Abstract
Background
Based on published data, we aimed to quantitatively elucidate the possible genetic influence of rs17561 G/T and rs1800587 C/T polymorphisms of the IL1A (interleukin 1 alpha) gene in the susceptibility to several autoimmune diseases.
Methods
A series of meta-analyses were carried out. After database searching, we utilized our inclusion/exclusion criteria to screen and include the eligible studies. Passociation (P value of association test), Bonferroni-corrected Passociation value; false discovery rate (FDR)-corrected Passociation, ORs (odd ratios), and 95% CI (confidence interval) were generated to assess the magnitudes of genetic relationships.
Results
A total of 35 eligible articles were included. Pooled analysis data of both rs17561 G/T and rs1800587 C/T in the overall population indicated a negative association between cases of autoimmune diseases and negative controls (all Passociation>0.05, Bonferroni-corrected Passociation>0.05, FDR-corrected Passociation>0.05). Similar results were found in most subgroup analyses (all Passociation>0.05, Bonferroni-corrected Passociation>0.05, FDR-corrected Passociation>0.05), apart from the rs1800587 in the Graves’ disease subgroup, which showed an increased risk in some cases, compared with controls, under the models of allele T vs. C, carrier T vs. C, CT+TT vs. CC, and CT vs. CC (all Passociation<0.05, Bonferroni-corrected Passociation<0.05, FDR-corrected Passociation>0.05, OR>1).
Conclusion
Based on the available data, C/T genotype of the rs1800587 polymorphism within IL1A gene may be associated with an increased Graves’ disease risk. We did not see evidence regarding a positive role for rs1800587 or rs17561 in the risk of other autoimmune diseases, such as systemic sclerosis or rheumatoid arthritis. These conclusions still merit further data support and molecular exploration.
Introduction
Human autoimmune diseases are a group of pathologies that cause clinical damage or destruction of body tissue due to an immune response to its own antigens [1, 2]. There are many types of autoimmune diseases, such as SSC (systemic sclerosis), JIA (juvenile idiopathic arthritis), BD (Behcet’s disease), RA (rheumatoid arthritis), MS (multiple sclerosis), GD (Graves’ disease), SLE (systemic lupus erythematosus), and TID (type 1 diabetes) [1, 2]. A few cytokine genes have been reported to be linked to the autoimmune disease [2–4].
Interleukin 1 (IL1), including interleukin 1 alpha (α), beta (β) and receptor antagonist (ra), is a family of cytokines implicated in regulation of the inflammatory response and the incidence of clinical immune disease [5, 6]. The human interleukin 1 alpha (IL1A) gene, located on chromosome 2q13 [7], contains some common single nucleotide polymorphisms (SNPs), including rs1800587 (NM_000575.4:c.-949C>T)and rs17561 (NM_000575.4:c.340G>T), which have been reported to be linked to several autoimmune diseases in some populations [8–11]. However, negative conclusions have also been obtained by some studies [12–15].
Several meta-analyses have reported an association between IL1A rs17561, rs1800587 polymorphisms and the presence of various autoimmune diseases, including systemic lupus erythematosus [16, 17], rheumatoid arthritis [18], multiple sclerosis [19] and Graves’ disease [20]. However, the genetic relationship between IL1A SNPs and the risk of other autoimmune diseases, including systemic sclerosis and type 1 diabetes, has not been reported. In the present study, we probed the genetic role of IL1A gene SNPs rs17561 and rs1800587 in the risk of autoimmune diseases using quantitative synthesis of overall meta-analysis followed by subgroup analyses.
Methods
The meta-analysis was conducted per the PRISMA (preferred reporting items for systematic reviews and meta-analyses) guidelines [21]. S1 File illustrates the meta-analysis on genetic association studies checklist, and S2 File shows the PRISMA 2009 checklist.
Database searching
We obtained potentially suitable articles by systematically searching three databases (up to April 2018): PubMed, WOS (Web of Science), and Embase (Excerpta Medica Database). The search terms were shown in S3 File.
Article screening
The following screening items were used to exclude publications: duplicates, reviews, letters, meta-analysis, abstracts or posters, and studies with unrelated data. Each study should have investigated an association between IL1A gene polymorphisms and autoimmune disease risk. The genotype frequency data could be extracted from both case and control groups. We also performed a chi-square-based Q-test to confirm that the genotype distribution of control group was consistent with HWE (Hardy-Weinberg Equilibrium).
Data extraction
Detailed data, including the first author name, publication year, SNP, disease type, genotype frequency, genotyping assay, and ethnicity, were extracted and summarized independently. Conflicting data were discussed with all authors, and missing data were requested by e-mail. We also used the Newcastle-Ottawa Scale (NOS) system to assess the study quality and generate an NOS score. An NOS score < 5 means the study was poor quality, and such studies were excluded.
Statistical association analysis
Stata/SE 12.0 software (StataCorp, USA) was used. To evaluate the strength of genetic relationships, Passociation, pooled ORs (odd ratios), and 95% CI (confidence interval) were generated referring to relevant publications [22–26]. The Passociation value was then adjusted by the Bonferroni and false discovery rate (FDR) correction method [27], using R software version 3.4.3. Bonferroni and FDR-corrected Passociation <0.05 from the association test was considered statistically significant. Six comparison models were utilized: allele T vs. G for rs17561, allele T vs. C for rs1800587 (allele); carrier T vs. G for rs17561, carrier T vs. C for rs1800587 (carrier); TT vs. GG for rs17561, TT vs. CC for rs1800587 (homozygote); GT vs. GG for rs17561, CT vs. CC for rs1800587 (heterozygote); GT+TT vs. GG for rs17561, CT+TT vs.CC for rs1800587 (dominant); TT vs. GG+GT for rs17561, and TT vs. CC+CT for rs1800587 (recessive). We also performed the subgroup analyses according to the characteristics of ethnicity, disease type, and control source.
Q statistics with Pheterogeneity (P value of heterogeneity) and I2 tests with I2 values were conducted to assess heterogeneity among the studies. When Pheterogeneity was >0.05 and the I2 value was <50%, the absence of high heterogeneity was inferred, and a fixed-effects model (Mantel-Haenszel method) was applied. Otherwise, a random-effects model (DerSimonian and Laird method) was utilized.
Sensitivity analysis and bias evaluation
We performed sensitivity analysis to test whether the pooled results were stable. In sensitivity analysis, the effect of each study on the pooled ORs was evaluated as each included study was excluded one-by-one. We also performed Begg’s test and Egger’s test to evaluate publication bias. P values of Begg’s test and Egger’s test, namely PBegg and PEgger, below 0.05 indicate the absence of publication bias.
Results
Study characteristics
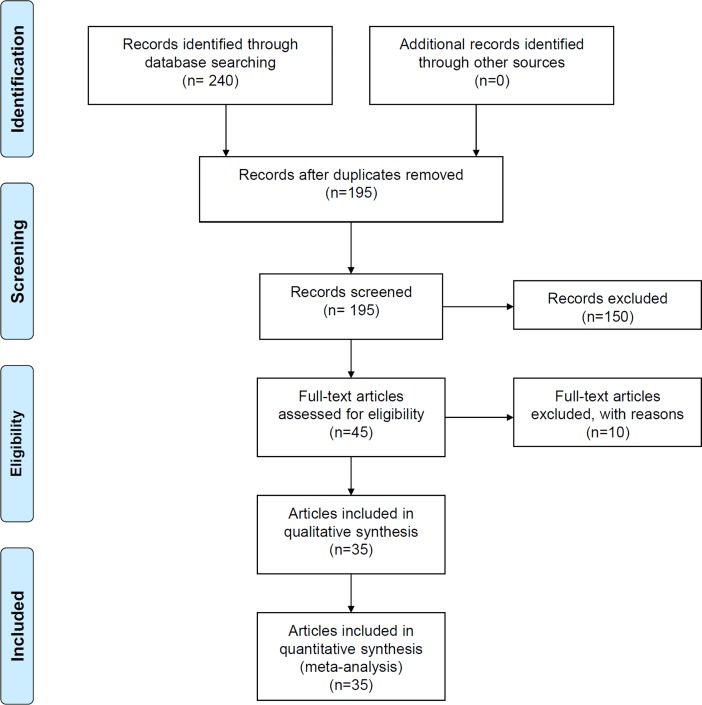
As shown in Fig 1, we searched three databases, identified a total of 240 articles [PubMed (n = 53), WOS (n = 81), Embase (n = 106)], and subsequently removed 45 duplicate articles. Then, 150 articles were excluded by our screening criteria. Assessing the eligibility of the remaining 45 articles, ten articles were removed, because seven did not contain complete genotype data and three were not consistent with HWE. Eventually, a total of 35 articles [8–15, 20, 28–53] were included, and none exhibited poor quality (all NOS score > 5). We list the characteristics of these studies in Table 1.
Fig 1. Flow diagram of database searching and article screening.
