Abstract
Introduction
CNTN6 is an immunoglobulin domain‐containing cell adhesion molecule that belongs to the contactin family. It is involved in the development of the nervous system. We aim to determine the effect of Cntn6 deficiency on the allocentric navigation in mice.
Methods
We recorded the travel distance and escape time of wild‐type and Cntn6 mutant male and female mice in the Morris water maze task according to the protocol.
Results
There was hardly any Cntn6 expression in the hippocampus of postnatal day 0 (P0) mice, while obvious Cntn6 expression was present in the hippocampal CA1 region of the P7 mice. During the acquisition period of Morris water maze task (Day 1 to 4), Cntn6 −/− male mice failed to shorten the escape time to reach platform on the third day, while the travel distance to platform was not significantly different. There was no significant difference in both escape time and travel distance to the platform among all female subjects. In the probe trial test (Day 5), spatial memory of the female mutant mice was mildly affected, while Cntn6 −/− male mice were normal. In the spatial relearning test (Day 7 to 10), Cntn6 −/− male mice showed no difference in escape time to the platform compared to the wild‐type male mice, while Cntn6 deficient female mice required shorter escape time to travel to the platform on day 7, day 8, and day 10.
Conclusions
Cntn6 is expressed in the developing hippocampus in mice. Cntn6 deficiency affects spatial learning and memory, indicating that Cntn6 plays a role in the development of hippocampus and affects allocentric navigation of the animals.
Keywords: allocentric navigation, CNTN6, hippocampus, Morris water maze, spatial learning, spatial memory
1. INTRODUCTION
Development of the central nervous system is dependent on the highly coordinated interactions between diverse cell types. Cell adhesion molecules (CAMs) are important signal molecules that mediate cell–cell and cell–extracellular matrix interactions in multiple neural developmental processes (Doving & Trotier, 1998; Schaal et al., 2003), including neuronal migration, neurite outgrowth, axon guidance, synaptogenesis, and synaptic connection (Dalva, McClelland, & Kayser, 2007; Geschwind & Levitt, 2007; Maness & Schachner, 2007; Murase & Schuman, 1999; Pardo & Eberhart, 2007; Rubenstein, 2011). Furthermore, CAMs may also function as receptors to regulate neuronal apoptosis and survival (Anderson et al., 2005; Naus et al., 2004).
Contactin‐6 (CNTN6), also termed NB‐3, is a member of the contactin family of immunoglobulin (Ig) domain‐containing cell adhesion molecules (IgCAMs). CNTN6 contains six N‐terminal Ig‐like and four fibronectin type III‐like (FNIII) domains and tethers to the cell membrane via a C‐terminal glycosylphosphatidylinositol (GPI)‐anchor (Maness & Schachner, 2007; Shimoda & Watanabe, 2009; Zuko et al., 2013). Cntn6 has been identified as a candidate risk gene of multiple psychiatric disorders including autism spectrum disorders (ASDs), schizophrenia, bipolar disorder, attention‐deficit hyperactivity disorder, intellectual disability, and Tourette syndrome (Guo et al., 2012; Hu et al., 2015; Huang et al., 2017; Kashevarova et al., 2014; Kerner, Lambert, & Muthen, 2011; Nava et al., 2014; Oguro‐Ando, Zuko, Kleijer, & Burbach, 2017; Okbay et al., 2016; Pinto et al., 2010; Van Daalen et al., 2011), suggesting the necessity of CNTN6 in neural development.
In mice, Cntn6 is exclusively expressed in the nervous system, such as cerebral cortex, accessory olfactory bulb, thalamus, and cerebellum (Huang, Yu, Shimoda, Watanabe, & Liu, 2012; Lee et al., 2000). However, the expression of Cntn6 displays distinct patterns in different regions in the mouse brain. The level of Cntn6 protein in the cerebrum reaches a maximum at P7 and thereafter declines to a constant low level in the adulthood (Huang et al., 2011; Lee et al., 2000). In contrast, the Cntn6 mRNA level in the cerebellum and the hippocampus increases until the adulthood (Lee et al., 2000). Plenty of studies using null mutant mice indicate that Cntn6 plays key roles in the developing and mature mouse brains (Mercati et al., 2013; Oguro‐Ando et al., 2017; Shimoda & Watanabe, 2009). In the visual cortex of one‐month‐old Cntn6 −/− mice, alterations in the orientation of apical dendrites of pyramidal neurons in layer V was observed (Ye et al., 2008). Cntn6 regulates neurite outgrowth in vitro, and this property was consistent with the finding that corticospinal tract formation was delayed in the Cntn6 −/− mice (Huang et al., 2011, 2012; Mercati et al., 2013). Moreover, Cntn6 contributes to glutamatergic synapse formation between parallel fibers and Purkinje cells during postnatal cerebellar development (Sakurai et al., 2009). In addition, behavioral studies have shown that Cntn6‐deficient mice display impaired motor coordination (Takeda et al., 2003).
In the hippocampus, a significant reduction in glutamatergic synapses was found in the Cntn6‐deficient mice in the postnatal stage (Sakurai, Toyoshima, Takeda, Shimoda, & Watanabe, 2010; Sakurai et al., 2009). Amila Zuko et al. found that Cntn6 deficiency in the dentate gyrus (DG) may impair the fasciculation of mossy fibers that innervate pyramidal cells in the hippocampus (Cremer, Chazal, Goridis, & Represa, 1997; Heyden, Angenstein, Sallaz, Seidenbecher, & Montag, 2008; Montag‐Sallaz, Schachner, & Montag, 2002; Zuko et al., 2016). Some studies showed that F3/Contactin, another member of the contactin family, promotes hippocampal neurogenesis in adult mice (Mercati et al., 2017; Puzzo et al., 2013; Sakurai et al., 2009, 2010). These studies suggest that Cntn6 may play an important role in the hippocampal development and function. However, the effect of Cntn6 deficiency on hippocampal‐related behavior is still unclear.
In this study, we found that there was hardly any Cntn6 expression in the hippocampus of P0 mice, but obvious Cntn6 expression in the hippocampal CA1 region of P7 mice. Morris water maze task (MWM) was used to determine whether Cntn6 deficiency in mice would affect allocentric navigation which involves hippocampus and its related brain structures. Our results suggest that deletion of Cntn6 leads to functional deficiency of the hippocampus, especially the spatial learning ability in mice.
2. MATERIALS AND METHODS
2.1. Animal
Cntn6‐deficient mice (Takeda et al., 2003) were maintained on a 12‐hour light/dark cycle with ad libitum food and water in a specific pathogen‐free (SPF) animal facility at the Capital Medical University, China. All animal procedures were approved by the university's Committee for Animal experiments and conformed to the guidelines for the care and use of laboratory animals of the Chinese Society for Neuroscience.
Cntn6 knockout mice were generated using 129/SVJ embryonic stem cells and then were backcrossed with C57BL/6J mice for more than 20 generations. In all experiments described in this article, homozygous and heterozygous mutants were compared with their wild‐type littermates.
