Abstract
Identifying regional effects of interest in MRI datasets usually entails testing a priori hypotheses across many thousands of brain voxels, requiring control for false positive findings in these multiple hypotheses testing. Recent studies have suggested that parametric statistical methods may have incorrectly modeled functional MRI data, thereby leading to higher false positive rates than their nominal rates. Nonparametric methods for statistical inference when conducting multiple statistical tests, in contrast, are thought to produce false positives at the nominal rate, which has thus led to the suggestion that previously reported studies should reanalyze their fMRI data using nonparametric tools.
To understand better why parametric methods may yield excessive false positives, we assessed their performance when applied both to simulated datasets of 1D, 2D, and 3D Gaussian Random Fields (GRFs) and to 710 real-world, resting-state fMRI datasets. We showed that both the simulated 2D and 3D GRFs and the real-world data contain a small percentage (< 6%) of very large clusters (on average 60 times larger than the average cluster size), which were not present in 1D GRFs. These unexpectedly large clusters were deemed statistically significant using parametric methods, leading to empirical familywise error rates (FWERs) as high as 65%: the high empirical FWERs were not a consequence of parametric methods failing to model spatial smoothness accurately, but rather of these very large clusters that are inherently present in smooth, high-dimensional random fields. In fact, when discounting these very large clusters, the empirical FWER for parametric methods was 3.24%. Furthermore, even an empirical FWER of 65% would yield on average less than one of those very large clusters in each brain-wide analysis. Nonparametric methods, in contrast, estimated distributions from those large clusters, and therefore, by construct rejected the large clusters as false positives at the nominal FWERs. Those rejected clusters were outlying values in the distribution of cluster size but cannot be distinguished from true positive findings without further analyses, including assessing whether fMRI signal in those regions correlates with other clinical, behavioral, or cognitive measures. Rejecting the large clusters, however, significantly reduced the statistical power of nonparametric methods in detecting true findings compared with parametric methods, which would have detected most true findings that are essential for making valid biological inferences in MRI data. Parametric analyses, in contrast, detected most true findings while generating relatively few false positives: on average, less than one of those very large clusters would be deemed a true finding in each brain-wide analysis. We therefore recommend the continued use of parametric methods that model nonstationary smoothness for cluster-level, familywise control of false positives, particularly when using a cluster defining threshold of 2.5 or higher, and subsequently assessing rigorously the biological plausibility of the findings, even for large clusters. Finally, because nonparametric methods yielded a large reduction in statistical power to detect true positive findings, we conclude that the modest reduction in false positive findings that nonparametric analyses afford does not warrant a re-analysis of previously published fMRI studies using nonparametric techniques.
Keywords: Random Fields, Cluster Level Inference, Permutation Testing, Parametric Testing, Family Wise Error Rates, Euler Characteristics
1. Introduction
Magnetic Resonance Imaging (MRI) has provided the opportunity to study the structure, function, and metabolism of the living brain[1–3], including its in vivo maturation, sex differences[4], illness-related effects[5–14], and the responses to pharmacological or behavioral interventions[15–20]. Advanced MRI techniques collect vast amounts of data at millimeter-level spatial resolution across many participants. Identifying biologically relevant effects in MRI data is challenging because of the presence of both individual variability and noise in the data, combined with the sheer number of voxels at which tissue characteristics of the brain are sampled. Hypothesis testing is conducted at each voxel to identify brain regions where MRI measures significantly associate with a condition or feature of interest. Real inter-individual variability or noise in the MRI measures can produce a statistically significant effect for that condition of interest, simply on the basis of chance, in the absence of a true effect. This is termed a false positive, or Type I, error. The probability of these errors increases as the number of statistical tests increases, especially in MRI data, where hundreds of thousands of tests are conducted at all voxels of the brain.
A number of statistical procedures have been proposed to reduce the probability of false positive findings in MRI datasets, including Bonferroni correction[21, 22], methods based on the Random Field Theory (RFT)[23–25] used in Statistical Parametric Mapping (SPM)[23], False Discovery Rate (FDR)[26, 27], and permutation testing[28, 29] and other nonparametric techniques[30, 31]. Bonferroni correction, which assumes the data are statistically independent across voxels, divides the nominal significance level by the number of statistical tests to yield the adjusted significance level for assessing significance of each individual statistical test. Although this correction reduces the likelihood of false positives, it also dramatically reduces statistical power, or the ability to detect a true finding when it is in fact present, because the intercorrelated values across voxels in brain images violates the Bonferroni assumption that data in neighboring voxels are statistically independent of one another. Consequently, methods based on the RFT have been developed that specifically model the correlations of values across voxels and that control false positives by minimizing the family-wise error rate (FWER) – i.e., the probability that the largest value of the random field at any one voxel among all voxels will be larger than a prespecified threshold. SPM, in particular, applies general linear model and parametric RFT to control for false positives when conducting multiple hypothesis testing. RFT-based methods, however, assume that the data vary sufficiently smoothly across voxels, an assumption that is generally satisfied in MRI data smoothed using a Gaussian filter. However, failure to accurately model heterogeneity in spatial smoothness and greater spatial autocorrelation function (SACF) than assumed in theory could possibly lead to greater FWER than those predicted by the theory[32]. Nonparametric methods of statistical inference, such as permutation testing, do not presuppose a given probability distribution but instead discovers the distribution from the data. Therefore, these methods can more accurately assess whether the data sufficiently support rejection of a specific null hypothesis, though they may have low statistical power (i.e., high Type II error rates), especially when the number of participants is small. Sensitivity of nonparametric as well as parametric methods can be improved by applying methods for threshold free cluster enhancement[33] of the imaging data. The False Discovery Rate (FDR)[27], in contrast, allows a pre-specified proportion of false positive findings, but with the aim of improving statistical power. Topological FDR[27] is more accurate than voxelwise procedure for FDR when controlling for false positive. The number of false positives that FDR permits depends on both the statistical procedure and whether the data are distributed as assumed. Assessing whether false positives are present at the nominal rate when any one of these statistical procedures is applied to real-world data is essential for making valid inferences about the brain from statistically significant findings.
Biologically valid MRI findings typically form clusters of voxels within the brain,[24] because the adjoining and interconnected cellular elements in tissues that support a given information processing function are considerably larger than a voxel[34, 35]. Their statistical significance is most commonly assessed using parametric methods, especially those based on GRF theory, as they are incorporated in freely available software packages such as SPM[36, 37], FSL[38], and AFNI[39]. Several studies[24, 40–44], especially the study that first proposed these methods[24], have assessed the validity of these parametric methods. These studies showed that parametric distributions accurately modeled the data and that the empirical FWERs were close to their nominal values. These studies, however, assessed the validity of their methods using only 1D[40] or 3D data,[24, 32, 42–45] and they typically performed the validation using only simulated datasets. Concerns have been raised as to whether their simulated data sufficiently modeled the degree of spatial smoothness in real-world data and whether the failure to model smoothness accurately may yield far more false positives than the nominal rate when these parametric procedures are applied to real-world data.[32, 43]
To assess whether the empirical FWERs equaled the nominal FWERs in real-world data, both parametric and nonparametric methods were applied[32] in more than 3 million statistical analyses applied to real-world fMRI datasets from 499 participants. This widely publicized study concluded that when SACF for data has longer tail than Gaussian distribution or when SACF is nonstationary across the image domain, parametric methods yielded an empirical FWER, or false positive rate, of 0.3 or higher -- much higher than the nominal FWER of 0.05.[32] This unacceptably high rate of false positive findings generated using parametric methods was suggested to derive from the greater smoothness in real-world data that generated clusters of sizes larger than were modeled parametrically under null hypothesis, i.e. in the absence of true effects in the data. The same study also reported, in contrast, that nonparametric methods generated false findings at exactly the nominal FWER of 0.05, presumably because those methods estimated the distributions from data and, therefore, accurately modeled smoothness of the data when calculating statistical significance of the clusters. The implications of these conclusions, if valid, were staggering, as a false positive rate of more than 30% suggested that previously reported findings about the brain in health and illness were neither reproducible nor biologically valid, and those concerns applied not only to fMRI studies, but to studies using any brain imaging modality that employed cluster-level parametric methods when correcting for multiple statistical comparisons. The study provocatively concluded that those datasets require reanalysis using nonparametric techniques.
Before undertaking that enormous, if not impossible task, it is important first to understand more deeply why parametric statistical procedures may have yielded high empirical FWERs, why the empirical FWERs varied from small (1%) to very large (70%), why in contrast the empirical FWERs for simulated data equaled their nominal FWERs, and how using parametric methods may have affected the ability to detect real effects in the data. At the same time, it is equally important to understand why nonparametric methods generated false positives at the nominal FWER for each and every analysis. Finally, it is important understand how the ability to detect real effects (statistical power) is influenced by various aspects of data processing and statistical analysis, especially the degree of smoothness these procedures introduce into the data, the Cluster Defining Threshold (CDT) employed, and the dimensionality of the random fields used for both parametric and nonparametric methods of statistical inference.
We therefore conducted experiments using both simulated and real-world datasets to understand why controlling for false positives with parametric methods during the testing of multiple hypothesis yielded much higher empirical FWERs than nominal values, whereas nonparametric methods generated false positives at the nominal FWE rates. The use of simulated data allowed us to control the experimental conditions precisely and, therefore, to evaluate how the performance of parametric methods varied with varying degrees of smoothness or statistical thresholds used to define spatial clusters and with variation in the dimensionality of the data. Varying performance across differing dimensionality data will allow us to understand better why parametric methods fail in one but not the other dimensional data. Simulated data, however, may fail to model adequately certain aspects of real-world random fields, particularly their degree of spatial smoothing, which may have produced high FWER when parametric methods are applied to real-world datasets. We therefore also assessed performance of both parametric and nonparametric methods when applied to a large, real-world resting-state fMRI dataset comprising 710 participants combined across 5 independent studies. We assessed whether varying cluster sizes, the mean cluster size, and the numbers of clusters affected accuracy when modeling the parametric probability distributions, whether parametric methods appropriately and sufficiently controlled for FWERs, and whether nonparametric methods generated false positives at the nominal FWERs. Our overarching aim was to assess whether nonparametric methods must be used to analyze fMRI datasets and whether indeed all previous fMRI studies should be reanalyzed using nonparametric techniques.
2. Materials and methods
2.1 Datasets
We assessed parametric and nonparametric methods for cluster-level inference using simulated as well as five large, real-world studies comprising resting state fMRI datasets from 710 participants.