Table 1. Characteristics of eligible studies in meta-analysis.
| First author, year | SNP | Disease | case | control | Source | Assay | NOS | Ethnicity | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | AA | AB | BB | |||||||
| Abtahi, 2015 | rs1800587 | SSc | 82 | 72 | 16 | 98 | 98 | 21 | PB | PCR-SSP | seven | Asian |
| Aggarwal, 2012 | rs1800587 | JIA | 42 | 47 | 5 | 93 | 78 | 14 | PB | PCR-RFLP | seven | Asian |
| Akman, 2008 | rs1800587 | BD | 32 | 17 | 4 | 19 | 22 | 7 | PB | PCR-SSP Tray/Minitray and String Kits | seven | Caucasian |
| Beretta, 2007 | rs1800587 | SSc | 117 | 70 | 17 | 112 | 76 | 16 | PB | PCR-SSP | eight | Caucasian |
| Crilly, 2000 | rs1800587 | RA | 45 | 47 | 7 | 33 | 22 | 5 | PB | PCR-RFLP | six | Caucasian |
| Dominguez, 2017 | rs1800587 | RA | 53 | 22 | 5 | 36 | 39 | 5 | PB | PCR | eight | Caucasian |
| rs17561 | RA | 55 | 21 | 4 | 47 | 29 | 4 | PB | PCR | eight | Caucasian | |
| Donn, 2001 | rs1800587 | JIA | 183 | 125 | 22 | 105 | 113 | 18 | PB | PCR-RFLP | six | Caucasian |
| Ferri, 2000 | rs1800587 | MS | 189 | 177 | 33 | 198 | 203 | 38 | PB | PCR-RFLP | eight | Caucasian |
| Genevay, 2002 | rs17561 | RA | 105 | 101 | 24 | 76 | 60 | 8 | PB | PCR | six | Caucasian |
| Harrison, 2008 | rs1800587 | RA | 355 | 321 | 63 | 286 | 269 | 49 | PB | PCR | eight | Caucasian |
| Havemose, 2007 | rs1800587 | JIA | 5 | 3 | 2 | 14 | 10 | 1 | PB | PCR-RFLP | seven | Caucasian |
| Havemose, 2007 | rs1800587 | RA | 10 | 7 | 6 | 14 | 10 | 1 | PB | PCR-RFLP | seven | Caucasian |
| Havemose, 2007 | rs17561 | JIA | 5 | 3 | 2 | 14 | 10 | 1 | PB | PCR-RFLP | seven | Caucasian |
| Havemose, 2007 | rs17561 | RA | 10 | 7 | 6 | 14 | 10 | 1 | PB | PCR-RFLP | seven | Caucasian |
| Hooper, 2003 | rs1800587 | MS | 189 | 239 | 64 | 102 | 105 | 21 | PB | PCR-RFLP | seven | Caucasian |
| Hutyrova, 2004 | rs1800587 | SSc | 17 | 23 | 6 | 87 | 49 | 14 | PB | PCR-SSP | seven | Caucasian |
| Johnsen, 2008 | rs1800587 | RA | 687 | 507 | 89 | 546 | 445 | 105 | PB | primer extension of multiplex products | eight | Caucasian |
| rs17561 | RA | 686 | 513 | 87 | 545 | 443 | 104 | PB | primer extension of multiplex products | eight | Caucasian | |
| Kaijzel, 2002 | rs17561 | RA | 194 | 171 | 31 | 117 | 79 | 22 | PB | PCR-RFLP | seven | Caucasian |
| Kammoun, 2007 | rs1800587 | GD | 89 | 42 | 0 | 188 | 37 | 0 | PB | PCR-RFLP | six | African |
| Karasneh, 2003 | rs1800587 | BD | 76 | 44 | 8 | 45 | 49 | 11 | PB | gene sequencing | six | Caucasian |
| Kawaguchi, 2003 | rs1800587 | SSc | 54 | 6 | 0 | 38 | 24 | 8 | PB | gene sequencing | seven | Asian |
| rs17561 | SSc | 54 | 6 | 0 | 30 | 30 | 10 | PB | gene sequencing | seven | Asian | |
| Khalilzadeh, 2009 | rs1800587 | GD | 23 | 57 | 27 | 62 | 62 | 12 | PB | PCR-SSP | seven | Asian |
| Kobayashi, 2007a | rs17561 | RA | 66 | 19 | 1 | 84 | 15 | 1 | PB | PCR-RFLP | seven | Asian |
| Kobayashi, 2007b | rs17561 | SLE | 24 | 1 | 0 | 37 | 7 | 0 | PB | PCR-RFLP | nine | Asian |
| Kobayashi, 2009 | rs17561 | RA | 116 | 20 | 1 | 91 | 16 | 1 | PB | PCR-RFLP | eight | Asian |
| Liu, 2010 | rs1800587 | GD | 617 | 137 | 5 | 638 | 92 | 3 | PB | GenomeLab SNPstream 12-plex Genotyping System | seven | Asian |
| Mann, 2002 | rs1800587 | MS | 169 | 152 | 39 | 68 | 64 | 11 | HB | PCR-RFLP | five | Caucasian |
| Mattuzzi, 2007 | rs1800587 | SSc | 43 | 28 | 7 | 364 | 275 | 50 | PB | Taqman MGB probes | seven | Caucasian |
| McDowell, 1995 | rs1800587 | RA | 108 | 127 | 34 | 51 | 37 | 11 | PB | gene sequencing | eight | Caucasian |
| Mirowska, 2011 | rs17561 | MS | 106 | 107 | 15 | 87 | 90 | 16 | PB | PCR-RFLP | six | Caucasian |
| Parks, 2004 | rs1800587 | SLE | 62 | 57 | 25 | 18 | 43 | 12 | PB | PCR-RFLP | seven | African |
| 43 | 32 | 11 | 68 | 109 | 25 | PB | PCR-RFLP | seven | Caucasian | |||
| Pehlivan, 2011 | rs1800587 | ITP | 53 | 18 | 0 | 67 | 4 | 0 | PB | PCR-RFLP | eight | Caucasian |
| Sánchez, 2006 | rs1800587 | SLE | 220 | 164 | 33 | 209 | 166 | 45 | PB | gene sequencing | seven | Caucasian |
| Sarial, 2008 | rs1800587 | MS | 33 | 66 | 1 | 62 | 62 | 12 | PB | PCR-SSP | six | Asian |
| Tahmasebi, 2013 | rs1800587 | SLE | 87 | 103 | 16 | 95 | 93 | 21 | PB | PCR-SSP | seven | Asian |
| Zhou, 2016 | rs1800587 | TID | 171 | 140 | 21 | 209 | 112 | 11 | PB | TaqMan allelic discrimination assay | seven | Asian |
| JIA | 23 | 27 | 3 | 62 | 62 | 12 | PB | PCR-SSP | six | Asian | ||
| Ziaee, 2014 | rs1800587 | SLE | 26 | 25 | 7 | 62 | 62 | 12 | PB | PCR-SSP | six | Asian |
Note: SNP, single nucleotide polymorphisms; SSC, systemic sclerosis; JIA, juvenile idiopathic arthritis; BD, Behcet’s disease; RA, rheumatoid arthritis; MS, multiple sclerosis; GD, Graves’ disease; SLE, systemic lupus erythematosus; ITP, immune thrombocytopenic purpura; TID, type 1 diabetes; AA, major allele/major allele; AB, major allele/minor allele; BB, minor allele/minor allele; PB, population-based; HB, hospital-based; PCR-SSP, polymerase chain reaction with sequence-specific primers; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; NOS, Newcastle-Ottawa Scale.
