Abstract
Although electrophiles are considered as detrimental to cells, accumulating recent evidence indicates that proliferating non-cancerous and particularly cancerous cells utilize these agents for pro-survival and cell cycle promoting signaling. Hence, the redox shift to mild oxidant release must be balanced by multiple defense mechanisms. Our latest findings demonstrate that cell cycle progression, which dictates oxidant level in stress-free conditions, determines PARP1 transcription. Growth modulating factors regulate CDK4/6-RBs-E2Fs axis. In cells arrested in G1 and G0, RB1-E2F1 and RBL2-E2F4 dimers recruit chromatin remodelers such as HDAC1, SWI/SNF and PRC2 to condense chromatin and turn off transcription. Release of retinoblastoma-based repressive complexes from E2F-dependent gene promoters in response to cell transition to S phase enables transcription of PARP1. This enzyme contributes to repair of oxidative DNA damage by supporting several strand break repair pathways and nucleotide or base excision repair pathways, as well as acting as a co-activator of transcription factors such as NRF2 and HIF1a, which control expression of antioxidant enzymes involved in removal of electrophiles and secondary metabolites. Furthermore, PARP1 is indispensible for transcription of the pro-survival kinases MAP2K6, ERK1/2 and AKT1, and for maintaining MAPK activity by suppressing transcription of the MAPK inhibitor, MPK1. In summary, cell cycle controlled PARP1 transcription helps cells to adapt to a pro-oxidant redox shift.
Keywords: Cell proliferation, Redox homeostasis, Poly-ADP-ribose polymerase 1 (PARP1), Gene transcription, Signaling, DNA repair
Highlights
-
•
Cell proliferation determines PARP1 transcription and production of electrophiles.
-
•
PARP1 contributes to cell protection against electrophiles.
-
•
PARP1 controls transcription of redox-sensitive kinases, antioxidants and detoxifying enzymes.
-
•
DNA repair machinery requires PARP1 to maintain genome integrity.
-
•
G1 and G0 arrest vulnerable cells during mild oxidative stress.
Graphical abstract
1. Pro-oxidant physiology of proliferating cells
Human cells proliferate in a variety of contexts. Controlled cell divisions play a particular function for development of the embryo, while in the adult organism mainly stem and some immune cells retain the ability to proliferate. Cancer cells, as a special type of transformed cells, are capable of unlimited and uncontrolled growth. Regardless of the type of dividing cell, proliferation imposes a requirement for energy and reducing power. Although mitochondrial oxidative phosphorylation is the most efficient source of ATP, it can cause extensive release of O2•-, which is dismutated to H2O2 either in mitochondria (by SOD2) or in the cytoplasm (by SOD1). Therefore, above some critical threshold value of this oxidant in cell compartments, aerobic glycolysis becomes more favorable than oxidative phosphorylation in order to limit the hazardous waste products resulting from the mitochondrial metabolic pathway [1]. During fatty acid oxidation, O2•- and H2O2 can also be produced by xanthine oxidase in peroxisomes, which duplicate and are segregated between progeny cells. Although metabolically unrelated, NADPH oxidases act as a primary source of oxidants in macrophages and some cancer cells [2].
Depending on the cell type, proliferation-inducing agents such as growth factors (platelet-derived, fibroblast, epidermal, insulin-like and transforming growth factor β), cytokines (type I interferons, granulocyte-macrophage colony-stimulating factor), mutant K-ras or small GTPase Rac-1 elevate intracellular O2•- through NADPH oxidase and/or mitochondria [3], [4]. Due to pressure induced by an elevated and sustained redox shift to a mild oxidative environment, cells have developed efficient mechanisms of adaptation and functional transformation of „bad” to „good” molecules, which promote cell proliferation and survival at different signaling levels [5].
