215 ± 4** | 34 ± 7 |
| Sci | 159 ± 8 | 185 ± 6* | 26 ± 9 |
| NREM Fragmentation (No. Episodes) | |||
| Baseline | Post-SD | Difference | |
| +/+ | 88 ± 8 | 90 ± 8 | 1 ± 8 |
| Sci | 104 ± 9 | 107 ± 6 | 3 ± 14 |
| NREM Mean Episodes Duration (min) | |||
| Baseline | Post-SD | Difference | |
| +/+ | 14 ± 1 | 19 ± 3 | 6 ± 3 |
| Sci | 10 ± 1 | 11 ± 1 | 1 ± 1 |
| NREM Delta Power | |||
| Baseline | Post-SD | Difference | |
| +/+ | 1.15 ± 0.04 | 1.6 ± 0.1* | 0.45 ± 0.2 |
| Sci | 1.12 ± 0.05 | 1.33 ± 0.08 | 0.1 ± 0.08 |
| REM (min) | |||
| Baseline | Post-SD | Difference | |
| +/+ | 25 ± 1 | 33 ± 2* | 7 ± 2 |
| Sci | 24 ± 2 | 26 ± 2 | 1 ± 3 |
Delta power was calculated over the first hour of the normalized values after SD (Zeitgeber Time [ZT] 7). The total NREM and REM duration, fragmentation, and mean episodes duration were calculated over 6 hr (ZT 7–12). The differences are taken as individual variation between Post-SD (ZT 7) and baseline (ZT 24). All the values are expressed as mean ± SEM. Statistical significance is reported as follows: one-way ANOVA, *p < 0.05, **p < 0.01.
A Faster Timer Runs in Zfhx3Sci/+ Mutants across Timescales
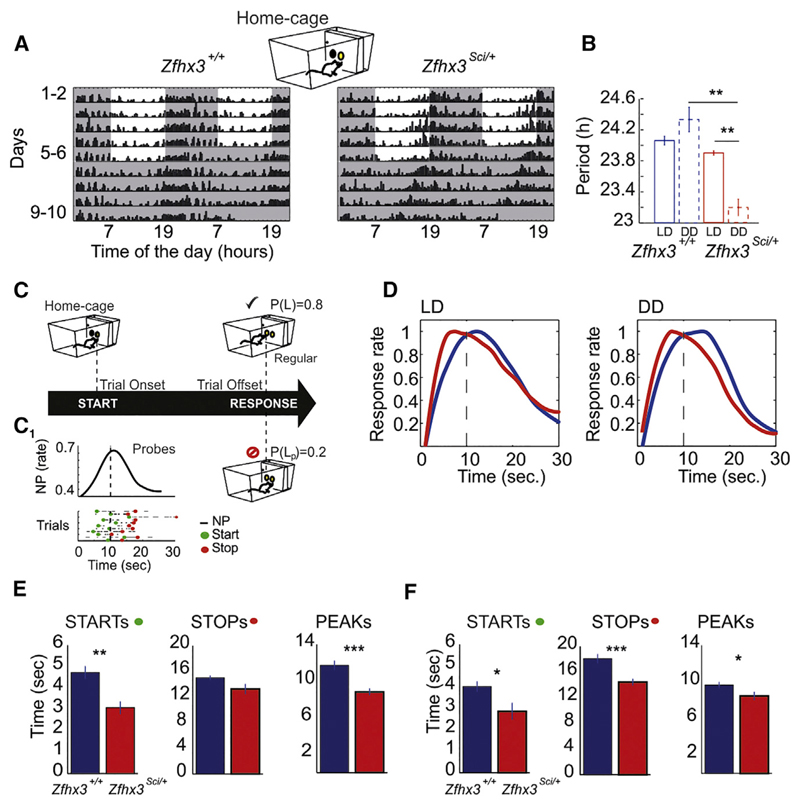
We conducted a behavioral investigation in Zfhx3Sci/+ mutant and wild-type littermate control mice to test whether the effects of the mutation on the circadian clock was distributed across timescales and influenced short-interval behaviors. From the circadian profiling of nose-poking activity (Figure 3A), we confirmed that Zfhx3Sci/+ mutant mice present a shortening of the circadian period in constant darkness (Figure 3B), as described earlier for wheel running (Parsons et al., 2015). In our study, while maintained in home cages, mice performed a work-for-food behavioral task that was designed to test interval timing. Specifically, mice were trained to estimate a 10-s interval, after which time they obtained a reward pellet. The ability to produce these timed behavioral responses is represented by an increase in response rate around the target time (10 s) during probe trials (Figure 3C). Probe trials are a subset of interpolated trials in which animals are not rewarded; therefore, the distribution of the behavioral response around the predicted time interval is a measure of the individual time perception. At any time during the 24 hr cycle, mice were allowed to self-initiate trials by simply nose poking in the central hopper of an operant wall mounted in the cage. Strikingly, we observed advanced timing behavior in Zfhx3Sci/+ mice, which was evident in both the light-dark (LD) condition and constant darkness (Figure 3D). Whereas the control mice showed a right-shifted (delayed) peak with respect to the 10-s target, the peak in the mutant mice was left shifted (advanced). A detailed analysis of trial-by-trial performance in LD (Figure 3E) and dark-dark (DD) (Figure 3F) confirmed that genotypic differences were reflected in advanced initiation, termination, and maximum response rate of the behavioral response.
