Abstract
It is widely accepted that obesity and associated metabolic diseases, including type 2 diabetes, are intimately linked to diet. However, the gut microbiota has also become a focus for research at the intersection of diet and metabolic health. Mechanisms that link the gut microbiota with obesity are coming to light through a powerful combination of translation-focused animal models and studies in humans. A body of knowledge is accumulating that points to the gut microbiota as a mediator of dietary impact on the host metabolic status. Efforts are focusing on the establishment of causal relationships in people and the prospect of therapeutic interventions such as personalized nutrition.
Worldwide, obesity has more than doubled since 1980 according to the World Health Organization. In 2014, more than 1.9 billion adults were overweight, and over 600 million of those people were obese. Obesity results from a positive energy balance, which occurs when the amount of energy ingested exceeds the amount expended, and it is a strong risk factor for other metabolic complications such as type 2 diabetes. Type 2 diabetes is increasing in prevalence in low-income countries, and in 2014, approximately 422 million adults worldwide had diabetes. The condition is characterized by high blood sugar, resistance to insulin and a relative lack of insulin. Insulin resistance is also associated with an increased flux of free fatty acids that contribute to diabetic dyslipidaemia, which is characterized by a high concentration of triglycerides in blood plasma, a low concentration of high-density lipoprotein (HDL) cholesterol and an increased concentration of small, dense low-density lipoprotein cholesterol particles1. Dyslipidaemia is one of the major risk factors for cardiovascular disease in people with diabetes. Accordingly, abnormal metabolism of glucose and lipids is the hallmark of metabolic syndrome, which is defined by central (abdominal) obesity and the presence of two or more of four factors — elevated triglycerides, reduced HDL cholesterol, high blood pressure, and increased fasting blood glucose. As governments and health organizations struggle to find solutions to these largely preventable health issues, a rapidly expanding area of research that is focused on the microbes that live within our digestive tract is offering fresh and interesting insights and potential avenues for intervention.
The human gut is a bioreactor with a microbiota that typically encompasses hundreds or thousands of bacterial taxa, which predominantly belong to two phyla: Firmicutes and Bacteroidetes2–5. Tremendous strides have been taken over the past decade towards mapping the composition and basic functional attributes of the gut microbiota of people from industrialized countries3,5. This ensemble of organisms has coevolved with the human host and complements the coding potential of our own genome with 500-fold more genes6. However, the annotation, and consequently the biological function, of many of these remain poorly defined.
The observation that germ-free mice, which lack a microbiota, have reduced adiposity and improved tolerance to glucose and insulin when compared with conventional (colonized) counterparts7 jump-started a decade of research that focused on the clarification of underlying mechanisms. Germ-free mice are protected from diet-induced obesity when fed a Western-style diet8–10, which further supports a link between the gut microbiota and the host metabolism. The altered microbiota that is observed in genetically obese mice11,12 is sufficient to promote increased adiposity in lean mice that receive a microbiota transplant12, demonstrating that the microbiota contributes to the regulation of adiposity. The importance and generalizability of these initial findings are strengthened by reports of alterations in the gut microbiota of obese people4,13–15, which confer the obese or adiposity phenotypes when transferred to mice14,16.
Here, we review the large body of data that is shaping our understanding of how the gut microbiota can alter the absorption, metabolism and storage of calories. Despite broad agreement that gut microbes modify how the human body responds to components of diet to influence metabolism, the mechanisms that underlie this process are exceptionally complex and the data can be difficult to reconcile. The picture that is emerging suggests that obesity is associated with reduced diversity of the gut microbiota13,17. Systemic inflammation and microbial metabolites, such as bile acids and short-chain fatty acids, are also commonly implicated. The ability to easily access and reprogramme the composition and function of the microbiota make it an attractive target for intervention.
Diet as an important modulator of the gut microbiota
Extensive research on the gut microbiota has shown that diet modulates the composition and function of this community of microbes in humans and other mammals18–25, with the earliest literature26 published almost 100 years ago. Human intervention studies from the past decade have revealed the extent to which different aspects of the microbiota can be influenced through dietary change; this can be summarized by three main themes.
The first theme is that the microbiota of the human gut responds rapidly to large changes in diet. The existence of these fast, diet-induced dynamics is supported by evidence from people who switch between plant- and meat-based diets, who add more than 30 grams per day of specific dietary fibres to their diet or who follow either a high-fibre–low-fat diet or a low-fibre–high-fat diet for 10 days; in all cases, the composition and function of the microbiota shifted over 1–2 days18,20,23. Such marked shifts in response to nutrient availability are perhaps unsurprising given that populations of microbes can double within an hour and the gut extensively purges the community every 24–48 hours. This responsiveness might represent an advantageous feature of enlisting microbes as part of the digestive structure — especially when considering the possible day-to-day variation in food that is available to foragers. It might also be an inescapable consequence of dealing with a complex and competitive microbial community that undergoes rapid turnover.
The second theme is that, despite these rapid dynamics, long-term dietary habits are a dominant force in determining the composition of an individual’s gut microbiota. Despite detectable responses of the microbiota within 24 hours of dietary intervention, a 10-day feeding study in 10 people20 failed to alter the major compositional features and the overall classification of each participant’s microbiota. Some, but not all, cross-sectional studies reported that long-term dietary trends are linked to features of microbiota composition20,25,27,28.
The third theme is that a particular change in diet can have a highly variable effect on different people owing to the individualized nature of their gut microbiota. For example, Ruminococcus bromii-related taxa bloomed in response to resistant-starch intervention in most of the 14 obese men in one study; the lack of response in the other individuals might reflect an absence of such taxa in those people23. A dietary intervention that includes a boosted intake of fibre and a decreased intake of energy can increase microbiota diversity — as defined by the gene content of the faecal metagenome — for individuals who start with a low microbiota gene content, but not those who start with a high gene content21. These individualized responses might fit into categories that enable a precision rather than a personalized approach to understanding responsiveness to diet.
The influence of diet on aspects of microbiota function might also help to explain how a specific metabolic input can alter microbiota composition over time. In a study that focused on the enzymatic activity of trimethylamine lyase, mice that harbour microbiotas with low production of trimethylamine (TMA) could be converted into high producers when their diet was supplemented with the TMA-containing compound L-carnitine for 10 weeks27. Similarly, a microbiota-encoded degradation system for porphyran, a polysaccharide that is found in certain species of edible seaweed, is rare in the microbiotas of Western people but prominent in those of populations that regularly consume seaweed29. This suggests that certain metabolic inputs can select for pathways as well as the organisms that harbour those pathways. One corollary of this interpretation is that there must be a reservoir of selectable functions — either present at low levels within the gut microbial community or able to invade from an environmental source. It is important to note that numerous other non-dietary mechanisms, such as interstrain killing that is mediated by the type VI secretion system, infection with bacteriophages and priority effects of colonization through which strains are able to exclude one another on the basis of relatedness of particular genetic loci, can underlie microbial community dynamics and might interact with or operate in parallel to dietary-mediated effects30–33.