2.1.1 Simulated Gaussian Random Fields
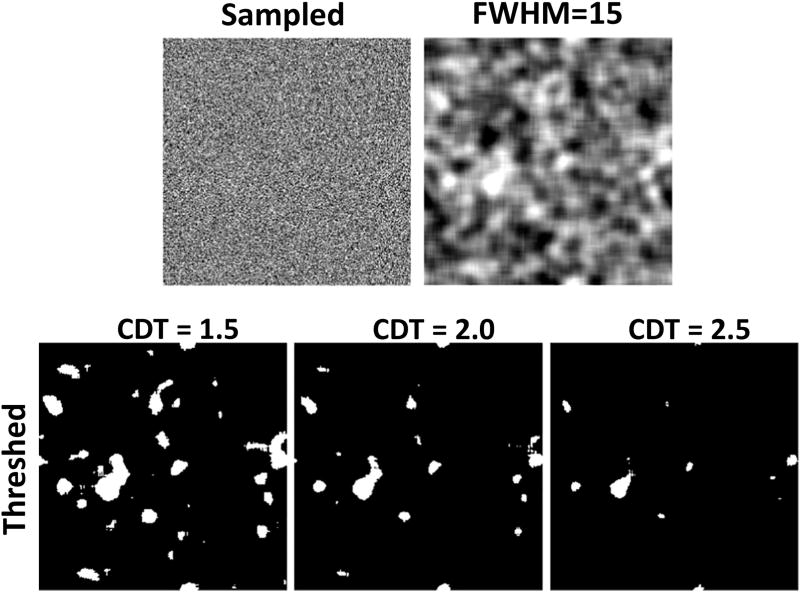
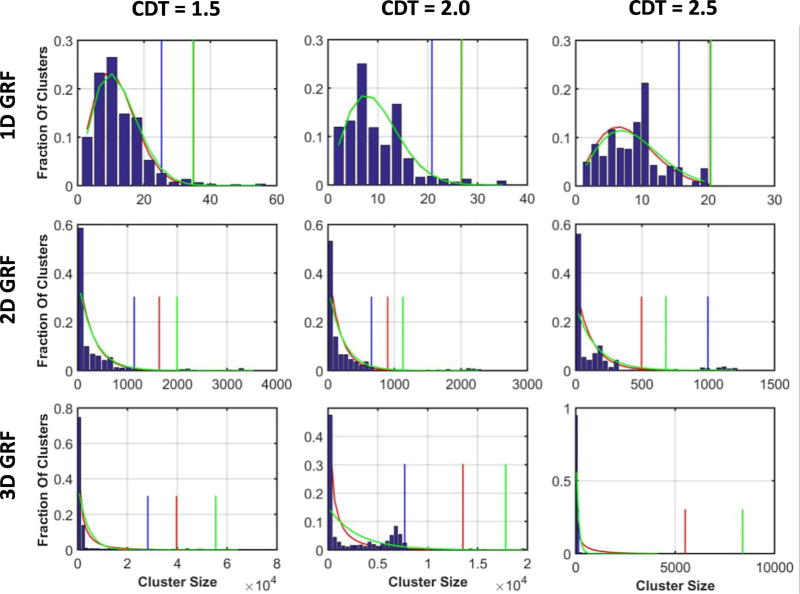
We simulated 50 realizations each of 1D, 2D, and 3D GRFs as follows. From a Gaussian distribution with mean zero and unit variance N(0,1), we sampled a sequence of 10,000 random numbers for each realization of a 1D GRF, a 2D array of 250 × 250 random numbers for each realization of a 2D GRF, and a 3D array of 90 × 90 × 90 random numbers for each realization of a 3D GRF. We smoothed each of those realizations using a Gaussian smoothing kernel with full width at half maximum (FWHM) of either 5, 10, 15, 20, 25, 30, or 35. We wrapped the smoothing kernel around the edges of the domain of the random field to ensure stationarity of the smoothed random fields. We then subtracted the residual mean and normalized the values, so that each smoothed random field had a zero mean and unit variance. We then defined clusters in the smoothed random fields by thresholding the smoothed fields at CDTs of Z=1.0, 1.5, 2.0, 2.5, 3.0, or 3.5 corresponding to uncorrected p-values 0.158, 0.0668, 0.0228, 0.0062, 0.0013, and 0.0002, respectively; we then computed the numbers and sizes of clusters in each thresholded realization of GRF (Figure 1).
Figure 1.
2.1.2 Real-World Datasets
Rationale for Use of Resting-State fMRI Datasets
Resting-state fMRI data are usually used to study functional brain connectivity[46] by measuring spatio-temporal correlations in spontaneously fluctuating fMRI signal[47, 48] across the brain. These data, therefore, naturally incorporate into their measures the spatial intercorrelations across voxels that derive in part from biologically driven fluctuations in the fMRI time series, in part from the point-spread function that inherently accompanies the low spatial resolution of fMRI data, and in part from the spatial smoothing of data performed during preprocessing. The spontaneous fluctuations in fMRI signal should not correlate with any arbitrary condition contrast (i.e., subtraction paradigm) to which the fMRI time series are subjected. We, therefore, used resting-state fMRI data to generate statistical contrasts under the null hypothesis -- i.e. for fMRI time series that contained no task-related brain activity but that had the spatial smoothness and noise characteristics of real-world fMRI data.
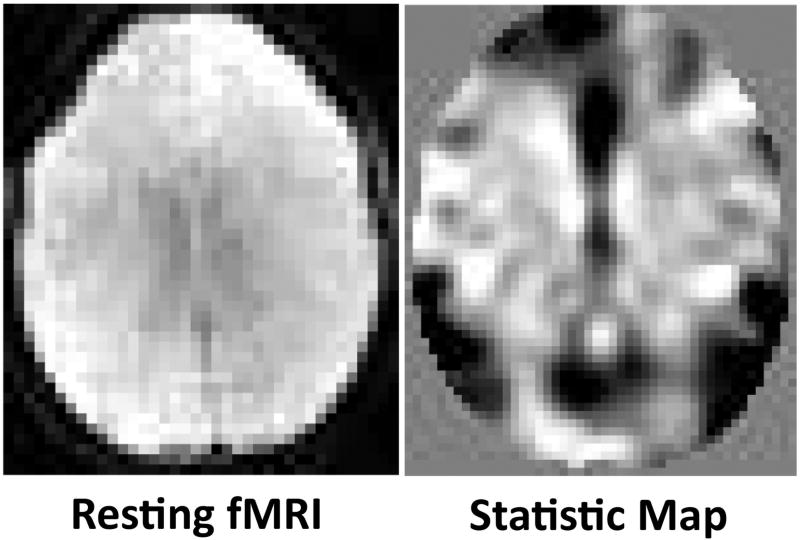
To generate these null hypothesis contrast maps, we preprocessed and modeled in the resting state data an event related design that alternated the falsely hypothesized activity (the ‘on’ condition) with rest (the ‘off’ condition) on consecutive epochs of twenty fMRI volumes throughout each run. We then performed a first-level statistical contrast across those conditions with a design matrix having 2 columns for each run of the resting state data: the first column incorporating 1’s for the ‘on’ condition and 0’s for ‘off’ condition, and the second column incorporating a constant term representing average fMRI signal intensity in the time series. The design matrix was convolved with hemodynamic response function of length 32.03 seconds. We computed the contrast for the first column in the design matrix. We visually assessed whether the contrast had large regions of similar values due to inherent correlations in resting state data. The statistical parametric map for the t-statistic of the contrast was used for the subsequent analyses. Because the falsely hypothesized task-related activity would correlate only with noise in the resting state fMRI signal, the estimated regression coefficient (i.e., beta value) and corresponding statistic at each voxel within each participant would represent only statistical noise within the fMRI signal. From these coefficients we then constructed a normalized z-statistic map across the entire brain for each participant that represented the null hypothesis of no task-related brain activity while at the same time containing the spatial correlations inherent in real-world fMRI data (Figure 2).
Figure 2.
Participants
We assessed the parametric and nonparametric approaches for their FWER using 710 resting state fMRI datasets acquired in 5 independent studies: (1) One of 61 autistic and 81 typically developing children and adults in the Autism Spectrum Disorder (ASD) study[49] (“Autism”); (2) Another of 123 children and young adults who were at either a low or high familial risk for depression[5, 50] (“High Risk”). (3) Another acquired 116 fMRI data longitudinally at three time points (pretreatment baseline, 10, and 12 weeks) during a clinical trial assessing the efficacy of an antidepressant medication in treating 40 participants with depressive disorder[51, 52] (“Depression”). Although within-subject, resting state fMRI data will be correlated across the 3 time points, maps for the test statistic under the null hypothesis derived from these data will be statistically independent. (4) Another acquired fMRI data at one time point in 39 children and adolescents with stuttering disorder and 30 age- and sex-matched healthy controls[53] (“Stuttering”). (5) Finally, one acquired cross-sectional fMRI data for assessing the effects of air pollution on the brains of 260 healthy children[54–56] (“Toxins”) who at the time of scan did not have any neuropsychiatric disorder even though they showed symptoms of anxiety, depression, or inattention[57] and had reduced full-scale and verbal IQ.[58] All resting state fMRI data were acquired in two runs, each comprising 140 images. Data for each study were processed and assessed independently; however, some of the statistics were averaged across all studies as indicated. All adult participants provided written informed consent. Child participants provided informed assent, and their legal guardian provided written informed consent. Study procedures were approved by the Institutional Review Board of the New York State Psychiatric Institute.
MRI Pulse Sequences
All MRI data were acquired on a 3T GE Signa whole body scanner using 8 channel head coil at New York State Psychiatric Institute.
Anatomical MRI: We acquired high resolution, T1-weighed anatomical MRI data using a 3D spoiled gradient recall (SPGR) sequence with spatial resolution = 0.98 × 0.98 × 1.0 mm3, repetition time (TR) =4.7 ms, echo time (TE) = 1.3 ms, inversion time (TI) = 500 ms, flip angle (FA) = 11°, matrix size = 256 × 256, field of view (FOV) = 25 × 25 cm2, slice thickness = 1.0 mm. Anatomical MRI data were used to spatially normalize all participant images into a common template space.
Resting-State fMRI: Resting state blood oxygen level dependent (BOLD) fMRI data were acquired using an axial echoplanar imaging sequence with TR = 2200 ms, TE = 30 ms, FA = 90°, receiver bandwidth = 62.5 kHz, single excitation per image, slice thickness = 3.5 mm, slice gap = 0 mm, FOV = 24 × 24 cm2, matrix size = 64 × 64. During image acquisition, participants were instructed to remain still with their eyes closed and to let their minds wander freely. Two 5 minutes 21 seconds resting-state scans with 140 volumes in each run were obtained for every participant.
Processing of the fMRI Data
All fMRI data were processed using the typical preprocessing methods in SPM8 software (http://www.fil.ion.ucl.ac.uk/spm/) as follows[59]: slice-time correction using the middle slice as the reference; motion correction by realigning functional volumes to the middle volume in each run; temporal smoothing using a Gaussian kernel; coregisterating to each participant’s anatomical scan; spatially normalizing coregistered fMRI data into the Montreal Neurological Institute (MNI) space; resampling a spatial resolution of 2mm3; and spatial smoothing using a Gaussian kernel with FWHM of 6mm.[60] A run with motion greater than one voxel between consecutive functional volumes were excluded from further analyses[59].
2.2 Empirical Mean Number of Clusters
We thresholded each realization of our simulated and real-world datasets at varying CDT (u). Thresholding generated a binary field with values equal to either 0 or 1 at locations where the smoothed field had values smaller or larger, respectively, than the threshold value. Thus, contiguous regions with a value of 1 defined clusters in which the smoothed random field had values greater than the CDT. We then counted the number m and the size n of clusters within each binary field. We also computed the empirical mean number of clusters by averaging the number of clusters across all random field realizations. We generated the histogram of cluster size for clusters in all random field realizations and then normalized the histogram such that weights of the bar in the histogram summed to 1.0.
2.3 Expected Euler Characteristic
For a D-dimensional, real-valued function , an excursion set Au(F, S) on any volume S ⊂ ℝD for a fixed number u is defined as the set of all locations t where the function F(t) is greater than u, i.e., Au(F, S) = {t ∈ S:F(t) ≥ u}. The Euler characteristic χ(Au) of an excursion set Au(F, S) equals the number of up-crossings (i.e. the number of clusters) of the function above a specified CDT u[61, 62]. The expected Euler characteristic E(χ) for a D-dimensional random field equals the expected number of up-crossings above a specified threshold. For a smooth random field X(t) with variance σ2 defined over a D-dimensional volume S, the E(χ) at a threshold u is defined as[61–65]
where L(S) is the Lebesgue measure of the volume S, A is the determinant of the covariance matrix for the first order partial derivatives of the random field X(t), and P(u) is the Hermite polynomial[63] defined as . The E(χu) simplifies to E(χu) = L(S)·(2π)−1·|A|1/2·e−u2/2 for a 1D random field with unit variance, E(χu) = L(S)·(2π)−3/2·|A|1/2·u·e−u2/2 for a 2D random field with unit variance, and E(χu) = L(S)·(2π)−2·|A|1/2·(u2 − 1)·e−u2/2 for a 3D random field with unit variance.