Meta-analysis of rs17561
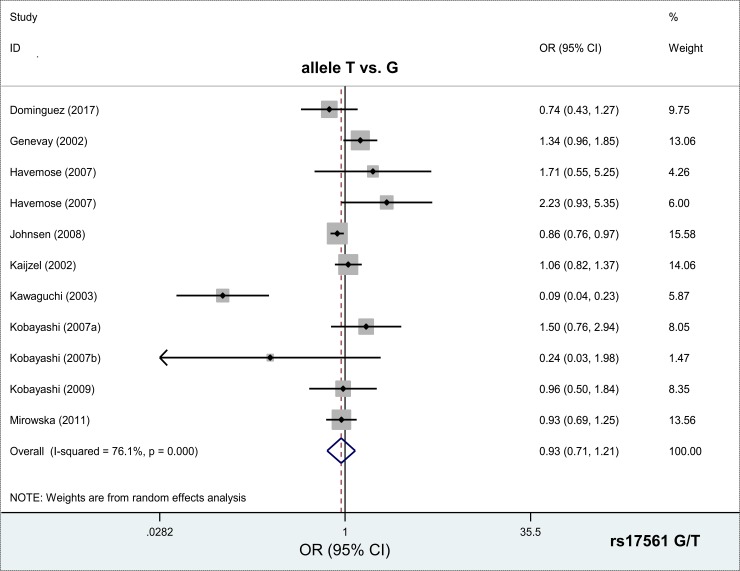
Eleven case-control studies with 2,561 cases and 2,099 controls were enrolled for the meta-analysis of the IL1A rs17561 G/T polymorphism. As shown in Table 2, compared with controls, no increased risk was detected in any of the cases under six comparison models, including allele T vs. G [Passociation (P value in test of association) = 0.576, Bonferroni-corrected Passociation = 1.000, FDR-corrected Passociation = 0.703]; carrier T vs. G (Passociation = 0.586, Bonferroni-corrected Passociation = 1.000, FDR-corrected Passociation = 0.703); TT vs. GG (Passociation = 0.909, Bonferroni-corrected Passociation = 1.000, FDR-corrected Passociation = 0.909); GT vs. GG (Passociation = 0.419, Bonferroni-corrected Passociation = 1.000, FDR-corrected Passociation = 0.703); GT+TT vs. GG (Passociation = 0.438, Bonferroni-corrected Passociation = 1.000, FDR-corrected Passociation = 0.703); TT vs. GG+GT (Passociation = 0.043, Bonferroni-corrected Passociation = 1.000, FDR-corrected Passociation = 0.258). Forest plot data of the meta-analyses under different models are provided in Fig 2 and S1–S5 Figs. We also performed subgroup analyses by ethnicity and disease types. Similar negative results were obtained under different comparison models (all Passociation>0.05, Bonferroni-corrected Passociation >0.05, FDR-corrected Passociation>0.05, Table 3), except for the Asian (Passociation = 0.024, Bonferroni-corrected Passociation = 0.144, FDR-corrected Passociation = 0.048) and PB (Passociation = 0.043, Bonferroni-corrected Passociation = 0.258, FDR-corrected Passociation = 0.043) subgroups under the TT vs. GG+GT model. These data suggested that the IL1A rs17561 G/T polymorphism seems not be related to a risk for autoimmune disease overall.
Table 2. Meta-analysis of IL1A rs17561 G/T and rs1800587 C/T polymorphism.
| SNP | Genetic models | N | Case/Control | Passociation | Passociation& | Passociation# | ORs (95% CIs) | I2 (%) | Pheterogeneity | F/R | PBegg | PEgger |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs17561 | allele T vs. G | 11 | 2,561/2,099 | 0.576 | 1.000 | 0.703 | 0.93 (0.71, 1.21) | 76.1 | <0.001 | R | 1.000 | 0.950 |
| carrier T vs. G | 11 | 2,561/2,099 | 0.586 | 1.000 | 0.703 | 0.94 (0.75, 1.18) | 56.2 | 0.011 | R | 0.876 | 0.724 | |
| TT vs. GGxs | 10 | 2,536/2,055 | 0.909 | 1.000 | 0.909 | 0.97 (0.59, 1.59) | 51.4 | 0.029 | R | 1.000 | 0.368 | |
| GT vs. GG | 11 | 2,561/2099 | 0.419 | 1.000 | 0.703 | 0.89 (0.67, 1.18) | 64.4 | 0.002 | R | 0.161 | 0.393 | |
| GT+TT vs. GG | 11 | 2,561/2,099 | 0.438 | 1.000 | 0.703 | 0.88 (0.65, 1.21) | 72.2% | <0.001 | R | 0.640 | 0.668 | |
| TT vs. GG+GT | 10 | 2,536/2,055 | 0.043 | 0.258 | 0.258 | 0.79 (0.64, 0.99) | 44.9 | 0.060 | F | 0.858 | 0.289 | |
| rs1800587 | allele T vs. C | 31 | 7,381/4,049 | 0.548 | 1.000 | 0.860 | 1.04 (0.92, 1.18) | 76.5 | <0.001 | R | 0.634 | 0.396 |
| carrier T vs. C | 31 | 7,381/4,049 | 0.546 | 1.000 | 0.860 | 1.03 (0.93, 1.14) | 54.7 | <0.001 | R | 0.683 | 0.502 | |
| TT vs. CC | 29 | 7.179/3,794 | 0.860 | 1.000 | 0.860 | 1.02 (0.81, 1.28) | 54.7 | <0.001 | R | 1.000 | 0.499 | |
| CT vs. CC | 31 | 7,381/4,049 | 0.747 | 1.000 | 0.860 | 1.03 (0.87, 1.21) | 74.7 | <0.001 | R | 0.919 | 0.915 | |
| CT+TT vs. CC | 31 | 7,381/4,049 | 0.672 | 1.000 | 0.860 | 1.04(0.88, 1.22) | 77.4 | <0.001 | R | 0.734 | 0.662 | |
| TT vs. CC+CT | 29 | 7.179/3794 | 0.698 | 1.000 | 0.860 | 1.04(0.86, 1.25) | 38.2 | 0.020 | R | 0.866 | 0.481 |
Note: SNP, single nucleotide polymorphisms; N, number of case-control study; ORs, odd ratios; CIs, confidence intervals; Passociation, P value of association test; &, Bonferroni-corrected Passociation value; #, FDR-corrected Passociation value; F, fixed; R, random.
Fig 2. Meta-analysis of the IL1A rs17561 G/T polymorphism and the risk of autoimmune diseases under allele T vs. G model.
Table 3. Subgroup analysis of IL1A rs17561 G/T polymorphism.
| Genetic models | subgroup | N | Case/Control | Passociation | Passociation& | Passociation# | ORs (95% CIs) | I2(%) | Pheterogeneity |
|---|---|---|---|---|---|---|---|---|---|
| Allele T vs. G | Caucasian | 7 | 2,253/1,777 | 0.811 | 1.000 | 0.811 | 1.02 (0.85, 1.24) | 53.4 | 0.045 |
| Asian | 4 | 308/322 | 0.258 | 1.000 | 0.516 | 0.94 (0.75, 1.18) | 88.9 | <0.001 | |
| RA | 7 | 2,238/1,767 | 0.593 | 1.000 | 0.625 | 1.06 (0.86, 1.32) | 55.9 | 0.035 | |
| PB | 11 | 2,561/2,099 | 0.576 | 1.000 | 0.576 | 0.93 (0.71, 1.21) | 76.1 | <0.001 | |
| carrier T vs. G | Caucasian | 7 | 2,253/1,777 | 0.493 | 1.000 | 0.493 | 0.96 (0.86, 1.07) | 0.0 | 0.570 |
| Asian | 4 | 308/322 | 0.279 | 1.000 | 0.493 | 0.54 (0.18, 1.64) | 81.9 | 0.001 | |
| RA | 7 | 2,238/1,767 | 0.612 | 1.000 | 0.790 | 0.97 (0.87, 1.09) | 0.0 | 0.448 | |
| PB | 11 | 2,561/2,099 | 0.586 | 1.000 | 0.586 | 0.94 (0.75, 1.18) | 56.2 | 0.011 | |
| TT vs. GG | Caucasian | 7 | 2,253/1,777 | 0.851 | 1.000 | 0.851 | 1.05 (0.65, 1.69) | 54.0 | 0.043 |
| Asian | 3 | 283.278 | 0.355 | 1.000 | 0.710 | 0.30 (0.02, 3.79) | 58.4 | 0.090 | |
| RA | 7 | 2,238/1,767 | 0.882 | 1.000 | 0.882 | 1.04 (0.63, 1.72) | 45.5 | 0.088 | |
| PB | 10 | 2,536/2,055 | 0.909 | 1.000 | 0.909 | 0.97 (0.59, 1.59) | 51.4 | 0.029 | |
| GT vs. GG | Caucasian | 7 | 2,253/1,777 | 0.805 | 1.000 | 0.805 | 0.98 (0.86, 1.12) | 0.0 | 0.444 |
| Asian | 4 | 308/322 | 0.280 | 1.000 | 0.560 | 0.50 (0.14, 1.77) | 85.3 | 0.000 | |
| RA | 7 | 2,238/1,767 | 0.694 | 1.000 | 0.904 | 1.04 (0.86, 1.25) | 18.7 | 0.287 | |
| PB | 11 | 2,561/2,099 | 0.419 | 1.000 | 0.419 | 0.89 (0.67, 1.18) | 64.4 | 0.002 | |
| GT+TT vs. GG | Caucasian | 7 | 2,253/1,777 | 0.984 | 1.000 | 0.984 | 1.00 (0.84, 1.19) | 24.9 | 0.239 |
| Asian | 4 | 308/322 | 0.262 | 1.000 | 0.524 | 0.45 (0.11, 1.82) | 88.3 | 0.000 | |
| RA | 7 | 2,238/1,767 | 0.657 | 1.000 | 0.772 | 0.45 (0.11, 1.82) | 37.8 | 0.141 | |
| PB | 11 | 2,561/2,099 | 0.438 | 1.000 | 0.438 | 0.88 (0.65, 1.21) | 72.2 | 0.000 | |
| TT vs. GG+GT | Caucasian | 7 | 2,253/1,777 | 0.125 | 0.750 | 0.125 | 0.84 (0.67, 1.05) | 52.2 | 0.051 |
| Asian | 3 | 283.278 | 0.024 | 0.144 | 0.048 | 0.21 (0.05, 0.81) | 40.9 | 0.184 | |
| RA | 7 | 2,238/1,767 | 0.123 | 0.738 | 0.220 | 0.83 (0.65, 1.05) | 41.2 | 0.116 | |
| PB | 10 | 2,536/2,055 | 0.043 | 0.258 | 0.043 | 0.79 (0.64, 0.99) | 44.9 | 0.060 |
Note: RA, rheumatoid arthritis; PB, population-based control; ORs, odd ratios; CIs, confidence intervals; Passociation, P value of association test; &, Bonferroni-corrected Passociation value; #, FDR-corrected Passociation value; Pheterogeneity, P value of heterogeneity.
