To the editor
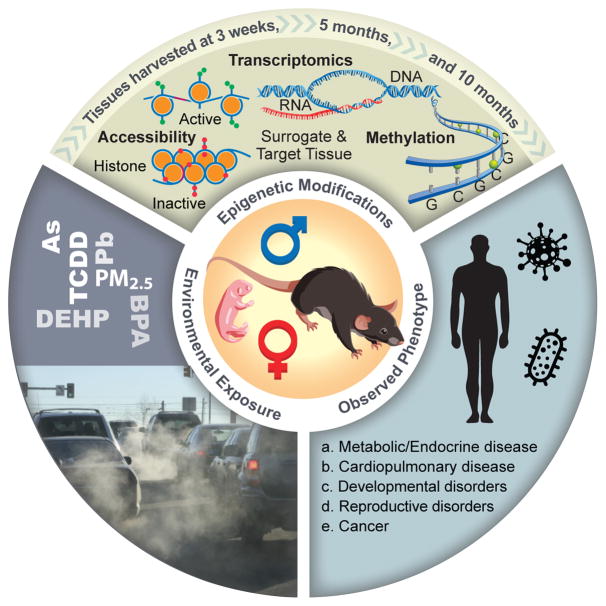
We describe here the aims and scope of one component of the US National Institute of Environmental Health Sciences’ (NIEHS; Durham, NC) multi-phased Toxicant Exposures and Responses by Genomic and Epigenomic Regulators of Transcription (TaRGET) Program to address the role of the environment in disease susceptibility as a function of changes to the epigenome. The TaRGET II Consortium was established in 2016 to explore the conservation of perturbations in epigenomic marks across target tissues/cells (those adversely affected by environmental exposures) and surrogate tissues/cells (those that are easily accessible and reflect the environmental exposures) using mouse models of environmentally relevant exposures (Fig. 1). Many human studies have begun to generate epigenetic data using surrogate tissues with the potential to aid in study design and interpretation. Data from TaRGET II will provide additional exposure-specific insights, and Next-Gen epigenetic signature data will enable further refinement of the design and analysis of human studies where target tissues are inaccessible. In addition, the side-by-side design using target and surrogate tissues from multiple animals will provide insights on the extent of inter-individual differences in response to these exposures.
Figure 1. TaRGET II summary diagram.
Shown are the environmental exposures and associated phenotypes. Tissues are harvested at 3 weeks, 5 months and 10 months and subjected to the indicated epigenomic assays. See text for abbreviation of exposures.
Environmental exposures can alter the epigenome, often referred to as ‘epigenetic reprogramming,’ including changes to DNA methylation, post-translational histone modifications, and chromatin accessibility. The resulting alterations in transcription, both immediately after exposure and revealed by later life events, have been associated with the development of environmentally induced diseases across the lifespan. For example, exposure to heavy metals, such as arsenic and nickel, is associated with epigenetic changes that may underlie the development of diseases, such as cancer, cardiovascular disease, neurological disorders and autoimmune disease1–5.
The human reference epigenome maps generated by consortia, such as the Roadmap Epigenomics Project and the International Human Epigenome Consortium6,7, set the stage for understanding cell type-specific epigenetic patterns and their dysregulation in disease. However, a similar understanding is lacking of how epigenetic patterns are perturbed by environmental exposures and in turn, influence susceptibility to environmental diseases. Thus, a major challenge for the environmental health community is to elucidate the mechanisms responsible for epigenome perturbation that drive pathogenesis of chronic diseases in response to pertinent environmental exposures.
Additionally, it is impossible to sample all relevant tissues involved in disease pathogenesis in human populations. To make direct connections between exposure-induced epigenetic changes and health outcomes, it is therefore critical to determine whether epigenetic alterations are conserved across tissues in such a way that easily sampled surrogate tissues could be used to assess the impact of environmental exposure on disease-relevant but inaccessible target tissues (Table 1). The correlation between exposure-induced epigenetic alterations in target and surrogate tissues is currently unclear and may not be straightforward, as it may depend on the normal epigenetic landscape of the tissues, the timing, route, and dose of exposure, as well as other variables. Determining the utility of surrogate tissue epigenomic analyses will enable more effective use of population-based studies to make connections between exposure, epigenetic changes, and the development of disease. Furthermore, it remains to be elucidated if cessation or elimination of pertinent exposure that may result in reversal of phenotype is associated with changes in the epigenome.
Table 1.
Environmental exposures, associated adverse health outcomes, and target and surrogate tissues.