Figure 3. Zfhx3Sci/+ Mutant Mice Shorten Both Circadian Period of Peak Nose-Poking Activity and Short-Interval Responses over 24 hr.
(A) Representative double-plotted actograms of nose-poking activity performed by Zfhx3+/+ and Zfhx3Sci/+ mice in home cages. Mouse nose-poking activity is collapsed into 15-min bins and is represented by black bars. Shaded regions indicate darkness in both the 12:12 light:dark (LD) and dark:dark (DD) conditions.
(B) Periodicities in LD and DD are shown for Zfhx3+/+ and Zfhx3Sci/+ mice.
(C) Representation of the short-interval task in home cages. The cartoon recapitulates how mice can initialize the trial by nose poking in the central hopper of an operant wall; after a FI, a nose poke at the left hopper is rewarded by the release of a food pellet with a probability 0.8. Unrewarded trials (probe trials) have probability of 0.2. (C1) An example of a raster plot and the distribution of nose-poke activity across the fixed-interval target during unrewarded probe trials. Green and red dots in the raster plot represent the “starts” and “stops” of responses extracted from trial-by-trial nose-poking activity (see Experimental Procedures).
(D) Temporal distribution of nose-poking responses in probe trials of Zfhx3Sci/+ and Zfhx3+/+ mice in LD and DD conditions. The FI target was 10 s.
(E and F) Mean values and SEMs of starts, stops, and peaks (start + stop/2) in LD (E) and DD (F) conditions. Significant differences between the groups are derived via one-way ANOVA. Significant differences are indicated as follows: ***p < 0.0005; **p < 0.005; *p < 0.05.
Overall, these behavioral data suggest that the timer of Zfhx3Sci/+ mice runs faster across 24-hr (circadian) and seconds-to-minutes (cognitive processes) timescales.
Light-Masking and Food-Seeking Behavior in Zfhx3Sci/+ Mutants Compared to Littermate Controls
During standard 12:12 LD cycles, Zfhx3Sci/+ mutant mice show decreased nose-poking activity (Figure S1A) and food intake (Figure S1B) compared to littermate control animals. The latter result in mutant behavior suggests an increased sensitivity to light-masking effects in Zfhx3Sci/+ mice, which could account for the rebound peak of both activity and food intake during the early phases of the dark period in these mice. Indeed, the peak of activity is missing under constant DD condition (Figures S1B and S1D). Nevertheless, Zfhx3Sci/+ mice also showed a reduction in both activity and food intake during the active phase both in LD and DD, indicating that mutants are poorly engaged in food seeking behavior during this phase independently from the light zeitgeber.