2.4 Parametric Distributions and Cluster Level Inference
The number of clusters m is assumed, asymptotically for a large CDT u, to be Poisson distributed[24, 66] as , where λ = E(χ) is the mean and variance of the number of clusters. Although a Poisson distribution is satisfying because, for λ > 5, it tends to a Gaussian distribution with mean and variance λ, the distribution is not validated using either simulated or real-world data. The distribution P(n = k) of the cluster size n is assumed[24, 67] to be , where , and N is the number of locations with values greater than the threshold u. For cluster-level inference while controlling for multiple statistical tests, the FWERs, or the probability of at least one cluster having size greater than k, is computed as . That is, for a large CDT u, cluster-level inference is equivalent to Bonferroni correction[68, 69], because the clusters likely are distributed independently in the volume S for large values of u.
2.5 Nonparametric Methods for Familywise Control of False Positives
Nonparametric methods, such as permutation testing, do not a priori assume a particular model for the distributions of either data or test statistics, but instead discover the distribution for the test statistic from the data. Nonparametric methods therefore require fewer untested assumptions of the data and can be applied to any test statistic. These methods all calculate a test statistic from the data, estimate the probability distribution of the test statistic under the assumption that the data satisfy the null hypothesis, and then use the estimated distribution to evaluate the probability value for the computed test statistic. Permutation testing, when applied to data from a single group of participants, specifically assumes that the data are distributed symmetrically around a mean value,[32, 70] and then flips the data about the mean for randomly selected participants to estimate the probability distribution of the test statistic under the null hypothesis. When applied to data from two groups of participants, permutation testing reassigns participants to one of the two groups by permuting their group labels. The nonparametric procedure that controls for false positive findings across multiple statistical tests of the null hypothesis (i.e. familywise control of false positives) calculates the size of the largest cluster for each of these permutations, and then forms a histogram of those cluster sizes across all of the permutations[32, 71]. The histogram is normalized so that it sums to 1.0 and thereby estimates the probability distribution for the size of the largest cluster under the null hypothesis. The P-value for any given cluster in the dataset (i.e. the probability of finding a cluster as large or larger) with true participant assignments, while controlling for multiple statistical tests, is calculated as the fraction of cluster with sizes greater than the size of that given cluster.
2.6 Experiments
We computed empirical FWERs for parametric method as the fraction of analyses with at least one cluster of size greater than the cluster size at its nominal FWER[32]. We conducted the following experiments to assess how well empirical FWERs for either parametric or nonparametric methods approximated their nominal values, and if they differed from those values then what the source of difference was likely to be.
-
Using the smoothed 1D, 2D, and 3D random fields as well as the real-world data, we computed E(χ) and assessed whether it differed significantly from the empirically identified average number of clusters Em. The number and size of clusters in simulated and real-world random fields were calculated by accounting for the wraping of clusters around the boundaries of random fields.
The smoothness of real-world random fields was estimated from the covariance matrix for the first order partial derivatives of a random field. We hypothesized that E(χ) would not differ from Em.
We generated histograms of the cluster sizes for varying CDTs and varying amounts of smoothness in both the simulated GRFs and real-world data, and then fitted an appropriate parametric distribution to the histogram. In the histogram we also plotted the distribution of the theoretically predicted cluster size P(n = k). We used the Kolmogorov-Smirnov statistic to compare the differences between the two distributions. The null hypothesis was that the fitted distribution would not differ from the theoretical one.
We also assessed whether the number of clusters was distributed according to the theoretically assumed Poisson distribution, and whether use of a delta distribution affected the calculated FWER for clusters. Using a narrower distribution for the number of clusters would allow us to assess sensitivity of the parametric methods to the assumed form of the distribution.
We derived an expression for the FWER P(nmax ≥ k) using E(χ) and Em as the mean number of clusters, and then, using these two different parametric formulations, calculated the cluster sizes and where the FWER equaled the nominal value of 0.05 using E(χ) and Em, respectively.
We also generated the histogram for the size of the largest cluster in each realization of the random fields, which we used for nonparametric inferences on cluster size. Using this histogram we calculated the cluster size such that a fraction of 0.05 largest clusters had a size greater than . We then numerically compared the cluster sizes , and as well as used them to assess statistical power for the parametric and nonparametric methods.
We assumed that in the presence of true effects, i.e. under an alternate hypothesis, cluster size was Gaussian distributed with varying mean μ and standard deviation σ. We systematically varied the mean from small to large values and then calculated statistical power for the cluster sizes (for parametric inference) and (for nonparametric inference) estimated at a FWER of 0.05. We expected that parametric methods would provide greater statistical power than would nonparametric methods.
3. Results
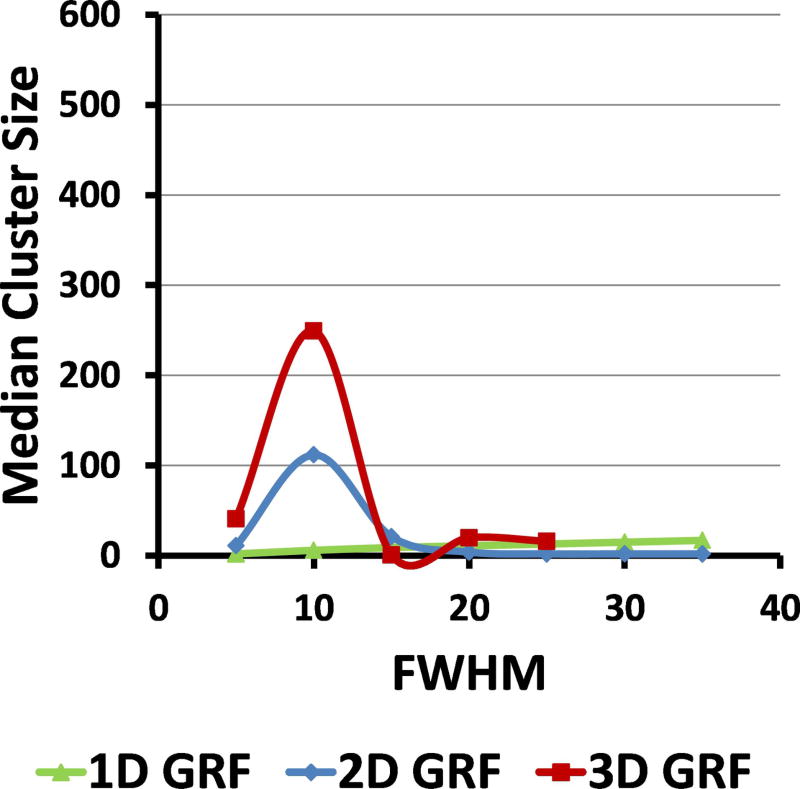
In our simulated GRFs the median size of clusters defined at CDT = 2.5 varied smoothly with varying amounts of smoothness: as the FWHM of the smoothing Gaussian kernel increased from 5 to 35 voxels, the median size of clusters first increased and then decreased to the smallest value for FWHM of 15 voxels (Figure 3). That is, FWHM = 15 voxels reduced the false positives clusters in simulated 3D GRFs and was nearly optimal in simulated 2D GRFs. We therefore presented our findings for simulated GRFs that were smoothed with a Gaussian kernel of FWHM = 15 voxels.
Figure 3.
3.1 Euler Characteristics and Average Number of Clusters
The E(χ) did not differ significantly from the empirically identified average number of clusters at any CDT in simulated 1D GRFs that were smoothed by Gaussian kernels of varying FWHMs (Table 1). For 2D GRFs, however, the empirically identified average number of clusters was significantly higher than the E(χ) (p < 0.0001, df = 49, one sample t test) (Table 1). Similar to the 2D GRFs, the number of clusters for the simulated 3D GRFs (Table 1) and for the real-world data (Table 2) was significantly higher than the E(χ) (p < 0.0001, df = 49, one sample t test). At a CDT of 3.0 or higher for the real-world data but not for the simulated 3D GRF, however, the number of clusters was significantly higher than the E(χ) (Table 1), suggesting greater excursions of random field values in real-world data than in the simulated random fields.
Table 1.
Comparing the Expected Euler Characteristic (EC) with the Average Number of Clusters in 50 simulated fields each in 1D, 2D, & 3D GRFs smoothed using a Gaussian kernel of increasing full width at half maximum (FWHM). We thresholded the smoothed fields with increasing Cluster Defining Thresholds (CDTs) and averaged the number of clusters across all 50 realizations of the smoothed GRFs. The expected ECs are shown in parentheses. For 1D GRFs, the average number of clusters did not differ significantly (one sample t test, df = 49) from the expected EC for any combination of CDT and FWHM. Furthermore, χ2 test for one population variance shows that the variance of the empirical variance for the number of clusters is statistically significantly smaller in general than the theoretically assumed variance (p<0.005, χ2 = 25.08, for FWHM=15, CDT = 2.5). For 2D & 3D GRFs, the average number of clusters was significantly higher (P-value < 0.0001, df = 49, one sample t test) than the expected EC, except at CDT = 3.0 in 3D GRFs. Therefore, the 2D & 3D random fields have much higher number of clusters under null hypothesis than predicted by the theory.
| GRF | Cluster Defining Threshold Average Number of Clusters (and Expected ECs) |
||||
|---|---|---|---|---|---|
| FWHM | 1.5 | 2.0 | 2.5 | 3.0 | |
| 1D | 5 | 169±0.73 (163.4) | 70±0.69 (68.1) | 22.2±0.63 (22.1) | 4.8±0.33 (5.9) |
| 10 | 85.1±0.93 (84.8) | 35.8±0.73 (35.3) | 11.2±0.47 (11.4) | 3.8±0.19 (2.9) | |
| 15 | 54.2±0.57 (57) | 25.2±0.50 (23.8) | 8.4±0.30 (7.7) | 2.66±0.13 (1.9) | |
| 20 | 41.2±0.65 (43) | 18.6±0.38 (17.9) | 7.26±0.22 (5.8) | 2±0.13 (1.5) | |
| 25 | 33.3±0.64 (34.5) | 15±0.31 (14.4) | 5.4±0.18 (4.7) | 1.4±0.14 (1.2) | |
| 2D | 5 | 210.5±0.93 (68.4) | 118.5±0.56 (38) | 53.5±0.38 (15.4) | 15.7±0.18 (4.7) |
| 10 | 55.3±0.35 (17.6) | 30±0.25 (9.8) | 14.8±0.21 (4) | 8.4±0.16 (1.2) | |
| 15 | 50.9±0.54 (14.4) | 29.6±0.50 (8) | 15.5±0.31 (3.2) | 4.7±0.13 (1) | |
| 20 | 94.3±0.97 (19.7) | 41.9±0.62 (10.9) | 17.8±0.34 (4.5) | 6.9±0.29 (1.3) | |
| 25 | 124.2±1.15 (24.9) | 57.8±0.98 (13.8) | 24.1±0.52 (5.6) | 7.8±0.26 (1.7) | |
| 3D | 5 | 259.6±1.14 (168.9) | 265.8±1.04 (168.9) | 175.2±0.78 (96) | 35.8±0.62 (37) |
| 10 | 47.18±1.12 (23.8) | 55.9±0.96 (23.8) | 33.1±0.46 (13.5) | 0.5±0.08 (5.2) | |
| 15 | 45.8±1.13 (11.7) | 35.1±1.32 (11.7) | 32.1±0.68 (6.6) | 0.5±0.10 (2.5) | |
| 20 | 116.5±3.46 (14) | 109±1.67 (14) | 27±0.66 (8) | 2.2±0.29 (3) | |
| 25 | 200.4±3.72 (18.3) | 97.1±3.0 (18.3) | 42.2±0.78 (10.4) | 3±0.30 (4) | |
Table 2.
