SUMMARY
Human APOBEC3H and homologous single-stranded DNA cytosine deaminases are unique to mammals. These DNA editing enzymes function in innate immunity by restricting the replication of viruses and transposons. APOBEC3H also contributes to cancer mutagenesis. Here we address the fundamental nature of RNA in regulating human APOBEC3H activities. APOBEC3H co-purifies with RNA as an inactive protein, and RNase A treatment enables strong DNA deaminase activity. RNA binding-defective mutants demonstrate clear separation-of-function by becoming DNA hypermutators. Biochemical and crystallographic data demonstrate a mechanism in which double-stranded RNA mediates enzyme dimerization. Additionally, APOBEC3H separation-of-function mutants show that RNA binding is required for cytoplasmic localization, packaging into HIV-1 particles, and antiviral activity. Overall, these results support a model in which structured RNA negatively regulates the potentially harmful DNA deamination activity of APOBEC3H, while at the same time positively regulating its antiviral activity.
Keywords: APOBEC3H, DNA cytosine deaminase, HIV-1, retrovirus restriction factor, ribonucleoprotein complex, RNA duplex, structural virology
INTRODUCTION
Human cells have the potential to express up to nine active DNA cytosine deaminase enzymes collectively called APOBECs (Conticello, 2008; Harris and Dudley, 2015). Activation-induced deaminase (AID) is expressed in antigen-responsive B lymphocytes, and it initiates the diversification of antibody genes through the distinct processes of somatic hypermutation and class switch recombination (Di Noia and Neuberger, 2007; Robbiani and Nussenzweig, 2013). APOBEC1 is expressed in gastrointestinal tissues and edits a range of RNA species including the APOB mRNA (inspiring the original family name) (Fossat and Tam, 2014; Koito and Ikeda, 2013). APOBEC3A (A3A), APOBEC3B (A3B), APOBEC3C (A3C), APOBEC3D (A3D), APOBEC3F (A3F), APOBEC3G (A3G), and APOBEC3H (A3H) are expressed at varying levels in different human tissues. These enzymes function broadly and inducibly in innate immunity by restricting the replication of DNA-based parasites, including common retrotransposons L1 and Alu and retroviruses such as HTLV-1, HIV-1, and HIV-2 (Harris and Dudley, 2015; Malim and Bieniasz, 2012; Simon et al., 2015).
Although single-stranded DNA cytosine deamination is the hallmark activity of the APOBEC enzyme family, RNA is important for several aspects of APOBEC biology. First, RNA editing is a bona fide function for at least two family members, APOBEC1 (Teng et al., 1993) and A3A (Sharma et al., 2015). Second, RNA has been implicated in governing the cytoplasmic localization of A3G and A3F, including accumulation in stress granules and RNA processing bodies (Gallois-Montbrun et al., 2007; Izumi et al., 2013; Kozak et al., 2006; Phalora et al., 2012; Stenglein et al., 2008; Wichroski et al., 2006). Third, an RNA-dependent interaction is required for A3G and A3F to interact with HIV-1 Gag, gain access to assembling viral particles, and exert antiviral activity (Apolonia et al., 2015; Bogerd and Cullen, 2008; Cen et al., 2004; Huthoff and Malim, 2007; Khan et al., 2005; Schafer et al., 2004; Svarovskaia et al., 2004; Wang et al., 2007; Wang et al., 2008; York et al., 2016; Zennou et al., 2004). A similar RNA-dependent packaging mechanism is proposed for the other HIV-1 restrictive APOBEC3 family members, A3D and A3H (Wang et al., 2011; York et al., 2016; Zhen et al., 2012). Fourth, interactions between many different cellular proteins, including but not limited to A3F, A3G, APOBEC1, and AID, are disrupted by RNase A treatment and thus likely RNA-mediated (Basu et al., 2011; Chiu et al., 2006; Gallois-Montbrun et al., 2008; Gallois-Montbrun et al., 2007; Kozak et al., 2006; Nowak et al., 2011). Finally, nearly all APOBEC biochemical studies include RNase A treatment in purification protocols suggesting that RNA may be a negative regulatory factor that prevents access to single-stranded DNA substrates (Ara et al., 2014; Bransteitter et al., 2003; Chelico et al., 2006; McDougall and Smith, 2011; Mitra et al., 2015; Shlyakhtenko et al., 2011; Starrett et al., 2016). However, despite an overarching importance for RNA in APOBEC biology, mechanistic details are lacking largely due to the absence of molecular and structural information on APOBEC-RNA complexes.
A3H is unique among APOBEC3 subfamily members for several reasons. First, the gene encoding A3H is phylogenetically distinct, shows high conservation, and invariably anchors the 3′-end of the APOBEC3 locus (LaRue et al., 2008; Münk et al., 2008). The encoded enzyme exists as a single zinc-coordinating domain Z3-type deaminase in higher primates including humans, or as a single- or double-domain enzyme, typically Z2–Z3 organization, in most other mammals including artiodactyls, carnivores, and rodents (LaRue et al., 2008; Münk et al., 2008). Second, unlike Z1 and Z2-type deaminase genes, the A3H or Z3-type genes show no copy number variation, existing in each species as a full gene or as the 3′ half of a gene encoding a double domain deaminase. Third, despite strict copy number conservation, A3H is the most polymorphic family member in humans with 7 reported haplotypes due to variations at 5 amino acid positions and 4 reported splice variants (Harari et al., 2009; OhAinle et al., 2008; Wang et al., 2011). These naturally occurring A3H variations are known to influence protein stability, antiviral activity, and subcellular localization, with at least two stable variants capable of potently suppressing Vif-deficient HIV-1 replication (Harari et al., 2009; Hultquist et al., 2011; Nakano et al., 2017; OhAinle et al., 2008; Ooms et al., 2013; Refsland et al., 2014; Wang et al., 2011). A3H is also implicated in restricting the replication of multiple other viruses and transposons (Bouzidi et al., 2016; Hultquist et al., 2011; Kock and Blum, 2008; Ooms et al., 2012; Tan et al., 2009). Moreover, mislocalized A3H (haplotype I) is a likely contributor to cancer mutagenesis (Starrett et al., 2016).
Here we investigate the role of RNA in regulating the activities of A3H. The most common, stable, active, and antiviral A3H variant in humans is the 183 amino acid haplotype II enzyme, hereafter called A3H. RNA invariably co-purifies with wild-type A3H and suppresses enzyme activity. RNase A treatment removes most, but not all, of the bound RNA and enables strong DNA deaminase activity. Mutagenesis of a positively charged patch predicted to bind RNA results in enzymes with DNA hypermutator activity. Wild-type A3H and hyperactive variants show dimeric and monomeric size exclusion profiles, respectively, suggesting a RNA-mediated dimerization mechanism. Indeed, crystal structures of a near wild-type human A3H-mCherry protein and a catalytic mutant derivative reveal a duplex RNA-bridged enzyme dimer involving the precise amino acid residues that yield DNA deaminase hyperactivity upon mutagenesis. Experiments with these separation-of-function mutants demonstrate that the RNA binding surface is required for A3H cytoplasmic localization and for packaging into HIV-1 particles and antiviral activity. Thus, RNA serves multiple regulatory roles in A3H biology.