Sleep Is a Poor Predictor of Circadian Times in Zfhx3Sci/+ Mutants
The distribution of sleep-wake cycles as well as behavioral performance over 24 hr results from the integration of the homeostatic regulatory process and the circadian clock. The former depends on the sleep drive that accumulates during wake, whereas the latter is a self-sustained mechanism that sets the time for sleep, wake, and behavioral processes. Here, we questioned whether the Zfhx3Sci/+ mutation affects the dynamics of sleep and behavior across the circadian cycle and whether the two functions are predictors of the circadian phase over 24 hr. To answer this, we analyzed group averages of sleep and behavioral measures from independent groups of animals. Using a linear discriminant analysis (LDA) approach, we employed sleep and behavioral measures to identify combinations of features that maximize the discrimination between light/resting and dark/active phases throughout the LD cycle (Figure S2; Tables S1, S2, S3, and S4). In particular, reaction times and error rates per hour were positively correlated with all sleep measures across different times of the day, confirming that sleep pressure affects behavioral performance. Moreover, LDA successfully separated between circadian phases in all mice (Figure S2) so that sleep and behavioral performance were good predictors of circadian times in wild-type littermate control mice. Interestingly, in Zfhx3Sci/+ mutants, the role of sleep to the linear discriminant was very little (see Supplemental Information) compared to the role of behavioral measures, indicating that sleep is a poor predictor of the circadian phase in mutant animals.
Discussion
We demonstrated that the SCN-enriched transcription factor Zfhx3 represents a significant player in the regulation of sleep homeostasis and important cognitive processes. Analyses of the SCN transcriptome of Zfhx3Sci/+ and Zfhx3+/+ mice revealed that Zfhx3-mediated targets significantly interact with proteins that control fundamental aspects of sleep physiology and cell-cell signaling. Sleep, like many biological systems, is homeostatically regulated, meaning that the pressure to sleep increases and decreases as wakefulness and sleep progress, respectively. Although circadian rhythms and sleep homeostasis have traditionally been considered to be independent biological processes, our study supports recent advances in the study of clock genes that have demonstrated that the circadian clock system plays a significant role in sleep homeostatic responses (Franken, 2013). Evidence now suggests that sleep and the circadian clock are linked by evolutionarily conserved motifs that are observed as recurrent patterns in DNA and have important biological functions. Indeed, the Zfhx3 gene directly controls transcription through interaction with AT motifs, working in concert with the circadian clock. Both the present study and the preceding study (Parsons et al., 2015) highlight the importance of specific DNA AT motifs of target genes in the regulation of the circadian clock and sleep homeostasis. In addition, a previous study reported that sleep loss modifies the specific DNA binding of clock genes to the E-box motif (Mongrain et al., 2011).
Moreover, our study indicates a link between the circadian clock and short-interval cognitive processes. Our work reports an example of a genetically determined defect that shortens the circadian clock, also anticipating interval timing, suggesting that the mutation affects the process that enables mice to predict the interval between two events over different timescales. Our findings will have a profound influence on the understanding of the relationship between long- and short-interval timers. In particular, we know that short-interval behavioral responses show scalar invariance (Allman et al., 2014); the variability of the response increases as the temporal interval increases, resulting in a stable coefficient of variation across timescales. The distribution of the behavioral response rate in such temporal cognitive processes is not linear, which has prompted the hypothesis that the underlying regulation is due to an oscillator-like mechanism (Crystal, 1999), and evidence suggests that there may be shared mechanisms existing between the circadian clock and interval-timing processes (Agostino et al., 2011). Here, we observed that the mean reaction times during trials and the error rate in completing a trial worsened during the light phase in the mice, when the animals sleep more. We recently observed this detrimental effect of behavioral performance on different inbred strains (Maggi et al., 2014), implying that sleep and/or circadian phases modulate interval timing performance in mice. Here, we show, in mice, that a specific sleep history is associated with a gradual increase in reaction times and error rate in timing behaviors.