Comparing Expected Euler Characteristic (EC) and Average Number of Clusters in real-world, resting-state fMRI studies from a total of 710 participants across 5 independent studies. We thresholded the smoothed fields at increasing Cluster Defining Thresholds (CDTs) and averaged the number of clusters across all participants in each study. The average numbers of clusters were significantly higher than the expected ECs shown in the brackets (P value < 0.0001, one sample t test, df=differed across studies).
| Study | Cluster Defining Threshold Average Number of Clusters (and Expected ECs) |
|||
|---|---|---|---|---|
| 1.5 | 2.0 | 2.5 | 3.0 | |
| Autism (N = 142) | 45.4±1.67 (12.6) | 34.8±1.37 (12.6) | 22.5±1.11 (7.2) | 12.8±0.92 (2.8) |
| High Risk (N = 123) | 51.2±1.81 (8.7) | 31±1.44 (8.7) | 15.8±1.23 (4.9) | 7.7±0.98 (1.9) |
| Depression (N = 116) | 44.9±1.69 (15.9) | 34.3±1.61 (15.9) | 22.8±1.27 (9.0) | 12.9±0.96 (3.5) |
| Stuttering (N =69) | 61.7±2.58 (13) | 42.9±2.1 (13) | 22.5±1.44 (7.4) | 11.6±1.25 (2.6) |
| Toxins (N=260) | 51.6±1.17 (10.9) | 38±0.99 (10.9) | 23.4±0.82 (6.2) | 12.4±0.63 (2.4) |
3.2 Distribution of the Number of Clusters
The empirically computed variance for the number of clusters was significantly smaller (p<0.005, χ2 test for one population variance; Table 1) than the variance of the theoretical Poisson distribution for the number of clusters for 1D GRFs, and for 2D and 3D GRFs smoothed by Gaussian kernels of FWHM = 5. In other words, the number of clusters across all realizations of the smoothed GRFs was distributed closer to the average number than assumed in the theoretical distribution. The distribution for the number of clusters determines the probability distribution for the size of the largest cluster, and therefore the distribution also determines the FWER corrected p-values for the clusters. Thus, we next assessed how a narrower distribution for the number of clusters may affect the FWER correction. We assumed that the number of clusters is a delta distribution – i.e., that the distribution has nonzero support only at the average number x0 and is zero at all other cluster sizes. Therefore, for P(m = x) = δx0 the probability P(nmax ≥ k) that the size of the largest cluster is greater than k is evaluated as
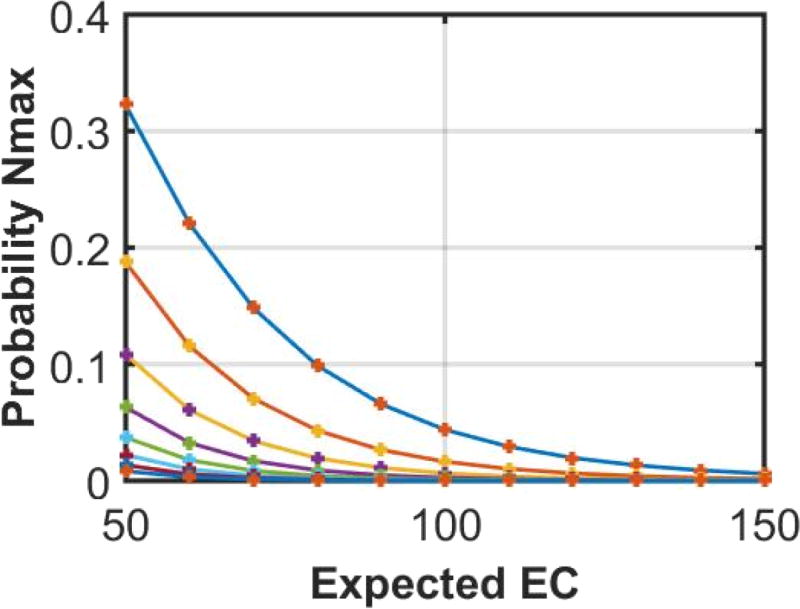
The plots of P(nmax ≥ k) and P(nmax ≥ k)|δ show that the two probabilities matched closely for varying values of x0 and P(n < k) (Figure 4), thereby indicating that the probability P(nmax ≥ k) is not sensitive to the analytic form of the parametric distribution for the number of clusters.
Figure 4.
3.3 Empirical and Parametric Distributions for Cluster Size
We generated histograms of cluster sizes and superimposed the parametric distribution predicted by theory, as well as the parametric distribution fitted to the histogram that maximized the likelihood of the observed data (Figures 5 & 6). The theoretical distribution did not differ significantly from the histogram of cluster sizes as assessed using the Kolmogorov-Smirnov test[72, 73] (Table 3), providing strong statistical evidence that cluster sizes in the simulated random fields are distributed according to the theoretical distribution predicted using E(χ), even though E(χ) differed significantly from the average number of clusters in the random fields. Although the random fields were distributed according to the null hypothesis and the empirical distributions for the cluster size matched closely the predicted and fitted parametric distributions, histograms showed that, on average, approximately 6% of the clusters were 60 times larger than the average cluster size (Table 4). These large clusters were not distributed according to P(n = k), as those clusters were in regions that constituted only 0.2% of the probability mass in P(n = k). For the parametric method, the presence of these large clusters raised the empirical FWER to 64% at CDT = 2.5, which decreased to 40% at a very high CDT = 3.5 (Table 4). When ignoring those large clusters, however, the empirical FWER was only 3.24% when using to calculate the parametric statistics (Table 4).
Figure 5.
Figure 6.
Table 3.
Kolmogorov-Smirnov Statistic Comparing the Theoretical Parametric Distribution to the histogram of cluster sizes. The KS statistic was computed from the histogram of cluster sizes and from the values of the parametric distribution at the centers of the bins in the histogram. The cluster size histograms were formed with 20 bins for 1D GRFs, 45 bins for 2D GRFs, and 200 bins for 3D GRFs. For computing p-value we selected degree of freedom equal to the number of bins.
| Dataset | Cluster Defining Threshold | ||
|---|---|---|---|
| 1.5 | 2.0 | 2.5 | |
| 1D GRFs | 0.025 | 0.081 | 0.125 |
| 2D GRFs | 0.247 | 0.187 | 0.280 |
| 3D GRFs | 0.36* | 0.264 | 0.711* |
| Toxins | 0.368* | 0.128 | 0.082 |
| Depression | 0.329* | 0.085 | 0.098 |
The star (*) denotes that the two distributions are significantly (p<0.05) different. The simulated GRFs were smoothed with a Gaussian kernel of FWHM = 15. For simulated 3D GRFs, the distributions differed because most (> 80%) of the clusters were of sizes 1 or 2 voxels, thereby deviating empirical distribution from theoretical distributions. In contrast, for the real-world data, the theoretical distributions did not differ from the histogram of cluster sizes for CDT > 1.5.
Table 4.
Average Number of False Positives under the Null Hypothesis when the parametric method for cluster-level inference is applied to our real-world datasets at a Cluster Defining Threshold (CDT) = 2.5. Second Column: The number of datasets analyzed in each real-world resting state fMRI study. The 2 runs each with 140 rsFMR images in each dataset were combined and then analyzed independently. Third Column: The number of datasets analyzed with a cluster of size greater than . Averaged across the 5 studies, the empirical FWER for the parametric method was 64% at CDT = 2.5. The empirical FWERs were 52% and 40% at CDT of 3.0 and 3.5, respectively (data not shown). Fourth Column: The number of datasets analyzed with a cluster of size greater than but of size smaller than a threshold size. The threshold size was calculated from the parametric probability distribution P(n = k) of cluster sizes under the null hypothesis, such that <0.2% of clusters had a size greater than the specified threshold. We identified this threshold to discount very large clusters present in 3D random fields. Discounting those large clusters, the empirical FWER averaged across the 5 studies was 29.5%. Fifth Column: The number of analyses with a cluster size greater than but of size smaller than the threshold size. Discounting those large clusters, the empirical FWER averaged across the 5 studies was 3.24%. Sixth Column: For each dataset, the 3 numbers represent: average cluster size discounting the very large clusters; threshold (in number of voxels) for defining the very large clusters; and average size of the very large clusters. On average the very large clusters are 60 times larger than the average cluster size. Seventh Column: For each study the 2 numbers are: the number of large clusters across all datasets analyzed; the total number of clusters in that dataset. Average across the 5 studies, 6% of all clusters are 60 times larger than the average cluster size; however, those large clusters constituted less than 0.2% of the probability mass of the parametric probability distribution P(n = k) for cluster size. Furthermore, averaged across all studies, there were 1.3 of the very large clusters in each analysis. A post hoc, independent analysis of the rsFMRI data for the 81 healthy participants (average age 22 years) showed similar rates of false positives, cluster size threshold, and the average cluster sizes as for all 142 participants in the Autism study.
| Study | # of Datasets |
# of Analyses with Clusters of Size > |
Ignoring clusters of size > a specified threshold |
Avg Size/ Threshold/ Largest Avg Size |
# of Large Clusters/Total # of Clusters |
|
|---|---|---|---|---|---|---|
| # of Analyses with Clusters of Size > |
# of Analyses with Clusters of Size > |
|||||
| Autism | 142 | 100 (70%) | 47 (33.1%) | 4 (2.8%) | 142/1546/6922 | 224/3193 (7%) |
| High Risk | 123 | 48 (39%) | 19 (15.4%) | 8 (6.5%) | 134/2243/10572 | 78/1942 (4%) |
| Depression | 116 | 88 (76%) | 42 (36.2%) | 4 (3.4%) | 116/1231/7623 | 231/2646 (8.7%) |
| Stuttering | 69 | 46 (67%) | 22 (31.9%) | 2 (2.9%) | 117/1494/6129 | 82/1553 (5.3%) |
| Toxins | 260 | 174 (67%) | 79 (30.4%) | 5 (1.9%) | 144/1798/7746 | 314/6076 (5.2%) |
3.4 Parametric Distribution for FWER Inference
Although the average number of clusters (Em) were significantly higher than the (Eχ), the theoretical parametric distribution accurately modeled the empirical distribution of cluster sizes. We therefore propose using the Poisson distribution , where λ̂ = Em is the average number of clusters, to model the distribution of the number of clusters in random fields. Using the parametric distribution P(n = k) predicted by theory for E(χ), the probability P(nmax ≥ k) that the largest cluster size will be ≥ k can be written as . We therefore calculated the corrected cluster sizes and at a FWER of 0.05 using P(nmax ≥ k) and P̂(nmax ≥ k), respectively, and the uncorrected cluster size at p-value = 0.05 using the empirical histogram of the cluster sizes (Figures 5 & 6). We located these cluster sizes using vertical lines in the histograms (Figure 5) and computed their p-values using the theoretical distribution P(n = k). The P-values indicated the fraction of clusters with size ≥ , or (Tables 5 & 6). For 1D GRFs the equaled and, as expected, for 2D and 3D GRFs the was larger than . The P-values for are 4 to 5 times smaller than those for and therefore 4 to 5 times fewer clusters will be considered statistically significant when FWER is computed using P̂(nmax ≥ k) than when using P(nmax ≥ k).
Table 5. Cluster Sizes and Their Associated P-Values for GRFs Smoothed with a Gaussian Kernel of FWHM = 15.