2.2. Colorimetric detection of LacZ expression
Cntn6 +/− mice at postnatal day 0 and 7 were perfused with PBS and then with 2% paraformaldehyde dissolved in PIPES, pH 6.9, containing 2 mM MgCl2 and 5 mM EGTA. Brains were removed and postfixed overnight at 4°C. The brains were then cryoprotected by incubation overnight in 20% sucrose containing 2 mM MgCl2. Floating sections (50 μm) were prepared using a cryostat. Sections were washed twice in PBS containing 2 mM MgCl2 and then incubated in PBS containing 2 mM MgCl2, 0.005% sodium deoxycholate and 0.01% NP‐40 for 10 min at 4°C. Colorimetric reaction was performed in the same solution containing 5 mM K3[Fe(CN)6], 5 mM K4[Fe(CN) 6], and 0.05% 5‐bromo‐4‐chloro‐3‐indolyl–D‐galactoside (X‐gal) at 37°C overnight. The sections were washed, mounted, air‐dried and were counterstained with 0.5% neutral red to visualize the brain architecture.
2.3. Morris water maze task
Learning and memory tasks of adult mice (2–4 months) were assessed using a Morris water maze task according to previous reports (Petravicz, Boyt, & McCarthy, 2014; Schenk & Morris, 1985). The stainless steel circular pool (150 cm in diameter, 51 cm in depth) was filled with white opaque water maintained at 21 ± 1°C. The platform (10 cm in diameter) was submerged 1 cm beneath water surface. The locations of the starting points were identified using different colors and dimensions visual extra‐maze cues attached to the room walls and were kept consistent during each experiment. The pool was divided into four quadrants using a computerized tracking/image analyzing system (video camcorder coupled with computational tracking system: Coulbourn Instrument). During the acquisition training trails, the platform was placed in the middle of the northwest (NW) quadrant and remained in the same position. Subjects were placed pseudorandomly with their heads facing the pool wall into each of four starting locations (northwest, northeast, southeast, and southwest) for each of four daily acquisition training trials. Trials lasted 60 s or until the subjects mounted the platform with a 30‐min intertrial interval. On the first day (Day 1) of training, the subjects were manually placed on the platform and allowed to stand on it for 15–20 s if they did not find the platform after 60 s. The escape time, travel distance and mean velocity to reach the platform were recorded during the four‐day training. A probe trial to test reference memory was conducted on day 5. Subjects were placed into the opposite quadrant of the platform quadrant and allowed to swim during 60 s in the absence of the platform. The number of platform crossings, the number of target quadrant crossings, and the proportion of swimming time spent in four quadrants were recorded and analyzed.
The reversal task (relearning training trial) was performed from day 7 to day 10 exactly as the acquisition training protocol, while the hidden platform was placed in the opposite quadrant (southeast). The escape time, travel distance, and mean velocity to reach the platform were recorded. The subjects were blind to the genotypes.
2.4. Statistical analysis
A two‐way ANOVA followed by the Bonferroni posttest was used to analyze escape time to platform and travel distance. The results are displayed as mean ± standard error of the mean (SEM). Multiple t test followed by the Sidak–Bonferroni method was used to analyze the time in quadrant. A one‐way ANOVA followed by the Bonferroni posttest was used to analyze and obtain statistics of the entries to target quadrant.
3. RESULTS
3.1. Expression of Cntn6 in the developing mouse hippocampus
To assess the potential role of Cntn6 in hippocampal development, the spatiotemporal expression of Cntn6 was analyzed in the developing mouse hippocampus. The segment between initiation codon of the second exon and the Bgl I site in the second intron of the Cntn6 gene was replaced by LacZ gene, so that the generated mutant mice were expected to produce β‐galactosidase instead of Cntn6 protein. The LacZ gene expression was driven by the promoter of the Cntn6 gene and accordingly reflected the expression of Cntn6 (Takeda et al., 2003). We first examine the expression of the LacZ in the whole brain (Figure 1a,b) and hippocampus (Figure 1c,d) of P0 and P7 Cntn6 +/− mice via X‐gal staining. The LacZ expression pattern was essentially the same as that observed in the Cntn6 in situ hybridization previously reported by Lee et al. (2000). In the hippocampus of P0 mice, there was hardly any Cntn6 expression in the CA1, CA3, and DG regions (Figure 1c). However, there was obvious Cntn6 expression in the CA1 but not in the CA3 and DG regions of P7 mice (Figure 1d). These results were indicating that Cntn6 is expressed in the developing hippocampus.
Figure 1.
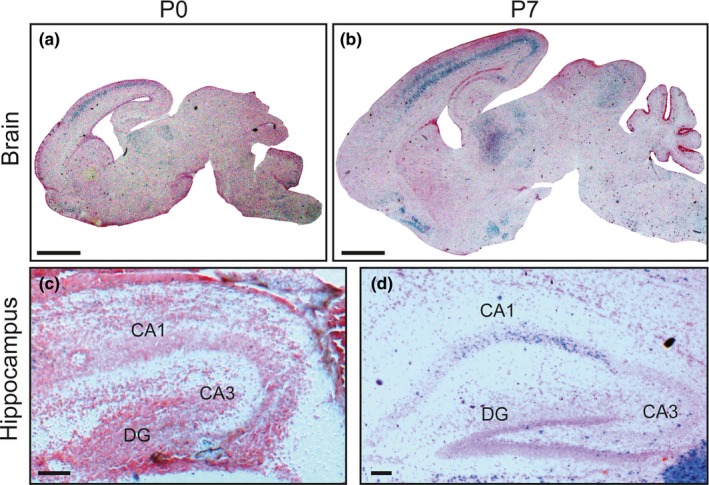
Expression of Cntn6 in the developing mouse hippocampus. Localization of cells expressing Cntn6 monitored by LacZ expression in the medial sagittal sections of the Cntn6 +/− brains at P0 and P7. (c,d) Higher magnification of the hippocampus in (a,b). Scale bars, (a,b) 1 mm, (c,d) 0.1 mm
3.2. Cntn6 deficiency affects spatial learning of male mice in the Morris water maze task
The hippocampal structure plays an important role in spatial learning and memory. It has been reported that the length and area size of the suprapyramidal bundle (SPB) in the hippocampus were significantly increased in Cntn6 −/− mice (Zuko et al., 2016). Here, we examined whether Cntn6 deficiency affected hippocampus‐related behavior in the Morris water maze task. Over the 4‐day acquisition training period, all animals improved their ability to find the submerged platform by exhibiting shorter escape time and travel distance to the platform (Figure 2a,b). There was no significant difference in performance among all female subjects (Figure 2d). However, although Cntn6 −/− male mice could swim as fast as wild‐type mice and willingly found a hidden platform, their escape time was significantly longer than their wild‐type and Cntn6 +/− littermates on the third day (Figure 2c). No significant difference in escape time was detected on the fourth day in Cntn6 −/− male mice (Figure 2c). These results indicated that spatial learning is mildly compromised in the Cntn6 −/− male mice.