RESULTS
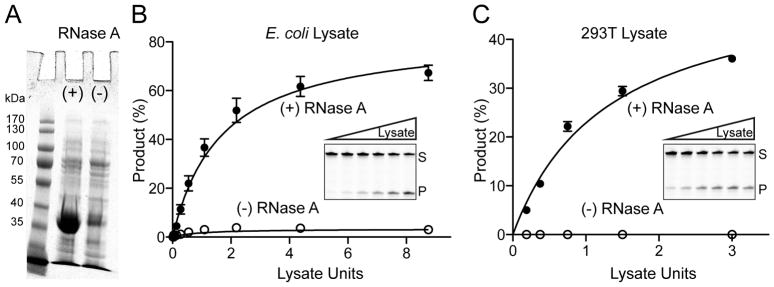
RNA Digestion Enhances APOBEC3H Purification and DNA Cytosine Deaminase Activity
Initial attempts to purify His6-SUMO-A3H from E. coli were unsuccessful because most protein failed to bind the affinity resin. We speculated that RNA may be promoting aggregation and preventing the hexahistidine tag from binding. We therefore treated lysates with RNase A, repeated metal affinity purifications, and compared fractions by SDS PAGE. This procedure enables a strong recovery of A3H but only with RNase A treatment (Figure 1A). Furthermore, using an oligo-based single-stranded DNA deamination activity assay, we found that A3H catalytic activity is only detected in E. coli lysates treated with RNase A suggesting that RNA inhibits deaminase activity (Figure 1B). A similar requirement for RNase A is evident in activity assays of A3H expressed in human 293T cells (Figure 1C). The necessity for RNase A treatment has been described previously for A3H, as well as for all other family members including A3G and AID [e.g., (Bransteitter et al., 2003; Chiu et al., 2006; Gu et al., 2016; Mitra et al., 2015; Shlyakhtenko et al., 2011)]. Our results extend prior studies and motivate the following experiments to delineate the RNA binding mechanism.
Figure 1. RNase A Digestion Enables APOBEC3H Purification and DNA Deaminase Activity.
(A) Coomassie-stained image showing His6-SUMO-A3H recovery from E. coli +/− RNase A treatment.
(B) DNA deaminase activity of His6-SUMO-A3H in extracts from E. coli +/− RNase A treatment (mean +/− SD; n = 3 experiments; inset gel image shows A3H-mediated conversion of a single-stranded DNA substrate to product, S to P, in the presence of RNase A). Lysate units (μl volumes) were chosen to include reactions with single-hit kinetics.
(C) DNA deaminase activity of untagged A3H in extracts from 293T cells, with experimental parameters similar to those in panel B.
Identification of Hyperactive APOBEC3H Variants by Altering a Predicted, Positively Charged RNA Binding Surface
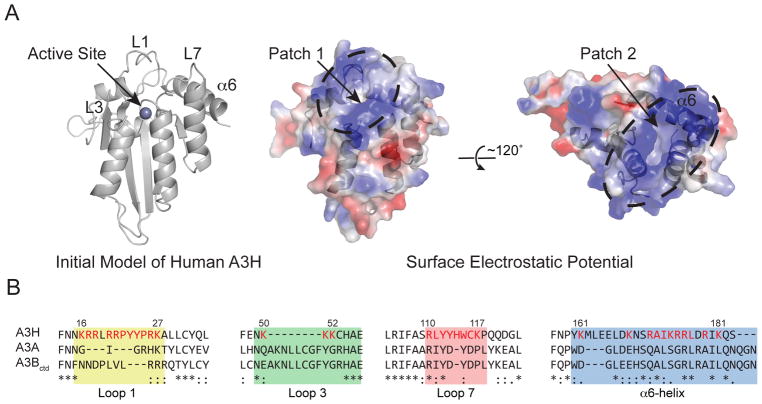
Since RNA inhibits A3H catalytic activity, we reasoned that RNA may bind the active site pocket and directly prevent the binding of single-stranded DNA. Alternatively, the RNA binding domain may be distinct and indirectly inhibit DNA deamination activity by forming inhibitory complexes. An A3H structural model was generated to help distinguish between these possibilities and inform mutagenesis experiments (Figure 2A). Based on recent A3A/B-ssDNA co-crystal structures (Kouno et al., 2017; Shi et al., 2017), loop 1, loop 7, and active site residues define a positively charged region that is likely essential for A3H to bind single-stranded DNA and potentially also RNA (i.e., in this model, the two activities will be inseparable by mutagenesis; patch 1 in Figure 2A).
Figure 2. Initial Structural Model Predicts Two Positively Charged Patches in Human APOBEC3H.
(A) Initial structural model of human A3H. A single zinc ion in the active site, loops 1, 3, and 7, and the α6-helix are labeled for orientation. The center and right space-filled schematics show the predicted electrostatic potential for A3H in the same orientation and rotated 120º, respectively. Patch 1 encompasses the active site and residues known to be required for DNA deamination. Patch 2 is distinct and includes several residues in the α6-helix.
(B) Alignment of human A3H, A3A, and A3B C-terminal domain (ctd) depicting loop 1, loop 3, loop 7, and α6-helix regions. Numbers correspond to the 183 amino acid splice variant of human A3H haplotype II, and residues highlighted in red were changed by site-directed mutagenesis.
See also Figures S2 and S7.
Interestingly, this structural model also predicts a second positively charged region centered upon α-helix 6, hereafter referred to as patch 2 (Figure 2A). Amino acids within this basic patch include Arg171, Lys174, Arg175, Arg176, and Arg179 (Figure 2B). We reasoned that if a residue located in this basic patch is important for binding RNA then substitutions at this site may impair the formation of RNA-inhibited complexes and simultaneously increase catalytic DNA deaminase activity.