To date, several examples in the literature propose a link between the circadian clock and neuromodulators that affect interval timing, for example, arguing that a disturbed clock can advance or delay the perception of timing (Merchant et al., 2013; Golombek et al., 2014). Nevertheless, until now, evidence that a single gene could affect both circadian and interval timing was still missing. For example, no evidence has been reported that canonical E-box-dependent circadian clock genes influence interval timing. For example, studies investigating the Clock (Cordes and Gallistel, 2008) and Cry (Papachristos et al., 2011) mouse mutants have shown no effects on interval timing. The Zfhx3/AT axis represents an important link between the circadian clock and interval timing. The axis is part of a process that is interdependent with the TTFL mechanism but also introduces an additional level of organization in the SCN along with the TTFL canonical clock. Consequently, it may support and sense biological mechanisms across different systems with respect to the classical TTFL clock mechanism.
Experimental Procedures
Mice
All Zfhx3Sci/+ and Zfhx3+/+ mice were bred at the “Istituto Italiano di Tecnologia.” Mice were genotyped as described (Parsons et al., 2015). The original mutant founder was on a C3H/HeH × BALB/cAnNCrl background. Animals were bred onto both C57BL/6J and C3H/HeH backgrounds and backcrossed for ten generations until congenic, ensuring that other functional N-ethyl-N-nitrosourea (ENU)-induced mutations were removed. Breeding difficulties, however, were encountered on both backgrounds. To circumvent these problems, the Sci animals were maintained on a mixed C3H/HeH × C57BL6/J F1 background. All experimental procedures were conducted with age-matched groups of male mice. Wild-type littermate controls and heterozygous mutant males were group housed in the experimental room for 1 week before the experiments with food and water provided ad libitum under a 12:12 LD cycle (lights on from 7:00 to 19:00). All procedures were conducted under the Italian Policy.
In Silico Analysis of RNA Sequencing Results
The subnetwork of differentially expressed transcripts was taken from the RNA sequencing dataset of our previous work (Parsons et al., 2015). Associations with sleep/wake Gene Ontology terms were determined using Amigo (Ashburner et al., 2000). gProfiler (Reimand et al., 2011) was used to find over-represented Kyto Encyclopedia of Genes and Genomes (KEGG) pathways. Associations with the nervous system and sleep/neurological Mammalian Phenotype Ontology terms were determined using the disease connection resource in the Mouse Genome Database (Eppig et al., 2015).
Wireless Sleep Experiment
Mice were implanted with a wireless EEG system to define wakefulness, REM, and NREM sleep. We subjected all Zfhx3Sci/+ and Zfhx3+/+ mice to 24-hr baseline recordings to assess the basal architecture of their sleep-wake profile. Subsequently, all animals were sleep deprived for 6 hr (from 7:00 to 13:00), starting at the time the lights were turned on, and we monitored the sleep recovery process for 18 additional hours to assess the homeostatic rebound of sleep. Seven Zfhx3+/+ and seven Zfhx3Sci/+ heterozygous mice (12–16 weeks of age) were used to investigate sleep patterns. All mice were anesthetized with ketamine/xylazine (90–150 mg/kg ketamine/7.5–16K/7.5–16 mg/kg xylazine) intra-peritoneally and implanted with telemetry transmitters (Data Sciences; F20-EET; Gold system) for recording electroencephalography, electromyography, and body temperature (°C), as described previously in Lassi et al. (2012). Ten days post-surgery recovery time was allowed to mice to ensure a full recovery of normal sleep.
Automated Home-Cage Behavioral Testing
Behavioral testing was conducted in automated home cages in which the animals performed spontaneous nose-poking behaviors throughout the day. Mice were housed singly and maintained for several days in a novel home cage (cognition and welfare [COWE]) apparatus developed by TSE Systems to monitor nose pokes by mice (Maggi et al., 2014). Each home cage was equipped with three holes/hoppers over a metal wall. The central hole was the control hopper, and the lateral holes were connected to two feeders that released 20 mg dustless precision pellets (BioServ). All three holes were equipped with infrared beams and a light-emitting diode (LED) for light signals. Software allowed the implementation of various behavioral protocols by setting combinations of LED stimuli and pellet dispensing.