Several issues can complicate the unravelling of mechanisms and the interpretion of data in dietary intervention studies in humans. People are notoriously poor at adhering to dietary regimes, and it is difficult to accurately measure the extent of their adherence because the self-assessment of food intake can be clouded by numerous factors. Budget limitations often mean that researchers must choose either tightly controlled studies of small cohorts, for example, in which food is provided, or larger cohort studies that could be confounded by the free will of the participants and by their self-assessment. Because dietary change often involves both the elimination and addition (that is, the substitution) of dietary components, even the most successful intervention studies can raise questions about which diet modification was responsible for the change in the microbiota. A further complication is that many of the dietary changes in such studies also have the potential to directly influence host metabolism in a microbiota-independent way.
As an alternative, animal models enable researchers to tightly control the diet of subjects and to have multiple biological replicates that represent the response of a single microbiota. Experimental models that lack a gut microbiota offer further power for determining whether the effects of diet in the host depend on the microbiota. For example, germ-free rats harvest less energy from a polysaccharide-rich diet34 and germ-free mice have a reduced adiposity despite an increased intake of food by comparison with their colonized counterparts7, which demonstrates that the microbiota helps to extract energy from food. These results are consistent with the fact that the fermentation of dietary fibre represents one of the dominant microbial metabolic activities in the colon, the region of the gut in which the microbiota is most dense35,36.
The short-chain fatty acid end-products of fermentation in the gut can be absorbed into the circulation to serve as both microbiota-generated calories and important regulatory molecules, and it has been estimated that people who consumed a typical British diet in the 1980s received 6–10% of their energy from short-chain fatty acids37. By contrast, people who eat large quantities of plants, the main source of dietary fibre38, such as those in certain African communities that consume up to sevenfold more fibre than people the industrialized world39, might generate considerably more short-chain fatty acids, which therefore probably contribute more to the whole-body energy requirement. This is in agreement with the increased abundance of taxa that ferment polysaccharides in the gut microbiota of African populations40. Certain recurrent physiological states in mammals, such as the non-hibernating period in bears41 and advanced pregnancy42, result in a markedly altered microbiota with an increased capacity to harvest energy from the diet without metabolic derangement. It should also be noted that the effects observed in animal models extend beyond a simple improvement in calorie harvest. The microbiota of mice suppress the expression of intestinal angiopoietin-like protein 4, an inhibitor of the enzyme lipoprotein lipase, which increases lipoprotein-lipase activity in adipose tissue and promotes the storage of fat7. Accordingly, mice that are deficient in Angptl4 have increased adiposity, even under germ-free conditions7.
Experiments that use a Western-style diet, which is devoid of fibres and rich in calories from saturated fat and sucrose, demonstrate that the gut microbiota regulates obesity through additional pathways8. For example, germ-free mice are protected from diet-induced obesity when fed high levels of sucrose and lard8, a diet that alters the composition of the gut microbiota. The presence of the microbiota is both necessary and sufficient for obesity: the transfer of microbiota from mice fed a Western diet to germ-free mice transfers the obese phenotype43. By contrast, germ-free mice that are fed a high-fat diet with less sucrose are only partly protected against obesity44, and all protection from obesity (that is, microbiota-dependent obesity) is lost when sucrose is omitted from the diet45. The molecular mechanisms that underpin this finding are unknown. The source of dietary fat also seems to be important. Saturated and unsaturated fats have profoundly different effects on the gut microbiota, and the altered microbiota that results from feeding unsaturated fats can offer protection from lard-induced weight gain44. These findings suggest that simple carbohydrates and fats could exert unexpected effects on the host metabolism through the microbiota. Further research is required to clarify how microbial taxa and ecosystems interact with specific macronutrients.
Emerging evidence suggests that the deleterious metabolic effects of processed foods might involve more than just macronutrients. Emulsifiers and artificial sweeteners have been shown to be involved in the development of metabolic syndrome features through their modulation of the microbiota in mice46,47. In a study in seven people, artificial sweeteners given at high doses resulted in insulin resistance after only 7 days47; however, this dramatic finding needs to be reproduced in a larger study. These data provide evidence that artificial food additives might contribute to metabolic disease through disruption of the microbiota. Notably, an important and unwavering commonality of Western dietary trends is the paucity of plant-based dietary fibre48, an important fuel for the microbiota. The absence of dietary fibre together with an abundance of nutrients that negatively affect the microbiota could be of considerable importance for understanding metabolic diseases.
Microbial ecology in metabolic disease
The interaction of numerous species, the allocation of resources and the dynamic response to perturbation within the gut provide many of the hallmarks of a complex ecosystem. The application of macroecological concepts to the gut microbiota might therefore be instructive in guiding scientific inquiry and understanding49, particularly when considering the associations between microbiota diversity and metabolic output (such as the link between short-chain fatty acids and obesity and metabolic disease). For example, many macroecology data suggest that the extent of biodiversity within an ecosystem can serve as an important measure of stability and robustness50, which are relevant to research that looks at the link between gut microbes and health.
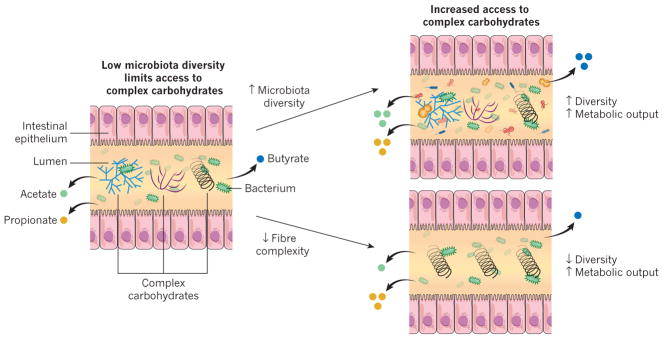
Three metagenomic studies13,21,51 have shown that improved metabolic health is associated with a relatively high microbiota gene content and with an increased microbial diversity. These data indicate that the extent of the diversity might be an important factor for metabolic health, which is consistent with findings from microbiota studies that have focused on traditional human societies. The gut microbiota of eight hunter–gatherer or rural farming populations in various parts of the world showed increased bacterial diversity compared with those of Western populations19,40,52–55. Notably, the microbial taxa that are absent from the Western gut are found in many populations of traditional people that have been separated for thousands of years on different continents. The parsimonious explanation for this is that industrialization has been accompanied by an overall decline in gut microbiota biodiversity as well as the loss of specific phylogenetic groups — a potential consequence of modern lifestyles, medical practices and processed foods. It is unclear whether certain taxa are keystones that promote diversity. It is also unknown whether the increased diversity is only a reflection of a healthy and varied diet or whether it directly contributes to protection from metabolic disease. One theory is that the microbiota of industrialized nations are experiencing a widespread change in functional capacity (for instance, altered production of short-chain fatty acids), which is contributing to modern health issues such as obesity56–58. Dietary reinforcement, and specifically the provision of diverse complex carbohydrates, could provide the key to sustaining, and perhaps recovering, a diverse resident ecosystem that is capable of the functions that the human body expects or requires (Fig. 1). A caveat is that diversity can be measured in many ways that include or exclude the relative abundances of species and the functions encoded within them. It is also important to note that a high level of biodiversity does not always correspond to a health-promoting ecosystem: for example, bacterial vaginosis is characterized by a diversity greater than that observed in a healthy state59. Undoubtedly, an understanding of diversity within the context of organism identity, location and function enriches the utility of measures that fail to capture important details when used alone.