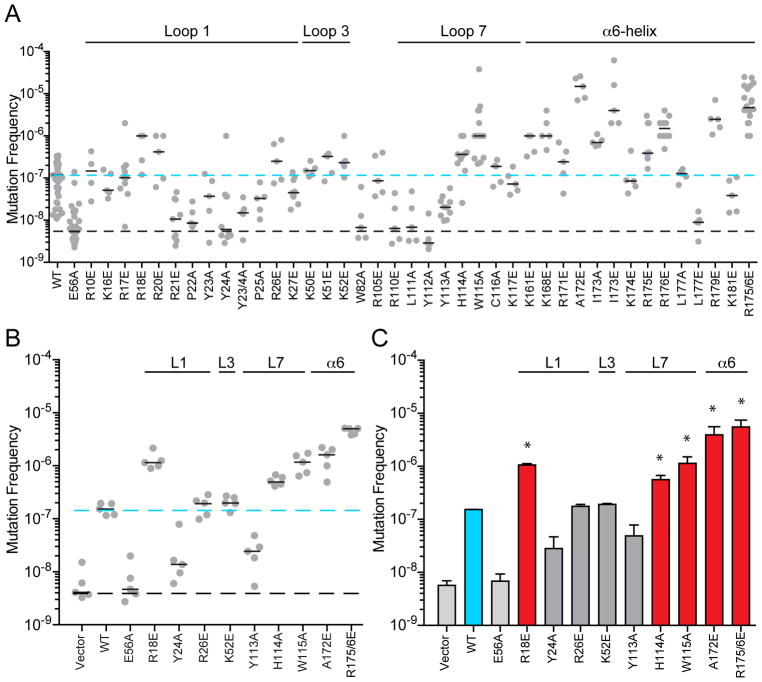
A panel of mutant A3H constructs covering both patch 1 and patch 2 was tested in rifampicin-resistance (RifR) mutation assays, which provide a quantitative measure of DNA mutator activity (Harris et al., 2002). Positively charged Arg and Lys residues are changed to Glu, other residues (Leu, Ile, Tyr, Trp) to Ala, and Ala residues to Glu (Figure 2B). The background RifR mutation frequency is approximately 1 x 10−8 (catalytic mutant E56A and empty vector controls; black dotted line in Figure 3A–B). Wild-type A3H causes a 10-fold increase in the median RifR mutation frequency (blue dotted line in Figure 3A–B). Several A3H amino acid substitutions have no significant effect, such as several loop 1 and 3 changes [R26E, K50E, K51E, K52E (important below for structural studies)], whereas other substitutions partially or completely abrogate mutator activity (catalytic E56A, loop 1 R21E, P22A, Y23A, Y24A, P25A, and loop 7 R110E, L111A, Y112A, Y113A). The latter group of mutants is most likely defective in DNA deamination.
Figure 3. Identification of Hyperactive Human APOBEC3H Mutants.
(A) RifR (rpoB) mutation frequency of E. coli expressing the indicated His6-Sumo-A3H constructs (WT, wild-type). Each dot represents data from an individual culture (n ≥ 5 per condition) with short horizontal lines depicting medians. The black dashed line represents the background mutation frequency of E. coli expressing an empty vector (not shown) or A3H catalytic mutant (E56A). The blue dashed line shows the median mutation frequency of E. coli expressing WT A3H.
(B) Data from a representative RifR mutation experiment comparing WT A3H and the indicated mutants including 5 hypermutators. Conditions and labels are similar to panel A.
(C) Quantification of key RifR mutation data. Each histogram bar reports the mean +/− SEM of the median mutation frequency from 3 independent experiments (panel B and 2 independent experiments not shown). The mutation frequencies induced by the 5 hypermutators (red bars) are significantly greater than that of WT A3H (blue bar; p<0.01 by Student’s t-test).
Strikingly, several substitutions cause much higher RifR mutation frequencies, including predicted loop 1, loop 7, and α6-helix residues (loop 1 R18E and R20E, loop 7 H114A and W115A, and α6-helix R171E, A172E, I173A/E, R175E, R176E, R179E). In a few instances, combinations of these amino acid substitutions lead to mutation frequencies nearly 100-fold higher than wild-type A3H (e.g., R175/6E; quantification in Figure 3C). One triple mutant combination is toxic to E. coli (W115A plus R175/6E), consistent with it eliciting the highest overall DNA mutator activity. Toxicity has been reported for wild-type A3A and A3B (Burns et al., 2013; Stenglein et al., 2010), but this is the first instance for A3H. These hypermutator phenotypes provide strong support for the model in which the RNA binding site in A3H is distinct from the single-stranded DNA binding and catalytic region.
DNA Deaminase Activity of APOBEC3H and Derivatives in Human Cell Extracts
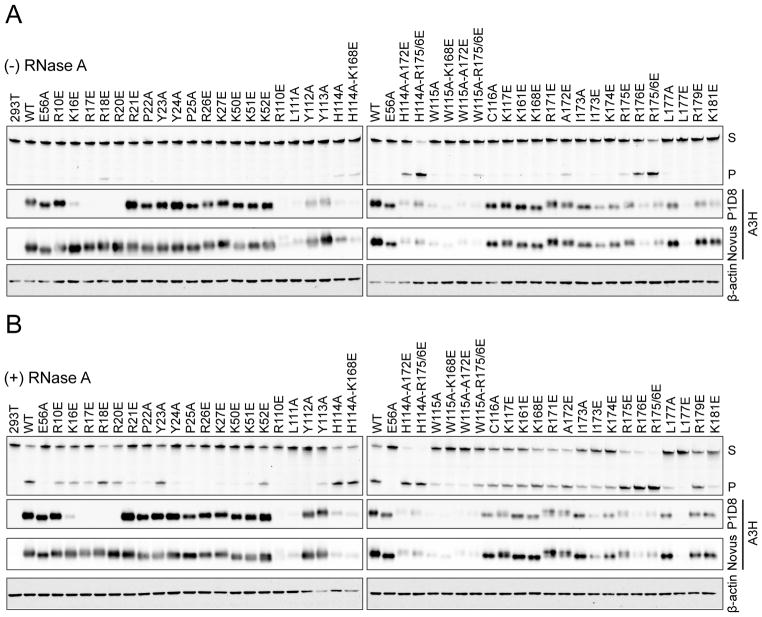
To extend the bacterial mutation results, the majority of the panel of A3H mutant constructs was expressed in human 293T cells and tested for single-stranded DNA cytosine deaminase activity +/− RNase A treatment. Human 293T cell lysates have no detectable single-stranded DNA cytosine deaminase activity unless transfected with an active APOBEC expression construct [e.g., (Carpenter et al., 2012; Stenglein et al., 2010; Thielen et al., 2007)]. As expected, wild-type A3H and most mutant derivatives show no activity in the absence of RNase A treatment (Figure 4A, upper panels). In contrast, most of the E. coli hypermutators display clear DNA cytosine deaminase activity under these repressive conditions, including loop 1 R18E, loop 7 H114A and W115A, and α6-helix A172E, R175E, and R176E, consistent with a RNA binding defect. Moreover, combinations of activating amino acid substitutions result in enzymes with further enhanced DNA deaminase activity (e.g., R175/6E). It is also notable that several of the hypermutators have low steady-state expression levels, suggesting they may be even more active than indicated in this series of comparative biochemical experiments (Figure 4A, lower panels).
Figure 4. DNA Deaminase Activity of APOBEC3H and Derivatives in 293T Extracts.
(A and B) Comparison of the DNA deaminase activities of untagged wild-type (WT) human A3H or the indicated amino acid substitution mutants in 293T extracts +/− RNase A treatment (S, substrate; P, product). Corresponding immunoblots show levels of A3H in cell lysates using a murine mAb (P1D8) or a rabbit pAb (Novus). β-actin is a loading control.