We thresholded the smoothed GRFs at varying Cluster Defining Thresholds (CDTs), and at those CDTs estimated the theoretical, uncorrected distribution P(n = k) of cluster size (3rd column) and the corrected probability P(nmax ≥ k) for the largest cluster having size greater than k, using either the expected Euler characteristic (4th column) or average number of clusters (5th column). We constructed the nonparametric distribution for the size of the largest cluster from the 50 random realizations of the smoothed GRFs thresholded at each CDT (6th column). We then computed the cluster size for a p-value of 0.05 without correcting for multiple statistical tests (uncorrected, 3rd column), cluster-level inference corrected using E(χ) (4th column), cluster-level inference corrected using the average number of clusters (5th column), and cluster-level inference corrected using nonparametric testing (6th column). In parentheses we present the associated p-values for those cluster sizes in the theoretical distribution P(n = k) of cluster size. The cluster size at p-value = 0.05 to control for multiple hypothesis testing in 1D simulated GRFs are equal for various procedures because the distribution P(n = k) is accurately modeled by parametric model. In 2D and 3D simulated GRFs the cluster size at p-value = 0.05 is much larger in nonparametric compared to that in parametric methods because of the presence of few very large clusters.
| GRF with FWHM = 15 |
Cluster Defining Threshold |
Cluster Size (P-value) | |||
|---|---|---|---|---|---|
| Uncorrected | Corrected, Expected EC |
Corrected, Average Number |
Corrected, Nonparametric |
||
| 1D | 1.5 | 25.26 (0.05) | 34.83 (9.5×10−4) | 34.97 (8.9×10−4) | 55.75 (1.8×10−8) |
| 2.0 | 20.83 (0.05) | 26.73 (2.2×10−3) | 26.84 (2.1×10−3) | 35.1 (2.5×10−5) | |
| 2.5 | 15.56 (0.05) | 20.31 (6.6×10−3) | 20.38 (6.4×10−3) | 19.55 (9.6×10−3) | |
| 2D | 1.5 | 1136.6 (0.05) | 1638 (3.6×10−3) | 2000 (1.0×10−3) | 3516.8 (5.6×10−6) |
| 2.0 | 654.3 (0.05) | 899 (6.4×10−3) | 1129 (1.8×10−3) | 2256 (3.1×10−6) | |
| 2.5 | 996.5 (0.05) | 496.5 (15.8 ×10−3) | 680.1 (3.4×10−3) | 1188.2 (4.9×10−5) | |
| 3D | 1.5 | 28,183 (0.05) | 39,711 (4.4×10−3) | 55,421 (1.1×10−3) | 63,162 (6.1×10−4) |
| 2.0 | 7693 (0.05) | 13,518 (4.4×10−3) | 17,824 (1.5×10−3) | 19,174 (1.1×10−3) | |
| 2.5 | 141 (0.05) | 5506 (7.7×10−3) | 8387 (1.6×10−3) | 4005 (19.6×10−3) | |
Table 6. Cluster Size and Its Associated P-Value for Real-World, Resting-State fMRI Data.
We thresholded the statistical maps at varying Cluster Defining Threshold (CDTs), and at those CDTs we estimated the theoretical, uncorrected distribution P(n = k) of cluster sizes (3rd column), the corrected probability P(nmax ≥ k) for the largest cluster having size greater than k using the expected Euler characteristic (4th column), and the average number of clusters (5th column). At each specified CDT we constructed the nonparametric distribution for the size of largest clusters in the statistical map for each participant in each study’s dataset (6th column). We then computed the cluster size for a p-value of 0.05 without correcting for multiple statistical tests (uncorrected, 3rd column), and for a cluster-level inference corrected using E(χ) (4th column), a cluster-level inference corrected using the average number of clusters (5th column), and a cluster-level inference corrected using nonparametric permutation testing (6th column), presenting in parentheses the p-values for those cluster sizes in the theoretical distribution P(n = k) of cluster sizes. Therefore, clusters comprising of 50,000 or more voxels will not be considered statistically significant when nonparametric methods for cluster level inference are applied to real-world data.
| Dataset | CDT | Cluster Size (P-value) | |||
|---|---|---|---|---|---|
| Uncorrected | Corrected, Expected EC |
Corrected, Average Number |
Corrected, Nonparametric |
||
| Autism | 1.5 | 3,629 (0.05) | 7,887 (4.1×10−3) | 10,764 (1.1×10−4) | 62,063 (3.4×10−10) |
| 2.0 | 3,613 (0.05) | 2,685 (4.1×10−3) | 3,438 (1.5×10−3) | 48,769 (2.9×10−17) | |
| 2.5 | 2,101 (0.05) | 1,096 (7.1×10−3) | 1,489 (2.3×10−3) | 28,355 (1.7×10−19) | |
| High Risk | 1.5 | 2,144 (0.05) | 10,303 (5.9×10−3) | 16,501 (1.0×10−3) | 54,708 (1.6×10−07) |
| 2.0 | 2,175 (0.05) | 3,507 (5.9×10−3) | 4,883 (1.7×10−3) | 48,925 (1.2×10−13) | |
| 2.5 | 1,706 (0.05) | 1,414 (10.4 ×10−3) | 1,958 (3.4×10−3) | 34,312 (2.3×10−17) | |
| Depression | 1.5 | 3,586 (0.05) | 6,674 (3.2×10−3) | 8,530 (1.2×10−3) | 61,345 (1.2×10−11) |
| 2.0 | 3,523 (0.05) | 2,272 (3.2×10−3) | 2,739 (1.5×10−3) | 52,897 (2.7×10−21) | |
| 2.5 | 2,178 (0.05) | 934 (5.7 ×10−3) | 1,185 (2.3×10−3) | 48,991 (3.4×10−32) | |
| Stuttering | 1.5 | 3,578 (0.05) | 7,693 (3.9×10−3) | 11,112 (0.8×10−3) | 54,909 (1.2×10−09) |
| 2.0 | 2,043 (0.05) | 2,619 (3.9×10−3) | 3,487 (2.3×10−3) | 52,081 (2.1×10−18) | |
| 2.5 | 1,373 (0.05) | 1,070 (6.9 ×10−3) | 1,439 (3.4×10−3) | 30,888 (4.6×10−21) | |
| Toxins | 1.5 | 3,638 (0.05) | 8,798 (4.7×10−3) | 12,867 (1.0×10−4) | 62,207 (2.7×10−9) |
| 2.0 | 2,159 (0.05) | 2,995 (4.7×10−3) | 4,102 (1.4×10−3) | 55,033 (6.3×10−17) | |
| 2.5 | 2,127 (0.05) | 1,217 (8.3×10−3) | 1,750 (2.2×10−3) | 35,081 (2.7×10−20) | |
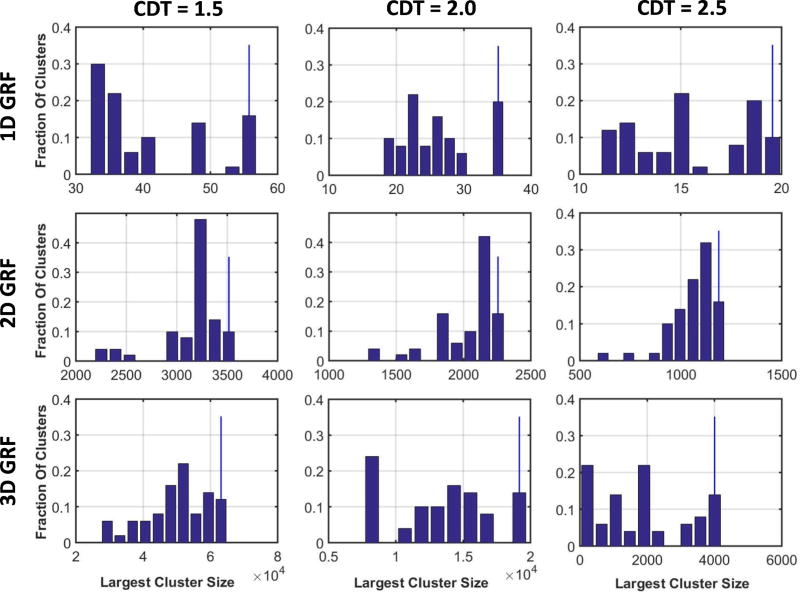
3.5 Nonparametric Distribution for Size of the Largest Cluster
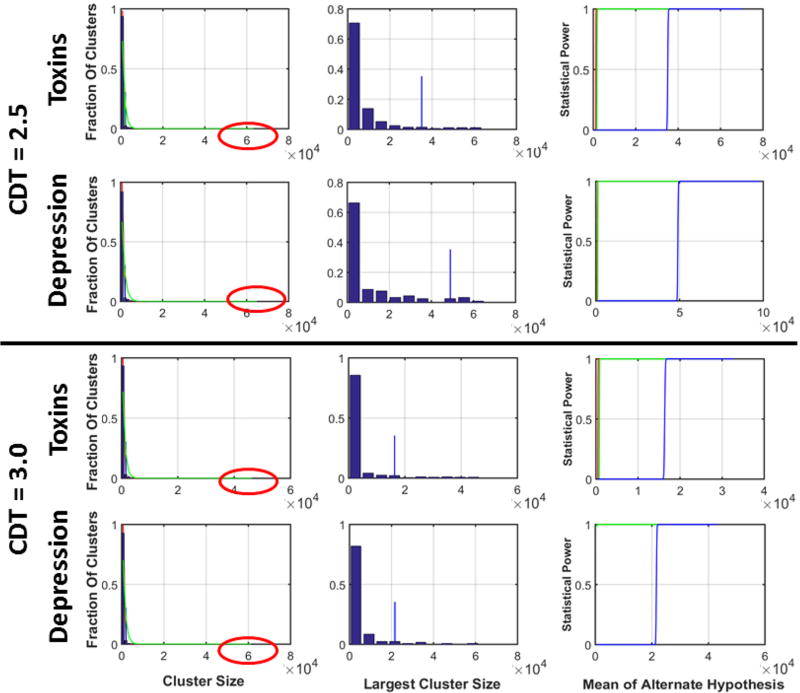
We generated histogram for the sizes of the largest cluster in each realization of 1D, 2D, and 3D GRFs and in the real-world data. We normalized the histogram such that the bins summed to 1.0, thereby generating the nonparametric distribution for the size of the largest cluster (Figures 6 & 7). We then computed the cluster size such that 5% of the largest clusters had size greater than . In other words, the probability of finding a cluster larger than across the entire family of random variables equaled 0.05. We located these cluster sizes by plotting vertical lines on the histograms, and we computed the p-value for this cluster size using the parametric distribution P(n = k) (Tables 5 & 6). The p-values we calculated for were up to 1000 times smaller than the p-values for , thus demonstrating that nonparametric methods yield far fewer false positives than do parametric methods when controlling for multiple comparisons. At a CDT = 2.5 simulated GRFs had very few clusters, leading to cluster size in 1D GRF approximately equal to, but in 3D GRF smaller than, the cluster sizes and . Therefore, in simulated GRFs, FWERs for the parametric methods will be smaller than that for the nonparametric methods at CDT of 2.5 or higher. However, in real-world data, even at CDT = 2.5, was 30 times smaller than and (Table 6 & Figure 6) because of a few very large clusters in the data.
Figure 7.
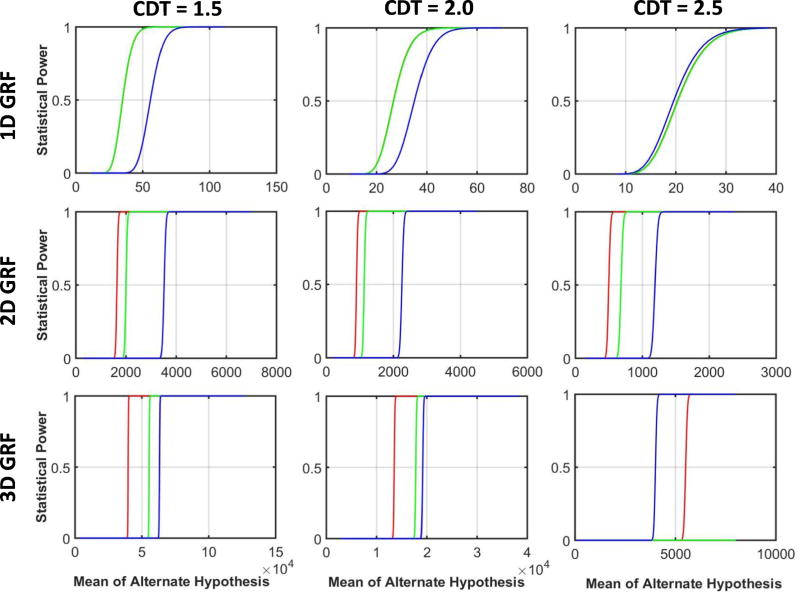
3.6 Statistical Power
We compared statistical power when using parametric methods, parametric methods with an expected number of clusters, and nonparametric methods at cluster sizes , and , for the alternate hypothesis in which cluster sizes of true effects were Gaussian distributed with varying means and variances. These plots (Figure 8) show that in general the parametric method (red curve) provides the greatest statistical power, the parametric method with the expected number of clusters is intermediate (green curve), and the nonparametric method provides the lowest power (blue curve) across all random fields. In simulated GRFs, the nonparametric method provided similar power as parametric methods for 1D GRFs but greater power than parametric methods for 3D GRFs at a CDT = 2.5 because simulated random fields at CDT = 2.5 or higher had only a few clusters most of which comprised of 1 voxel (Tables 3 & 7). That is, a CDT = 2.5 was a very high threshold for defining clusters in simulated GRFs. In contrast, although the number of clusters were small even in the real-world data at CDT = 2.5 or higher, the cluster sizes were large (Figure 8), thereby leading to much larger than and and significantly lower statistical power for nonparametric methods than that for parametric methods.