Figure 1. Interactions between the diet and the gut microbiota dictate the production of short-chain fatty acids.
Dietary fibre is a source of complex carbohydrates, which are required for the production of short-chain fatty acids such as acetate, butyrate and propionate. When the diversity of the microbiota is high and the diet contains many types of complex carbohydrates (top right), a relatively high percentage of complex carbohydrates will be accessible to the microbiota. But when the diversity of the microbiota is low and the diet contains many types of complex carbohydrates (left), only a low percentage of these complex carbohydrates are accessible to the microbiota. If the fibre composition of the diet is matched to the needs of a low-diversity microbiota (bottom right) by limiting the types of complex carbohydrate that are available, the levels of production of certain short-chain fatty acids, such as propionate, might increase. However, the diversity of the microbiota will probably remain low and it might not be able to provide as many functions as a diverse microbiota. Consumption of a complex diet (top right) might result in increased levels of production of multiple types of short-chain fatty acids and helps to recruit additional diversity to the gut microbiota. The level of propionate production is correlated with the abundance of Bacteroides species in the gut, which is consistent with the involvement of these bacteria in the production of propionate125. Fermentation of fibre in the colon has been shown to decrease pH levels, which can help to increase the diversity of the gut microbiota or results in the reinforcement by certain taxa of a pH that favours their own growth126–129.
Fuel for the microbial ecosystem
Many of the plant polysaccharides that are found within dietary fibre are structurally complex. It is therefore unsurprising that the numerous enzymes that are required to de-modify, liberate, transport and metabolize component monosaccharides are not encoded within the human genome60. Furthermore, the time that would be required to perform these steps is probably not compatible with the rapid transit that occurs in the small intestine, the region of the gut in which simple carbohydrates are digested and absorbed. Consequently, complex carbohydrates travel to the distal gut for fermentation by its dense community of microbes.
Many complex plant carbohydrates qualify as dietary fibre, according to laboratory tests. However, the amount of fibre that can be metabolized (for example, through the enzymatic degradation of glycosidic linkages and the fermentation of liberated monosaccharides into short-chain fatty acids) will depend on many factors, including the composition of the microbiota. Carbohydrates that can be metabolized by the microbiota are known as microbiota-accessible carbohydrates61 and can be contrasted with those that pass through the digestive tract without undergoing metabolic transformation. This metabolic accessibility is an important distinguishing characteristic: it defines a carbohydrate as a resource that drives the interspecies economy within the gut and it implies that metabolic products, such as short-chain fatty acids, will be generated.
Notably, high diversity in the microbiota corresponds with high levels of short-chain fatty acid production in rural farmers in Burkina Faso19, as well as with the enrichment of genes in the microbiome of hunter–gatherers that are associated with the metabolism of complex carbohydrates62. In a multigenerational study in mice, the consumption of a Western-style diet exacerbated the loss of microbiota diversity compared with a diet that was rich in microbiota-accessible carbohydrates, and the extinction of taxa corresponded with a predicted loss in diversity of glycoside hydrolases63. Several studies in humans indicate that there is a population-specific ‘ceiling’ on microbiota diversity and metabolic output. For example, following a vegan diet for at least 6 months or a high-fibre–low-fat diet for 10 days were insufficient to substantially increase microbiota diversity or production of faecal short-chain fatty acids28. A plant-based diet could significantly alter the composition of the gut microbiota, although a change in diversity was not observed18. When fed high levels of resistant starch, individuals who fail to show a bloom in Ruminococcus bromii and its relatives also have the highest levels of undigested starch in their stool, which supports the idea that the composition of the microbiota determines whether a carbohydrate is accessible to the microbiota23. Overall, these data suggest that the production of short-chain fatty acids is affected by the existing diversity within a microbiota.
Eating whole grains for just 3 days can improve tolerance to glucose in some people, and these ‘responders’ show an increased representation of specific glycoside hydrolases within the gut microbiome compared with non-responders who received the same dietary intervention22. This indicates that the microbiota might need to already have the capacity to degrade certain complex carbohydrates in the diet to reap the potential benefits of microbiota-accessible carbohydrates. Notably, individuals whose microbiota and glucose tolerance respond to a whole-grain intervention tend to consume diets that are higher in fibre. The complex carbohydrates that are associated with whole grains and that were metabolically accessible to the microbiotas of the responders might therefore have been inaccessible to non-responders who also did not routinely consume high-fibre diets.
Microbial metabolites
Microbes that live in the gut continually produce numerous small molecules through primary and secondary metabolic pathways64, many of which are dependent on the diet of the host. Although some of these compounds are retained within the gut ecosystem, others will be absorbed into the circulation and then chemically modified (that is, co-metabolized) by the host, and eventually secreted in the urine65. Much research has focused on short-chain fatty acids, which have been implicated in diverse roles in obesity and metabolic syndrome. Pathways that generate short-chain fatty acids were found to be enriched in metagenomic studies of obesity, and levels of short-chain fatty acids were elevated in overweight or obese people and animal models12,66,67, which is consistent with these products of microbial fermentation providing extra calories to the host. By contrast, increased levels of the short-chain fatty acid propionate promoted intestinal gluconeogen-esis68 or were associated with the microbiota following gastric bypass69, which conferred protection from diet-induced obesity on transfer to germ-free recipient mice. The direct delivery of propionate to the colon through propionate-esterified carbohydrate reduced weight gain in a randomized 24-week study of 60 overweight adults70.