Figure 2.
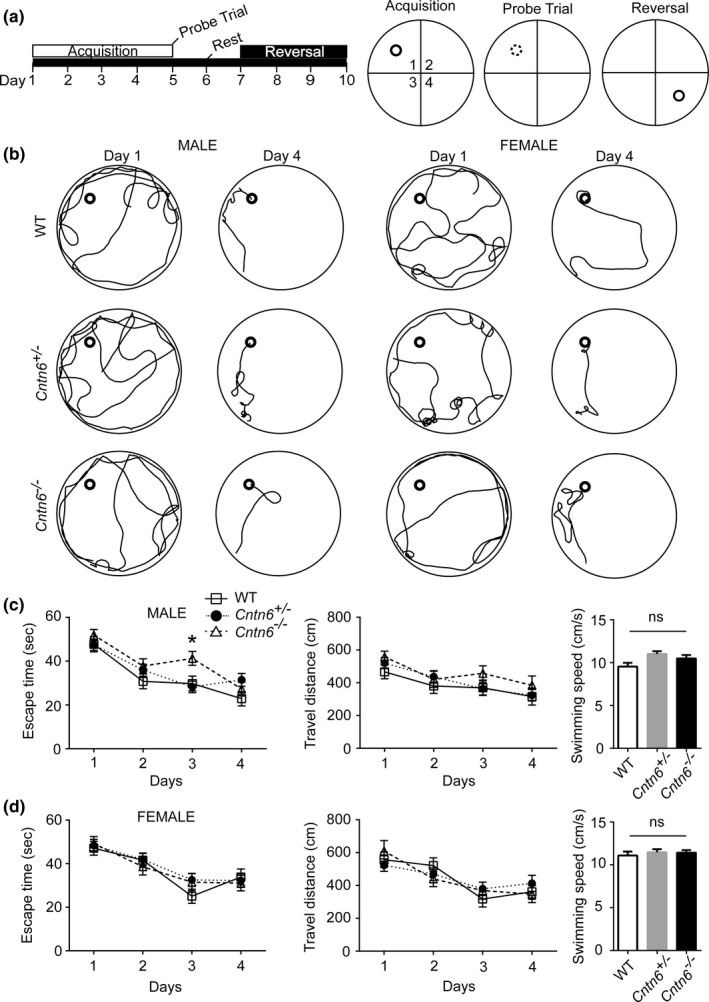
Cntn6 deficiency affects spatial learning of male mice in the Morris water maze task. (a) A schematic representation of the Morris water maze training protocol. Mice were trained for 4 days to locate a hidden platform (acquisition trials). A probe trial was performed on the fifth day, when the platform was removed. The hidden platform was moved to the opposite quadrant during reversal training. (b) Representative traces of swimming plot in Morris water maze task. (c) Quantitative analyses of the Morris water maze. Performance of the Cntn6 −/− male mice (2–4 months) in the spatial learning phases of the Morris water maze task, measured by escape time to platform. n = 10 (wild‐type, WT), 15 (Cntn6 +/−), 10 (Cntn6 −/−). Right panel, the swimming speed of male mice on the first day. (d) Performance of Cntn6 −/− female mice in the spatial learning. n = 9 (WT), 12 (Cntn6 +/−), 8 (Cntn6 −/−). Data represent as mean ± SEM. Two‐way ANOVA followed by Bonferroni posttest for escape time and travel distance and One‐way ANOVA for swimming speed. *, p < .05; ns, not significant
3.3. Cntn6 deficiency affects the spatial memory of female mice, but not male mice
After the 4‐day successive acquisition training period, we measured the time of movement of all the experimental groups in the 60‐second probe trial test on day 5 (Figure 3a). We calculated the time the mice spent in the target quadrant and the opposite quadrant after entering the pool in the last 40 s of the probe trial. Similar with the wild‐type male mice, Cntn6 −/− mutant male mice spent significant shorter time in the opposite quadrant than in the target quadrant, indicating that the Cntn6 deficiency has no serious effect on male mice's ability of recalling the previously learned spatial strategy (Figure 3b). Although Cntn6 +/− and Cntn6 −/− female mice also spent shorter time in the opposite quadrant, the change was not significant, (Figure 3c). We further analyzed the number of times the mice crossed the target platform location. There was no significant difference in the entries to target quadrant among all experimental subjects (Figure 3d). Together, these results indicated that Cntn6 deficiency of leads to mild deficits in the spatial memory of female mice.
Figure 3.
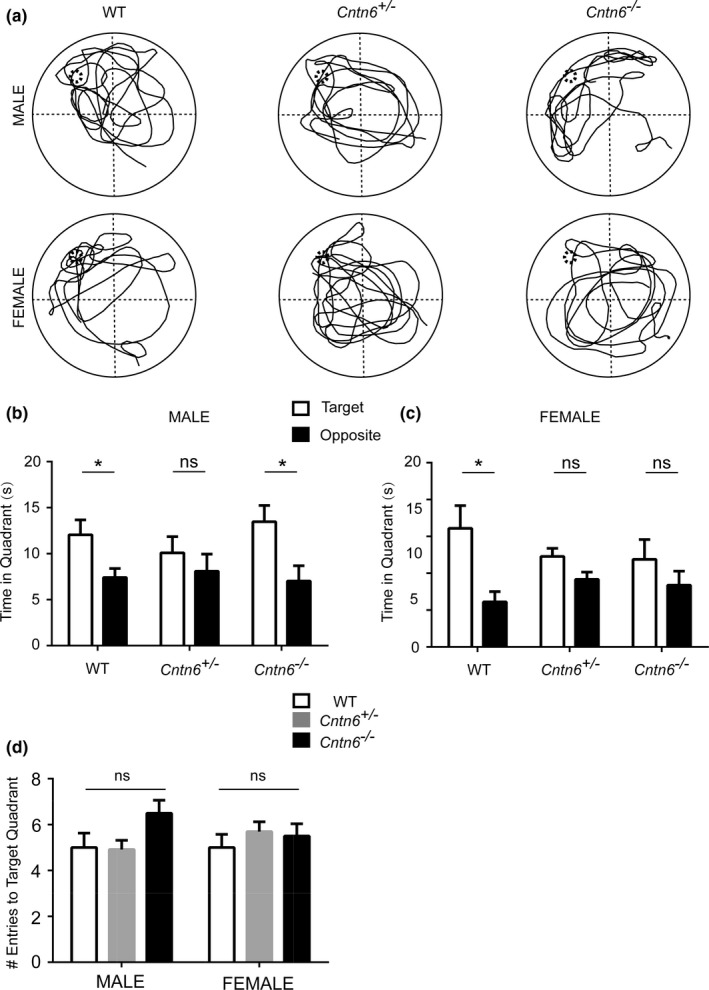
Cntn6 deficiency affects spatial memory of female mice. (a–d), A probe trial of the Morris water maze. (a) Representative trajectories of WT, Cntn6 +/−, and Cntn6 −/− mice during the probe trial. (b) The time male mice spent in the target quadrant and the opposite quadrant during the probe trial in which the target platform is removed. n = 10 (WT), 10 (Cntn6 +/−), 9 (Cntn6 −/−). Multiple t test followed by Sidak–Bonferroni posttest. (c) The time female mice spent in the target quadrant and the opposite quadrant during the probe trial. n = 7 (WT), 10 (Cntn6 +/−), 8 (Cntn6 −/−). Multiple t test followed by Sidak–Bonferroni posttest. (d) Number of entry to the target quadrant in the 60 s probe trial. Two‐way ANOVA. Data represent as mean ± SEM. *, p < .05; ns, not significant
3.4. Improved spatial relearning in Cntn6 deficient female mice
To investigate the effect of Cntn6 deficiency on spatial relearning, we performed a reversal task in the Morris water maze. Mice were trained for 4 additional days (day 7 to day 10) with the hidden platform placed in the opposite quadrant (Figure 4a). There was no significant difference in travel distance between wild‐type and mutants mice in both sexes (Figure 4b,c). Cntn6 −/− and Cntn6 +/− male mice showed no difference in escape time to the platform in the reversal task (Figure 4b). Interestingly, compares with the wild‐type female mice, both Cntn6 +/− and Cntn6 −/− female mice spent shorter time to reach the platform, and the change was significant between the wild‐type and the Cntn6 +/− female mice on day 7 (wild‐type vs. Cntn6 +/−, p = .031), day 8 (wild‐type vs. Cntn6 –/−, p = .0288; wild‐type vs. Cntn6 +/−, p = .0002), and day 10 (wild‐type vs. Cntn6 +/−, p = .0228) (Figure 4c). These results indicate that Cntn6 deficiency improves spatial relearning in female mice.