2. Cell cycle progression regulates PARP1 transcription
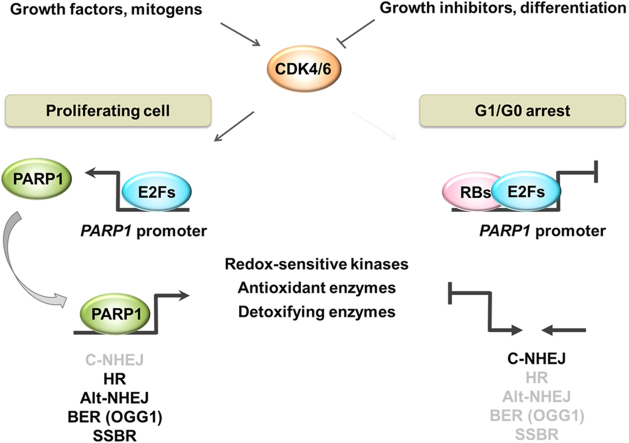
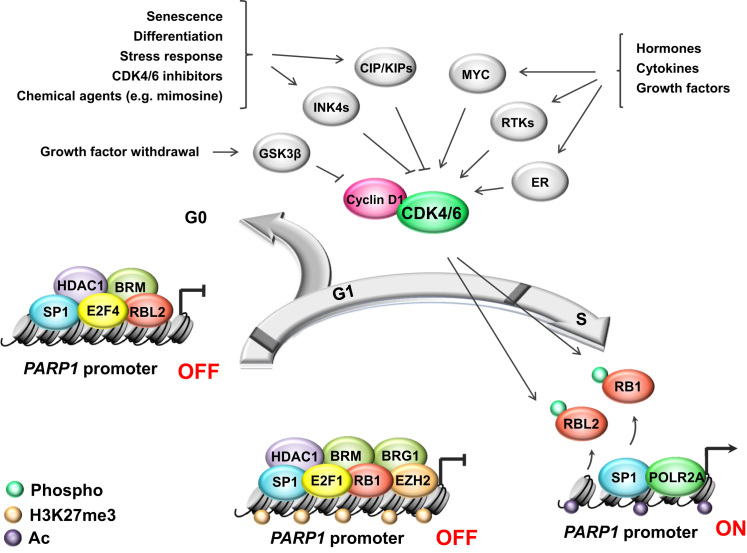
Poly-ADP-ribose polymerase 1 (PARP1) is a multitasking enzyme that regulates many intracellular processes, including DNA repair, metabolism, signaling and transcription, by direct interaction with other proteins and DNA, involving their ADP-ribosylation and auto-ADP-ribosylation of PARP1. The data acquired and published in the EMBL-EBI Expression Atlas indicate high PARP1 abundance in proliferating cancerous and non-cancerous cells (e.g. macrophages) [6]. In search for the link between PARP1 transcription and cell cycle progression, we recently revealed that cell arrest in G1 or exit to G0 lead to PARP1 repression by retinoblastoma-based multiprotein complexes, which are also known to repress transcription of E2F-dependent genes encoding proteins responsible for cell transition to S phase [7]. The mode of growth inhibition determines the composition of the repressive complex at the PARP1 promoter, giving priority to E2F1-RB1 dimers under G1 arrest in cancer, as well as in CD34 + hematopoietic progenitor/stem cells treated with cyclin-dependent kinases 4 and 6 (CDK4/6) pharmacological inhibitors or depleted of nucleotides by mimosine. E2F4-RBL2-based complexes were found to be prevalent in differentiated cells (Fig. 1). Since PARP1 is involved in cell protection against oxidants, one may think that PARP1 repression in response to proliferation arrest may sensitize cells to agents that challenge redox homeostasis. Some ongoing and recruiting clinical trials have been testing FDA approved CDK4/6 inhibitors Palbociclib (IBRANCE®, PD0332991) and Ribociclib (LEE011, Kisqali) in combination with drugs such as doxorubicin, carboplatin and paclitaxel, which trigger acute redox imbalance [8].
Fig. 1.
Cell cycle progression dictatesPARP1transcription via growth factors/inhibitors-G1/G0-CDK4/6-RBs axis. Cell cycle machinery is controlled by external signals in order to adapt cells to environmental requirements and conditions. Stimulation of receptor tyrosine kinases (RTKs), MYC protooncogene or estrogen receptor (ER) in response to peptide and non-peptide growth-promoting agents activates cyclin-dependent kinase 4 and 6 (CDK4/6), which associate with cyclin D1 and phosphorylate retinoblastoma proteins (RB1, RBL2). This modification keeps retinoblastoma proteins released from promoters of PARP1 and cell cycle promoting genes, thereby allowing active gene transcription and enabling cell transition from G1 to S phase. Upon cell growth arrest in G1 or cell cycle exit to G0, CDK4/6 inhibition results in hypophosphorylation of retinoblastoma proteins, their binding to E2F-driven gene promoters and recruitment of chromatin remodelers, which are capable of inactivating gene expression by removing transcription-promoting indicators and/or inserting transcription-inhibiting histone modification(s). It leads to an increase in nucleosome density and chromatin condensation. Notably, composition of the repressive complex varies between cells arrested in G1 and in G0. Limiting PARP1 expression in G0 is achieved solely by histone deacetylase 1 (HDAC1) for histone deacetylation, while in G1 HDAC1 additionally requires PRC2 (polycomb repressor complex 2) activity and trimethylation of H3K27 by enhancer of zeste homolog 2 (EZH2) to repress PARP1 transcription. Cell cycle arrest in G2 does not affect the mRNA and protein levels of PARP1.
Furthermore, PARP1 enhances cell proliferation. Hormone-activated cyclin-dependent kinase 2 (CDK2) phosphorylates and activates PARP1, thereby facilitating H1 displacement and transcription of the majority of hormone-responsive genes in breast cancer [9]. In urinary bladder carcinoma cells, PARP1 regulates cyclin E expression, cell cycle re-entry and G1/S progression [10]. Thus, high levels of PARP1 in cancer cells promote cell cycle progression, which is associated with an increased level of oxidants, thereby maintaining PARP1 transcription and creating a self-promoting cycle.