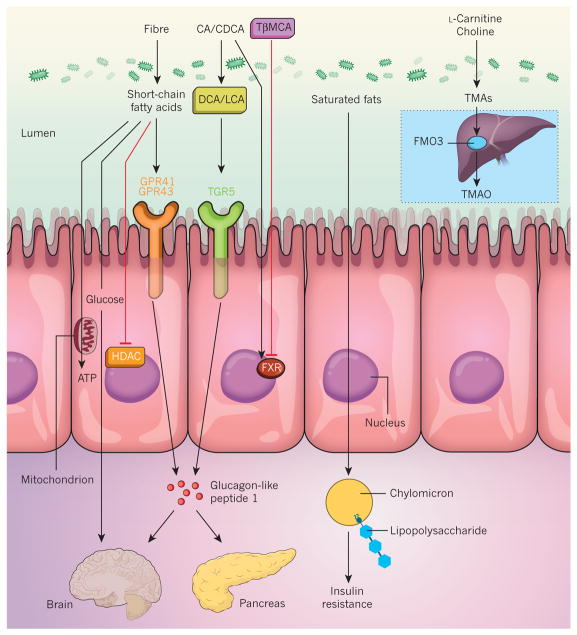
Short-chain fatty acids can signal to the host through at least four distinct pathways (Fig. 2). First, short-chain fatty acids, particularly butyrate, are an energy substrate for colonocytes71,72, and in response to reduced energy availability, germ-free mice slow down the transit through the small intestine to allow more time for nutrient absorption73. Second, propionate is a substrate for gluconeogenesis and can induce intestinal gluconeogenesis, which signals through the central nervous system to protect the host from diet-induced obesity and associated glucose intolerance68. Third, butyrate and acetate, another short-chain fatty acid, can act as histone deacetylase inhibitors74,75. (Acetate acts in peripheral tissues, in which the concentration of butyrate might not be high enough to exert an effect.) Fourth, short-chain fatty acids signal through G-protein-coupled receptors such as GPR41 (also known as FFAR3) and GPR43 (also known as FFAR2), which affects several important processes that include inflammation76 and enteroendocrine regulation77. However, the generation of short-chain fatty acids is only one aspect of microbial metabolism in the gut.
Figure 2. Mechanisms of signalling from the gut microbiota to the host.
The gut microbiota interacts with dietary components and metabolites to form bioactive metabolites that signal to the host through distinct mechanisms. Short-chain fatty acids that are produced by the fermentation of fibre are an important source of energy (ATP) for colonocytes. They are also a substrate for gluconeogenesis, which modulates central metabolism, and are involved in signalling to the host by inhibiting histone deacetylase (HDAC) or by activating G-protein-coupled receptors such as GPR41 and GPR43, which triggers the release of the hormone glucagon-like peptide-1. The primary bile acids cholic acid (CA) and chenodeoxycholic acid (CDCA) are metabolized into the secondary bile acids deoxycholic acid (DCA) and lithocholic acid (LCA), which activates signalling to the host through the G-protein-coupled bile acid receptor 1 (GPBAR1; also known as TGR5). Tauro-β-muricholic acid (TβMCA) is deconjugated into β-muricholic acid (βMCA; not shown), which alleviates the inhibition of the farnesoid X-activated receptor (FXR; also known as the bile acid receptor) by TβMCA. Microbially produced endotoxins (also known as lipopolysaccharides) are taken up into chylomicrons that are formed from dietary saturated fats and subsequently they promote inflammation in the host that induces insulin resistance. L-Carnitine and choline, compounds that are found in red meat, are metabolized into TMAs that are oxidized further into TMAO by the enzyme flavin-containing monooxygenase 3 (FMO3) in the liver (inset).
The microbial metabolism of phosphatidylcholine78, a phospholipid that is abundant in cheese, seafood eggs and meat, and of L-carnitine27, an amino acid that is abundant in red meat, produce high levels of TMA. Once it has been absorbed from the gut into the bloodstream, TMA circulates to the liver and is enzymatically oxidized to TMA N-oxide (TMAO), a compound that has been associated with poor cardiovascular outcomes in humans and the acceleration of atherosclerosis in mice27,78,79 (Fig. 2). TMA production serves as an excellent example of the interaction between the diet and the microbiota. For example, microbiotas that are capable of producing TMA make the metabolite only when compounds that contain trimethyl ammonium are present in the diet, and some microbiotas (such as those of vegans) are poor producers of TMA27, even when precursor compounds are transiently provided through the diet. Together, these data suggest that the microbiota evolves to adapt to specific macronutrients. Many of the experiments that demonstrated the atherogenic nature of TMAO involved supplementing the low-fat diets of animals with the compound. Other metabolites probably contribute to metabolic disease — as supported by evidence from people who have undergone bariatric surgery, a procedure that produces long-term weight loss and improved metabolism and reduces the risk of cardiovascular disease and death80,81 but that is associated with elevated levels of circulating TMAO16. The increased levels of TMAO in such patients might reflect the creation of a more aerobic gut environment that is conducive to generation of this metabolite. It is therefore essential to determine the conditions under which TMAO promotes cardiovascular disease and whether TMAO directly affects cardiovascular disease in humans.
Bile acids, formed by the microbiota from host cholesterol, are another group of metabolites with a profound effect on human health82. They are metabolized by the microbiota in the lower part of the small intestine and the colon to generate secondary bile acids83. They were originally thought only to act as soaps that solubilize dietary fats to promote their absorption, but over the past two decades, it has become clear that they serve as signalling molecules and bind to distinct receptors such as G-protein-coupled bile acid receptor 1 (also known as TGR5) and the bile acid receptor FXR84 (Fig. 2). The microbiota regulates TGR5 signalling by producing agonists85 and FXR signalling by metabolizing antagonists86. TGR5 and FXR both have a major impact on host metabolism84 and, accordingly, an altered microbiota might affect host physiology by modulating the signals that pass through these receptors. The capacity to metabolize tauro-β-muricholic acid, a naturally occurring FXR antagonist86,87, is essential for the microbiota to induce obesity and steatosis, as well as impaired tolerance to glucose and insulin87–89. At least some of this effect is mediated by an altered microbiota89. Bariatric surgery is associated with an altered microbiota and metabolism of bile acids16,90. Mechanistic links between bile acids and bariatric surgery demonstrate that functional FXR signalling is required for a reduction in body weight and an improvement in glucose tolerance after vertical sleeve gastrectomy90. Similarly, TGR5 is required for the improved metabolism of glucose following this procedure. Germ-free mice that received a faecal transplant from people who had undergone Roux-en-Y gastric bypass 10 years earlier gained less fat than did mice that were colonized by microbiota from obese people16. Some of the beneficial effects of bariatric surgery might therefore be mediated by the altered microbial metabolism of bile acids, which affects their capacity for signalling. Other mechanisms and metabolites might have equally important roles.