Meta-analysis of rs1800587
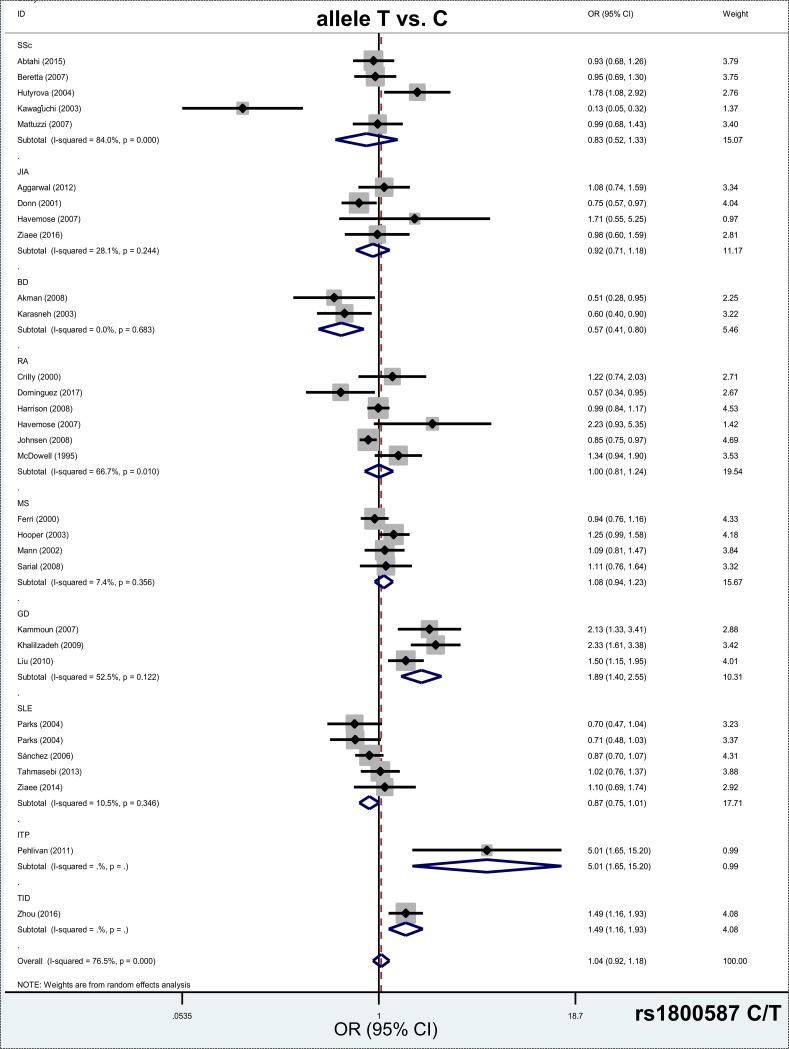
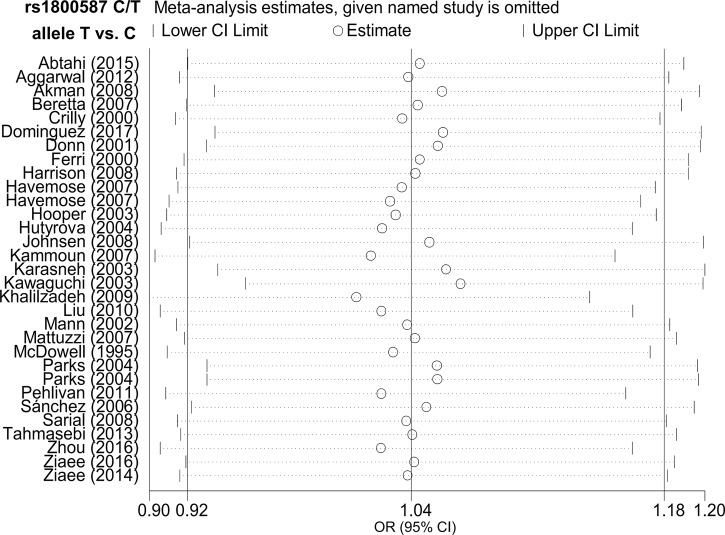
A total of 31 case-control studies with 7,381 cases and 4,049 controls were used for meta-analysis of the IL1A rs1800587 C/T Polymorphisms. Pooled data from the overall population (Table 2) presented the negative results under all comparison models (all Passociation >0.05, Bonferroni-corrected Passociation >0.05, FDR-corrected Passociation>0.05). Nevertheless, the data from the GD (Graves’ disease) subgroup analysis (Table 4), comprising three studies, showed an increased risk in cases of autoimmune diseases compared with controls under the genetic models of allele T vs. C (Passociation<0.001, Bonferroni-corrected Passociation <0.006, FDR-corrected Passociation <0.006, OR = 1.89, 95% CIs = 1.40, 2.55), carrier T vs. C (Passociation<0.001, Bonferroni-corrected Passociation <0.006, FDR-corrected Passociation <0.006, OR = 1.60, 95% CIs = 1.30, 1.98), CT vs. CC (Passociation<0.001, Bonferroni-corrected Passociation <0.006, FDR-corrected Passociation <0.006, OR = 1.94, 95% CIs = 1.38, 2.72), CT+TT vs.CC (Passociation = 0.001, Bonferroni-corrected Passociation = 0.006, FDR-corrected Passociation = 0.006, OR = 2.12, 95% CIs = 1.38, 3.25). We did not observe a positive association between case and control groups in other subgroup analyses (Table 4, all Passociation>0.05). Fig 3 and S6–S8 Figs show the forest plots of the subgroup analysis by disease type under the models of allele T vs. C, carrier T vs. C, CT+TT vs.CC and CT vs. CC, respectively. S9 and S10 Figs show the forest plot data of subgroup analysis by ethnicity and control source under allele models. Based on the above data, the C/T genotype of IL1A rs1800587 C/T polymorphism is more likely to be statistically associated with an increased risk of Graves’ disease, but not other autoimmune diseases, such as systemic sclerosis, juvenile idiopathic arthritis, rheumatoid arthritis, multiple sclerosis and systemic lupus erythematosus.
Table 4. Subgroup analysis of IL1A rs1800587 C/T polymorphism.