3. PARP1 co-activates expression of proteins that enzymatically decompose oxidants and remove secondary metabolites
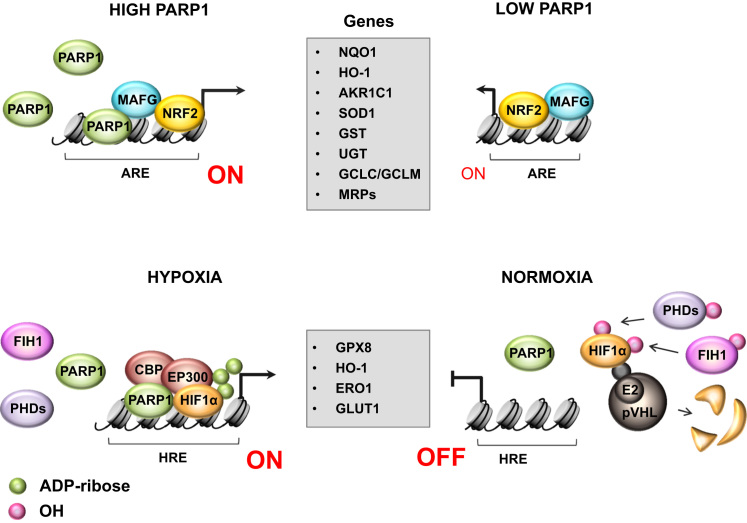
The primary role in antioxidant defense and in cell adaptation to excessive oxidant or electrophile production is fulfilled by enzymatic antioxidant defense, which comprises direct scavengers of electrophiles, but also enzymes that detoxify the secondary metabolites. Many such enzymes are under transcriptional control of nuclear factor erythroid 2 (NFE2)-related factor 2 (NRF2), a basic leucine zipper (bZIP) protein, which dissociates from its repressor Keap1 and translocates to nucleus in response to a physiological shift in redox homeostasis towards oxidant production. NRF2 requires PARP1 for full transcriptional activity, because PARP1 facilitates interaction of NRF2 and NRF2-partner (small MAF protein; MAFG) with the antioxidant response element (ARE) (Fig. 2) [11]. An inhibitory effect of PARP1 knockdowns was found in breast cancer cells and proliferating mouse fibroblasts. Although in normal cells NRF2 suppresses tumor promotion and progression, this pathway is constitutively activated in various cancers by mutation and transcriptional repression of Keap-1, accumulation of Keap-1-NRF2 disruptors, transcriptional and post-translational NRF2 induction. In view of NRF2 targets, this transcription factor provides chemoresistance and, like PARP1, has become a target for anticancer interventions [12].
Fig. 2.
PARP1 contributes to antioxidant cell defense by enhancing transcription of enzymatic scavengers of electrophiles and secondary metabolites. Under normal oxygen conditions, PARP1 determines intracellular redox homeostasis by intensifying nuclear factor erythroid 2 (NFE2)-related factor 2 (NRF2)-dependent transcription of enzymatic redox-balancing enzymes (NAD(P)H quinone oxidoreductase 1, NQO1; heme oxygenase-1, HO-1; aldo-keto reductase family 1, member C1, AKR1C1; superoxide dismutase 1, SOD1), as well as phase II detoxifying enzymes (glutathione S-transferase, GST; UDP-glucuronosyltransferase, UGT; catalytic and modifier subunits of glutamate cysteine ligase, GCLC and GCLM respectively) and drug transporters (multidrug resistance-associated proteins, MRPs). In the absence of PARP1, transcription of the above-mentioned genes is restricted as NRF2 moderately associates with small MAF proteins (in this case with MAFG) and the antioxidant response element (ARE), which is localized within the promoter of NRF2 target genes. When abundant, PARP1 enhances the interaction among NRF2, MAFG and ARE, thereby acting as a co-activator of NRF2-dependent gene transcription. PARP1 also functions actively in cell adaptive responses to match O2 supply under hypoxia by supporting hypoxia-inducible factor 1-alpha (HIF1α) at different signaling levels. This protein, together with PARP1, acts as a key modulator of the transcription response in cells that experience a low O2 level. Under normal oxygen condition, factor inhibiting HIF1α (FIH1; asparaginyl hydroxylase) and propyl hydroxylase domain-containing enzymes (PHDs) hydroxylate HIF1α, thereby preventing transcription factor interaction with EP300/CBP coactivators and marking HIF1α for proteasomal degradation by E3 ubiquitin ligase, the von Hippel-Lindau (pVHL) complex. Low O2 concentration inhibits hydroxylases and stabilizes HIF1α. PARP1 forms complex and co-activates HIF1α in a PARP1 enzymatic activity-dependent manner, therefore enabling expression of genes controlled by hypoxia response element (HRE)-positive promoters. This group comprises antioxidant defense enzymes such as heme oxygenase-1 (HO-1), glutathione peroxidase 8 (GPX8), ER oxidoreductin 1 (ERO1) and glucose transporter 1 (GLUT1), the activity of which helps to maintain glutathione homeostasis.