Figure 4.
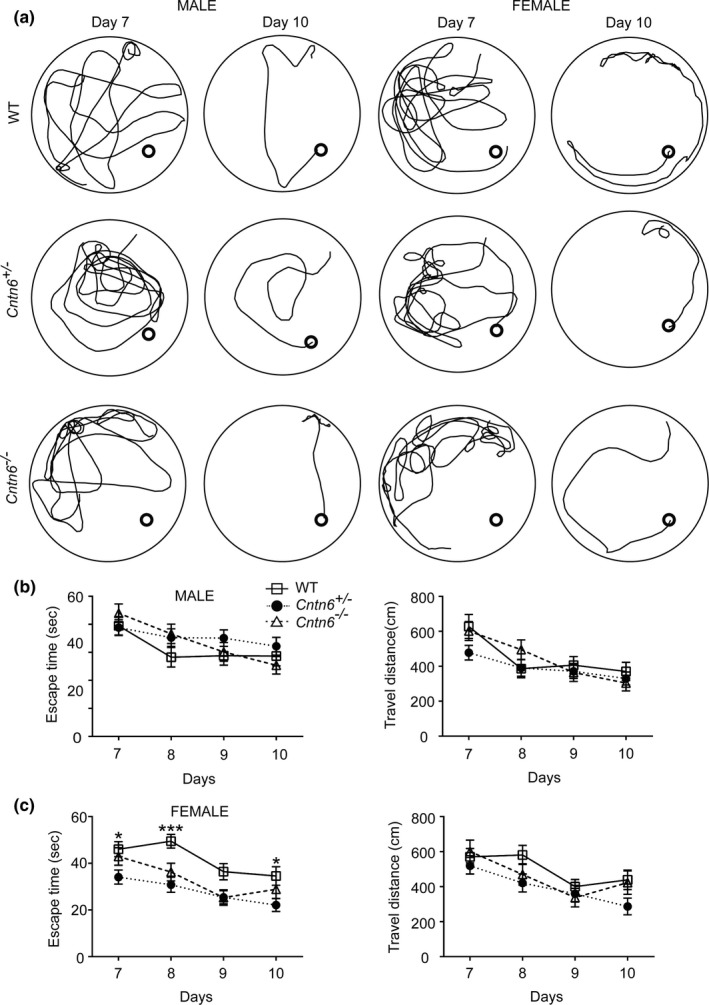
Cntn6 deficiency improves spatial relearning of female mice. (a) Representative traces of swimming plot in Morris water maze reversal task. (b) Quantitative analyses of the Morris water maze. Performance of Cntn6 −/− male mice (2–4 months) in the spatial relearning phase of the Morris water maze task, as measured by escape time to platforms. n = 10 (WT), 14 (Cntn6 +/−), 9 (Cntn6 −/−). (c) Performance of Cntn6 −/− female mice in spatial relearning. n = 8 (WT), 12 (Cntn6 +/−), 8 (Cntn6 −/−). Data represent mean ± SEM. Two‐way ANOVA followed by Bonferroni posttest. *, p < .05; ***, p < .001
4. DISCUSSION
Previous studies have shown that CNTN6 is important for the normal development and stability of the a few brain regions (Hu et al., 2015; Kashevarova et al., 2014; Lee et al., 2000; Sakurai et al., 2009). Here, we found that the expression of Cntn6 in the hippocampal CA1 region increases during early postnatal stage, which is consistent with the data set provided by Allen Brain database (http://developingmouse.brain-map.org/gene/show/33165), suggesting that Cntn6 is necessary for hippocampal structural formation and function. In the Morris water maze task, we found Cntn6 −/− male mice failed to reduce the escape time to reach the hidden platform on day 3 of the acquisition trials. Interestingly, although female Cntn6 mutant mice exhibited similar performance as the wild‐type mice in the acquisition trials, their spatial memory was mildly affected in the following probe trial. Moreover, female Cntn6 mutant mice also showed a decreased escape time to reach the platform in the spatial relearning test.
The structural integrity of hippocampus is crucial for spatial learning and memory (Daugherty, Bender, Yuan, & Raz, 2016; Guderian et al., 2015; Penner & Mizumori, 2012). The so‐called “trisynaptic loop” in hippocampus conducts synaptic transmission and consists of three major excitatory pathways: perforant path (from entorhinal cortex to DG), mossy fiber (from DG to CA3), and Schaffer collateral (from CA3 to CA1) (Andersen, Bliss, Lomo, Olsen, & Skrede, 1969; Inoue & Watanabe, 2014; Kesner, Lee, & Gilbert, 2004; Knierim, 2015; Lee et al., 2017; Okada & Okaichi, 2009; Piatti, Ewell, & Leutgeb, 2013; Rolls & Kesner, 2006; Rongo, 2002). The CA1 region is also thought to help encode memory into a form that can be sent back to the entorhinal cortex via the subiculum for subsequent longer‐term spatial memory and consolidation, but not short‐term acquisition or encoding processes (Lassalle, Bataille, & Halley, 2000; Lee & Kesner, 2004; Rolls, 2015; Rolls, Dempere‐Marco, & Deco, 2013; Rolls & Treves, 1994; Rolls & Xiang, 2006; Treves & Rolls, 1992). We found that Cntn6 is not expressed in the hippocampus of P0 mice, but is expressed in the CA1 region of P7 mice (Figure 1). Consistent with the expression pattern of Cntn6, the length and area size of mossy fiber projections in the SPB were significantly increased in the hippocampus of Cntn6 −/− mice, indicating that Cntn6 deficiency may impair the fasciculation of mossy fibers (Zuko et al., 2016). We therefore used the Morris water maze task to check whether the loss of Cntn6 affects hippocampus‐regulated spatial learning and memory.