| Genetic models | subgroup | N | Case/Control | Passociation | Passociation& | Passociation# | ORs (95% CIs) | I2(%) | Pheterogeneity |
|---|---|---|---|---|---|---|---|---|---|
| allele T vs. C | Asian | 10 | 1,939/1,419 | 0.465 | 1.000 | 0.733 | 1.10 (0.85, 1.43) | 80.5 | <0.001 |
| Caucasian | 19 | 5,167/2,424 | 0.612 | 1.000 | 0.733 | 0.97 (0.85, 1.10) | 65.5 | <0.001 | |
| SSc | 5 | 558/699 | 0.441 | 1.000 | 0.593 | 0.83 (0.52, 1.33) | 84.0 | <0.001 | |
| JIA | 4 | 487/274 | 0.494 | 1.000 | 0.593 | 0.92 (0.71, 1.18) | 28.1 | 0.244 | |
| RA | 6 | 2,493/966 | 0.980 | 1.000 | 0.980 | 1.00 (0.81, 1.24) | 66.7 | 0.010 | |
| MS | 4 | 1,351/430 | 0.284 | 1.000 | 0.568 | 1.11 (0.76, 1.64) | 7.4 | 0.356 | |
| GD | 3 | 997/888 | <0.001 | <0.006 | <0.006 | 1.89 (1.40, 2.55) | 52.5 | 0.122 | |
| SLE | 5 | 911/452 | 0.071 | 0.426 | 0.213 | 0.87 (0.75, 1.01) | 10.5 | 0.346 | |
| PB | 30 | 7,021/3,981 | 0.582 | 1.000 | 0.582 | 1.04 (0.91, 1.18) | 77.2 | <0.001 | |
| carrier T vs. C | Asian | 10 | 1,939/1,419 | 0.316 | 1.000 | 0.677 | 1.11 (0.90, 1.37) | 61.8 | 0.005 |
| Caucasian | 19 | 5,167/2,424 | 0.451 | 1.000 | 0.677 | 0.96 (0.87, 1.06) | 30.6 | 0.101 | |
| SSc | 5 | 558/699 | 0.512 | 1.000 | 0.614 | 0.88 (0.59, 1.30) | 71.5 | 0.007 | |
| JIA | 4 | 487/274 | 0.385 | 1.000 | 0.614 | 0.91 (0.73, 1.13) | 0.0 | 0.576 | |
| RA | 6 | 2,493/966 | 0.680 | 1.000 | 0.680 | 0.97 (0.84, 1.12) | 24.3 | 0.252 | |
| MS | 4 | 1,351/430 | 0.494 | 1.000 | 0.614 | 1.05 (0.91, 1.22) | 0.0 | 0.721 | |
| GD | 3 | 997/888 | <0.001 | <0.006 | <0.006 | 1.60 (1.30, 1.98) | 0.0 | 0.518 | |
| SLE | 5 | 911/452 | 0.236 | 1.000 | 0.614 | 0.91 (0.78, 1.06) | 0.0 | 0.718 | |
| PB | 30 | 7,021/3,981 | 0.568 | 1.000 | 0.787 | 1.03 (0.93, 1.15) | 56.2 | <0.001 | |
| TT vs. CC | Asian | 10 | 1,939/1,419 | 0.746 | 1.000 | 0.746 | 1.11 (0.60, 2.04) | 69.5 | 0.001 |
| Caucasian | 18 | 5,096/2,357 | 0.651 | 1.000 | 0.746 | 0.95 (0.77, 1.18) | 36.5 | 0.061 | |
| SSc | 5 | 558/699 | 0.893 | 1.000 | 0.893 | 1.04 (0.58, 1.86) | 44.6 | 0.125 | |
| JIA | 4 | 487/274 | 0.334 | 1.000 | 0.668 | 0.78 (0.46, 1.30) | 0.0 | 0.505 | |
| RA | 6 | 2,493/966 | 0.876 | 1.000 | 0.893 | 0.97 (0.66, 1.43) | 46.0 | 0.099 | |
| MS | 4 | 1,351/430 | 0.660 | 1.000 | 0.893 | 1.13 (0.66, 1.91) | 52.6 | 0.097 | |
| GD | 2 | 866/700 | 0.032 | 0.192 | 0.192 | 3.72 (1.12, 12.39) | 54.8 | 0.137 | |
| SLE | 5 | 911/452 | 0.082 | 0.492 | 0.246 | 0.75 (0.55, 1.04) | 0.0 | 0.773 | |
| PB | 28 | 6,819/3,726 | 0.963 | 1.000 | 0.963 | 1.01 (0.80, 1.27) | 36.5 | 0.061 | |
| CT vs. CC | Asian | 10 | 1,939/1,419 | 0.120 | 0.720 | 0.360 | 1.24 (0.94, 1.64) | 69.2 | 0.001 |
| Caucasian | 19 | 5,167/2,424 | 0.327 | 1.000 | 0.491 | 0.92 (0.78, 1.09) | 64.4 | <0.001 | |
| SSc | 5 | 558/699 | 0.513 | 1.000 | 0.770 | 0.84 (0.50, 1.42) | 77.5 | 0.001 | |
| JIA | 4 | 487/274 | 0.795 | 1.000 | 0.883 | 0.94 (0.60, 1.48) | 54.3 | 0.087 | |
| RA | 6 | 2,493/966 | 0.883 | 1.000 | 0.883 | 0.98 (0.74, 1.29) | 64.0 | 0.016 | |
| MS | 4 | 1,351/430 | 0.351 | 1.000 | 0.702 | 1.15 (0.86, 1.53) | 57.9 | 0.068 | |
| GD | 3 | 997/888 | <0.001 | <0.006 | <0.006 | 1.94 (1.38, 2.72) | 42.1 | 0.178 | |
| SLE | 5 | 911/452 | 0.166 | 1.000 | 0.498 | 0.76 (0.51, 1.12) | 70.9 | 0.008 | |
| PB | 30 | 7,021/3,981 | 0.733 | 1.000 | 0.826 | 1.03 (0.87, 1.22) | 75.5 | <0.001 | |
| CT+TT vs. CC | Asian | 10 | 1,939/1,419 | 0.237 | 1.000 | 0.650 | 1.20 (0.89, 1.63) | 77.0 | <0.001 |
| Caucasian | 19 | 5,167/2,424 | 0.433 | 1.000 | 0.650 | 0.94 (0.79, 1.11) | 67.3 | <0.001 | |
| SSc | 5 | 558/699 | 0.473 | 1.000 | 0.710 | 0.81 (0.46, 1.43) | 82.6 | <0.001 | |
| JIA | 4 | 487/274 | 0.730 | 1.000 | 0.876 | 0.93 (0.62, 1.40) | 48.4 | 0.121 | |
| RA | 6 | 2,493/966 | 0.963 | 1.000 | 0.963 | 0.99 (0.75, 1.31) | 66.8 | 0.010 | |
| MS | 4 | 1,351/430 | 0.294 | 1.000 | 0.588 | 1.14 (0.89, 1.45) | 45.5 | 0.138 | |
| GD | 3 | 997/888 | 0.001 | 0.006 | 0.006 | 2.12 (1.38, 3.25) | 63.7 | 0.063 | |
| SLE | 5 | 911/452 | 0.139 | 1.000 | 0.417 | 0.78 (0.55, 1.09) | 63.5 | 0.027 | |
| PB | 30 | 7,021/3,981 | 0.680 | 1.000 | 0.902 | 1.04 (0.88, 1.23) | 78.1 | <0.001 | |
| TT vs. CC+CT | Asian | 10 | 1,939/1,419 | 0.948 | 1.000 | 0.948 | 1.02 (0.60, 1.73) | 62.9 | 0.004 |
| Caucasian | 18 | 5,096/2,357 | 0.684 | 1.000 | 0.948 | 0.97 (0.82, 1.14) | 13.0 | 0.299 | |
| SSc | 5 | 558/699 | 0.786 | 1.000 | 0.794 | 1.06 (0.69, 1.63) | 14.0 | 0.325 | |
| JIA | 4 | 487/274 | 0.503 | 1.000 | 0.794 | 0.84 (0.51, 1.39) | 0.0 | 0.444 | |
| RA | 6 | 2,493/966 | 0.723 | 1.000 | 0.794 | 0.94 (0.67, 1.31) | 35.0 | 0.174 | |
| MS | 4 | 1,351/430 | 0.794 | 1.000 | 0.794 | 1.08 (0.62, 1.86) | 58.6 | 0.065 | |
| GD | 2 | 866/700 | 0.001 | 0.006 | 0.006 | 2.97 (1.54, 5.72) | 0.0 | 0.349 | |
| SLE | 5 | 911/452 | 0.356 | 1.000 | 0.794 | 0.87 (0.64, 1.17) | 0.0 | 0.688 | |
| PB | 28 | 6,819/3,726 | 0.823 | 1.000 | 0.794 | 1.02 (0.85, 1.24) | 38.8 | 0.020 |
Note: SSC, systemic sclerosis; JIA, juvenile idiopathic arthritis; RA, rheumatoid arthritis; MS, multiple sclerosis
GD, Graves’ disease; SLE, systemic lupus erythematosus; PB, population-based control; ORs, odd ratios; CIs, confidence intervals.
Passociation, P value of association test; &, Bonferroni-corrected Passociation value; #, FDR-corrected Passociation value; Pheterogeneity, P value of heterogeneity.
Fig 3. Subgroup analysis by disease type of the association between IL1A rs1800587 C/T polymorphism and the risk of autoimmune diseases under allele T vs. C model.