Figure 8.
Table 7.
Empirical FWER for Parametric Methods for Increasing CDT for real-world data.
| Studies | # of Analyses with Clusters of Size >
at increasing CDT |
||
|---|---|---|---|
| 2.5 | 3.0 | 3.5 | |
| Autism (142) | 100 (70%) | 88 (62%) | 67 (47%) |
| High Risk (123) | 48 (39%) | 40 (33%) | 33 (27%) |
| Depression (116) | 88 (76%) | 72 (62%) | 58 (50%) |
| Stuttering (69) | 46 (67%) | 31 (45%) | 22 (32%) |
| Toxins (260) | 174 (67%) | 145 (56%) | 118 (45%) |
| Empirical FWER | 64% | 52% | 40% |
CDT = Cluster Defining Threshold
4. Discussion
We have shown, using both our simulated and real-world resting state fMRI data, that random fields under the null hypothesis have a small fraction (< 6%) of clusters with a very large spatial extent. Parametric methods will deem these large clusters to be statistically significant, which cannot be distinguished from true positive, biologically valid clusters under the alternative hypothesis. The presence of these unpredictable, large clusters yields empirical FWERs as high as 70%, as reported previously[32].
We have also shown that although the Expected Euler characteristic, E(χ), did not differ significantly from the empirically identified expected (mean) number of clusters, Em, for 1D random fields, E(χ) was significantly smaller than Em for both 2D and 3D GRFs and in real-world data. In other words, the empirically observed numbers of positive findings for cluster-based statistical inference equaled the theoretical number for 1D fields, but were far greater for 2D and 3D fields than predicted by the theoretical distributions. The practical consequence of this finding is that an increasing number of statistical tests in imaging data will generate increasing number of false positive findings. More stringent control than predicted by theory therefore is necessary when conducting multiple statistical tests based on cluster-level inference. Theory predicts[24] that the parametric FWER P(nmax ≥ k) (the probability of finding a cluster size nmax greater than k across the entire family of random variables) approximately equals the product of E(χ) and the probability P(n ≥ k). Under the null hypothesis and at higher CDTs (≥2.5), cluster location and size are independently distributed, and therefore the parametric FWER approximates Bonferroni correction in controlling false positive rates across E(χ) clusters. Because Em is significantly larger than E(χ) for 2D and 3D random fields, however, the FWER P(nmax ≥ k) should account for an Em rather than for an E(χ) number of clusters in the data.
The probability P(m = x) for the number of clusters m, therefore, should be a Poisson distribution[74] , with mean λ = Em, and variance Em. However, our simulated 1D GRFs showed that the variance of m was significantly smaller than Em, (p < 0.005, Table 1), suggesting the presence of a narrower spread of the distribution around the mean Em than assumed in the Poisson distribution. We assessed whether a smaller spread in the distribution P(m = x) affected the empirical FWER by assuming a delta distribution for m around its mean, i.e. P(m = x) = δεm, which has nonzero support only at the mean Em. Our results showed that the FWERs computed under assumption of a delta distribution closely matched those calculated when assuming a Poisson distribution for P(m = x), thus suggesting that the FWERs were robust to the parametric form assumed for the probability P(m = x). Furthermore, the probability P(n = k) of the cluster size n predicted by theory did not differ significantly from the empirical distribution when tested using the Kolmogorov-Smirnov statistic[73, 75]. Thus, we suggest a modified FWER to control for multiple statistical tests when using cluster-level inference, which will increase P(nmax ≥ k) for each k, increase the cluster size for a FWER of 0.05, and consequently reduce the number of false positives. Although use of this empirically derived distribution will likely reduce the number of false positive findings, it will also likely reduce the statistical power to detect true findings in the data.
The Presence of Large Clusters in High Dimensional Random Fields
Our data, simulated under the null hypothesis, showed that each realization of the smoothed 2D and 3D GRFs thresholded at CDT = 2.5 or 3.0 had a small fraction (< 6%) of clusters that were 2–3 times larger than the cluster sizes and for a FWER = 0.05 when calculated using E(χ) and Em, respectively. Because these large clusters were present in all realizations of simulated GRFs, an empirical FWER would equal 1.0 if it were calculated as a fraction of all brain-wide analyses that yielded false positive findings. In other words, parametric methods for cluster-level inference yield at least one false positive finding in every brain-wide analysis. These large clusters in simulated random fields smoothed with a Gaussian kernel of specified FWHM and thresholded at CDT = 2.5 or 3.0 show that large clusters are inherent property of smoothed random fields rather than due to a larger SACF than that of a Gaussian distribution. This was true not only for simulated data, but equally so in our 710 real-world, resting-state fMRI datasets as well as in a subset of 81 healthy participants from the Autism study (Table 4).
Several published studies using real-world datasets under the null hypothesis also reported clusters as large as 55,000 voxels (supplementary Tables 3, 4, & 5)[32] or more (Figures 6 & 7)[32, 43]. A practical consequence of having large clusters under the null hypothesis was that when controlling for familywise false positives at a nominal FWER of 0.05, nonparametric methods estimated that a cluster should be larger than 12,000 contiguous voxels to considered statistically significant, whereas for the same dataset, parametric methods estimated that clusters should be larger than 3,000 (Supplementary figure 16)[32]. These extremely large clusters were an inherent property of the random fields because these large clusters were present in our simulated datasets that we smoothed only by an a priori specified kernel. Therefore, the previously reported[32] high empirical FWERs associated with parametric methods were not a consequence of the failure to model sufficiently the amount of spatial smoothness (SACF) in the data[32], but instead derived from an inherent property of smoothed random fields that generates a small fraction of very large clusters. Because the numbers of these clusters is small, the number and size of these large clusters are unpredictable and cannot be modeled using parametric or nonparametric distributions. Even if their distributions could be learned from data, these large clusters under null hypothesis cannot be distinguished from clusters that form true positive findings. Moreover, the large variability in the empirical FWERs reported in previous studies[32, 43–45] likely derives from the presence of the unpredictable numbers and sizes of these large clusters. This unpredictability likely in turn derives from differences in how the data were acquired and processed -- including differences in MRI scanner performance, differences in the imaging pulse sequences employed, differences in the degree of motion artifact and other structured noise present in the data that increases correlations among neighboring voxels,[76] and differences in image processing methods employed -- with the empirical FWERs being closer to the nominal value of 0.05 for some platforms and datasets but not for others.[32]
The permutation-based, nonparametric method for cluster-level inference first estimated the distribution of the sizes of the largest cluster and then used that distribution to compute the cluster size at a FWER = 0.05, such that the probability of the largest cluster having a size greater than equaled 0.05. Because the nonparametric distribution for the largest cluster was estimated from the largest clusters in the data under the null hypothesis, empirical FWER equaled the nominal FWER, irrespective of the software platform or how the data were acquired and processed[32]. The cluster size estimated from the largest clusters in the dataset was much larger than the cluster sizes and calculated using parametric methods. Because of these large, unpredictable clusters, the empirical FWERs for parametric methods varied from as small as 1% to as large as 80%, whereas the empirical FWERs for nonparametric methods were always close to the nominal FWER of 5%. The nonparametric methods are more influenced by these large clusters whose numbers and sizes cannot be modelled: nonparametric methods learn the distribution of the largest clusters empirically from the clusters in the data; because the distributions of the large clusters are unpredictable, so too are the performances of the nonparametric techniques that depends on them. The performance of parametric methods, however, are generally robust to the presence or absence of these large clusters in the random fields, because these large clusters constitute only a small fraction (< 6%) of all clusters in the dataset, and parametric distributions model well the other 94% of clusters. The use of nonparametric methods that are robust to small perturbations in the probability distributions[77] possibly can reduce the influence of these large clusters on statistical inference.
Statistical Power
Statistical power (detecting real effect or sensitivity) is generally equal in importance to specificity (rejecting false positive findings) when testing hypotheses. Optimizing these two capabilities typically involves a trade-off in which improving one necessarily compromises the other. Prior studies of cluster-based statistical inference were concerned primarily with the specificity of parametric methods -- whether those analyses rejected false positives at the nominal FWER. Although these studies found that empirical FWERs for parametric, but not nonparametric, methods were much higher than the nominal FWERs, they did not assess the relative statistical power of these two general statistical approaches when applied to real-world datasets. The inherent trade-off in sensitivity and specificity necessitates that a method for statistical inference that yields low rates of false positives will also inherently tend to have low statistical power. When undertaking cluster-level inference, false positive rates can be made arbitrarily small by requiring clusters to be of sufficiently large size or by increasing the Z-score of the Cluster Defining Threshold (CDT). Those practices, however, will also increasingly reject true positive findings and therefore will have low statistical power. Ideally, a statistical procedure would permit only a few false positives while still detecting most or all real effects in the data. Use of the False Discovery Rate (FDR)[26, 78], for example, permits false positives at a pre-specified rate so as to detect most true findings while trying to contain the rate of false positive findings to an acceptable level. In most neuroimaging studies, detecting most of the true findings at the cost of a few false positives is preferable to overly conservative control of false positives that miss true effects; moreover, false positives are unlikely to replicate across independent studies[79] and therefore will be rejected as false in meta-analyses of those studies. For example, a meta-analysis[80] of 13 task-based fMRI studies showed that although findings varied across studies possibly due to several factors, including presence of false positives, their findings when combined across all studies showed that ADHD participants had reduced activity across several regions of the brain. That is, meta analysis may lead to accurate understanding of the pathophysiology provided statistical procedures detect most true findings even in the presence of few false positives.
Parametric and nonparametric methods have similar statistical power when the data are well behaved and are available from a sufficient number of participants.[81] If the distributions of data cannot be modeled accurately, then nonparametric methods may be more accurate -- i.e. they may have both greater sensitivity and specificity -- than parametric approaches to cluster-based statistical inference. In our simulated 1D random fields, distributions were well behaved because of the absence of very large clusters, meaning that those distributions could be modeled accurately using a priori specified parametric models. Therefore, parametric and nonparametric methods provided similar statistical power and similar cluster size thresholds for rejecting clusters as false positives. For 2D and 3D random fields, however, the distributions of cluster sizes were not as well behaved, as approximately 6% of the clusters in those distributions were very large. Consequently, the empirical FWERs for parametric methods were as high as 70%, in contrast to nonparametric methods, where the empirical FWERs equaled the nominal FWERs. Statistical power, however, was generally substantially lower for nonparametric than for parametric methods; nonparametric methods will risk failing to detect real effects when in fact they are present.
Practical Implications and Recommendations
Statistical analyses of imaging data must first carefully evaluate whether the data and test statistics follow assumed distributions and, in particular, are devoid of outlying values. Evaluating the validity of these assumptions is especially critical for parametric techniques, as they employ a presupposed parametric function to model the distributions of data. Deviations in the distributions of real data from the assumed models can have drastic consequences that range from, most commonly, allowing too many false positive findings, to failing to detect real effects. For example, SACF for real-world data is nonstationary and have longer tail than Gaussian distribution over the domain of the data. Recently proposed methods[82] minimize the effects of these deviations on statistical parametric mapping, thereby controlling for their effects on the false positive findings in the analyses. Nonparametric methods overcome this limitation by learning distributions from the data, and therefore they can be applied to any data that meets exchangeability criteria[83, 84], without specifying an a priori model and when evaluating the significance of any test statistic. Consequently, nonparametric methods are generalizable, easy to implement in diverse situations, and can be as statistically powerful as parametric methods when sufficient data are available. Nonparametric methods, however, may have lower statistical power to detect real effects when assumptions in parametric methods are valid.[85] Even when using nonparametric methods, however, one must still carefully evaluate distributions of data for outlying values in order to avoid making incorrect inferences from those distributions.