At first acquisition of spatial learning was evaluated via repetitive training during which the mice use distinct spatial cues to swim from the starting position to the submerged platform. On day 3 of the acquisition training trails, the Cntn6 −/− male mice took longer time to find the hidden platform than the wild‐type male mice, indicating that Cntn6 −/− male mice learn more slowly but catch up at a later stage of the acquisition training trials (Figure 2). This increase in escape time on day 3 in Cntn6 −/− male mice is not due to impaired motor coordination as their swimming speed was comparable with the wild‐type male mice, and they performed equally well on day 1, 2, and 4 of the acquisition trials (Figure 2). After the acquisition training, a single probe trial was performed on day 5 with the platform withdrawn from the water tank to assess their spatial memory. The Cntn6 −/− male mice performed similar as the wild‐type mice, while the spatial memory in female mutant mice was mildly compromised (Figure 3). Interestingly, in the relearning/reversal phase (day 7 to 10) when mice were forced to find the submerged platform at a different location, Cntn6 +/− and Cntn6 −/− female mice performed better than their wild‐type littermates (Figure 4), while no difference was detected in the male mice, suggesting that female Cntn6 mutant mice are less perseverative for the previous acquisition platform location and are more readily to adapt to the changed contingencies.
Contactin family belongs to immunoglobulin (Ig) domain‐containing cell adhesion molecules (IgCAMs) and contains six members, CNTN1 (Contactin), CNTN2 (TAG‐1), CNTN3 (BIG‐1), CNTN4 (BIG‐2), CNTN5 (NB‐2), and CNTN6 (NB‐3) (Shimoda & Watanabe, 2009). CNTN6 is structurally and functionally similar to the other five family members. CNTN4 and CNTN6 followed by the close homologue of L1 (CHL1) are located on chromosome 3p25‐pter in the human genome (Kamei, Tsutsumi, Taketani, & Watanabe, 1998; Wei et al., 1998; Zeng et al., 2002). The deletion of this locus will cause 3p deletion syndrome with symptoms of microcephaly, growth retardation, intellectual disability, and distinctive facial features (Dijkhuizen et al., 2006; Fernandez et al., 2004, 2008). These three genes are closely located on chromosome 6p~ in the mouse genome and exhibit similar expression pattern. Thus, we speculate that the mild effect of Cntn6 deficiency on learning and memory may be due to the compensational effects of other contactin family members for the in the Cntn6 −/− brain.
Our results show that Cntn6 mutant mice exhibit sexual difference in spatial learning and memory impairments. The selection of female mice was random and did not exclude the factors of the menstrual cycle. Cntn6 −/− male mice show slower spatial learning, while female mutant mice may be compromised in long‐term memory retention. No sexual difference in hippocampus morphology or architecture has been discovered in the Cntn6 mutant mice. Sex hormones are involved in the cognitive differences between men and women, and sex‐selective effects were also detected with regard to spatial learning and memory (Piber, Nowacki, Mueller, Wingenfeld, & Otte, 2018). Young males rodents also have an advantage in spatial learning in Morris water maze tasks (Brandeis, Brandys, & Yehuda, 1989). Male and female mice perform the same when they are 6 months old, suggesting that the sex difference in young animals may reflect a difference in maturation rate (Bucci, Chiba, & Gallagher, 1995). We also found that Cntn6 −/− female mice have an advantage in spatial relearning during the reversal task compared with the wild‐type female mice. Reversal learning is a form of cognitive flexibility, an executive process that allows the adaptive modification of behavior in response to changes (Rygula, Walker, Clarke, Robbins, & Roberts, 2010). It has been reported that abnormal hippocampal structure leads to inflexible behaviors in women (Vilà‐Balló et al., 2017). We therefore speculate that the Cntn6 deficiency may specifically increase cognitive flexibility in female mice.
In conclusion, Cntn6 is expressed during postnatal hippocampal development. The absence of Cntn6 affects hippocampal spatial learning and memory. However, its cellular and molecular mechanism need further study.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
ACKNOWLEDGMENTS
We thank Dr. Lei Liu from Center of Stroke, Beijing Institute for Brain Disorder and Xuanwu Hospital, Capital Medical University, Beijing, China, for providing the venue, equipment, and technical support. This study was supported by the National Natural Science Foundation of China (31571486, 31270033), the Beijing Natural Science Foundation (5132003), and the Importation and Development of High‐Caliber Talents Project of Beijing Municipal Institutions (CIT&TCD20130339).