The microbiota produces a vast number of metabolites and much work remains to be done to investigate fully their functions in physiology and pathophysiology. Examples of such metabolites include: ethylphenyl sulfate, which is connected to the exacerbation of autistic behaviour in a mouse model91; indole propionic acid, which is linked to improved function of the epithelial barrier in the gut92; and indoxyl sulfate and p-cresyl sulfate, both of which are associated with poor cardiovascular outcomes in people with uraemia (p-cresyl sulfate is also associated with insulin resistance)93–95. These metabolites undoubtedly give a glimpse of how this poorly explored universe of molecules can affect the host. The relevance of some of these metabolites in humans is yet to be established. Although several bioactive metabolites are the derivatives of amino acids, neither the effect of the quantity and quality of protein in the diet on metabolite synthesis nor the ensembles of microbial genes that are responsible for metabolite production are well understood.
Inflammation and diet
Obesity and insulin resistance are associated with the increased infiltration of macrophages into and the inflammation of adipose tissue96,97. Because the gut microbiota is known to contribute to the obese phenotype, at least in mice, it might also contribute to increased adipose inflammation. A model of adipose inflammation that is dependent on the microbiota but independent of diet is supported by evidence from Swiss Webster mice. While consuming a standard diet, these animals develop a similar amount of adiposity to C57Bl6 common laboratory mice that are fed a high-fat diet for 8 weeks. When germ-free Swiss Webster and C57Bl6 mice are fed their respective adiposity-inducing diets, both exhibit reduced adiposity, lower levels of endotoxins (known as lipopolysaccharides) in the circulation and decreased macrophage infiltration into white adipose tissue, as well as improved metabolism of glucose44,98. Obesity in mice is also associated with increased numbers of T cells99,100 and mast cells101 and reduced numbers of regulatory T cells102. In mouse models, the fermentation of fibre and the generation of short-chain fatty acids seem to promote anti-inflammatory responses both within the gut and systemically through regulatory T cells103–106. Although dietary fibre and the production of short-chain fatty acids exert a positive metabolic impact through non-immunological mechanisms in a mouse model of diet-induced obesity68, it is unclear whether similar interactions that are mediated through the immune compartment contribute to metabolic changes. The supplementation of high-fat diets with fermentable fibres protects mice from obesity and associated diseases107 but the mechanism that underlies this action remains unclear.
The gut microbiota also interacts with the innate immune system to induce adipose inflammation, and mice that lack Toll-like receptor signalling, through loss of either of the adaptor proteins MyD88 or TRIF (also known as TICAM1), have reduced levels of inflammation in adipose tissue and are protected from insulin resistance that is induced by saturated fatty acids44. Mice that are deficient in the gene Myd88, but not the gene Trif, are protected from diet-induced obesity, which therefore separates obesity from insulin resistance and suggests that they are controlled by different mechanisms. Mice raised in conventional conditions that are fed saturated fatty acids exhibit increased levels of endotoxins in the circulation in comparison to mice that consume polyunsaturated fatty acids. Dietary fat has been demonstrated to increase the amount of endotoxins in the blood plasma of both mice108 and humans109, probably by allowing endotoxins to be transported across the epithelium on chylomicrons110. These higher levels of endotoxins activate Toll-like receptors in adipose tissue that, in turn, induce the expression of the chemokine CCL2, which is required for macrophage infiltration44. The source of dietary fat might therefore have specific interactions with the microbiota that lead to altered interactions with the innate immune system and contribute to metabolic diseases. Mice fed a diet supplemented with fish oil are protected from obesity and insulin resistance. Furthermore, mice that consume lard and receive the microbiota of those fed fish oil are protected against obesity44, which demonstrates that the modified microbiotas themselves have a protective effect.
A switch to a diet rich in saturated fatty acids shifts the composition of the microbiota44. Levels of the bacterium Bilophila wadsworthia increase when mice are fed a diet rich in milk fat or supplemented with the bile acid taurocholic acid111. Similarly, increased levels of Bilophila and a reduced abundance of Desulfovibrio were observed in mice that were fed lard compared with fish oil as a source of fat44. B. wadsworthia increases gut inflammation in mice that lack in the anti-inflammatory cytokine interleukin (IL)-10. Insulin resistance that is induced through a high-fat diet is associated with reduced levels of T helper 17 (TH17) cells that are positive for IL-17 and retinoic acid receptor-related orphan receptor γt (RORγt)112. It is tempting to speculate that one of the underlying mechanisms involves the fat-induced restriction of a specific taxon known as the segmented filamentous bacteria, which induce the expression of IL-23 in enterocytes. IL-23 causes the release of IL-22 from innate lymphoid cells in the ileum, which subsequently induces the production of the proteins serum amyloid A1 and serum amyloid A2 from the epithelium in a paracrine fashion — a process that is required for the activation of TH17 cells in the ileum113. In mice, IL-22 has been shown to protect against metabolic disease, which further suggests a link between the altered gut microbiota, TH17 cells and IL-22 signalling and the mediation of metabolic disease. However, it is unknown whether taxa that induce specific immune responses, such as the segmented filamentous bacteria in mice, protect against metabolic disease. Despite efforts, there are no reports on the role of segmented filamentous bacteria in people, but other bacteria in the human microbiota might have developed similar functions.
Dietary interventions and diet-based therapeutics
The gut microbiota provides a powerful route to influencing human health. It has many attributes with biomedical potential, such as a connection to multiple facets of human biology, malleability and accessibility for therapeutic targeting or diagnostics. The microbes of the gut can therefore be likened to an easily accessible control centre for the modulation of human physiology. However, owing to the complexity and individuality of each microbiota, the rate at which this potential can be realized is unknown.
Diet and, in particular, polysaccharides serve as primary modulators of the composition and function of the microbiota. Polysaccharides, which are widely consumed components of human food, are therefore functionally analogous to small-molecule drugs. Because of their relative safety (that is, their lack of acute toxicity), availability and low cost, it might be feasible systematically and empirically to determine which dietary polysaccharides, alone or in combinations, can improve human health in different situations.
Such an empirical approach is compatible with emerging concepts in precision health22,114. Although the dietary interventions affect the metabolic responses of hosts in an individualized manner, elements of the microbiome can help to predict the response. One study used continuous blood-glucose monitoring to follow postprandial glycaemic responses in 800 people114. The responses of individuals to particular foods were highly variable. However, when compared with microbiome profiles and with measurements of metabolism and behaviour, using a machine-learning approach, the response of an individual to a given food could be predicted — even in an independent cohort. Similarly, individuals show large differences in glucose metabolism in response to an intervention that is based on whole grains22. Improved tolerance to glucose could be explained largely through enrichment of the genus Prevotella within the microbiota. Prevotella could also improve the glucose metabolism of mice that were fed carbohydrate-rich diets but not a high-fat diet that was devoid of fermentable polysaccharides. These findings point to the possibility of a mechanism-free, empirical approach for determining a dietary intervention that is appropriate for a given individual or group. They also highlight the potential of a next generation of probiotics (sets of microbiota-derived living microbes that will be tailored to interact with a given diet) as a method for converting non-responders into responders. A further outcome of this approach might be the use of predictive elements of metadata to guide the generation of hypotheses and to determine priorities for investigation into underlying mechanisms.