It is important to emphasize that a FWER as high as 70% in parametric methods should not be interpreted as meaning that 70% of all findings in a given statistical analysis are false positives, but rather as meaning that 70% of independent analyses each likely will have one false positive cluster. That is, on average each analysis will have fewer than one false positive finding. In our real-world data, for example, the empirical FWER for parametric methods was 65% because of the presence of a few very large clusters: on average there were 1.3 very large clusters in each analysis (Table 4). If those large clusters were discounted, the empirical FWER was only 3.24% when average across all studies (Table 4) – i.e., only 3% of all independent analyses would contain at least one false positive finding. Although it is comforting to know that these FWERs will yield very few false positive clusters, it is at the same time disconcerting to know that a large cluster that otherwise appears convincing by virtue of its very size could in fact be a false positive finding. In real-world settings where the data are generated under an alternative hypothesis, these false positive findings cannot be distinguished on any statistical basis from true positive findings, although perhaps they can be distinguished in individual studies on the basis of their location and biological plausibility if fMRI signal significantly correlates with other measures of interest. It is also important to emphasize that parametric methods for cluster-based inference that allow less than one false positive finding per study will be far more conservative, as well as lower statistical power, than an FDR-based analysis that by fiat permits up to 5% of false positive findings. Thus, even at an empirical FWER of 70%, parametric methods for cluster-based inference likely permit less than one false positive finding on average in each analysis, while at the same time having much higher statistical power than nonparametric methods for discovering most of the true positive effects. These considerations strongly suggest that reanalysis of data in the previously published fMRI studies is unwarranted as nonparametric methods would have reduced statistical power to detect true findings. We therefore recommend using parametric techniques for cluster-level statistical inference with CDT of 2.5 or higher, thereby minimizing false positives while providing better statistical power than nonparametric methods.
Future Directions
Our simulated data showed that, on average, the number of clusters Em in random fields is significantly higher than predicted by E(χ). We also showed that the FWER for parametric methods can be approximated as a product of the number of clusters and the probability P(n ≥ k), thus approximating Bonferroni correction for multiple statistical comparisons. Because the number of clusters in real-world data is higher than predicted by theory, we suggest using the average number of clusters Em rather than E(χ) to reduce the number of false positive findings. However, unlike E(χ), which can be computed using a simple formula, Em is not known a priori and must be estimated from the data under the null hypothesis. The average number of clusters Em could possibly be estimated from the data using procedures similar to Monte Carlo[86, 87] or permutation testing, in which all data are assumed to be distributed according to the null hypothesis; then maps for the test statistic could be computed and thresholded at a specified CDT, and the number of clusters in the thresholded map counted. Repeating this procedure for several permutations and averaging the number of clusters across those permutations would generate an estimate for Em that could be used subsequently within parametric methods for statistical inference. This hybrid approach, whose validity needs to be established in independent studies, to cluster-based statistical inference would first apply a nonparametric tool to estimate Em and then use that estimate within a parametric framework to estimate the probability P(n ≥ k). Although computationally more expensive, our results suggest strongly that this approach would yield lower false positive rates than would a purely parametric approach, but it would provide much greater statistical power than would a purely nonparametric approach.
Acknowledgments
This research was made possible by the provision of data by New York State Psychiatric Institute and Columbia University. This study was supported by Institute for the Developing Mind, Children’s Hospital Los Angeles, and the University of Southern California. This study was supported by National Institute of Mental Health (grants MH36197, MH068318, MH089582, K02-74677), National Institute of Environmental Health Sciences (grants 5P01ES09600, P50ES015905, 5R01ES08977), an investigator-initiated grant from Eli Lilly company to Drs. Hellerstein and Peterson, Milhiser family fund, and the Murphy endowment at Columbia University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
Authors report no financial relationships with commercial interests.
References
- 1.Sowell ER, Peterson BS, Thompson PM, Welcome SE, Henkenius AL, Toga AW. Mapping Cortical Change Across the Human Life Span. Nature Neuroscience. 2003:309–15. doi: 10.1038/nn1008. [DOI] [PubMed] [Google Scholar]
- 2.Sowell ER, Thompson PM, Toga AW. Mapping changes in the human cortex throughout the span of life. Neuroscientist. 2004;10(4):372–92. doi: 10.1177/1073858404263960. [DOI] [PubMed] [Google Scholar]
- 3.Sowell ER, Jernigan TL. Further MRI evidence of late brain maturation: Limbic volume increases and changing asymmetries during childhood and adolescence. Dev Neuropsychol. 1998;14(4):599–617. [Google Scholar]
- 4.Sowell E, Peterson BS, Bansal R, Xu D, Zhu H. Sex Differences in Cortical Thickness Mapped in 176 Healthy Individuals Between 7 and 87 Years of Age. Cerebral Cortex. 2006 doi: 10.1093/cercor/bhl066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bansal R, Peterson BS, Gingrich J, Hao XJ, Odgerel Z, Warner V, et al. Serotonin signaling modulates the effects of familial risk for depression on cortical thickness. Psychiat Res-Neuroim. 2016;248:83–93. doi: 10.1016/j.pscychresns.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterson BS, Choi HA, Hao X, Amat J, Zhu H, Whiteman R, et al. Morphology of the Amygdala and Hippocampus in Children and Adults with Tourette Syndrome. Archives General Psychiatry. 2007 doi: 10.1001/archpsyc.64.11.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson BS, Skudlarski P, Gatenby JC, Zhang H, Anderson AW, Gore JC. An fMRI study of Stroop word-color interference: evidence for cingulate subregions subserving multiple distributed attentional systems. Biol Psychiatry. 1999;45(10):1237–58. doi: 10.1016/s0006-3223(99)00056-6. [DOI] [PubMed] [Google Scholar]
- 8.Peterson BS, Staib L, Scahill L, Zhang H, Anderson C, Leckman JF, et al. Regional brain and ventricular volumes in Tourette syndrome. Arch Gen Psychiatry. 2001;58:427–40. doi: 10.1001/archpsyc.58.5.427. [DOI] [PubMed] [Google Scholar]
- 9.Peterson BS, Thomas P, Kane MJ, et al. Basal ganglia volumes in patients with Gilles de la Tourette syndrome. Arch Gen Psychiatry. 2003;60:415–24. doi: 10.1001/archpsyc.60.4.415. [DOI] [PubMed] [Google Scholar]
- 10.Plessen KJ, Bansal R, Peterson BS. Imaging evidence for anatomical disturbances and neuroplastic compensation in persons with Tourette syndrome. J Psychosom Res. 2009;67(6):559–73. doi: 10.1016/j.jpsychores.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plessen KJ, Bansal R, Zhu H, Whiteman R, Quackenbush GA, Hugdahl K, et al. Hippocampus and amygdala morphology in Attention-Deficit/Hyperactivity Disorder. Arch Gen Psychiatry. 2006;63:795–807. doi: 10.1001/archpsyc.63.7.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sowell ER, Thompson PM, Welcome SE, Henkenius AL, Toga AW, Peterson BS. Cortical abnormalities in children and adolescents with attention-deficit hyperactivity disorder. Lancet. 2003;362(9397):1699–707. doi: 10.1016/S0140-6736(03)14842-8. [DOI] [PubMed] [Google Scholar]
- 13.Amat JA, Whiteman R, Bansal R, Davies M, Haggerty R, Peterson BS. The cognitive correlates of amygdala and hippocampus volumes in healthy adults. Brain Cognit. 2008;66:105–14. doi: 10.1016/j.bandc.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold SJM, Ivleva EI, Gopal TA, Reddy AP, Jeon-Slaughter H, Sacco CB, et al. Hippocampal Volume Is Reduced in Schizophrenia and Schizoaffective Disorder But Not in Psychotic Bipolar I Disorder Demonstrated by Both Manual Tracing and Automated Parcellation (FreeSurfer) Schizophrenia Bull. 2015;41(1):233–49. doi: 10.1093/schbul/sbu009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bansal R, Hellerstein DJ, Peterson BS. Evidence for Neuroplastic Compensation in the Cerebral Cortex of Persons with Dysthymia. Mol Psychiatr. 2017 doi: 10.1038/mp.2017.34. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolb B, Whishaw IQ. Brain plasticity and behavior. Annu Rev Psychol. 1998;49:43–64. doi: 10.1146/annurev.psych.49.1.43. [DOI] [PubMed] [Google Scholar]
- 17.Kolb B. Brain development, plasticity, and behavior. Am Psychol. 1989;44(9):1203–12. doi: 10.1037//0003-066x.44.9.1203. [DOI] [PubMed] [Google Scholar]
- 18.Zilles K. Neuronal Plasticity as an Adaptive Property of the Central-Nervous-System. Annals of Anatomy-Anatomischer Anzeiger. 1992;174(5):383–91. doi: 10.1016/s0940-9602(11)80255-4. [DOI] [PubMed] [Google Scholar]
- 19.Chang Y. Reorganization and plastic changes of the human brain associated with skill learning and expertise. Front Hum Neurosci. 2014;8:35. doi: 10.3389/fnhum.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobel LJ, Bansal R, Maia TV, Sanchez J, Mazzone L, Durkin K, et al. Basal ganglia surface morphology and the effects of stimulant medications in youth with attention deficit hyperactivity disorder. Am J Psychiatry. 2010;167(8):977–86. doi: 10.1176/appi.ajp.2010.09091259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonferroni CE. Il calcolo delle assicurazioni su gruppi di teste. Studi in Onore del Professore Salvatore Ortu Carboni. 1935:13–60. [Google Scholar]
- 22.Bonferroni CE. Teoria statistica delle classi e calcolo delle probabilita. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze. 1936;8:3–62. [Google Scholar]
- 23.Statistical Parametric Mapping: The Analysis of Functional Brain Images. Statistical Parametric Mapping: The Analysis of Functional Brain Images. 2007:1–680. [Google Scholar]
- 24.Friston KJ, Worsley KJ, Frackowiak RS, Mazziotta JC, Evans AC. Assessing the significance of focal activations using their spatial extent. Hum Brain Mapp. 1994;1(3):210–20. doi: 10.1002/hbm.460010306. [DOI] [PubMed] [Google Scholar]
- 25.Bansal R, Staib LH, Xu D, Zhu H, Peterson BS. Statistical Analysis of Brain Surfaces Using Gaussian Random Fields on 2D Manifold. IEEE Transactions on Medical Imaging. 2007;26(1):46–57. doi: 10.1109/TMI.2006.884187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
- 27.Chumbley J, Worsley K, Flandin G, Friston K. Topological FDR for neuroimaging. NeuroImage. 2010;49(4):3057–64. doi: 10.1016/j.neuroimage.2009.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: A primer with examples. Human Brain Mapping. 2002;15(1):1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mielke PW, Berry KJ. Permutation methods : a distance function approach. 2. New York: Springer; 2007. [Google Scholar]
- 30.Efron B, Tibshirani R. An introduction to the bootstrap. New York: Chapman & Hall; 1993. [Google Scholar]
- 31.Efron B. The jackknife, the bootstrap, and other resampling plans. Philadelphia, Pa: Society for Industrial and Applied Mathematics; 1982. [Google Scholar]