| Exposures | Outcomes | Target | Surrogate |
|---|---|---|---|
| PM2.5 | Cardiopulmonary and Metabolic effects | Brain, Liver, Adipose tissue, Lung, Macrophages, and Heart | Blood, Skin, and Nasal epithelial cells |
| Lead and Arsenic | Neurodevelopmental toxicity, Metabolic effects, Cancer | Brain, Lung, Heart, Skeletal muscle, Kidney, Liver, and Adipose tissue | Blood/peripheral monocytes, Skin, and Teeth |
| BPA and TBT | Metabolic and Reproductive effects, Cancer | Brain, Uterus, Liver, and Placenta | Blood, Skin, Placenta, and Teeth |
| Dioxin | Neurodevelopmental toxicity | Brain and Liver | Blood and Skin |
| Phthalates | Metabolic and Reproductive effects | Brain, Kidney, and Liver/liver progenitor cells | Blood, Skin, Hair, and Teeth |
The TaRGET II Consortium takes advantage of next-generation sequencing technologies to produce epigenomic maps resulting from environmental exposures, interrogating a broad class of epigenomic features (DNA methylation, histone modifications, and chromatin accessibility) and transcriptomic alterations (RNA-seq) in both target tissues/cells and surrogate tissues/cells under well-defined exposure paradigms (Fig. 1). Across the consortium studies, target and surrogate tissues will be harvested over the life-course following perinatal, peri-adolescent or adult exposure to arsenic, lead, bisphenol A (BPA), tributyltin (TBT), the phthalate di-2-ethylhexyl phthalate (DEHP), the dioxin tetrachlorodibenzo-p-dioxin (TCDD), or air pollution in the form of particulate matter <2.5μ (PM2.5) (Table 1). These tissue- and exposure-specific epigenomic maps will be produced by five consortium groups across the country (Data Production Centers) and supported by a Data Coordination Center, which will coordinate experimental and analytical efforts to maximize consistency, data quality, and overall coverage of the exposure reference epigenomic landscape. Collectively, the consortium will determine changes in target tissues/cells with those of surrogate tissue/cells to identify predictive locus-specific or genome-wide epigenetic alterations, and to dissect the epigenomic marks that precede and after development of the phenotype.
The consortium will also investigate a variety of factors, including timing of exposure and gender, that may in turn influence whether environmentally induced epigenetic changes occur and additionally to what extent these changes are conserved across tissues or persist throughout the lifespan. These will ultimately provide the context with which to interpret the utility and admissibility of surrogate tissues as representative of changes that occur in target tissues (Fig. 1). Together, the TaRGET II research program will enhance our understanding of the relationship between exposure-induced perturbations of epigenetic marks in target versus surrogate tissues; determine exposure conditions and life-course events where surrogate-tissue epigenetic biomarkers are useful; and aid in the design and interpretation of human environmental health studies.
Key to developing a conceptual framework that describes the impact of specific environmental exposures on epigenetic marks in target and surrogate tissue/cell types, and how the conservation of epigenomic signatures is influenced by variables such as timing of exposure, will be the production of comprehensive, high resolution, integrative epigenome maps for both target and surrogate tissues. Importantly, delivery of environmental epigenomic data sets generated by the TaRGET II Consortium to the broader scientific community will facilitate further data analyses by integrating additional data from specific target:surrogate pairs and for different epigenetic marks, as well as promoting ancillary studies to develop data on the functional impact of these epigenetic alterations.
As part of its mission, the TaRGET II Consortium will provide the scientific community with a plethora of high-resolution, integrative epigenome profiles in response to environmental exposure. Raw sequencing data will be quality controlled and processed using state-of-the-art metrics and pipelines developed by the Consortium for each experimental assay. Furthermore, each file will be densely annotated with multiple layers of informative metadata that will enable comparisons across experiments and studies. The Consortium will facilitate integrative analysis through the development of software and algorithms, such as tools to model correlated epigenome changes between target and surrogate tissues or to transfer epigenomic signatures from mouse to orthologous human (epi)genomes. Like the data, these tools will be publically available to support future complementary analyses by the environmental health research community.
Descriptions of the consortium and its goals and protocols detailing target/surrogate tissue collection, epigenomic assays, and exposure paradigms are available at the consortium website (http://targetepigenomics.org). Additionally, raw and processed datasets will be publicly accessible through a data portal created by the Data Coordination Center, which will offer tools for in-browser analysis and visualization of datasets, including links to the Washington University Epigenome Browser8.
The TaRGET II Consortium will greatly advance our knowledge of the epigenetic mechanisms linking environmental exposure to disease, how they are influenced by the timing and dosage of exposure, and their persistence across the life-course. Specifically, the integrative datasets we generate will determine the utility of surrogate tissues to detect epigenomic changes in tissues that are disease-relevant but inaccessible in humans, facilitating future human population-based studies.
References
- 1.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nature Reviews Genetics. 2012;(13):97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 2.Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. Int J Epidemiol. 2012;41(1):79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Somers EC, Richardson BC. Environmental Exposures, Epigenetic Changes and the Risk of Lupus. Lupus. 2014;23(6):568–576. doi: 10.1177/0961203313499419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baccarelli A, Ghosh S. Environmental Exposures, Epigenetics and Cardiovascular Disease. Current opinion in clinical nutrition and metabolic care. 2012;15(4):323–329. doi: 10.1097/MCO.0b013e328354bf5c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perera F, Herbstman J. Prenatal environmental exposures, epigenetics, and disease. Reproductive toxicology. 2011;31(3):363–373. doi: 10.1016/j.reprotox.2010.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stunnenberg HG, Hirst M The International Human Epigenome Consortium. A Blueprint for Scientific Collaboration and Discovery. Cell. 2016;167(5):1145–1149. doi: 10.1016/j.cell.2016.11.007. doi.org/10.1016/j.cell.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA, Koebbe BC, Nielsen C, Hirst M, Farnham P, Kuhn RM, Zhu J, Smirnov I, Kent WJ, Haussler D, Madden PA, Costello JF, Wang T. The Human Epigenome Browser at Washington University. Nat Methods. 2011;8(12):989–90. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]