Perspective
It is becoming clear that an altered gut microbiota is associated with metabolic diseases in humans that range from obesity to type 2 diabetes and cardiovascular disease. Causality has also been demonstrated in animal models. To move forwards, it will be essential to understand whether the gut microbiota is causally linked to host metabolism in humans. Prospective studies should be performed to determine whether the gut microbiota is altered before or after the onset of disease. This will require large cohorts that allow considerable numbers of participants to develop the disease under investigation, and it will probably involve the high-resolution monitoring of host and microbial parameters to determine the progression of derangements.
Another approach is to transfer microbiotas from humans to mice, and this is particularly powerful when focused on twin cohorts to control for human genetics14,15,51. In one-such study, transplantation of the microbiota from obese individuals to germ-free mice transfers the obese phenotype, as determined by increased weight gain, whereas administration of Christensenella minuta prevents weight gain15. In a separate study, bacterial representatives from the microbiota of lean individuals were associated with an increased production of short-chain fatty acids, whereas the microbiota of obese individuals had an increased abundance of genes that are involved in biosynthesis of branched-chain amino acids, which are associated with impaired sensitivity to insulin14. Importantly, the lean microbiota could only invade and prevent increased adiposity when the recipient mice consumed a diet that was low in fat and high in fruits and vegetables. Consistent with the idea that the microbiota reinforces the diet, supplementation with Prevotella produces an improved tolerance to glucose only when mice are fed a standard diet that is rich in fibre, and not a Western-style diet, which is devoid of fibre22.
A similar dependency on diet was observed in children with a type of malnutrition known as kwashiorkor115. Twins that are discordant for kwashiorkor have distinct microbiotas, and germ-free mice that have been colonized with a ‘kwashiorkor’ microbiota experience weight loss when they are fed a typical Malawian diet, which is based on tomatoes and corn. However, when the mice are fed a peanut-based, ‘ready-to-use’ therapeutic food, their weight transiently increases and their microbiota normalize115. It is becoming increasingly important to consider how the diet can modify microbiota-linked disease states in mice to generate hypotheses about underlying molecular mechanisms that can then be tested and validated in people. Faecal microbiota transplantation, which has been shown to cure recurrent infection with Clostridium difficile116, has also been used to directly address whether the gut microbiota can affect the metabolism of the host. Eighteen insulin-resistant obese men were randomly designated to receive either an autologous (control) faecal microbiota transplant or a similar transplant from a lean, insulin-sensitive donor. Insulin clamps that were performed before and after the intervention revealed that the insulin sensitivity of a subset of the participants had significantly improved 6 weeks after the transplant117. It is unclear whether the positive responses of these individuals are dependent on characteristics of the donors or the recipients as well as what the duration of the responses should be. Research in larger cohorts is required to verify the effects of faecal microbiota transplantion and to answer remaining questions. For example, experiments could be performed using specific bacteria from lean microbiotas with the aim of developing next-generation probiotics. It is clear that stratification might be required to identify groups that are likely to respond to such interventions22,114.
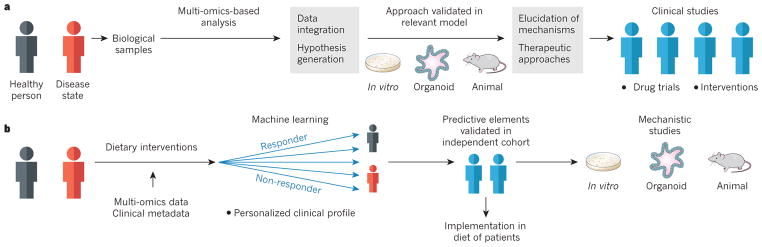
To improve the understanding of how the microbiota affects the metabolism in humans, metagenomics, transcriptomics, proteomics and metabolomics data from key target tissues and the microbiota during various disease states and interventions should be combined to provide a map of co-occurrences. These data enable the formation of testable hypotheses that can be pursued in validated animal models, and they will form the foundation for precision interventions (Fig. 3).
Figure 3. Strategies for modulating the gut microbiota to improve human health.
a, The collection and comparison of multi-omics data from healthy people and those who are affected by metabolic disorders will implicate various genes, pathways and molecules as potential targets for intervention. Relevant experimental models (in vitro, organoid or animal models) are then used to elucidate underlying mechanisms and to pilot therapeutic approaches to modulating the gut microbiota, which lay the foundations for intervention studies or drug trials in humans. b, Studies in humans can also be a starting point for the identification of strategies to modulate the gut microbiota through components of the diet, which are generally considered to be ‘safe’ interventions. Data-processing algorithms, such as machine learning, can be used to identify aspects of the clinical profile of individuals (including data on the microbiota) that help to predict the response of others to dietary interventions. After validation of these predictive elements in independent cohorts, the best intervention can be determined and then implemented to improve human health. Such predictive elements can also be used to guide mechanistic studies in experimental models.
It will also be important to gain a more nuanced understanding of the foundational principles of the microbiota, such as the cross-sectional or longitudinal spatial organization of interactions between the host and its microbes in the intestine118. The majority of studies in humans and mice rely on faecal samples, which provide some representation of what is occurring throughout the digestive tract; however, aspects of microbial communities and host responses that are specific to the small intestine might be obscured by faecal sampling119,120. For example, it could miss information on how the microbiota affects nutrient absorption in the small intestine through its impact on glucose transporters and bile acids, which are essential for the absorption of lipids and fat-soluble vitamins.
Microbial metabolites probably act as mediators for the host metabolism and can be either beneficial (for example, butyrate) or detrimental (TMAO). Such molecules might therefore provide fresh therapeutic approaches in which beneficial metabolites could be supplemented pharmacologically or the bacteria that produce them are developed into probiotics. And receptor antagonists could be developed from detrimental metabolites if the relevant receptor has been identified. Another possibility is to target the microbial enzymes that produce metabolites with inhibitors. An inhibitor of TMA lyase that stops the microbial synthesis of TMA and therefore reduces the levels of circulating TMAO prevents the development of atherosclerosis in mice121. However, such inhibitors are yet to be tested in humans, and it is unlikely that one metabolite acts alone to promote or prevent metabolic diseases. Strategies that promote or prevent suites of metabolites are more likely to have wider applicability and larger effects on host metabolism.