During this experiment, only two hoppers were available: the central and the left hopper. This experiment was run using six Zfhx3Sci/+ heterozygous and six wild-type littermate Zfhx3+/+ mice at 12–16 weeks of age.
FI and PI Task
This behavioral task involves the timing of a fixed interval (FI) associated with a reward pellet. FI refers to the training of repeated associations of a FI (in this case, 10 s) with the reward whereas the peak interval (PI) refers to the same FI training procedure except for the interpolation of 20% of probe (unrewarded) trials. Mice had access to food over the entire 24-hr period, and no food restriction was implemented.
Pre-training
The pre-training phase of the task was aimed at training the mice to associate the light/hopper with food pellet delivery. The mice self-initiated the trial by poking the central hopper. This caused a 2-s illumination (L = 2 s) of the two hoppers. If the mice poked the lateral hopper after the light signal and before the time limit (30 s), they received a pellet reward. The inter-trial interval (ITI) was defined as a 30-s FI plus a random delay drawn from a geometric distribution with a mean of 60 s. The pre-training phase lasted 3 days.
Training
In this phase, we implemented the FI and PI tasks at once. Unlike in pre-training, mice had to wait 10 s (L = 10 s) before receiving a pellet that was delivered only corresponding to a nose poke in the left location and within the time limit (30 s). In the training phase, 20% of the trials were unreinforced probe trials. An ITI followed each trial.
Data Analyses
Circadian Periodicities
Periodicity and amplitude were extracted by fitting nose-poking data with a sine function using repeated runs of the Levenberg-Marquardt (LM) algorithm (Levenberg, 1944; Marquardt, 1963).
The function used for the fit was as follows:
where translation (τ), amplitude (α), period (T), and phase (Φ) were the parameters that were optimized. In the first iteration of the LM algorithm, we choose an initial set of parameters according to the characteristics of the nose-poking profile.
Analyses of Sleep Data
EEG data recordings were analyzed using SleepSign software. A combination of automatic sleep scoring and manual correction was used. Scoring and extrapolation of the power spectra were performed in 4-s epochs. Each epoch was subjected to fast Fourier transformation (FFT) and classified as wakefulness (W), NREM, or REM.
The sleep scoring was analyzed in 1-hr bins, and various parameters were extracted, such as the total sleep time, which combined the durations of NREM and REM sleep. Fragmentation referred to the number of episodes/hour, where an episode is a period of one or more equal, consecutive, 4-s epochs.
The EEG delta power during NREM sleep epochs was expressed as a percentage of the average values of delta power during the last 4 hr of the light phase of the baseline profile (Franken et al., 2001).
Statistical Analysis
Data were analyzed with paired t tests and one-way ANOVAs. We used the Statistics Toolbox of the Matlab package for statistical analysis. Significant differences were considered for p values < 0.05 and indicated as follows: * p < 0.05; ** p < 0.005; *** p < 0.001.
Supplemental Information
Supplemental Information includes Supplemental Experimental Procedures, two figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.06.017.
Highlights.
The Zfhx3Sci/+ mutation leads to a defect in sleep homeostasis
This mutation accelerates biological timers across timescales
The Zfhx3Sci/+-dependent gene network contains a significant number of sleep targets
Sleep and the circadian clock are predictors of behavioral performance in mice
Footnotes
Author Contributions
E.B. conducted the computational work; S.S. and M.S. conducted mRNA-seq analyses; G.L. conducted the sleep work; S.M. conducted the behavioral/computational work; M.J.P. and P.M.N. developed the mouse model; and V.T. conceived, designed, and supervised all of the experiments and analyses and wrote the manuscript.