Heterogeneity, bias and sensitivity
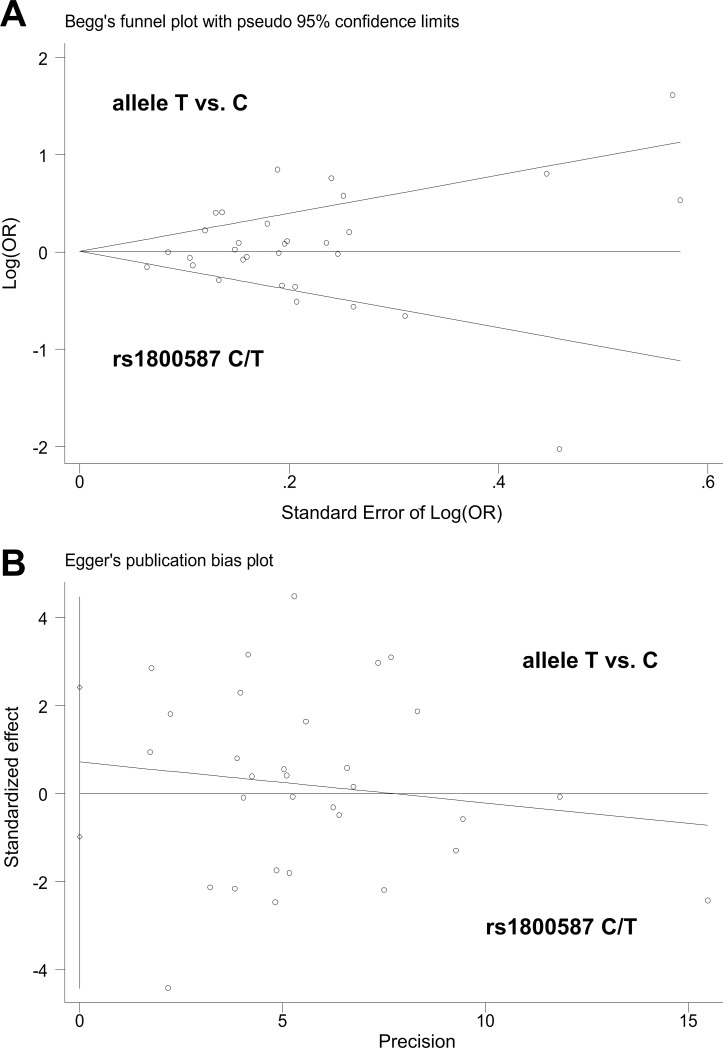
Apart from the TT vs. GG+GT comparison of rs17561, larger heterogeneity was detected (Table 2, I2 >50.0% or Pheterogeneity >0.05), and random effect models were utilized. In addition, as shown in Table 2, P value of Begg’s test and Egger’s test were >0.05 in all genetic models, indicating the absence of large publication bias. The plot data are shown in Fig 4 and S11 Fig. Furthermore, we believe our data are stable, because we did not observe any remarkable change of pooled ORs under any genetic models. The data for the allele T vs. C model of rs1800587 are shown in Fig 5, and other data are not shown.
Fig 4. Begg’s test and Egger’s test for the allele T vs. C model of IL1A rs1800587 C/T polymorphism.
(A) Begg’s test; (B) Egger’s test.
Fig 5. Sensitivity analysis for the allele T vs. C model of IL1A rs1800587 C/T polymorphism.
Discussion
Previously, the rs1800587 C/T SNP of IL1A gene was reported to not be linked to the risk or severity of systemic lupus erythematosus in a Spanish population [12], juvenile idiopathic arthritis in an Iranian population [15], and juvenile idiopathic arthritis in the UK [13]. IL1A rs17561 SNP was not associated with rheumatoid arthritis susceptibility in a Mexican population [14]. However, the IL1A rs1800587 and rs17561 SNPs were also reported to be associated with the risk of systemic sclerosis in a Japanese population [8]. The rs1800587 C/T SNP of IL1A gene has been related to susceptibility to systemic sclerosis in a Slovak Caucasian population [9], Graves’ ophthalmopathy in an Iranian population [10], and Graves’ disease in a Tunisian population [11]. Therefore, we first comprehensively explored the association between IL1A rs17561 and rs1800587 SNPs and the risk of overall autoimmune diseases using meta-analysis and subgroup analyses by characteristics of ethnicity, disease type and source of control.
In 2013, a meta-analysis was reported [17], investigating the genetic relationship between IL1A rs1800587 and rs17561 SNPs and the risk of systemic lupus erythematosus based on four case-control studies from three articles [12, 42, 48], which did not provide strong evidence for an association. In 2014, data from another meta-analysis containing four studies from three articles [12, 48, 51] supported a potential association for rs1800587 in Europeans [16]. In this study, we added another case-control study [53] to the subgroup meta-analysis of systemic lupus erythematosus for rs1800587, and observed a negative association.
In one meta-analysis of rheumatoid arthritis susceptibility[18], there were four case-control studies [32, 35, 38, 54] for rs1800587 and three case-control studies [34, 39, 43] for rs17561. No positive association between IL1A rs1800587 and rs17561 SNPs and the risk of rheumatoid arthritis was observed [18]. Here, we included more data for our updated meta-analysis and removed one case-control study [54], in which the genotype distribution of control group did not fulfill Hardy-Weinberg equilibrium. Seven case-control studies [14, 34, 36, 38, 39, 41, 43] were enrolled for the subgroup analysis of rs17561, and six case-control studies [14, 32, 35, 36, 38, 46] were used for rs1800587. Our pooled data with enhanced statistical power also indicated that the IL1A rs1800587 and rs17561 SNPs were not linked to the risk of rheumatoid arthritis, which was consistent with previous data [18].
Regarding multiple sclerosis susceptibility, in 2013, Huang et al. enrolled five case-control studies [33, 37, 44, 50, 55] for a meta-analysis of rs1800587 SNP and two case-control studies [47, 56] for meta-analysis of rs17561 SNP. However, negative association was reported for both s1800587 and rs17561 [19]. Here, due to the limitation of Hardy-Weinberg equilibrium, one case-control study [55] was excluded from our subgroup meta-analysis of rs1800587. We also found that the rs1800587 SNP was not linked to the risk of multiple sclerosis.
In 2010, Liu et al. investigated the genetic relationship between IL1A rs1800587 SNP and risk of Graves’ disease via a meta-analysis and found a positive association in an Asian population [20]. Here, our data in the subgroup meta-analysis of Graves’ disease showed similar results. It is possible that the rs1800587 SNP within the 5'-flanking regulatory region of IL1A gene affects the normal production, secretion or function of interleukin-1.
Some limitations exist in our meta-analysis. First, we did not obtain strong evidence regarding the effect of rs1800587 and rs17561 SNPs for the risk of different types of autoimmune diseases, due to the limited number of included independent case-control studies. Only two case-control studies [30, 40] were included in the subgroup of Graves’ disease under the homozygote and recessive models. Second, even though no remarkable publication bias was detected by our Begg’s test and Egger’s test, larger heterogeneity existed in the majority of comparisons. We observed a decreased level of heterogeneity in some subgroup analyses by disease type, such as the “rheumatoid arthritis, RA” subgroup of rs17561 and “multiple sclerosis, MS” subgroup of rs1800587. The factor of specific disease type may be involved in the source of heterogeneity. Further relevant researches with larger sample sizes were required. Third, we only acquired suitable case-control studies published in English. The outcome may be affected by the inclusion of unpublished articles, or articles published in another language. Fourth, it is worth analyzing the combined influence of different SNPs or cytokine genes, when more case-control studies become available.
Taken together, based on published articles in databases, our meta-analysis suggested that the rs1800587 polymorphism, rather than rs17561, within the IL1A gene, may be a genetic risk factor for Graves’ disease. However, IL1A rs17561 or rs1800587 polymorphism seems not to be statistically linked to the risk of other analyzed autoimmune diseases, such as systemic sclerosis, juvenile idiopathic arthritis, rheumatoid arthritis, multiple sclerosis and systemic lupus erythematosus.
Supporting information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(A) Begg’s test; (B) Egger’s test.