In comparison, RNase A treatment enables most constructs to elicit DNA cytosine deaminase activity, except those predicted to be defective in binding single-stranded DNA or catalyzing C-to-U deamination (e.g., catalytic E56A, loop 1 R21E, P22A, Y24A, and P25A, and loop 7 R110E, L111A, Y112A, and Y113A; Figure 4B). Immunoblots of the same cell extracts show that most constructs, apart from several hyperactive mutants (as above), are expressed at near wild-type A3H levels. The use of two different antibodies enables analyses of untagged constructs, which overcomes issues associated with epitope alterations. For instance, constructs with changes spanning residues Lys16 to Arg20 were detected readily using a rabbit polyclonal reagent, but showed no signal with our mouse anti-A3H monoclonal antibody P1D8, consistent with its binding site mapping to the N-terminal 30 residues (Refsland et al., 2014). Overall, the results of the RifR mutation experiments and in vitro DNA deaminase studies correlate strongly, with several hyperactive mutants clearly standing-out.
Human APOBEC3H Purification and X-ray Crystal Structure Determination
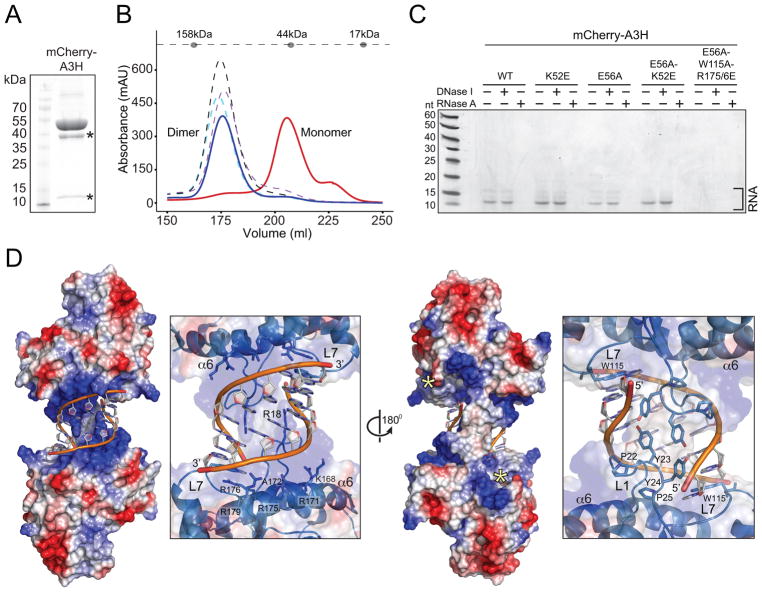
To directly compare the RNA binding activities of A3H and representative hyperactive mutants, we expressed His6-mCherry-tagged A3H constructs in E. coli and performed metal affinity purification. Again, wild-type A3H, a fully functional derivative (K52E), and a catalytic mutant derivative (E56A) show requirements for RNase A treatment during purification, whereas hyperactive mutants do not (Figure 5A and data not shown). Further purification of wild-type A3H using an ion exchange column and size exclusion chromatography results in a ~100 kDa species consistent with dimerization (Figure 5B). Several derivative proteins (K52E, E56A, and E56A/K52E) profile similarly and, like wild-type A3H, show high 260/280 ratios (>1) suggesting that some RNA is still bound. In contrast, hyperactive mutants elute as monomeric species with low 260/280 ratios (~0.6) consistent with near-pure protein (e.g., Figure 5B). To characterize the bound nucleic acid, a panel of purified A3H proteins was proteinase K-treated and the remaining material was heat-denatured, treated with RNase A or DNase I, and analyzed by polyacrylamide gel electrophoresis (Figure 5C). The resulting gel image reveals a range of short RNase A-sensitive RNA oligomers with a major species of ~10 nucleotides long. In contrast, no RNA is apparent in hyperactive enzyme preparations (e.g., Figure 5C). These results support a model in which RNA somehow mediates A3H dimerization.
Figure 5. Structure of a Human APOBEC3H Dimer Bound to a RNA Double Helix.
(A) Coomassie-stained gel image showing mCherry-A3H-K52E recovery from E. coli. The asterisks represent known heat-induced mCherry degradation products (Gross et al., 2000).
(B) SEC profiles of wild-type mCherry-A3H (solid blue line), mCherry-A3H-K52E (dashed cyan line), mCherry-A3H-E56A (dashed purple line), mCherry-A3H-E56A-K52E (dashed black line), and a representative RNA binding-defective mutant (W115A/R175/6E with E56A to prevent E. coli toxicity; solid red line). Molecular weight standards are shown on the dashed line above chromatograms.
(C) Polyacrylamide-urea gel image of A3H preparations after proteinase K treatment. Wild-type, K52E, E56A, and E56A-K52E preparations contain multiple small RNA species that are sensitive to RNase A but not DNase I. A representative RNA binding defective mutant lacks RNA (W115A/R175/6E with E56A to prevent E. coli toxicity).
(D) X-ray structure of human A3H in complex with duplex RNA. Left: a crystallographic pose showing the A3H electrostatic surface potential and double-stranded RNA sandwiched between the positively charged surfaces (patch 2) of two A3H monomers. Right: a partially transparent zoom-in of the duplex RNA-binding region highlighting positively charged residues in loop 1 (L1 R18) and α6-helix (K168, R171, A172, R175, R176, and R179). A 180º rotation shows the back side of the positively charged cradle bound to double-stranded RNA (loop 7, L7, labeled for orientation). This orientation also shows the active site pocket (patch 1, asterisked). The zoom-in depicts the observed loop 1 contacts between the two A3H monomers.
See also Figures S1 and S3.
We next sought to determine the structure of wild-type human A3H to provide a molecular explanation for the aforementioned genetic and biochemical data. During purification from E. coli we found that wild-type A3H is prone to aggregation and precipitation. We therefore tried multiple N-terminal solubility tags (Sumo, MBP, GST, GFP, mCherry) and found that mCherry-A3H is soluble but fails to crystallize. We then optimized the linker region between mCherry and A3H, and this resulted in non-diffracting crystals. As discussed above, extensive portions of the A3H surface are predicted to be charged positively with Lys and Arg residues projecting into solution (Figure 2A). Because an excessive concentration of positive charge may hinder crystallization, loop 3 lysine residues were systematically changed to glutamate to help overcome this issue while simultaneously preserving the integrity of patch 2. A single amino acid A3H variant, K52E, was identified, which has wild-type activities in multiple systems described above indicating fully intact DNA and RNA binding activities (Figures 3, 4 and 5B–C).