Mu D, Xu Y, Zhao T, Watanabe K, Xiao Z‐C, Ye H. Cntn6 deficiency impairs allocentric navigation in mice. Brain Behav. 2018;8:e00969 https://doi.org/10.1002/brb3.969
Funding information
Importation and Development of High‐Caliber Talents Project of Beijing Municipal Institutions, (Grant/Award Number: CIT&TCD20130339); National Natural Science Foundation of China, (Grant/Award Number: 31571486, 31270033); Beijing Natural Science Foundation, (Grant/Award Number: 5132003)
REFERENCES
- Andersen, P. , Bliss, T. V. , Lomo, T. , Olsen, L. I. , & Skrede, K. K. (1969). Lamellar organization of hippocampal excitatory pathways. Acta Physiologica Scandinavica, 76(1), 4A–5A. [DOI] [PubMed] [Google Scholar]
- Anderson, A. A. , Kendal, C. E. , Garcia‐Maya, M. , Kenny, A. V. , Morris‐Triggs, S. A. , Wu, T. , … Saffell, J. L. (2005). A peptide from the first fibronectin domain of NCAM acts as an inverse agonist and stimulates FGF receptor activation, neurite outgrowth and survival. Journal of Neurochemistry, 95(2), 570–583. https://doi.org/10.1111/j.1471-4159.2005.03417.x [DOI] [PubMed] [Google Scholar]
- Brandeis, R. , Brandys, Y. , & Yehuda, S. (1989). The use of the Morris Water Maze in the study of memory and learning. International Journal of Neuroscience, 48(1–2), 29–69. https://doi.org/10.3109/00207458909002151 [DOI] [PubMed] [Google Scholar]
- Bucci, D. J. , Chiba, A. A. , & Gallagher, M. (1995). Spatial learning in male and female Long‐Evans rats. Behavioral Neuroscience, 109(1), 180–183. https://doi.org/10.1037/0735-7044.109.1.180 [DOI] [PubMed] [Google Scholar]
- Cremer, H. , Chazal, G. , Goridis, C. , & Represa, A. (1997). NCAM is essential for axonal growth and fasciculation in the hippocampus. Molecular and Cellular Neurosciences, 8(5), 323–335. https://doi.org/10.1006/mcne.1996.0588 [DOI] [PubMed] [Google Scholar]
- Dalva, M. B. , McClelland, A. C. , & Kayser, M. S. (2007). Cell adhesion molecules: Signalling functions at the synapse. Nature Reviews Neuroscience, 8(3), 206–220. https://doi.org/10.1038/nrn2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty, A. M. , Bender, A. R. , Yuan, P. , & Raz, N. (2016). Changes in search path complexity and length during learning of a virtual water maze: Age differences and differential associations with hippocampal subfield volumes. Cerebral Cortex, 26(6), 2391–2401. https://doi.org/10.1093/cercor/bhv061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkhuizen, T. , van Essen, T. , van der Vlies, P. , Verheij, J. B. , Sikkema‐Raddatz, B. , van der Veen, A. Y. , … Kok, K. (2006). FISH and array‐CGH analysis of a complex chromosome 3 aberration suggests that loss of CNTN4 and CRBN contributes to mental retardation in 3pter deletions. American Journal of Medical Genetics. Part A, 140(22), 2482–2487. [DOI] [PubMed] [Google Scholar]
- Doving, K. B. , & Trotier, D. (1998). Structure and function of the vomeronasal organ. Journal of Experimental Biology, 201(Pt 21), 2913–2925. [DOI] [PubMed] [Google Scholar]
- Fernandez, T. , Morgan, T. , Davis, N. , Klin, A. , Morris, A. , Farhi, A. , & Lifton, R. P. (2004). Disruption of contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. American Journal of Human Genetics, 74(6), 1286–1293. https://doi.org/10.1086/421474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, T. , Morgan, T. , Davis, N. , Klin, A. , Morris, A. , Farhi, A. , & Lifton, R. P. (2008). Disruption of Contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. American Journal of Human Genetics, 82(6), 1385 https://doi.org/10.1016/j.ajhg.2008.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind, D. H. , & Levitt, P. (2007). Autism spectrum disorders: Developmental disconnection syndromes. Current Opinion in Neurobiology, 17(1), 103–111. https://doi.org/10.1016/j.conb.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Guderian, S. , Dzieciol, A. M. , Gadian, D. G. , Jentschke, S. , Doeller, C. F. , Burgess, N. , … Vargha‐Khadem, F. (2015). Hippocampal volume reduction in humans predicts impaired allocentric spatial memory in virtual‐reality navigation. Journal of Neuroscience, 35(42), 14123–14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H. , Xun, G. , Peng, Y. , Xiang, X. , Xiong, Z. , Zhang, L. , … Long, Z. (2012). Disruption of Contactin 4 in two subjects with autism in Chinese population. Gene, 505(2), 201–205. [DOI] [PubMed] [Google Scholar]
- Heyden, A. , Angenstein, F. , Sallaz, M. , Seidenbecher, C. , & Montag, D. (2008). Abnormal axonal guidance and brain anatomy in mouse mutants for the cell recognition molecules close homolog of L1 and NgCAM‐related cell adhesion molecule. Neuroscience, 155(1), 221–233. https://doi.org/10.1016/j.neuroscience.2008.04.080 [DOI] [PubMed] [Google Scholar]
- Hu, J. , Liao, J. , Sathanoori, M. , Kochmar, S. , Sebastian, J. , Yatsenko, S. A. , & Surti, U. (2015). CNTN6 copy number variations in 14 patients: A possible candidate gene for neurodevelopmental and neuropsychiatric disorders. Journal of Neurodevelopmental Disorders, 7(1), 26 https://doi.org/10.1186/s11689-015-9122-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Sun, J. , Zhao, T. , Wu, K. W. , Watanabe, K. , Xiao, Z. C. , … Fan, M. (2011). Loss of NB‐3 aggravates cerebral ischemia by impairing neuron survival and neurite growth. Stroke, 42(10), 2910–2916. [DOI] [PubMed] [Google Scholar]
- Huang, A. Y. , Yu, D. , Davis, L. K. , Sul, J. H. , Tsetsos, F. , Ramensky, V. , … McGrath, L. M. (2017). Rare copy number variants in NRXN1 and CNTN6 increase risk for tourette syndrome. Neuron, 94(6), 1101–1111e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Z. , Yu, Y. , Shimoda, Y. , Watanabe, K. , & Liu, Y. (2012). Loss of neural recognition molecule NB‐3 delays the normal projection and terminal branching of developing corticospinal tract axons in the mouse. Journal of Comparative Neurology, 520(6), 1227–1245. https://doi.org/10.1002/cne.22772 [DOI] [PubMed] [Google Scholar]
- Inoue, N. , & Watanabe, S. (2014). Effects of reversible deactivation of mossy fibers in the dentate‐CA3 system on geometric center detection task in mice: Functional separation of spatial learning and its generalization to new environment. Physiology & Behavior, 131, 75–80. https://doi.org/10.1016/j.physbeh.2014.04.015 [DOI] [PubMed] [Google Scholar]
- Kamei, Y. , Tsutsumi, O. , Taketani, Y. , & Watanabe, K. (1998). cDNA cloning and chromosomal localization of neural adhesion molecule NB‐3 in human. Journal of Neuroscience Research, 51(3), 275–283. https://doi.org/10.1002/(ISSN)1097-4547 [DOI] [PubMed] [Google Scholar]