Another oxidant-counteracting mechanism that involves PARP1 is represented by its interaction with the transcription factor hypoxia-inducible factor 1-alpha (HIF1α), which undergoes activation during hypoxia and hypoxia-triggered redox imbalance [13], [14]. PARP1 co-activates HIF1α-dependent transcription of genes, which promotes cell survival. In murine embryonic fibroblasts, PARP1 caused accumulation of HIF1α via upregulation of NO and oxidant production in cells treated with deferoxamine [15]. In addition to PARP1, HIF1α binds EP300/CBP acetylase(s) for full transcriptional activity. A similar observation was made for nuclear factor kappa B (NF-κB), activation of which required synergistic interaction with PARP1 and EP300/CBP. However, for HIF1α mutual dependence between these two types of co-activators has not been documented yet.
Promoters of some antioxidant enzymes such as catalase, SOD1 or SOD2 carry the binding motif for NF-κB, but the role of PARP1 in transcription activation of these genes has not been confirmed. Instead, PARP1 level negatively correlates with mitochondrial SOD in cancer cells (EMBL-EBI Expression Atlas), where SOD2 overexpression causes a growth inhibitory effect by shifting the O2•-/H2O2 balance towards H2O2 accumulation [16]. If PARP1 is involved in SOD2 repression, this enzyme could be capable of defining the intracellular repertoire of growth promoting or inhibiting oxidants. Furthermore, low SOD2 level is known to stabilize HIF1α [17].
4. PARP1 regulates redox-sensitive signaling pathways
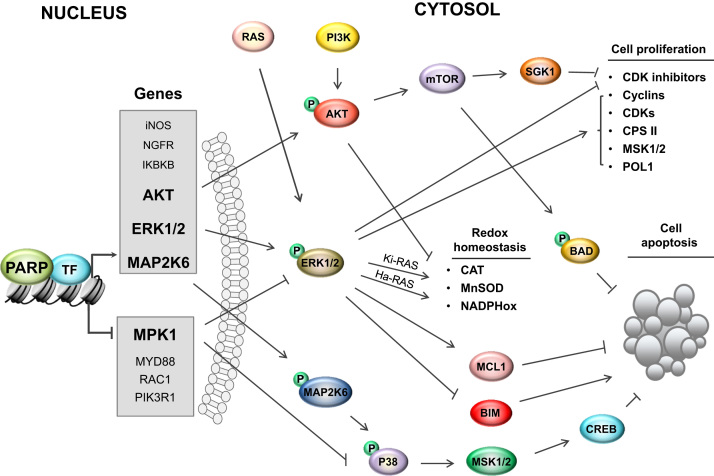
The roles that small species of oxidizing nature play in cellular signaling are becoming increasingly appreciated. Redox-sensitive pathways allow cells to adapt to mild oxidant/antioxidant imbalance and promote survival by linking redox shifts to post-translational modifications of proteins and to their interactome [5], [18]. According to our and previous findings, PARP1 regulates transcription of numerous genes encoding redox sensors and mediators transmitting signals upstream or downstream of electrophile sources (Fig. 3). Redox-sensitive MAP kinase signaling serves as a good example of the PARP1-MAPK-ROS-cell survival axis since PARP1 couples transcription of MAP2K6, ERK1/2 and AKT1 with the cell cycle progression, thereby assuring active transcription of kinases that have a pro-survival function under mild redox imbalance [19], [20], [21], [22], [23], [24], [25], [26]. Furthermore, oxidative stress-induced PARP1 activation represses transcription of MPK1, a known MAPK inhibitor [27]. Although in this particular case JNK and p38 were shown to act as pro-death kinases, the beneficial or detrimental activity of kinases and MAPK pathways is determined by the type of intracellular or extracellular stimuli that challenges redox homeostasis, for example whether the action is acute or prolonged, mild or severe, but is also dependent on the cell type. All these aspects also apply to PARP1. In addition to conditions listed above, PARP1-dependent cell life or death fate is determined by the pathway to which PARP1 contributes (pro-survival or suicidal), the direction (inhibition or stimulation) and mode of mutual interdependence with its interacting partner (direct protein-protein binding or covalent modification, i.e. ADP-ribosylation). Oxidants are one of major agents triggering mono- or poly-ADP-ribosylation; the severity of oxidative stress determines the length of ADP polymer synthesized, thus also impinging on the beneficial or detrimental effect of PARP1 activation, since NAD+ is utilized as a substrate for ADP-ribosylation. High doses of H2O2 cause metabolic catastrophe, parthanatos, and activation of detrimental signaling pathways, while moderate PARylation protects cells from mild oxidative stress by attracting DNA repair complexes, clearing and removal of oxidized or damaged proteins, and re-establishing homeostasis [28], [29].