This optimized construct – mCherry-A3H-K52E – is amenable to purification, crystallization, and structure determination. The resulting crystal structure reveals two A3H monomers bridged by a RNA double helix (Figure 5D; Table 1 and Figure S1). Each A3H monomer has a cytidine deaminase fold with a central 5-strand β-sheet and 6 surrounding α-helices (Supplementary Figure S2). A single zinc ion is coordinated in the active site of each monomer. A 7 bp RNA duplex with an additional 1 nucleotide overhang is nestled between the same region of each A3H monomer, anchored by positively charged α6-helix residues (Arg171, Arg175, Arg176, and Arg179). These residues make direct interactions with the RNA phosphate backbone and encompass a total surface area of ~650 Å2 (Figure 5D). Notably, Arg to Glu substitutions at these positions cause increased activity in both the RifR mutation assay and the in vitro DNA deamination assay, with combinations often eliciting the highest activities (Figures 3 and 4). In addition, although Ala172 does not appear to interact directly with the RNA, an A172E substitution yields a hyperactive protein (RNA binding-defective), likely due to repulsive electrostatic effects.
Table 1.
Data Collection and Refinement Statistics.
| 6B0B | 6BBO (E56A) | |
|---|---|---|
| Data collection | ||
| Resolution range | 49.2 - 3.28 (3.54 - 3.28) | 81.4 – 3.45 (3.78 – 3.45) |
| Space group | P 3112 | P 3112 |
| Unit cell | ||
| a,b,c (Å) | 101.29 101.29 211.22 | 101.97 101.97 210.00 |
| Total reflections | 226613 (21387) | 78256 (18359) |
| Unique reflections | 19319 (1672) | 16797 (3988) |
| Multiplicity | 11.6 (11.4) | 4.7 (4.6) |
| Completeness (%) | 99.9 (99.4) | 99.9 (99.9) |
| I/σ(I) | 12.6 (0.7) | 5.5 (1.1) |
| R-merge | 0.132 (3.82) | 0.188 (1.55) |
| R-meas | 0.144 (4.19) | 0.241 (1.98) |
| R-pim | 0.043 (1.23) | 0.111 (0.92) |
| CC1/2 | 1.000 (0.499) | 0.997 (0.494) |
|
| ||
| Refinement | ||
|
| ||
| Reflections | 18933 (1672) | 16395 (1353) |
| Reflections for R-free | 967 (109) | 866 (74) |
| R-work (%) | 35.3 | 36.3 |
| R-free (%) | 36.2 | 39.2 |
| # non-H atoms | 6932 | 6934 |
| Macromolecules | 6928 | 6920 |
| Ligands | 2 | 14 |
| Solvent | 2 | N/A |
| Protein residues | 770 | 770 |
| r.m.s.d. | ||
| Bond lengths (Å) | 0.004 | 0.003 |
| Bond angles (˚) | 1.05 | 0.94 |
| Ramachandran plot | ||
| Favored (%) | 92.49 | 95.44 |
| Allowed (%) | 7.51 | 4.56 |
| Outliers (%) | 0.00 | 0.00 |
| Average B-factor (Å2) | 203.82 | 142.03 |
| Macromolecules (Å2) | 203.82 | 142.15 |
| Ligands (Å2) | 156.83 | 85.32 |
| Solvent (Å2) | 129.89 | N/A |
Statistics for the highest-resolution shell are shown in parentheses.
Additional A3H-RNA contacts are made by residues in loops 1 and 7, as anticipated by E. coli and in vitro activity data. In particular, Trp115 of loop 7 is base stacked with the 5′ end of the RNA, and a portion of loop 1, including Arg18, of each monomer projects into the major groove of the RNA helix (180º view in Figure 5D). Tyr residues in the loop 1 PYYP motif make direct contacts with the RNA phosphate backbone, and a Tyr23 interaction may also facilitate a monomer-monomer contact. Overall, loop 1, loop 7, and α6-helix residues cradle the RNA duplex through extensive interactions. Identical contacts occur in an independent crystallographic data set using a catalytic mutant derivative (mCherry-A3H-K52E-E56A) (Table 1 and Figure S3). These structural results demonstrate a molecular mechanism in which the prime mediator of A3H dimerization is duplex RNA.
RNA-binding Is Required for APOBEC3H Cytoplasmic Localization
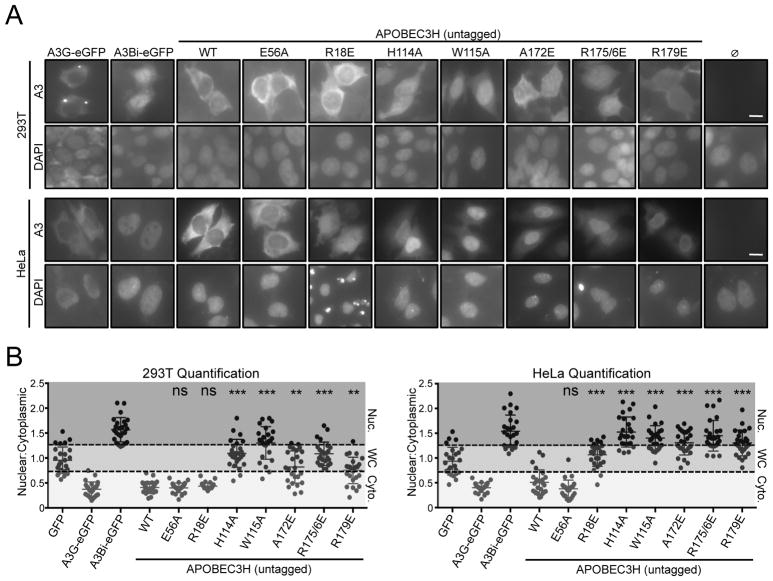
As summarized in the Introduction, an APOBEC-RNA interaction is thought to have roles in subcellular localization and packaging into HIV-1 particles, which is an essential step in the overall retrovirus restriction mechanism (Harris and Dudley, 2015; Malim and Bieniasz, 2012; Simon et al., 2015). Previous studies have shown that A3H is predominantly cytoplasmic, and that this activity is conserved between the human and rhesus macaque enzymes (Hultquist et al., 2011; Li and Emerman, 2011). To test whether the RNA binding domain of A3H is required for cytoplasmic localization, we performed a series of fluorescent microscopy experiments with untagged, wild-type human A3H and a panel of RNA binding mutants in two different cell types (293T and HeLa). Cytoplasmic A3G and nuclear A3B were scored in parallel as controls. As expected, wild-type A3H is predominantly cytoplasmic. In contrast, 5/6 RNA binding domain mutants show disrupted cytoplasmic localization in 293T cells and 6/6 in HeLa cells (representative images in Figure 6A and quantification in Figure 6B). The different localization phenotype of A3H R18E, as well as A172E and R179E, between the two cell types may be due to a partial loss of RNA binding activity and/or to altered constellations of potential RNA partners. In comparison, A3H H114A, W115A, and R175/6E enzymes are similarly and strongly mislocalized in both cell types suggesting a complete loss of RNA binding activity. Similar subcellular localization phenotypes are apparent for mCherry-tagged constructs in living 293T and HeLa cells (Supplementary Figure S4). Taken together with the aforementioned genetic, biochemical, and structural data, we conclude that RNA binding is essential for A3H cytoplasmic localization.