(TIF)
(DOCX)
(DOC)
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Wang L, Wang FS, Gershwin ME. Human autoimmune diseases: a comprehensive update. J Intern Med. 2015;278(4):369–395. Epub 2015/07/28. doi: 10.1111/joim.12395 . [DOI] [PubMed] [Google Scholar]
- 2.Kunz M, Ibrahim SM. Cytokines and cytokine profiles in human autoimmune diseases and animal models of autoimmunity. Mediators Inflamm. 2009;2009:979258 Epub 2009/11/04. doi: 10.1155/2009/979258 ; PubMed Central PMCID: PMCPmc2768824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kochi Y. Genetics of autoimmune diseases: perspectives from genome-wide association studies. Int Immunol. 2016;28(4):155–161. Epub 2016/02/10. doi: 10.1093/intimm/dxw002 ; PubMed Central PMCID: PMCPmc4889885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Gorp H, Van Opdenbosch N, Lamkanfi M. Inflammasome-Dependent Cytokines at the Crossroads of Health and Autoinflammatory Disease. Cold Spring Harb Perspect Biol. 2017. Epub 2017/10/19. doi: 10.1101/cshperspect.a028563 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roerink ME, van der Schaaf ME, Dinarello CA, Knoop H, van der Meer JW. Interleukin-1 as a mediator of fatigue in disease: a narrative review. J Neuroinflammation. 2017;14(1):16 Epub 2017/01/22. doi: 10.1186/s12974-017-0796-7 ; PubMed Central PMCID: PMCPmc5251329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mistry A, Savic S, van der Hilst JCH. Interleukin-1 Blockade: An Update on Emerging Indications. BioDrugs. 2017;31(3):207–221. Epub 2017/05/13. doi: 10.1007/s40259-017-0224-7 . [DOI] [PubMed] [Google Scholar]
- 7.Lafage M, Maroc N, Dubreuil P, de Waal Malefijt R, Pebusque MJ, Carcassonne Y, et al. The human interleukin-1 alpha gene is located on the long arm of chromosome 2 at band q13. Blood. 1989;73(1):104–107. Epub 1989/01/01. . [PubMed] [Google Scholar]
- 8.Kawaguchi Y, Tochimoto A, Ichikawa N, Harigai M, Hara M, Kotake S, et al. Association of IL1A gene polymorphisms with susceptibility to and severity of systemic sclerosis in the Japanese population. Arthritis and Rheumatism. 2003;48(1):186–192. doi: 10.1002/art.10736 [DOI] [PubMed] [Google Scholar]
- 9.Hutyrová B, Lukác J, Bosák V, Buc M, Du Bois RM, Petrek M. Interleukin 1α Single-Nucleotide Polymorphism Associated with Systemic Sclerosis. Journal of Rheumatology. 2004;31(1):81–84. [PubMed] [Google Scholar]
- 10.Khalilzadeh O, Anvari M, Esteghamati A, Mahmoudi M, Tahvildari M, Rashidi A, et al. Graves' ophthalmopathy and gene polymorphisms in interleukin-1α, interleukin-1β, interleukin-1 receptor and interleukin-1 receptor antagonist. Clinical and Experimental Ophthalmology. 2009;37(6):614–619. doi: 10.1111/j.1442-9071.2009.02093.x [DOI] [PubMed] [Google Scholar]
- 11.Kammoun-Krichen M, Bougacha-Elleuch N, Makni K, Rebai M, Peraldi-Roux S, Rebai A, et al. Association analysis of interleukin-1 gene polymorphisms in autoimmune thyroid diseases in the Tunisian population. European Cytokine Network. 2007;18(4):196–200. doi: 10.1684/ecn.2007.0104 [DOI] [PubMed] [Google Scholar]
- 12.Sánchez E, Sabio JM, Callejas JL, de Ramón E, Garcia-Portales R, García-Hernández FJ, et al. Association study of genetic variants of pro-inflammatory chemokine and cytokine genes in systemic lupus erythematosus. BMC Medical Genetics. 2006;7 doi: 10.1186/1471-2350-7-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donn RP, Barrett JH, Farhan A, Stopford A, Pepper L, Shelley E, et al. Cytokine gene polymorphisms and susceptibility to juvenile idiopathic arthritis. British Paediatric Rheumatology Study Group. Arthritis Rheum. 2001;44(4):802–810. Epub 2001/04/24. doi: 10.1002/1529-0131(200104)44:4<802::AID-ANR136>3.0.CO;2-G . [DOI] [PubMed] [Google Scholar]
- 14.Dominguez-Perez RA, Loyola-Rodriguez JP, Abud-Mendoza C, Alpuche-Solis AG, Ayala-Herrera JL, Martinez-Martinez RE. Association of cytokines polymorphisms with chronic peridontitis and rheumatoid arthritis in a Mexican population. Acta Odontol Scand. 2017;75(4):243–248. Epub 2017/03/31. doi: 10.1080/00016357.2017.1280846 . [DOI] [PubMed] [Google Scholar]
- 15.Ziaee V, Maddah M, Harsini S, Rezaei A, Sadr M, Zoghi S, et al. Association of interleukin-1 family gene polymorphisms with juvenile idiopathic arthritis in Iranian population. Allergol Immunopathol (Madr). 2016;44(6):542–546. Epub 2016/10/09. doi: 10.1016/j.aller.2016.07.002 . [DOI] [PubMed] [Google Scholar]
- 16.Song GG, Kim JH, Seo YH, Choi SJ, Ji JD, Lee YH. Associations between interleukin 1 polymorphisms and susceptibility to systemic lupus erythematosus: a meta-analysis. Hum Immunol. 2014;75(1):105–112. Epub 2013/09/24. doi: 10.1016/j.humimm.2013.09.002 . [DOI] [PubMed] [Google Scholar]
- 17.Wang B, Zhu JM, Fan YG, Feng CC, Chen GM, Chen H, et al. The association of IL1alpha and IL1beta polymorphisms with susceptibility to systemic lupus erythematosus: a meta-analysis. Gene. 2013;527(1):95–101. Epub 2013/06/12. doi: 10.1016/j.gene.2013.05.059 . [DOI] [PubMed] [Google Scholar]
- 18.Lee YH, Bae SC. Associations between interleukin-1 and IL-1 receptor antagonist polymorphisms and susceptibility to rheumatoid arthritis: A meta-analysis. Cell Mol Biol (Noisy-le-grand). 2015;61(8):105–111. Epub 2016/01/01. . [PubMed] [Google Scholar]
- 19.Huang J, Xie ZK, Lu RB, Xie ZF. Association of interleukin-1 gene polymorphisms with multiple sclerosis: a meta-analysis. Inflamm Res. 2013;62(1):97–106. Epub 2012/10/12. doi: 10.1007/s00011-012-0556-1 . [DOI] [PubMed] [Google Scholar]
- 20.Liu N, Li X, Liu C, Zhao Y, Cui B, Ning G. The association of interleukin-1alpha and interleukin-1beta polymorphisms with the risk of Graves' disease in a case-control study and meta-analysis. Hum Immunol. 2010;71(4):397–401. Epub 2010/02/02. doi: 10.1016/j.humimm.2010.01.023 . [DOI] [PubMed] [Google Scholar]
- 21.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097 Epub 2009/07/22. doi: 10.1371/journal.pmed.1000097 ; PubMed Central PMCID: PMCPmc2707599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Wang T, Wu Z, Zhang K, Li W, Yang J, et al. Association between TERT rs2853669 polymorphism and cancer risk: A meta-analysis of 9,157 cases and 11,073 controls. 2018;13(3):e0191560 doi: 10.1371/journal.pone.0191560 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hua J, Huang W. Peptidylarginine deiminase 4 -104C/T polymorphism and risk of rheumatoid arthritis: A pooled analysis based on different populations. PLoS One. 2018;13(3):e0193674 Epub 2018/03/14. doi: 10.1371/journal.pone.0193674 10.1371/journal.pone.0193674. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Niu T. A meta-analysis of associations of LEPR Q223R and K109R polymorphisms with Type 2 diabetes risk. 2018;13(1):e0189366 doi: 10.1371/journal.pone.0189366 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Hu F, Liang H, Liu Y, Yang J, Zhou W. Association between a miRNA-146a polymorphism and susceptibility to head and neck squamous cell carcinoma in Chinese patients: A meta-analysis of 8 case-control studies. 2017;12(10):e0186609 doi: 10.1371/journal.pone.0186609 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siokas V, Dardiotis E, Tsironi EE, Tsivgoulis G, Rikos D, Sokratous M, et al. The Role of TOR1A Polymorphisms in Dystonia: A Systematic Review and Meta-Analysis. PLoS One. 2017;12(1):e0169934 Epub 2017/01/13. doi: 10.1371/journal.pone.0169934 ; PubMed Central PMCID: PMCPmc5231385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Wu J, Peng X, Song J, Wang J, Dong W. Associations between STAT3 rs744166 polymorphisms and susceptibility to ulcerative colitis and Crohn's disease: a meta-analysis. PLoS One. 2014;9(10):e109625 Epub 2014/10/07. doi: 10.1371/journal.pone.0109625 ; PubMed Central PMCID: PMCPmc4186844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abtahi S, Farazmand A, Mahmoudi M, Ashraf-Ganjouei A, Javinani A, Nazari B, et al. IL-1A rs1800587, IL-1B rs1143634 and IL-1R1 rs2234650 polymorphisms in Iranian patients with systemic sclerosis. Int J Immunogenet. 2015;42(6):423–427. Epub 2015/09/30. doi: 10.1111/iji.12212 . [DOI] [PubMed] [Google Scholar]
- 29.Aggarwal A, Srivastava R, Singh S, Dubey PK. IL1 gene polymorphisms in enthesitis related arthritis category of juvenile idiopathic arthritis (ERA-JIA). Clinical Rheumatology. 2012;31(4):607–611. doi: 10.1007/s10067-011-1898-8 PubMed PMID: WOS:000303449200004. [DOI] [PubMed] [Google Scholar]