Fig. 3.
PARP1 contributes to regulation of redox-related signaling pathways. PARP1 regulates both positively and negatively transcription of numerous enzymes involved in the transmitting signals to and from oxidant-releasing intracellular systems or extracellular sources. Expression of MAP2K6 and ERK1 are, like PARP1, controlled by cell cycle progression and RB-based repressive complexes. In proliferating cells, PARP1 is indispensable for their transcription, because it mediates EP300 recruitment to their promoters. Although AKT and ERK2 are not directly repressed by RBs, PARP1 maintains their expression. All these kinases were shown to protect proliferating cells facing mild or physiological increases in the electrophile abundance from death. In the AKT pathway, PI3 acts as a redox sensor and after activation phosphorylates AKT, which in turn activates mTOR kinase. This enzyme stimulates cell proliferation via the SGK1-FOXO3 pathway, which represses transcription of CDK inhibitors. Moreover, mTOR phosphorylates and inactivates BAD, thus blocking the release of cytochrome c from mitochondria. AKT contributes to H2O2 accumulation by stimulating oxidative metabolism and FOXO-dependent repression of catalase. Growth factors and oxidants switch on ERK signaling via RAS, which, depending on the isoform expressed in a particular cell type, shifts up or down the intracellular level of oxidants. Ha-RAS isoform promotes O2•- accumulation by activating NADPH oxidase, while the Ki-RAS pathway upregulates transcription of SOD2. There are numerous ERK targets, which implicate this enzyme in cell proliferation: carbamoyl phosphate synthetase (CPS II, source of pyrimidine nucleotides), MSK1/2 (chromatin remodeling and induction of cell cycle-related gene transcription), RNA polymerase I (transcription of the ribosomal RNA genes), CDK inhibitors or MYC (transcription of cyclin D1). ERK protects cells from death by repressing pro-apoptotic BIM and stabilizing anti-apoptotic MCL-1 protein. PARP1-MAP2K6 functional cross-talk at the genomic level is not limited to upregulation of kinase transcription. MAP2K6 links PARP1 with transcription of anti-apoptotic genes indirectly by starting the phosphorylation cascade MAP2K6-p38-MSK1/2-CREB. The last component of this axis controls transcription of HO-1 and PGC-1α; the latter promotes mitochondrial biogenesis. Furthermore, PARP1 regulates activity of MAPKs independently of their promoters by regulating transcription of JNK, ER1/2 and the p38 inhibitor MKP1. Downregulation of MKP1 expression by PARP1 blocks dephosphorylation of tyrosine and threonine residues of MAPKs, which undergo activation upon acute cell exposure to H2O2.
5. Cell cycle determines DNA repair mechanisms
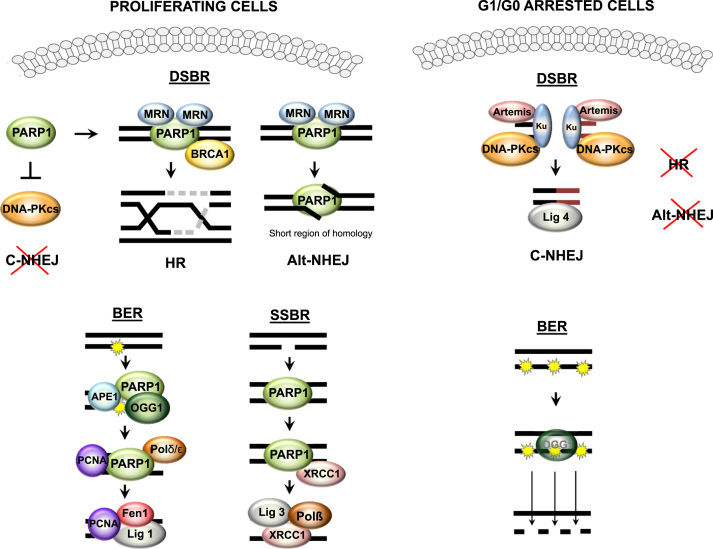
PARP1 actively contributes to numerous repair pathways of oxidative DNA lesions, which comprise covalent modifications of nucleobases as well as single and double strand breaks (SSB and DSB, respectively). The deformability of DNA within SSB is recognized by two flexibly linked N-terminal zinc fingers, and initiates self-assembly of remaining PARP1 domains leading to activation of the C-terminal catalytic domain [30]. In case of DSB induced by oxidative stress, JNK 6 phosphorylates SIRT6. This enzyme is rapidly mobilized to break sites, where it potentiates recruitment and activation of PARP1, which in turn stimulates DSB repair [31]. In proliferating cells all repair mechanisms are active, and PARP1 is highly expressed to support SSB repair (SSBR), base excision repair (BER), homologous recombination (HR), and alternative non-homologous end joining (Alt-NHEJ), but inhibits classical non-homologous end joining (C-NHEJ) (Fig. 4). Thus PARP1 has become a target for anticancer interventions. G1/G0 arrest shifts error-free HR and Alt-NHEJ to error-prone C-NHEJ, but low PARP1 impairs also BER and SSBR [32], [33], [34]. According to our new data, PARP1 repression by CDK4/6 inhibitors reduces PARP1-dependent 8-oxoguanine glycosylase (OGG1) activity, causing accumulation of single strand breaks and thereby increasing cell vulnerability to anticancer drugs and H2O2-induced oxidative stress [35]. The direct binding of OGG1 to PARP1 is stimulated by increased oxidant level, and for full activity OGG1 requires acetylation by EP300, which physically interacts with PARP1 and is recruited to some genomic loci by PARP1 [36], [37].