It is reasonable to consider what proportion of metabolic problems in humans could be addressed by properly caring for the gut microbiota. The use of antibiotics in early life is associated with obesity in both people and mice, which suggests that the disruption of microbial ecosystems at crucial points in time might affect physiology in later life and also that the amendment of medical practices could have a substantial impact66,122. However, changes in the diet might be more important for reaping the health benefits that the microbiota can provide. Increased levels of polysaccharides are likely to be of benefit to people who follow a typical Western-style diet, most of whom consume far below the recommended amounts of dietary fibre48; meta-analyses show that the increased consumption of fibre significantly decreases the risk of mortality123,124. Controlled dietary interventions that document the utility of various supplements, probiotics, nutrients and foods in modulating aspects of the gut microbiota and human health are required. The measurement of multiple aspects of individuality, including the microbiota, will provide insight into the characteristics of people who respond beneficially to a given intervention and will pave the way for microbiota-focused precision nutrition. A deeper understanding of the gut microbiota, an important aspect of failing health, has the potential to contribute big gains in our understanding of metabolic health and weight loss.
Acknowledgments
The authors thank members of the Sonnenburg and Bäckhed laboratories for discussions. This work was funded by a grant from the US National Institute of Diabetes and Digestive and Kidney Diseases NIDDK (R01-DK085025 to J.L.S.) and grants from the Swedish Research Council and the Novo Nordisk Foundation to F.B. F.B. is a recipient of a European Research Council Consolidator Grant (615362-METABASE).
Footnotes
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this paper at go.nature.com/28j4ikq.
References
- 1.Mooradian AD. Dyslipidemia in type 2 diabetes mellitus. Nature Clin Pract Endocrinol Metab. 2009;5:150–159. doi: 10.1038/ncpendmet1066. [DOI] [PubMed] [Google Scholar]
- 2.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 5.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, et al. An integrated catalog of reference genes in the human gut microbiome. Nature Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 7.Bäckhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabot S, et al. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010;24:4948–4959. doi: 10.1096/fj.10-164921. [DOI] [PubMed] [Google Scholar]
- 10.Ding S, et al. High-fat diet: bacteria interactions promote intestinal inflammation which precedes and correlates with obesity and insulin resistance in mouse. PLoS ONE. 2010;5:e12191. doi: 10.1371/journal.pone.0012191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. The first study to show that the microbiota from an obese mouse could confer increased weight gain to a germ-free recipient mouse. [DOI] [PubMed] [Google Scholar]
- 13.Le Chatelier E, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 14.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. This study showed that a microbiota from a lean individual could invade the microbiota of an obese individual and provide protection from weight gain, but that the invasion and protection was dependent on diet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tremaroli V, et al. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab. 2015;22:228–238. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. The first of several studies to show that the gut microbiota of a traditional rural population is more diverse than and contains distinct taxa in comparison to the microbiotas of Western populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cotillard A, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 22.Kovatcheva-Datchary P, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22:971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Walker AW, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muegge BD, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torrey JC. The regulation of the intestinal flora of dogs through diet. J Med Res. 1919;39:415–447. [PMC free article] [PubMed] [Google Scholar]
- 27.Koeth RA, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu GD, et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut. 2016;65:63–72. doi: 10.1136/gutjnl-2014-308209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hehemann JH, et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 30.Wexler AG, et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci USA. 2016;113:3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci USA. 2016;113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reyes A, Wu M, McNulty NP, Rohwer FL, Gordon JI. Gnotobiotic mouse model of phage–bacterial host dynamics in the human gut. Proc Natl Acad Sci USA. 2013;110:20236–20241. doi: 10.1073/pnas.1319470110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wostmann BS, Larkin C, Moriarty A, Bruckner-Kardoss E. Dietary intake, energy metabolism, and excretory losses of adult male germfree Wistar rats. Lab Anim Sci. 1983;33:46–50. [PubMed] [Google Scholar]
- 35.Lozupone CA, et al. The convergence of carbohydrate active gene repertoires in human gut microbes. Proc Natl Acad Sci USA. 2008;105:15076–15081. doi: 10.1073/pnas.0807339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nature Rev Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 37.McNeil NI. The contribution of the large intestine to energy supplies in man. Am J Clin Nutr. 1984;39:338–342. doi: 10.1093/ajcn/39.2.338. [DOI] [PubMed] [Google Scholar]
- 38.Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 39.Bingham S, Cummings JH. In: Medical Aspects of Dietary Fiber. Spiller GA, Kay RM, editors. Plenum; 1980. pp. 261–2884. [Google Scholar]
- 40.Schnorr SL, et al. Gut microbiome of the Hadza hunter–gatherers. Nature Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sommer F, et al. The gut microbiota modulates energy metabolism in the hibernating brown bear Ursus arctos. Cell Rep. 2016;14:1655–1661. doi: 10.1016/j.celrep.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 42.Koren O, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Backhed F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 2015;22:658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fleissner CK, et al. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br J Nutr. 2010;104:919–929. doi: 10.1017/S0007114510001303. [DOI] [PubMed] [Google Scholar]
- 46.Chassaing B, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suez J, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514:181–186. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- 48.McGill CR, Fulgoni VL, III, Devareddy L. Ten-year trends in fiber and whole grain intakes and food sources for the United States population: National Health and Nutrition Examination Survey 2001–2010. Nutrients. 2015;7:1119–1130. doi: 10.3390/nu7021119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cardinale BJ, et al. Biodiversity loss and its impact on humanity. Nature. 2012;486:59–67. doi: 10.1038/nature11148. [DOI] [PubMed] [Google Scholar]
- 51.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Obregon-Tito AJ, et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nature Commun. 2015;6:6505. doi: 10.1038/ncomms7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martínez I, et al. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep. 2015;11:527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 55.Clemente JC, et al. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1:e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forslund K, et al. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013;23:1163–1169. doi: 10.1101/gr.155465.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karlsson FH, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 58.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 59.Srinivasan S, et al. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS ONE. 2012;7:e37818. doi: 10.1371/journal.pone.0037818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martens EC, Kelly AG, Tauzin AS, Brumer H. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol. 2014;426:3851–3865. doi: 10.1016/j.jmb.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 2014;20:779–786. doi: 10.1016/j.cmet.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rampelli S, et al. Metagenome sequencing of the Hadza hunter-gatherer gut microbiota. Curr Biol. 2015;25:1682–1693. doi: 10.1016/j.cub.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 63.Sonnenburg ED, et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donia MS, Fischbach MA. Small molecules from the human microbiota. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meyer TW, Hostetter TH. Uremic solutes from colon microbes. Kidney Int. 2012;81:949–954. doi: 10.1038/ki.2011.504. [DOI] [PubMed] [Google Scholar]