References
- Agostino PV, do Nascimento M, Bussi IL, Eguía MC, Golombek DA. Circadian modulation of interval timing in mice. Brain Res. 2011;1370:154–163. doi: 10.1016/j.brainres.2010.11.029. [DOI] [PubMed] [Google Scholar]
- Allman MJ, Teki S, Griffiths TD, Meck WH. Properties of the internal clock: first- and second-order principles of subjective time. Annu Rev Psychol. 2014;65:743–771. doi: 10.1146/annurev-psych-010213-115117. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes S, Gallistel CR. Intact interval timing in circadian CLOCK mutants. Brain Res. 2008;1227:120–127. doi: 10.1016/j.brainres.2008.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crystal JD. Systematic nonlinearities in the perception of temporal intervals. J Exp Psychol Anim Behav Process. 1999;25:3–17. [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE, Mouse Genome Database Group The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43:D726–D736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken P. A role for clock genes in sleep homeostasis. Curr Opin Neurobiol. 2013;23:864–872. doi: 10.1016/j.conb.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Franken P, Chollet D, Tafti M. The homeostatic regulation of sleep need is under genetic control. J Neurosci. 2001;21:2610–2621. doi: 10.1523/JNEUROSCI.21-08-02610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golombek DA, Bussi IL, Agostino PV. Minutes, days and years: molecular interactions among different scales of biological timing. Philos Trans R Soc Lond B Biol Sci. 2014;369:20120465. doi: 10.1098/rstb.2012.0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WP, Li JD, Colwell CS, Zhou QY. Decreased REM sleep and altered circadian sleep regulation in mice lacking vasoactive intestinal polypeptide. Sleep. 2011;34:49–56. doi: 10.1093/sleep/34.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassi G, Ball ST, Maggi S, Colonna G, Nieus T, Cero C, Bartolomucci A, Peters J, Tucci V. Loss of Gnas imprinting differentially affects REM/NREM sleep and cognition in mice. PLoS Genet. 2012;8:e1002706. doi: 10.1371/journal.pgen.1002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenberg K. A method for the solution of certain non–linear problems in least squares. Q J Appl Math. 1944;II:164–168. [Google Scholar]
- Maggi S, Garbugino L, Heise I, Nieus T, Balci F, Wells S, Tocchini-Valentini GP, Mandillo S, Nolan PM, Tucci V. A cross-laboratory investigation of timing endophenotypes in mouse behavior. Timing & Time Perception. 2014;2:25–50. [Google Scholar]
- Marquardt DW. An algorithm for least-squares estimation of nonlinear parameters. J Soc Ind Appl Math. 1963;11:431–441. [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O’Neill JS, O’Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Merchant H, Harrington DL, Meck WH. Neural basis of the perception and estimation of time. Annu Rev Neurosci. 2013;36:313–336. doi: 10.1146/annurev-neuro-062012-170349. [DOI] [PubMed] [Google Scholar]
- Mongrain V, La Spada F, Curie T, Franken P. Sleep loss reduces the DNA-binding of BMAL1, CLOCK, and NPAS2 to specific clock genes in the mouse cerebral cortex. PLoS ONE. 2011;6:e26622. doi: 10.1371/journal.pone.0026622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papachristos EB, Jacobs EH, Elgersma Y. Interval timing is intact in arrhythmic Cry1/Cry2-deficient mice. J Biol Rhythms. 2011;26:305–313. doi: 10.1177/0748730411410026. [DOI] [PubMed] [Google Scholar]
- Parsons MJ, Brancaccio M, Sethi S, Maywood ES, Satija R, Edwards JK, Jagannath A, Couch Y, Finelli MJ, Smyllie NJ, et al. The regulatory factor ZFHX3 modifies circadian function in SCN via an AT motif-driven axis. Cell. 2015;162:607–621. doi: 10.1016/j.cell.2015.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggins HD, Cutler DJ. The roles of vasoactive intestinal polypeptide in the mammalian circadian clock. J Endocrinol. 2003;177:7–15. doi: 10.1677/joe.0.1770007. [DOI] [PubMed] [Google Scholar]
- Reimand J, Arak T, Vilo J. g:Profiler–a web server for functional interpretation of gene lists (2011 update) Nucleic Acids Res. 2011;39:W307–W315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.