Fig. 4.
PARP1 regulates repair of oxidative DNA damages. The cell decision on involvement of particular repair system depends on the type of DNA damage and is strongly related to cell cycle progression. For double strand breaks, the cell is equipped with three repair systems: homologous recombination (HR), classical non-homologous end joining (C-NHEJ) or alternative non-homologous end joining (Alt-NHEJ). Proliferating cells, with high PARP1 levels, make use mainly of two mechanisms: HR and/or Alt-NHEJ, which assure accurate and error-free repair of double strand breaks since they use a replicated DNA template to reconstruct the missing fragment with high fidelity. These two pathways rely on the recognition of detrimental lesions by PARP1, which recruits other proteins to the affected sites: first MRN complex (consisting of MRE11, Rad50, Nbs1), an initiator of repair, then MRE11 determines the composition of proteins for each repair mechanism (HR or NHEJ). The low C-NHEJ involvement in repair is achieved by high expression of PARP1, an inhibitor of DNA-dependent protein kinases (DNA-PKcs), which is crucial for C-NHEJ progression. In G1/G0 arrested cells (deficient in HR and Alt-NHEJ), low expression of PARP1 allows recruitment of the Ku70/80 heterodimer to double strand breaks and sites of DNA-PKcs activation. Repair machinery such as nuclease Artemis, DNA ligase IV and XRCC4 further process the damaged DNA and directly ligate DNA ends, leading to the irreversible loss of genetic material. Base excision repair (BER) is a substantial pathway to repair oxidized bases. The damaged base is recognized and removed by OGG1 glycosylase, which requires PARP1 for proper and efficient functioning. AP endonuclease (APE1) is binds to apurynic sites and produces single-strand breaks (SSBs), which again involve PARP1 in the BER machinery at a later repair step. DNA nick-induced poly-ADP-ribosylation facilitates PARP1 interaction with DNAPδ/ε and PCNA, which further govern repair machinery. Single strand breaks resulting from direct oxidant action also need PARP1 activity to be repaired. Poly-ADP-ribose polymers recruit XRCC1, then the remaining SSBR machinery comprising DNA polymerase (DNAP) and DNA ligase III, which also can be PARylated by PARP1. PARP1 deficiency in growth-arrested cells substantially impairs both pathways. Of note, inhibition of OGG1 in combination with G1-blockade leads to further accumulation of single strand breaks.
PARP1 has been postulated to cooperate with transcription factor(s) that activate expression of genes encoding proteins contributing to DNA repair [38]. Although such a premise must be experimentally confirmed, PARP1 is a bona fide co-regulator of NF-κB, p53, AP-1, E2F1, and BRCA1, which control promoter activation, epigenetic landscape and miRNA transcription of DNA repairing machinery. The confirmation for likely contribution of PARP1 in regulation of DNA repair gene transcription comes from observations in human growth arrested monocytes differentiating to proliferating macrophages [39]. This process was associated with increased PARP1 expression, but also with transcriptional activation of XRCC1, ligase IIIα, OGG1 and catalytic subunit of DNA-dependent protein kinase (DNA-PKcs). Severe DNA repair defects that impacted base excision repair and double-strand break repair in monocytes sensitized these cells to death by t-butyl hydroperoxide and irradiation with γ-rays, while macrophages revealed almost complete resistance to these redox-challenging agents.
To conclude, PARP1 provides cell with protection against oxidants at different levels: by activating expression of proteins setting up antioxidant defense and redox sensitive signaling pathways, and by fine tuning of DNA repair machinery. Therefore PARP1 expression, which is defined by the proliferative status of cells, can determine cell resistance to oxidants, even though an adaptive response is only apparent within a narrow dose window.
Acknowledgments
AR acknowledges grants from Polish National Science Center (DEC-2013/11/D/NZ2/00033) and Ministry of Science and Higher Education (776/STYP/11/2016); CMS acknowledges funding from the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement number 675132; LV acknowledges grants from the National Research, Development and Innovation Office (GINOP-2.3.2-15-2016-00020-TUMORDNS, GINOP-2.3.2-15-2016-00048-STAYALIVE, OTKA K112336).