- 66.Cho I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. This study demonstrated that the use of antibiotics in early life might cause metabolic disease in later life. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwiertz A, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 68.De Vadder F, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 69.Liou AP, et al. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med. 2013;5:178ra41. doi: 10.1126/scitranslmed.3005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chambers ES, et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut. 2015;64:1744–1754. doi: 10.1136/gutjnl-2014-307913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Donohoe DR, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Donohoe DR, Wali A, Brylawski BP, Bultman SJ. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PLoS ONE. 2012;7:e46589. doi: 10.1371/journal.pone.0046589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wichmann A, et al. Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe. 2013;14:582–590. doi: 10.1016/j.chom.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 74.Thorburn AN, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nature Commun. 2015;6:7320. doi: 10.1038/ncomms8320. [DOI] [PubMed] [Google Scholar]
- 75.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 76.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Samuel BS, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang WH, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sjöström L, et al. Effects of bariatric surgery on mortality in Swedish obese subjects. N Engl J Med. 2007;357:741–752. doi: 10.1056/NEJMoa066254. [DOI] [PubMed] [Google Scholar]
- 81.Sjöström L, et al. Bariatric surgery and long-term cardiovascular events. J Am Med Assoc. 2012;307:56–65. doi: 10.1001/jama.2011.1914. [DOI] [PubMed] [Google Scholar]
- 82.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 83.Midtvedt T. Microbial bile acid transformation. Am J Clin Nutr. 1974;27:1341–1347. doi: 10.1093/ajcn/27.11.1341. [DOI] [PubMed] [Google Scholar]
- 84.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nature Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 85.Kawamata Y, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 86.Sayin SI, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR Antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 87.Li F, et al. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nature Commun. 2013;4:2384. doi: 10.1038/ncomms3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiang C, et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest. 2015;125:386–402. doi: 10.1172/JCI76738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parsfus A, et al. Microbiota-induced obesity requires farnesoid X receptor. Gut. 2016 doi: 10.1136/gutjnl-2015-310283. http://dx.doi.org/10.1136/gutjnl-2015-310283. [DOI] [PMC free article] [PubMed]
- 90.Ryan KK, et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014;509:183–188. doi: 10.1038/nature13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Venkatesh M, et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meijers BK, et al. p-Cresol and cardiovascular risk in mild-to-moderate kidney disease. Clin J Am Soc Nephrol. 2010;5:1182–1189. doi: 10.2215/CJN.07971109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koppe L, et al. p-Cresyl sulfate promotes insulin resistance associated with CKD. J Am Soc Nephrol. 2013;24:88–99. doi: 10.1681/ASN.2012050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barreto FC, et al. Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin J Am Soc Nephrol. 2009;4:1551–1558. doi: 10.2215/CJN.03980609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weisberg SP, et al. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu H, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Caesar R, et al. Gut-derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut. 2012;61:1701–1707. doi: 10.1136/gutjnl-2011-301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Winer S, et al. Normalization of obesity-associated insulin resistance through immunotherapy. Nature Med. 2009;15:921–929. doi: 10.1038/nm.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nishimura S, et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nature Med. 2009;15:914–920. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- 101.Liu J, et al. Genetic deficiency and pharmacological stabilization of mast cells reduce diet-induced obesity and diabetes in mice. Nature Med. 2009;15:940–945. doi: 10.1038/nm.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Feuerer M, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nature Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 105.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nature Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 106.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cani PD, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 108.Cani PD, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. The first study to demonstrate that the presence of endotoxin is sufficient to alter glucose metabolism in mice. [DOI] [PubMed] [Google Scholar]
- 109.Erridge C, Attina T, Spickett CM, Webb DJ. A high-fat meal induces low-grade endotoxemia: evidence of a novel mechanism of postprandial inflammation. Am J Clin Nutr. 2007;86:1286–1292. doi: 10.1093/ajcn/86.5.1286. [DOI] [PubMed] [Google Scholar]
- 110.Ghoshal S, Witta J, Zhong J, de Villiers W, Eckhardt E. Chylomicrons promote intestinal absorption of lipopolysaccharides. J Lipid Res. 2009;50:90–97. doi: 10.1194/jlr.M800156-JLR200. [DOI] [PubMed] [Google Scholar]
- 111.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Garidou L, et al. The gut microbiota regulates intestinal CD4 T cells expressing RORγt and controls metabolic disease. Cell Metab. 2015;22:100–112. doi: 10.1016/j.cmet.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 113.Sano T, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell. 2015;163:381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zeevi D, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. This study used a machine-learning approach to mine personal health profiles that included microbiome data to predict the postprandial glycaemic response. [DOI] [PubMed] [Google Scholar]
- 115.Smith MI, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van Nood E, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 117.Vrieze A, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–916. doi: 10.1053/j.gastro.2012.06.031. This study demonstrated that sensitivity to insulin could be changed by directly altering the gut microbiota through faecal microbiota transplantation. [DOI] [PubMed] [Google Scholar]
- 118.Earle KA, et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe. 2015;18:478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lichtman JS, et al. The effect of microbial colonization on the host proteome varies by gastrointestinal location. ISME J. 2016;10:1170–1181. doi: 10.1038/ismej.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang Z, et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163:1585–1595. doi: 10.1016/j.cell.2015.11.055. The first example of inhibiting microbial enzymes (or ‘drugging the bug’) to prevent atherosclerosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ajslev TA, Andersen CS, Gamborg M, Sorensen TI, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes. 2011;35:522–529. doi: 10.1038/ijo.2011.27. [DOI] [PubMed] [Google Scholar]
- 123.Kim Y, Je Y. Dietary fiber intake and total mortality: a meta-analysis of prospective cohort studies. Am J Epidemiol. 2014;180:565–573. doi: 10.1093/aje/kwu174. [DOI] [PubMed] [Google Scholar]
- 124.Yang Y, Zhao LG, Wu QJ, Ma X, Xiang YB. Association between dietary fiber and lower risk of all-cause mortality: a meta-analysis of cohort studies. Am J Epidemiol. 2015;181:83–91. doi: 10.1093/aje/kwu257. [DOI] [PubMed] [Google Scholar]
- 125.Salonen A, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014;8:2218–2230. doi: 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microbiol. 2005;71:3692–3700. doi: 10.1128/AEM.71.7.3692-3700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chung WS, et al. Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC Biol. 2016;14:3. doi: 10.1186/s12915-015-0224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bown RL, Gibson JA, Sladen GE, Hicks B, Dawson AM. Effects of lactulose and other laxatives on ileal and colonic pH as measured by a radiotelemetry device. Gut. 1974;15:999–1004. doi: 10.1136/gut.15.12.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kettle H, Louis P, Holtrop G, Duncan SH, Flint HJ. Modelling the emergent dynamics and major metabolites of the human colonic microbiota. Environ Microbiol. 2015;17:1615–1630. doi: 10.1111/1462-2920.12599. [DOI] [PubMed] [Google Scholar]