Figure 6. RNA-binding Is Required for Human APOBEC3H Cytoplasmic Localization.
(A) Representative images of untagged A3H (WT) and mutant derivatives (E56A, R18E, H114A, W115A, A172E, R175/6E, R179E) in 293T cells and HeLa cells (top and bottom panels, respectively). A3B nuclear localization and A3G cytoplasmic localization provide positive controls, and the no APOBEC reaction provides a negative control. DAPI staining defines the nuclear compartment (10 μm scale bar).
(B) Quantification of the ratio of nuclear/cytoplasmic staining of the indicated constructs in 293T and HeLa cells (n = 25 cells represented by individual dots with mean +/− SEM superimposed; ns, not significant; **, p<0.001; ***, p<0.0001 by 2-sided Student’s t-test; Nuc, nuclear; WC, whole cell; Cyto, cytoplasmic).
See also Figure S4.
RNA-binding Is Required for HIV-1 Restriction
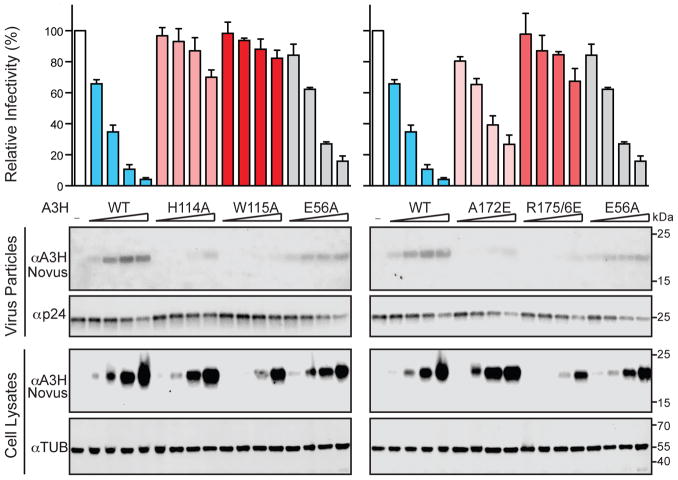
We next asked whether the RNA binding domain of A3H is required for HIV-1 encapsidation and restriction. Single-cycle infectivity experiments were performed using 293T cells to produce Vif-deficient HIV-1 (LAI) in the presence of wild-type A3H, a catalytic mutant E56A, or several of the RNA binding-defective constructs used above in localization studies (Figure 7). As expected, increasing amounts of wild-type A3H cause large reductions in viral infectivity (>10-fold at highest concentrations) compared to the vector only control, and the catalytic mutant E56A exerts a similar but slightly lower suppressive effect. This result suggests that most of the anti-HIV-1 activity of A3H is independent of DNA deaminase activity. In contrast, the RNA binding-deficient A3H mutants elicit considerably less than wild-type antiviral activity. For instance, in comparison to the vector only control, the H114A and W115A single mutants and the R175/6E double mutant have very little antiviral activity (less than 2-fold reduction at highest protein concentrations tested). Interestingly, the A172E single mutant has partially (not fully) compromised antiviral activity, suggesting retention of some RNA binding activity and consistent with the partial mislocalization phenotype described above. Corresponding immunoblots indicate that most of the compromised antiviral activity of these RNA binding mutants is due to defective packaging into viral particles. A different HIV-1 isolate (IIIB) shows very similar results (Supplementary Figure S5). Moreover, the defective packaging and antiviral activities of H114A, W115A, and R175/6E are not exacerbated by combining these amino acid changes suggesting that each is already completely defective in RNA binding activity (Supplementary Figure S6; further consistent with these mutants showing the strongest mislocalization phenotypes in Figure 6 and Supplementary Figure S4). We conclude that A3H binds duplex RNA as part of a HIV-1 packaging mechanism, and that this fundamental activity is required for virus restriction.
Figure 7. RNA Binding Is Required for HIV-1 Restriction by Human APOBEC3H.
Relative levels of infectivity for Vif-null HIV-1LAI produced in 293T cells in the presence of 12.5 ng, 50 ng, 200 ng, 400 ng of the indicated untagged, human A3H expression constructs (WT, wild-type; average +/− SD of 3 biologically independent experiments; average values for each condition normalized to those of the no A3 control). Corresponding immunoblots are shown below for proteins in viral particles and virus-producing 293T cells. See also Figures S5 and S6.
DISCUSSION
Human A3H is unique among APOBEC family members with HIV-1 restriction activity because it is the only enzyme with a single zinc-coordinating domain (A3D, A3F, and A3G have double-domain architectures). Thus, it has to pack more innate immune and biochemical activities into a smaller protein unit. Here, we show that A3H binds RNA through a RNA duplex-mediated dimerization mechanism. The RNA binding domain was initially identified through mutagenesis experiments, which yielded a group of A3H mutants with E. coli hypermutator activity, RNase A-independent DNA deaminase activity in 293T cell extracts, and monomeric size exclusion profiles. Two independent x-ray crystal structures of human A3H, a fully active K52E enzyme and an E56A derivative, demonstrate that all of the amino acid substitutions that confer these unique properties map to a positively charged protein surface that binds duplex RNA. The physiological importance of the RNA binding domain of A3H is evidenced by separation-of-function mutants showing deficiencies in cytoplasmic localization and HIV-1 encapsidation, whilst clearly retaining robust DNA deaminase activity. A related surprise is the observation that A3H RNA binding activity is more important than DNA deaminase activity for the overall HIV-1 restriction mechanism. Our results therefore provide molecular and structural explanations for a previously unclear step of the HIV-1 restriction mechanism in which interactions with duplex RNA are essential for both cytoplasmic localization and packaging into HIV-1 particles. Moreover, once inside HIV-1 cores, the duplex RNA binding activity of A3H may contribute to a reported block in reverse transcription (Mitra et al., 2015) and provide a mechanistic explanation for the strong deaminase-independent antiviral activity of this enzyme (Figure 7; Supplementary Figures 5 and 6). Deaminase-independent antiviral activity is likely to be mediated by binding to specific structured viral genomic RNA elements, consistent with recent CLIP-seq data (York et al., 2016). Human A3H shows similar deaminase-independent genetic requirements for restricting the replication of the related retrovirus HTLV-1 (Ooms et al., 2012) and transposition of the endogenous retroelement L1 (Feng et al., 2017), suggesting broad conservation of the mechanism detailed here.