References
- 1.Molavian H.R., Kohandel M., Sivaloganathan S. High concentrations of H2O2 make aerobic glycolysis energetically more favorable for cellular respiration. Front. Physiol. 2016;7:362. doi: 10.3389/fphys.2016.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambeth J.D., Kawahara T., Diebold B. Regulation of Nox and Duox enzymatic activity and expression. Free Radic. Biol. Med. 2007;43:319–331. doi: 10.1016/j.freeradbiomed.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liou G.Y., Storz P. Reactive oxygen species in cancer. Free Radic. Res. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24:453–462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espinosa-Diez C., Miguel V., Mennerich D., Kietzmann T., Sanchez-Perez P., Cadenas S., Lamas S. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015;6:183–197. doi: 10.1016/j.redox.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EMBL-EBI 〈https://www.ebi.ac.uk/services/gene-expression〉, 2018.
- 7.Wiśnik E., Płoszaj T., Robaszkiewicz A. Downregulation of PARP1 transcription by promoter-associated E2F4-RBL2-HDAC1-BRM complex contributes to repression of pluripotency stem cell factors in human monocytes. Sci. Rep. 2017;7:9483. doi: 10.1038/s41598-017-10307-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.NIH: US National Library of Medicine 〈https://clinicaltrials.gov/〉, 2018. [DOI] [PubMed]
- 9.Wright R.H., Castellano G., Bonet J., Le Dily F., Font-Mateu J., Ballare C., Nacht A.S., Soronellas D., Oliva B., Beato M. CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes Dev. 2012;26:1972–1983. doi: 10.1101/gad.193193.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leger K., Hopp A.K., Fey M., Hottiger M.O. ARTD1 regulates cyclin E expression and consequently cell-cycle re-entry and G1/S progression in T24 bladder carcinoma cells. Cell Cycle. 2016;15:2042–2052. doi: 10.1080/15384101.2016.1195530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu T., Wang X.J., Tian W., Jaramillo M.C., Lau A., Zhang D.D. Poly(ADP-ribose) polymerase-1 modulates Nrf2-dependent transcription. Free Radic. Biol. Med. 2014;67:69–80. doi: 10.1016/j.freeradbiomed.2013.10.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kansanen E., Kuosmanen S.M., Leinonen H., Levonen A.L. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majmundar A.J., Wong W.J., Simon M.C. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen T.L., Duran R.V. Prolyl hydroxylase domain enzymes and their role in cell signaling and cancer metabolism. Int. J. Biochem. Cell Biol. 2016;80:71–80. doi: 10.1016/j.biocel.2016.09.026. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Romero R., Martinez-Lara E., Aguilar-Quesada R., Peralta A., Oliver F.J., Siles E. PARP-1 modulates deferoxamine-induced HIF-1alpha accumulation through the regulation of nitric oxide and oxidative stress. J. Cell Biochem. 2008;104:2248–2260. doi: 10.1002/jcb.21781. [DOI] [PubMed] [Google Scholar]
- 16.Oberley L.W. Mechanism of the tumor suppressive effect of MnSOD overexpression. Biomed. Pharmacother. 2005;59:143–148. doi: 10.1016/j.biopha.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Movafagh S., Crook S., Vo K. Regulation of hypoxia-inducible factor-1a by reactive oxygen species: new developments in an old debate. J. Cell Biochem. 2015;116:696–703. doi: 10.1002/jcb.25074. [DOI] [PubMed] [Google Scholar]
- 18.Sies H. Oxidative stress: a concept in redox biology and medicine. Redox Biol. 2015;4:180–183. doi: 10.1016/j.redox.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robaszkiewicz A., Wisnik E., Regdon Z., Chmielewska K., Virag L. PARP1 facilitates EP300 recruitment to the promoters of the subset of RBL2-dependent genes. Biochim. Biophys. Acta. 2017 doi: 10.1016/j.bbagrm.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Mori S., Nada S., Kimura H., Tajima S., Takahashi Y., Kitamura A., Oneyama C., Okada M. The mTOR pathway controls cell proliferation by regulating the FoxO3a transcription factor via SGK1 kinase. PLoS One. 2014;9:e88891. doi: 10.1371/journal.pone.0088891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chambard J.C., Lefloch R., Pouyssegur J., Lenormand P. ERK implication in cell cycle regulation. Biochim. Biophys. Acta. 2007;1773:1299–1310. doi: 10.1016/j.bbamcr.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Kim E.K., Choi E.J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta. 2010;1802:396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Lee B., Cao R., Choi Y.S., Cho H.Y., Rhee A.D., Hah C.K., Hoyt K.R., Obrietan K. The CREB/CRE transcriptional pathway: protection against oxidative stress-mediated neuronal cell death. J. Neurochem. 2009;108:1251–1265. doi: 10.1111/j.1471-4159.2008.05864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glorieux C., Auquier J., Dejeans N., Sid B., Demoulin J.B., Bertrand L., Verrax J., Calderon P.B. Catalase expression in MCF-7 breast cancer cells is mainly controlled by PI3K/Akt/mTor signaling pathway. Biochem. Pharmacol. 2014;89:217–223. doi: 10.1016/j.bcp.2014.02.025. [DOI] [PubMed] [Google Scholar]