- 32.Eklund A, Nichols TE, Knutsson H. Cluster failure: Why fMRI inferences for spatial extent have inflated false-positive rates. P Natl Acad Sci USA. 2016;113(28):7900–5. doi: 10.1073/pnas.1602413113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith SM, Nichols TE. Threshold-free cluster enhancement: Addressing problems of smoothing, threshold dependence and localisation in cluster inference. NeuroImage. 2009;44(1):83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- 34.Fedorenko E, Behr MK, Kanwisher N. Functional specificity for high-level linguistic processing in the human brain. P Natl Acad Sci USA. 2011;108(39):16428–33. doi: 10.1073/pnas.1112937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanwisher N. Functional specificity in the human brain: A window into the functional architecture of the mind. P Natl Acad Sci USA. 2010;107(25):11163–70. doi: 10.1073/pnas.1005062107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friston KJ ebrary Inc. Statistical parametric mapping the analysis of functional brain images. vii, 647. London: Academic; 2007. p. 32. p. of plates ill. (some col.) [Google Scholar]
- 37.Ashburner J. SPM: a history. NeuroImage. 2012;62(2):791–800. doi: 10.1016/j.neuroimage.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. NeuroImage. 2012;62(2):782–90. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 39.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29(3):162–73. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 40.Pataky T. rft1d: Smooth One-Dimensional Random Field Upcrossing Probabilities in Python. Journal of Statistical Software. 2016;71(7):1–22. [Google Scholar]
- 41.Ashburner J, Friston KJ. Voxel-based morphometry - The methods. NeuroImage. 2000;11(6):805–21. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 42.Silver M, Montana G, Nichols TE, Neuroimaging AD. False positives in neuroimaging genetics using voxel-based morphometry data. NeuroImage. 2011;54(2):992–1000. doi: 10.1016/j.neuroimage.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eklund A, Andersson M, Josephson C, Johannesson M, Knutsson H. Does parametric fMRI analysis with SPM yield valid results?-An empirical study of 1484 rest datasets. NeuroImage. 2012;61(3):565–78. doi: 10.1016/j.neuroimage.2012.03.093. [DOI] [PubMed] [Google Scholar]
- 44.Hayasaka S, Nichols TE. Validating cluster size inference: random field and permutation methods. NeuroImage. 2003;20(4):2343–56. doi: 10.1016/j.neuroimage.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Meyer-Lindenberg A, Nicodemus KK, Egan MF, Callicott JH, Mattay V, Weinberger DR. False positives in imaging genetics. NeuroImage. 2008;40(2):655–61. doi: 10.1016/j.neuroimage.2007.11.058. [DOI] [PubMed] [Google Scholar]
- 46.Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional Connectivity in the Motor Cortex of Resting Human Brain Using Echo-Planar Mri. Magnetic Resonance in Medicine. 1995;34(4):537–41. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 47.Duyn J. Spontaneous fMRI activity during resting wakefulness and sleep. Slow Brain Oscillations of Sleep, Resting State and Vigilance. 2011;193:295–305. doi: 10.1016/B978-0-444-53839-0.00019-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fransson P, Skiold B, Engstrom M, Hallberg B, Mosskin M, Aden U, et al. Spontaneous Brain Activity in the Newborn Brain During Natural Sleep-An fMRI Study in Infants Born at Full Term. Pediatric Research. 2009;66(3):301–5. doi: 10.1203/PDR.0b013e3181b1bd84. [DOI] [PubMed] [Google Scholar]
- 49.Goh S, Dong ZC, Zhang YD, DiMauro S, Peterson BS. Mitochondrial Dysfunction as a Neurobiological Subtype of Autism Spectrum Disorder Evidence From Brain Imaging. Jama Psychiat. 2014;71(6):665–71. doi: 10.1001/jamapsychiatry.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weissman MM, Wickramaratne P, Nomura Y, Warner V, Verdeli H, Pilowsky DJ, et al. Families at high and low risk for depression - A 3-generation study. Arch Gen Psychiat. 2005;62(1):29–36. doi: 10.1001/archpsyc.62.1.29. [DOI] [PubMed] [Google Scholar]
- 51.Hellerstein DJ, Stewart JW, McGrath PJ, Deliyannides DA, Batchelder ST, Black SR, et al. A Randomized Controlled Trial of Duloxetine Versus Placebo in the Treatment of Nonmajor Chronic Depression. Journal of Clinical Psychiatry. 2012;73(7):984–91. doi: 10.4088/JCP.11m07230. [DOI] [PubMed] [Google Scholar]
- 52.Bansal R, Hellerstein DJ, Peterson BS. Evidence for neuroplastic compensation in the cerebral cortex of persons with depressive illness. Mol Psychiatry. 2017 doi: 10.1038/mp.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desai J, Huo Y, Wang Z, Bansal R, Williams SC, Lythgoe D, et al. Reduced perfusion in Broca's area in developmental stuttering. Hum Brain Mapp. 2016 doi: 10.1002/hbm.23487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rauh VA, Perera FP, Horton MK, Whyatt RM, Bansal R, Hao X, et al. Brain Abnormalities in Children Exposed to a Common Organophosphate Pesticide. Proceedings of the National Academy of Sciences. 2012;109(20):7871–6. doi: 10.1073/pnas.1203396109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peterson BS, Rauh VA, Bansal R, Hao XJ, Toth Z, Nati G, et al. Effects of Prenatal Exposure to Air Pollutants (Polycyclic Aromatic Hydrocarbons) on the Development of Brain White Matter, Cognition, and Behavior in Later Childhood. Jama Psychiat. 2015;72(6):531–40. doi: 10.1001/jamapsychiatry.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Posner J, Rauh V, Gruber A, Gat I, Wang Z, Peterson BS. Dissociable attentional and affective circuits in medication-naive children with attention-deficit/hyperactivity disorder. Psychiatry Res. 2013;213(1):24–30. doi: 10.1016/j.pscychresns.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perera FP, Tang D, Wang S, Vishnevetsky J, Zhang B, Diaz D, et al. Prenatal Polycyclic Aromatic Hydrocarbon (PAH) Exposure and Child Behavior at Age 6–7 Years. Environmental Health Perspectives. 2012;120(6) doi: 10.1289/ehp.1104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perera FP, Li Z, Whyatt R, Hoepner L, Wang S, Camann D, et al. Prenatal airborne polycyclic aromatic hydrocarbon exposure and child IQ at age 5 years. Pediatrics. 2009;124(2):e195–202. doi: 10.1542/peds.2008-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterson BS, Wang ZS, Horga G, Warner V, Rutherford B, Klahr KW, et al. Discriminating Risk and Resilience Endophenotypes From Lifetime Illness Effects in Familial Major Depressive Disorder. Jama Psychiat. 2014;71(2):136–48. doi: 10.1001/jamapsychiatry.2013.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Friston KJ, Holmes AP, Worsley KJ, Polime JB, Frith C, Frackwiak RSJ. Statistical parametric maps in functional imaging: A general linear approach. Human Brain Mapping. 1995;2:189–210. [Google Scholar]
- 61.Adler RJ. Geometry of Random Fields. Geometry of Random Fields. 2010;62:1–280. [Google Scholar]
- 62.Adler RJ, Hasofer AM. Level-Crossings for Random Fields. Annals of Probability. 1976;4(1):1–12. [Google Scholar]
- 63.Hasofer AM. Upcrossings of Random Fields. Advances in Applied Probability. 1978:14–21. [Google Scholar]
- 64.Adler RJ. Generalizing Notion of Upcrossings to Random Fields. Advances in Applied Probability. 1977;9(2):226-. [Google Scholar]
- 65.Adler RJ. Excursions above a Fixed Level by N-Dimensional Random Fields. Journal of Applied Probability. 1976;13(2):276–89. [Google Scholar]
- 66.Worsley KJ, Marrett S, Neelin P, Vandal AC, Friston KJ, Evans AC. A unified statistical approach for determining significant signals in images of cerebral activation. Hum Brain Mapp. 1996;4(1):58–73. doi: 10.1002/(SICI)1097-0193(1996)4:1<58::AID-HBM4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 67.Nosko VP. Proceedings of the USSR-Japan Symposium on Probability. Harbarovsk, Novosibirak; 1969. The characteristics of excursions of Gaussian homogeneous random fields above a high level; pp. 216–22. [Google Scholar]
- 68.Dunn OJ. Multiple Comparisons among Means. J Am Stat Assoc. 1961;56(293):52&. [Google Scholar]
- 69.Dunn OJ. Estimation of the Medians for Dependent-Variables. Annals of Mathematical Statistics. 1959;30(1):192–7. [Google Scholar]
- 70.Winkler AM, Ridgway GR, Webster MA, Smith SM, Nichols TE. Permutation inference for the general linear model. NeuroImage. 2014;92:381–97. doi: 10.1016/j.neuroimage.2014.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eklund A, Dufort P, Villani M, Laconte S. BROCCOLI: Software for fast fMRI analysis on many-core CPUs and GPUs. Front Neuroinform. 2014;8:24. doi: 10.3389/fninf.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smirnov N. Table for Estimating the Goodness of Fit of Empirical Distributions. Annals of Mathematical Statistics. 1948;19(2):279-. [Google Scholar]
- 73.Kolmogorov AN. Sulla determinazione empirica di una legge di distribuzione. G Ist Ital Attuari. 1933;4:83–91. [Google Scholar]
- 74.Adler RJ. The Geometry of Random Fields. John Wiley and Sons; 1981. [Google Scholar]
- 75.Kolmogorov AN. Grundbegriffe der Wahrscheinlichkeitsrechnung. Berlin: J. Springer; 1933. [Google Scholar]
- 76.Power JD, Schlaggar BL, Petersen SE. Recent progress and outstanding issues in motion correction in resting state fMRI. NeuroImage. 2015;105:536–51. doi: 10.1016/j.neuroimage.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hettmansperger TP, McKean JW. Robust nonparametric statistical methods. 2. Boca Raton, FL: CRC Press; 2011. [Google Scholar]
- 78.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics. 2001;29(4):1165–88. [Google Scholar]
- 79.Lieberman MD, Cunningham WA. Type I and Type II error concerns in fMRI research: re-balancing the scale. Social Cognitive and Affective Neuroscience. 2009;4(4):423–8. doi: 10.1093/scan/nsp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dickstein SG, Bannon K, Castellanos FX, Milham MP. The neural correlates of attention deficit hyperactivity disorder: an ALE meta-analysis. Journal of child psychology and psychiatry, and allied disciplines. 2006;47(10):1051–62. doi: 10.1111/j.1469-7610.2006.01671.x. [DOI] [PubMed] [Google Scholar]
- 81.Mumby PJ. Statistical power of non-parametric tests: a quick guide for designing sampling strategies. Mar Pollut Bull. 2002;44(1):85–7. doi: 10.1016/s0025-326x(01)00097-2. [DOI] [PubMed] [Google Scholar]
- 82.Cox RW, Chen G, Glen DR, Reynolds RC, Taylor PA. FMRI Clustering in AFNI: False-Positive Rates Redux. Brain Connect. 2017;7(3):152–71. doi: 10.1089/brain.2016.0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Winkler AM, Webster MA, Brooks JC, Tracey I, Smith SM, Nichols TE. Non-parametric combination and related permutation tests for neuroimaging. Human Brain Mapping. 2016;37(4):1486–511. doi: 10.1002/hbm.23115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Flandin G, Friston KJ. Analysis of family-wise error rates in statistical parametric mapping using random field theory. Wellcome Trust Centre for Neuroimaging. 2016 doi: 10.1002/hbm.23839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Colquhoun D. Lectures on biostatistics: an introduction to statistics with applications in biology and medicine. Oxford: Clarendon Press; 1971. [Google Scholar]
- 86.Forman SD, Cohen JD, Fitzgerald M, Eddy WF, Mintun MA, Noll DC. Improved Assessment of Significant Activation in Functional Magnetic-Resonance-Imaging (Fmri) - Use of a Cluster-Size Threshold. Magnetic Resonance in Medicine. 1995;33(5):636–47. doi: 10.1002/mrm.1910330508. [DOI] [PubMed] [Google Scholar]
- 87.Goebel R, Esposito F, Formisano E. Analysis of Functional Image Analysis Contest (FIAC) data with BrainVoyager QX: From single-subject to cortically aligned group general linear model analysis and self-organizing group independent component analysis. Human Brain Mapping. 2006;27(5):392–401. doi: 10.1002/hbm.20249. [DOI] [PMC free article] [PubMed] [Google Scholar]