- Kashevarova, A. A. , Nazarenko, L. P. , Schultz‐Pedersen, S. , Skryabin, N. A. , Salyukova, O. A. , Chechetkina, N. N. , … Romeo, G. (2014). Single gene microdeletions and microduplication of 3p26.3 in three unrelated families: CNTN6 as a new candidate gene for intellectual disability. Molecular Cytogenetics, 7(1), 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner, B. , Lambert, C. G. , & Muthen, B. O. (2011). Genome‐wide association study in bipolar patients stratified by co‐morbidity. PLoS ONE, 6(12), e28477 https://doi.org/10.1371/journal.pone.0028477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesner, R. P. , Lee, I. , & Gilbert, P. (2004). A behavioral assessment of hippocampal function based on a subregional analysis. Reviews in the Neurosciences, 15(5), 333–351. [DOI] [PubMed] [Google Scholar]
- Knierim, J. J. (2015). The hippocampus. Current Biology, 25(23), R1116–R1121. https://doi.org/10.1016/j.cub.2015.10.049 [DOI] [PubMed] [Google Scholar]
- Lassalle, J. M. , Bataille, T. , & Halley, H. (2000). Reversible inactivation of the hippocampal mossy fiber synapses in mice impairs spatial learning, but neither consolidation nor memory retrieval, in the Morris navigation task. Neurobiology of Learning and Memory, 73(3), 243–257. https://doi.org/10.1006/nlme.1999.3931 [DOI] [PubMed] [Google Scholar]
- Lee, I. , & Kesner, R. P. (2004). Encoding versus retrieval of spatial memory: Double dissociation between the dentate gyrus and the perforant path inputs into CA3 in the dorsal hippocampus. Hippocampus, 14(1), 66–76. https://doi.org/10.1002/(ISSN)1098-1063 [DOI] [PubMed] [Google Scholar]
- Lee, S. H. , Lutz, D. , Mossalam, M. , Bolshakov, V. Y. , Frotscher, M. , & Shen, J. (2017). Presenilins regulate synaptic plasticity and mitochondrial calcium homeostasis in the hippocampal mossy fiber pathway. Molecular Neurodegeneration, 12(1), 48 https://doi.org/10.1186/s13024-017-0189-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Takeda, Y. , Kawano, H. , Hosoya, H. , Nomoto, M. , Fujimoto, D. , … Watanabe, K. (2000). Expression and regulation of a gene encoding neural recognition molecule NB‐3 of the contactin/F3 subgroup in mouse brain. Gene, 245(2), 253–266. [DOI] [PubMed] [Google Scholar]
- Maness, P. F. , & Schachner, M. (2007). Neural recognition molecules of the immunoglobulin superfamily: Signaling transducers of axon guidance and neuronal migration. Nature Neuroscience, 10(1), 19–26. https://doi.org/10.1038/nn1827 [DOI] [PubMed] [Google Scholar]
- Mercati, O. , Danckaert, A. , André‐Leroux, G. , Bellinzoni, M. , Gouder, L. , Watanabe, K. , … Cloëz‐Tayarani, I. (2013). Contactin 4, ‐5 and ‐6 differentially regulate neuritogenesis while they display identical PTPRG binding sites. Biology Open, 2(3), 324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercati, O. , Huguet, G. , Danckaert, A. , André‐Leroux, G. , Maruani, A. , Bellinzoni, M. , … Amsellem, F. (2017). CNTN6 mutations are risk factors for abnormal auditory sensory perception in autism spectrum disorders. Molecular Psychiatry, 22(4), 625–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag‐Sallaz, M. , Schachner, M. , & Montag, D. (2002). Misguided axonal projections, neural cell adhesion molecule 180 mRNA upregulation, and altered behavior in mice deficient for the close homolog of L1. Molecular and Cellular Biology, 22(22), 7967–7981. https://doi.org/10.1128/MCB.22.22.7967-7981.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase, S. , & Schuman, E. M. (1999). The role of cell adhesion molecules in synaptic plasticity and memory. Current Opinion in Cell Biology, 11(5), 549–553. https://doi.org/10.1016/S0955-0674(99)00019-8 [DOI] [PubMed] [Google Scholar]
- Naus, S. , Richter, M. , Wildeboer, D. , Moss, M. , Schachner, M. , & Bartsch, J. W. (2004). Ectodomain shedding of the neural recognition molecule CHL1 by the metalloprotease‐disintegrin ADAM8 promotes neurite outgrowth and suppresses neuronal cell death. Journal of Biological Chemistry, 279(16), 16083–16090. https://doi.org/10.1074/jbc.M400560200 [DOI] [PubMed] [Google Scholar]
- Nava, C. , Keren, B. , Mignot, C. , Rastetter, A. , Chantot‐Bastaraud, S. , Faudet, A. , … Whalen, S. (2014). Prospective diagnostic analysis of copy number variants using SNP microarrays in individuals with autism spectrum disorders. European Journal of Human Genetics, 22(1), 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro‐Ando, A. , Zuko, A. , Kleijer, K. T. , & Burbach, J. P. H. (2017). A current view on contactin‐4, ‐5, and ‐6: Implications in neurodevelopmental disorders. Molecular and Cellular Neurosciences, 81, 72–83. https://doi.org/10.1016/j.mcn.2016.12.004 [DOI] [PubMed] [Google Scholar]
- Okada, K. , & Okaichi, H. (2009). Functional differentiation and cooperation among the hippocampal subregions in rats to effect spatial memory processes. Behavioral Brain Research, 200(1), 181–191. https://doi.org/10.1016/j.bbr.2009.01.011 [DOI] [PubMed] [Google Scholar]
- Okbay, A. , Beauchamp, J. P. , Fontana, M. A. , Lee, J. J. , Pers, T. H. , Rietveld, C. A. , … Oskarsson, S. (2016). Genome‐wide association study identifies 74 loci associated with educational attainment. Nature, 533(7604), 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo, C. A. , & Eberhart, C. G. (2007). The neurobiology of autism. Brain Pathology, 17(4), 434–447. https://doi.org/10.1111/j.1750-3639.2007.00102.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner, M. R. , & Mizumori, S. J. (2012). Neural systems analysis of decision making during goal‐directed navigation. Progress in Neurobiology, 96(1), 96–135. https://doi.org/10.1016/j.pneurobio.2011.08.010 [DOI] [PubMed] [Google Scholar]
- Petravicz, J. , Boyt, K. M. , & McCarthy, K. D. (2014). Astrocyte IP3R2‐dependent Ca(2 + ) signaling is not a major modulator of neuronal pathways governing behavior. Frontiers in Behavioural Neurosciences, 8, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti, V. C. , Ewell, L. A. , & Leutgeb, J. K. (2013). Neurogenesis in the dentate gyrus: Carrying the message or dictating the tone. Frontiers in Neuroscience, 7, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piber, D. , Nowacki, J. , Mueller, S. C. , Wingenfeld, K. , & Otte, C. (2018). Sex effects on spatial learning but not on spatial memory retrieval in healthy young adults. Behavioral Brain Research, 336, 44–50. https://doi.org/10.1016/j.bbr.2017.08.034 [DOI] [PubMed] [Google Scholar]