- 25.Ismail N., Ismail M., Imam M.U., Azmi N.H., Fathy S.F., Foo J.B., Abu Bakar M.F. Mechanistic basis for protection of differentiated SH-SY5Y cells by oryzanol-rich fraction against hydrogen peroxide-induced neurotoxicity. BMC Complement Altern. Med. 2014;14:467. doi: 10.1186/1472-6882-14-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyrsch P., Blenn C., Bader J., Althaus F.R. Cell death and autophagy under oxidative stress: roles of poly(ADP-Ribose) polymerases and Ca(2+) Mol. Cell Biol. 2012;32:3541–3553. doi: 10.1128/MCB.00437-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Racz B., Hanto K., Tapodi A., Solti I., Kalman N., Jakus P., Kovacs K., Debreceni B., Gallyas F., Jr., Sumegi B. Regulation of MKP-1 expression and MAPK activation by PARP-1 in oxidative stress: a new mechanism for the cytoplasmic effect of PARP-1 activation. Free Radic. Biol. Med. 2010;49:1978–1988. doi: 10.1016/j.freeradbiomed.2010.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Luo X., Kraus W.L. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hocsak E., Szabo V., Kalman N., Antus C., Cseh A., Sumegi K., Eros K., Hegedus Z., Gallyas F., Jr., Sumegi B., Racz B. PARP inhibition protects mitochondria and reduces ROS production via PARP-1-ATF4-MKP-1-MAPK retrograde pathway. Free Radic. Biol. Med. 2017;108:770–784. doi: 10.1016/j.freeradbiomed.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 30.Eustermann S., Wu W.F., Langelier M.F., Yang J.C., Easton L.E., Riccio A.A., Pascal J.M., Neuhaus D. Structural basis of detection and signaling of dna single-strand breaks by human PARP-1. Mol. Cell. 2015;60:742–754. doi: 10.1016/j.molcel.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Meter M., Simon M., Tombline G., May A., Morello T.D., Hubbard B.P., Bredbenner K., Park R., Sinclair D.A., Bohr V.A., Gorbunova V., Seluanov A. JNK phosphorylates SIRT6 to stimulate DNA double-strand break repair in response to oxidative stress by recruiting PARP1 to DNA breaks. Cell Rep. 2016;16:2641–2650. doi: 10.1016/j.celrep.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiruvella K.K., Liang Z., Wilson T.E. Repair of double-strand breaks by end joining. Cold Spring Harb. Perspect. Biol. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dean J.L., Mcclendon A.K., Knudsen E.S. Modification of the DNA damage response by therapeutic CDK4/6 inhibition. J. Biol. Chem. 2012;287:29075–29087. doi: 10.1074/jbc.M112.365494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei H., Yu X. Functions of PARylation in DNA damage repair pathways. Genom. Proteom. Bioinforma. 2016;14:131–139. doi: 10.1016/j.gpb.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tempka D., Tokarz P., Chmielewska K., Kluska M., Pietrzak J., Rygielska Z., Virag L., Robaszkiewicz A. Downregulation of PARP1 transcription by CDK4/6 inhibitors sensitizes human lung cancer cells to anticancer drug-induced death by impairing OGG1-dependent base excision repair. Redox Biol. 2018;15:316–326. doi: 10.1016/j.redox.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang L., Zhao W., Zhang G., Wu J., Guan H. Acetylated 8-oxoguanine DNA glycosylase 1 and its relationship with p300 and SIRT1 in lens epithelium cells from age-related cataract. Exp. Eye Res. 2015;135:102–108. doi: 10.1016/j.exer.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Noren Hooten N., Kompaniez K., Barnes J., Lohani A., Evans M.K. Poly(ADP-ribose) polymerase 1 (PARP-1) binds to 8-oxoguanine-DNA glycosylase (OGG1) J. Biol. Chem. 2011;286:44679–44690. doi: 10.1074/jbc.M111.255869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christmann M., Kaina B. Transcriptional regulation of human DNA repair genes following genotoxic stress: trigger mechanisms, inducible responses and genotoxic adaptation. Nucleic Acids Res. 2013;41:8403–8420. doi: 10.1093/nar/gkt635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bauer M., Goldstein M., Christmann M., Becker H., Heylmann D., Kaina B. Human monocytes are severely impaired in base and DNA double-strand break repair that renders them vulnerable to oxidative stress. Proc. Natl. Acad. Sci. USA. 2011;108:21105–21110. doi: 10.1073/pnas.1111919109. [DOI] [PMC free article] [PubMed] [Google Scholar]