- 30.Akman A, Ekinci NC, Kacaroglu H, Yavuzer U, Alpsoy E, Yegin O. Relationship between periodontal findings and specific polymorphisms of interleukin-1α and -1β in Turkish patients with Behçet's disease. Archives of Dermatological Research. 2008;300(1):19–26. doi: 10.1007/s00403-007-0794-1 [DOI] [PubMed] [Google Scholar]
- 31.Beretta L, Bertolotti F, Cappiello F, Barili M, Masciocchi M, Toussoun K, et al. Interleukin-1 gene complex polymorphisms in systemic sclerosis patients with severe restrictive lung physiology. Human Immunology. 2007;68(7):603–609. doi: 10.1016/j.humimm.2007.03.005 [DOI] [PubMed] [Google Scholar]
- 32.Crilly A, Maiden N, Capell HA, Madhok R. Predictive value of interleukin 1 gene polymorphisms for surgery. Ann Rheum Dis. 2000;59(9):695–699. Epub 2000/09/08. doi: 10.1136/ard.59.9.695 ; PubMed Central PMCID: PMCPmc1753257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferri C, Sciacca FL, Grimaldi LE, Veglia F, Magnani G, Santuccio G, et al. Lack of association between IL-1A and IL-1B promoter polymorphisms and multiple sclerosis. J Neurol Neurosurg Psychiatry. 2000;69(4):564–565. Epub 2001/02/24. doi: 10.1136/jnnp.69.4.564 ; PubMed Central PMCID: PMCPmc1737125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Genevay S, Di Giovine FS, Perneger TV, Silvestri T, Stingelin S, Duff G, et al. Association of interleukin-4 and interleukin-1B gene variants with Larsen score progression in rheumatoid arthritis. Arthritis Rheum. 2002;47(3):303–309. Epub 2002/07/13. doi: 10.1002/art.10394 . [DOI] [PubMed] [Google Scholar]
- 35.Harrison P, Pointon JJ, Chapman K, Roddam A, Wordsworth BP. Interleukin-1 promoter region polymorphism role in rheumatoid arthritis: a meta-analysis of IL-1B-511A/G variant reveals association with rheumatoid arthritis. Rheumatology (Oxford). 2008;47(12):1768–1770. Epub 2008/10/08. doi: 10.1093/rheumatology/ken374 . [DOI] [PubMed] [Google Scholar]
- 36.Havemose-Poulsen A, Sorensen LK, Bendtzen K, Holmstrup P. Polymorphisms within the IL-1 gene cluster: effects on cytokine profiles in peripheral blood and whole blood cell cultures of patients with aggressive periodontitis, juvenile idiopathic arthritis, and rheumatoid arthritis. J Periodontol. 2007;78(3):475–492. Epub 2007/03/06. doi: 10.1902/jop.2007.060135 . [DOI] [PubMed] [Google Scholar]
- 37.Hooper-van Veen T, Schrijver HM, Zwiers A, Crusius JB, Knol DL, Kalkers NF, et al. The interleukin-1 gene family in multiple sclerosis susceptibility and disease course. Mult Scler. 2003;9(6):535–539. Epub 2003/12/11. doi: 10.1191/1352458503ms974oa . [DOI] [PubMed] [Google Scholar]
- 38.Johnsen AK, Plenge RM, Butty V, Campbell C, Dieguez-Gonzalez R, Gomez-Reino JJ, et al. A broad analysis of IL1 polymorphism and rheumatoid arthritis. Arthritis Rheum. 2008;58(7):1947–1957. Epub 2008/06/26. doi: 10.1002/art.23592 ; PubMed Central PMCID: PMCPmc2533126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaijzel EL, van Dongen H, Bakker AM, Breedveld FC, Huizinga TW, Verweij CL. Relationship of polymorphisms of the Interleukin-1 gene cluster to occurrence and severity of rheumatoid arthritis. Tissue Antigens. 2002;59(2):122–126. Epub 2002/05/25. . [DOI] [PubMed] [Google Scholar]
- 40.Karasneh J, Hajeer AH, Barrett J, Ollier WER, Thornhill M, Gul A. Association of specific interleukin 1 gene cluster polymorphisms with increased susceptibility for Behçet's disease. Rheumatology. 2003;42(7):860–864. doi: 10.1093/rheumatology/keg232R [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi T, Ito S, Kuroda T, Yamamoto K, Sugita N, Narita I, et al. The interleukin-1 and Fcgamma receptor gene polymorphisms in Japanese patients with rheumatoid arthritis and periodontitis. J Periodontol. 2007a;78(12):2311–2318. Epub 2007/12/07. doi: 10.1902/jop.2007.070136 . [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi T, Ito S, Yasuda K, Kuroda T, Yamamoto K, Sugita N, et al. The combined genotypes of stimulatory and inhibitory Fc gamma receptors associated with systemic lupus erythematosus and periodontitis in Japanese adults. J Periodontol. 2007b;78(3):467–474. doi: 10.1902/jop.2007.060194 . [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi T, Murasawa A, Ito S, Yamamoto K, Komatsu Y, Abe A, et al. Cytokine gene polymorphisms associated with rheumatoid arthritis and periodontitis in Japanese adults. J Periodontol. 2009;80(5):792–799. Epub 2009/05/02. doi: 10.1902/jop.2009.080573 . [DOI] [PubMed] [Google Scholar]
- 44.Mann CL, Davies MB, Stevenson VL, Leary SM, Boggild MD, Ko Ko C, et al. Interleukin 1 genotypes in multiple sclerosis and relationship to disease severity. J Neuroimmunol. 2002;129(1–2):197–204. Epub 2002/08/06. . [DOI] [PubMed] [Google Scholar]
- 45.Mattuzzi S, Barbi S, Carletto A, Ravagnani V, Moore PS, Bambara LM, et al. Association of polymorphisms in the IL1B and IL2 genes with susceptibility and severity of systemic sclerosis. Journal of Rheumatology. 2007;34(5):997–1004. [PubMed] [Google Scholar]
- 46.McDowell TL, Symons JA, Ploski R, Forre O, Duff GW. A genetic association between juvenile rheumatoid arthritis and a novel interleukin-1 alpha polymorphism. Arthritis Rheum. 1995;38(2):221–228. Epub 1995/02/01. . [DOI] [PubMed] [Google Scholar]
- 47.Mirowska-Guzel D, Gromadzka G, Mach A, Czlonkowski A, Czlonkowska A. Association of IL1A, IL1B, ILRN, IL6, IL10 and TNF-alpha polymorphisms with risk and clinical course of multiple sclerosis in a Polish population. J Neuroimmunol. 2011;236(1–2):87–92. Epub 2011/05/31. doi: 10.1016/j.jneuroim.2011.04.014 . [DOI] [PubMed] [Google Scholar]
- 48.Parks CG. Systemic lupus erythematosus and genetic variation in the interleukin 1 gene cluster: a population based study in the southeastern United States. Annals of the Rheumatic Diseases. 2004;63(1):91–94. doi: 10.1136/ard.2003.007336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pehlivan M, Okan V, Sever T, Balci Oguzkan S, Yilmaz M, Babacan T, et al. Investigation of TNF-alpha, TGF-beta 1, IL-10, IL-6, IFN-gamma, MBL, GPIA, and IL1A gene polymorphisms in patients with idiopathic thrombocytopenic purpura. Platelets. 2011;22(8):588–595. doi: 10.3109/09537104.2011.577255 [DOI] [PubMed] [Google Scholar]
- 50.Sarial S, Shokrgozar MA, Amirzargar A, Shokri F, Radfar J, Zohrevand P, et al. IL-1, IL-1R and TNFalpha gene polymorphisms in Iranian patients with multiple sclerosis. Iran J Allergy Asthma Immunol. 2008;7(1):37–40. Epub 2008/03/07. doi: 07.01/ijaai.3740 . [PubMed] [Google Scholar]
- 51.Tahmasebi Z, Akbarian M, Mirkazemi S, Shahlaee A, Alizadeh Z, Amirzargar AA, et al. Interleukin-1 gene cluster and IL-1 receptor polymorphisms in Iranian patients with systemic lupus erythematosus. Rheumatology International. 2013;33(10):2591–2596. doi: 10.1007/s00296-013-2784-2 . [DOI] [PubMed] [Google Scholar]
- 52.Zhou X, Ca JG, Peng H, Wang JL, Li GM. Association of IL-1alpha gene polymorphism with susceptibility to type 1 diabetes in Chinese children. Genet Mol Res. 2016;15(3). Epub 2016/10/06. doi: 10.4238/gmr.15038025 . [DOI] [PubMed] [Google Scholar]
- 53.Ziaee V, Tahghighi F, Moradinejad MH, Harsini S, Mahmoudi M, Rezaei A, et al. Interleukin-6, interleukin-1 gene cluster and interleukin-1 receptor polymorphisms in Iranian patients with juvenile systemic lupus erythematosus. Eur Cytokine Netw. 2014;25(2):35–40. Epub 2014/08/12. doi: 10.1684/ecn.2014.0352 . [DOI] [PubMed] [Google Scholar]
- 54.Trajkov D, Mishevska-Perchinkova S, Karadzova-Stojanoska A, Petlichkovski A, Strezova A, Spiroski M. Association of 22 cytokine gene polymorphisms with rheumatoid arthritis in population of ethnic Macedonians. Clinical Rheumatology. 2009;28(11):1291–1300. doi: 10.1007/s10067-009-1238-4 [DOI] [PubMed] [Google Scholar]
- 55.Luomala M, Lehtimaki T, Elovaara I, Wang X, Ukkonen M, Mattila K, et al. A study of interleukin-1 cluster genes in susceptibility to and severity of multiple sclerosis. J Neurol Sci. 2001;185(2):123–127. Epub 2001/04/20. . [DOI] [PubMed] [Google Scholar]
- 56.Borzani I, Tola MR, Caniatti L, Collins A, De Santis G, Luiselli D, et al. The interleukin-1 cluster gene region is associated with multiple sclerosis in an Italian Caucasian population. Eur J Neurol. 2010;17(7):930–938. Epub 2010/03/03. doi: 10.1111/j.1468-1331.2010.02952.x . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(A) Begg’s test; (B) Egger’s test.
(TIF)
(DOCX)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.