- Pinto, D. , Pagnamenta, A. T. , Klei, L. , Anney, R. , Merico, D. , Regan, R. , … Almeida, J. (2010). Functional impact of global rare copy number variation in autism spectrum disorders. Nature, 466(7304), 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzzo, D. , Bizzoca, A. , Privitera, L. , Furnari, D. , Giunta, S. , Girolamo, F. , … Palmeri, A. (2013). F3/Contactin promotes hippocampal neurogenesis, synaptic plasticity, and memory in adult mice. Hippocampus, 23(12), 1367–1382. [DOI] [PubMed] [Google Scholar]
- Rolls, E. T. (2015). Diluted connectivity in pattern association networks facilitates the recall of information from the hippocampus to the neocortex. Progress in Brain Research, 219, 21–43. https://doi.org/10.1016/bs.pbr.2015.03.007 [DOI] [PubMed] [Google Scholar]
- Rolls, E. T. , Dempere‐Marco, L. , & Deco, G. (2013). Holding multiple items in short term memory: A neural mechanism. PLoS ONE, 8(4), e61078 https://doi.org/10.1371/journal.pone.0061078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls, E. T. , & Kesner, R. P. (2006). A computational theory of hippocampal function, and empirical tests of the theory. Progress in Neurobiology, 79(1), 1–48. https://doi.org/10.1016/j.pneurobio.2006.04.005 [DOI] [PubMed] [Google Scholar]
- Rolls, E. T. , & Treves, A. (1994). Neural networks in the brain involved in memory and recall. Progress in Brain Research, 102, 335–341. https://doi.org/10.1016/S0079-6123(08)60550-6 [DOI] [PubMed] [Google Scholar]
- Rolls, E. T. , & Xiang, J. Z. (2006). Spatial view cells in the primate hippocampus and memory recall. Reviews in the Neurosciences, 17(1–2), 175–200. [DOI] [PubMed] [Google Scholar]
- Rongo, C. (2002). A fresh look at the role of CaMKII in hippocampal synaptic plasticity and memory. BioEssays, 24(3), 223–233. https://doi.org/10.1002/(ISSN)1521-1878 [DOI] [PubMed] [Google Scholar]
- Rubenstein, J. L. (2011). Annual research review: Development of the cerebral cortex: Implications for neurodevelopmental disorders. Journal of Child Psychology and Psychiatry, 52(4), 339–355. https://doi.org/10.1111/j.1469-7610.2010.02307.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rygula, R. , Walker, S. C. , Clarke, H. F. , Robbins, T. W. , & Roberts, A. C. (2010). Differential contributions of the primate ventrolateral prefrontal and orbitofrontal cortex to serial reversal learning. Journal of Neuroscience, 30(43), 14552–14559. https://doi.org/10.1523/JNEUROSCI.2631-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai, K. , Toyoshima, M. , Takeda, Y. , Shimoda, Y. , & Watanabe, K. (2010). Synaptic formation in subsets of glutamatergic terminals in the mouse hippocampal formation is affected by a deficiency in the neural cell recognition molecule NB‐3. Neuroscience Letters, 473(2), 102–106. https://doi.org/10.1016/j.neulet.2010.02.027 [DOI] [PubMed] [Google Scholar]
- Sakurai, K. , Toyoshima, M. , Ueda, H. , Matsubara, K. , Takeda, Y. , Karagogeos, D. , … Watanabe, K. (2009). Contribution of the neural cell recognition molecule NB‐3 to synapse formation between parallel fibers and Purkinje cells in mouse. Developmental Neurobiology, 69(12), 811–824. [DOI] [PubMed] [Google Scholar]
- Schaal, B. , Coureaud, G. , Langlois, D. , Ginies, C. , Sémon, E. , & Perrier, G. (2003). Chemical and behavioural characterization of the rabbit mammary pheromone. Nature, 424(6944), 68–72. https://doi.org/10.1038/nature01739 [DOI] [PubMed] [Google Scholar]
- Schenk, F. , & Morris, R. G. (1985). Dissociation between components of spatial memory in rats after recovery from the effects of retrohippocampal lesions. Experimental Brain Research, 58(1), 11–28. [DOI] [PubMed] [Google Scholar]
- Shimoda, Y. , & Watanabe, K. (2009). Contactins: Emerging key roles in the development and function of the nervous system. Cell Adhesion and Migration, 3(1), 64–70. https://doi.org/10.4161/cam.3.1.7764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, Y. , Akasaka, K. , Lee, S. , Kobayashi, S. , Kawano, H. , Murayama, S. , … Sudo, K. (2003). Impaired motor coordination in mice lacking neural recognition molecule NB‐3 of the contactin/F3 subgroup. Journal of Neurobiology, 56(3), 252–265. [DOI] [PubMed] [Google Scholar]
- Treves, A. , & Rolls, E. T. (1992). Computational constraints suggest the need for two distinct input systems to the hippocampal CA3 network. Hippocampus, 2(2), 189–199. https://doi.org/10.1002/(ISSN)1098-1063 [DOI] [PubMed] [Google Scholar]
- van Daalen, E. , Kemner, C. , Verbeek, N. E. , van der Zwaag, B. , Dijkhuizen, T. , Rump, P. , … Beemer, F. A. (2011). Social Responsiveness Scale‐aided analysis of the clinical impact of copy number variations in autism. Neurogenetics, 12(4), 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilà‐Balló, A. , Mas‐Herrero, E. , Ripollés, P. , Simó, M. , Miró, J. , Cucurell, D. , … Rodríguez‐Fornells, A. (2017). Unraveling the role of the hippocampus in reversal learning. Journal of Neuroscience, 37(28), 6686–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, M. H. , Karavanova, I. , Ivanov, S. V. , Popescu, N. C. , Keck, C. L. , Pack, S. , … Lerman, M. I. (1998). In silico‐initiated cloning and molecular characterization of a novel human member of the L1 gene family of neural cell adhesion molecules. Human Genetics, 103(3), 355–364. [DOI] [PubMed] [Google Scholar]
- Ye, H. , Tan, Y. L. J. , Ponniah, S. , Takeda, Y. , Wang, S. Q. , Schachner, M. , … Xiao, Z. C. (2008). Neural recognition molecules CHL1 and NB‐3 regulate apical dendrite orientation in the neocortex via PTP alpha. EMBO Journal, 27(1), 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L. , Zhang, C. , Xu, J. , Ye, X. , Wu, Q. , Dai, J. , … Mao, Y. (2002). A novel splice variant of the cell adhesion molecule contactin 4 (CNTN4) is mainly expressed in human brain. Journal of Human Genetics, 47(9), 497–499. [DOI] [PubMed] [Google Scholar]
- Zuko, A. , Kleijer, K. T. , Oguro‐Ando, A. , Kas, M. J. , van Daalen, E. , van der Zwaag, B. , & Burbach, J. P. H. (2013). Contactins in the neurobiology of autism. European Journal of Pharmacology, 719(1–3), 63–74. https://doi.org/10.1016/j.ejphar.2013.07.016 [DOI] [PubMed] [Google Scholar]
- Zuko, A. , Oguro‐Ando, A. , van Dijk, R. , Gregorio‐Jordan, S. , van der Zwaag, B. , & Burbach, J. P. H. (2016). Developmental role of the cell adhesion molecule Contactin‐6 in the cerebral cortex and hippocampus. Cell Adhesion and Migration, 10(4), 378–392. https://doi.org/10.1080/19336918.2016.1155018 [DOI] [PMC free article] [PubMed] [Google Scholar]