In contrast to A3H, the majority of HIV-1 restriction activity is deaminase-dependent for the related restriction factor A3G (Browne et al., 2009; Miyagi et al., 2007; Schumacher et al., 2008). Therefore, an important future question is what encapsidated viral RNA species is being bound preferentially by A3H. For example, the abundant Alu-like 7SL RNA has been shown to interact with A3H and may contribute to packaging into HIV-1 particles (Zhen et al., 2012). Additionally or alternatively, A3H may bind to viral genomic RNA directly (York et al., 2016). Considering that catalytically dead A3H-E56A still strongly restricts HIV-1, it is likely that HIV-1 genomic RNA is a bona fide biological target of A3H. The precise nucleobase sequences of these RNA structures will also be of future interest because, despite clear A-form RNA in our A3H x-ray structures (Supplementary Figure S1), the crystal lattice has a mixed population of RNA species derived from E. coli making nucleobase identities unclear.
The human A3H-duplex RNA binding mechanism is mediated in large part by electrostatic contacts between the enzyme and the RNA phosphodiester backbone, as opposed to sequence-specific contacts. This binding mechanism may be favorable as an innate immune defense against virus and transposon replication because A3H would be able to engage and survey many different structured RNA species and then be strategically positioned to directly suppress parasite replication (here HIV-1 reverse transcription) and also, in the case of some retroviruses and transposons, deaminate single-stranded DNA replication intermediates. Moreover, such a general RNA superhighway patrol mechanism would prevent parasites from evolving resistance by, for instance, simply changing the nucleobase sequence of the binding site. This structure-guided mechanism is unlikely to affect cellular RNA functions at physiological protein levels because macromolecular machines such as the ribosome have powerful mechanisms to evict bound proteins and other mechanisms exist to remodel RNA pre- and post-translationally. Nevertheless, it will be interesting to identify the precise viral and cellular RNA structures recognized by A3H, and determine whether selectivity may be conferred by a higher-order, 3-dimensional features such as duplex intercalation or capping by Trp115. Interestingly, short duplex RNA structures can be substrates for RIG-I innate immune signaling (Jiang et al., 2011) and, perhaps, recognition of short RNA duplex structures is a general protective strategy used by innate immune factors.
Amino acid alignments indicate that the human A3H-RNA binding mechanism is conserved among primate A3H enzymes, and likely also for homologous mammalian Z3-type deaminases (Supplementary Figure S7). In particular, residues homologous to human A3H loop 1 Arg 18, loop 7 His114 and Trp115, and α-helix 6 Arg171, Ala172, Arg175, Arg176, and Arg179 are present in all primate A3H enzymes. Mammalian A3Z3 homologs also have near-identical α-helix 6 and loop 7 residues, as well as positively charged residues situated toward the N-terminal end of loop 1. More distantly related APOBEC family members may also have similar duplex RNA binding mechanisms. For instance, human A3C (Z2-type enzyme) and human AID have analogous Arg-rich α6-helices and loop 1 motifs. Interestingly, AID was shown recently to bind G-quadraplex DNA structures using this basic region (Qiao et al., 2017). Taken together with an analogous role for RNA in negatively regulating the DNA deaminase activity of AID (Bransteitter et al., 2003), it is conceivable that the same region may bind structured RNA, perhaps as part of a R-loop directed DNA targeting mechanism relevant to antibody diversification. In support of this idea, a variety of AID α6 mutants are defective in class switch recombination, including naturally occurring hyper-IgM disease alleles (Durandy et al., 2007). However, a homologous basic patch is lacking in single domain enzymes such as APOBEC1 and A3A, which suggests that these family members may bind both RNA and single-stranded DNA using active site residues and surrounding loop regions (especially loops 1 and 7). This inference is supported by the fact that both of these enzymes are RNA editing enzymes, potent DNA deaminases, and, at least for A3A, largely monomeric in single molecule studies in vitro (Shlyakhtenko et al., 2014) and in living human cells where RNA is abundant (Li et al., 2014). The double domain enzymes, A3B, A3D, A3F, and A3G, also interact with RNA (Introduction) and further work will be needed to assess conservation with the mechanism described here.
The roles of RNA in regulating the multifaceted functions of the APOBEC/AID family of polynucleotide editing enzymes have been challenging to define mechanistically, in large part due to a lack of separation-of-function mutants and structural information. The studies presented here with human A3H demonstrate a duplex RNA binding mechanism through identification and analyses of separation-of-function mutants and the determination of A3H-duplex RNA co-crystal structures. Both the negative regulatory role of RNA in suppressing the A3H DNA deaminase activity and the positive regulatory role of RNA in governing cytoplasmic localization are also likely to be relevant to preventing the accumulation of somatic mutations in development, ageing, and cancer.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Reuben S. Harris (rsh@umn.edu).
Method Details
Plasmids for Expression in E. coli
6xHis-SUMO-A3H was generated by PCR amplifying A3H HapII 1-183 cDNA (GenBank: ACK77775.1) from pcDNA3.1-A3H (Hultquist et al., 2011) using primers 5′-gcgcGGTCTCTAGGTGGCGGCGGCATGGCTCTGTTAACAGCCG and 5′-gcgcTCTAGATTAGGACTGCTTTATCCTC. The PCR product was digested with BsaI and XbaI and ligated into similarly digested pE-SUMO (Life Sensors). This forward primer also encodes a triple Gly linker. A large panel of mutant derivatives was made by site-directed mutagenesis and verified by Sanger sequencing (primer sequences available on request). This collection of pE-SUMO based constructs was used for experiments in Figures 1A–B and Figure 3.
His-tagged mCherry(1-232)-3xAla linker-HapII(1-183) was generated by overlap PCR using primers 5′-GGATCCGAATTCGCTGGAAGTTCTGTTCCAGGGGATGGTGAGCAAGGGCGAGGAGG, 5′-CTCCACCGGCGGCATGGACGCCGCTGCCATGGCTCTGTTAACAGCCGAAACATTCC, 5′-GGAATGTTTCGGCTGTTAACAGAGCCATGGCAGCGGCGTCCATGCCGCCGGTGGAG, and 5′-GCGCGCGGCCGCTCAGGACTGCTTTATCC. The PCR product was digested with EcoRI and NotI and ligated into MCS1 of the prSFDuet-1 (Novagen). A HRV rhinovirus 3C protease site was engineered prior to the mCherry ATG start site (sequence included in the forward primer used during overlapping PCR). The mCherry coding sequence was amplified from pRSETB-mCherry (Shaner et al., 2004) and A3H from 6x-His-SUMO-A3H (above). A3H-K52E, A3H-E56A, A3H-E56A-K52E, and A3H-E56A-W115A-R175/6E were made by site-directed mutagenesis and verified by Sanger sequencing. These prSFDuet-1 based constructs were used for experiments in Figure 5 including crystallography (Figure 5; Table 1 and Supplementary Figures S1–S3).
E. coli-based RifR Mutation Experiments
Experiments were done as described (Harris et al., 2002; Shi et al., 2015), with a minimum of 5 colonies tested for each condition in each independent experiment. His6-SUMO-A3H constructs were transformed into E. coli C43 (DE3) cells, and single colonies were grown to saturation at 37ºC in 2 mL LB plus ampicillin (100 μg/ml). A 100 μl aliquot of undiluted culture was plated directly onto LB-agar plates containing rifampicin (100 μg/ml) to select RifR mutants. Cultures were diluted serially in 1X M9 salts (sodium phosphate dibasic, potassium phosphate dibasic, sodium chloride, ammonium chloride) and plated on LB to determine viable cell counts. Mutation frequencies were calculated by dividing the number of colonies on the rifampicin plate by the total number of cells in the overnight culture (total number of colonies on LB plates multiplied by dilution factor). Key RifR mutation data were quantified by comparing mean +/− SEM of the median mutation frequency from 3 independent experiments and assessing statistical significance using the Student’s t-test. RifR mutation experiments are presented in Figure 3.
Plasmids for APOBEC Expression in Human Cell Lines
Coding sequences of A3H exon 2 and exons 3–5 were amplified from pcDNA3.1-A3H (Hultquist et al., 2011) using primer pairs 5′-NNNNGGTACCACCATGGCTCTGTTAACAG/ 5′-AAACATCTCCTGGACTCACCTTGTTTTCAAAGTAGCCTC and 5′-GTCTCCTTTCATCTCAACAGAAAAAGTGCCATGCGGAAAT/ NNNNGCGGCCGCTCAGGACTGCTTTATCCT, respectively. Human HGB2 intron 2 was amplified from an A3Bi containing plasmid (Burns et al., 2013) using primer pair 5′-GAGGCTACTTTGAAAACAAGGTGAGTCCAGGAGATGTTT/5′-ATTTCCGCATGGCACTTTTTCTGTTGAGATGAAAGGAGAC. The amplified fragments were fused together by overlap extension PCR, and inserted into the KpnI-NotI cloning site of pcDNA3.1(+) (ThermoFisher Scientific). Mutant derivatives of pcDNA3.1-A3Hi were generated using site-directed mutagenesis (primers available on request) and verified by Sanger sequencing. Plasmids pEGFP-N3 (Clontech Laboratories), pEGFP-N3-A3Bi-eGFP, and pEGFP-N3-A3G-eGFP have been reported in prior localization and HIV-1 restriction studies (Hultquist et al., 2011). These APOBEC expression constructs were used in experiments in Figures 1C, 4A–B, 6, and 7 and Supplementary Figures S5 and S6.
N-terminal mCherry-tagged, intron disrupted A3H HapII (1-183) was generated by overlap PCR using primers 5′-TGATCCGCGGCCGCACCACCATGGTGAGCAAGGGCGAGGA-3′ (mCherry Forward), 5′-CGGCTGTTAACAGAGCCATGGCAGCGGCGTCCATGCCACCGGTGGAGTG-3′ (overlap reverse, mCherry), 5′-CACTCCACCGGTGGCATGGACGCCGCTGCCATGGCTCTGTTAACAGCCG-3′ (overlap forward, A3H HapII), and 5′-ATTAAGCGTACGTCAGGACTGCTTTATCCTGTCAAG-3′ (A3H HapII reverse). The PCR product was digested with NotI and BsiWI and ligated into a pQCXIH retrovirus-based vector (Clontech Laboratories). Designated mutants were PCR-subcloned from pcDNA3.1 expression constructs (above) using the same set of A3H HapII primers. PCR products were digested with AgeI and BsiWI and subcloned into similarly digested parental vector (pQCXIH-mCherry-A3Hi). Sanger sequencing was used to verify the integrity of all constructs. These pQCXIH-based expression constructs were used in experiments in Supplementary Figures S4.
APOBEC3H DNA Deamination Experiments
293T cells were plated in 6-well plates at 400,000 cell density. The following day, the cells were transfected with 1 μg of each pcDNA3.1-A3Hi expression construct and 50 ng of an eGFP expression plasmid (transfection control). 48 hrs later, cells were harvested and resuspended in 200 μL HED buffer (25 mM HEPES pH 7.4, 15 mM EDTA, 1 mM DTT, Roche Complete EDTA-free protease inhibitors, 10% glycerol) and frozen at −80ºC. Cells were thawed, vortexed, sonicated for 20 min in a water bath sonicator (Branson), and centrifuged 10 min at 16,000 g, and then soluble extracts were transferred to a new tube. Half of each extract was treated with RNase A (100 μg/ml) at RT for 1 hr. An oligo master mix containing 1.6 μM 5′-ATTATTATTATTCTAATGGATTTATTTATTTATTTATTTATTT-fluorescein in HED buffer was made and mixed 1:1 with cell extracts (10 μL). Reactions were gently mixed, spun down, then incubated at 37ºC for 60 min, heated to 95ºC for 10 minutes to stop the reactions, then incubated with 0.1 U/rxn UDG for 10 min at 37ºC. Sodium hydroxide was added to a final concentration of 100 mM and reactions were heated to 95ºC to cleave the DNA at abasic sites. Reactions were mixed with 2x DNA PAGE loading dye (80% formamide, 1x TBE, bromophenol blue, xylene cyanol). Reaction products were separated by 15% denaturing PAGE (or TBE-Urea PAGE) and visualized by scanning on a Typhoon FLA-7000 scanner on fluorescence mode. A fraction of each extract was separated by 12% SDS-PAGE and transferred to low-fluorescence PVDF overnight at 20 V. Membranes were blocked with 4% milk in PBST then incubated with mouse anti-A3H mAb P1D8 (Refsland et al., 2014) or rabbit anti-β-actin mAb (Cell Signaling: 13E5), at dilutions of 1:1,000 or 1:10,000, respectively. After washing in PBST, membranes were probed with the goat anti-mouse 680 antibody (Life Technologies: A21057) or the goat anti-rabbit 800CW (Li-COR Biosciences: 926-32211), each 1:20,000. Blots were stripped and reprobed with rabbit anti-A3H pAb at 1:1,000 (Novus Biologicals NBP1-91682), followed by Li-COR anti-rabbit 800CW secondary at 1:20,000. Imaging was done with a Li-COR Odyssey. Results from these methods are shown in Figures 1C and 4.
APOBEC3H purification from E. coli
His6x-mCherry-A3H HapII 1-183 was transformed into OneShot BL21(DE3) competent cells (Thermo-Fischer Scientific) and, after overnight incubation, colonies were directly inoculated into 2XYT media containing 50 μg/ml kanamycin. Approximately 30 min. prior to induction, cultures were supplemented with 100 μM zinc sulfate and cooled to 16ºC. Protein expression was induced overnight by addition of 0.5 mM IPTG at ~1 OD600. Cells were centrifuged at 3800g for 20 min and resuspended in lysis buffer containing 50 mM Tris pH 8, 500 mM NaCl, 5 mM imidazole, lysozyme, and RNase A (10 mg). Cells were then sonicated (Branson Sonifer) and cell debris was removed by centrifugation (13000g, 45 min). The supernatant was added to Talon Cobalt Resin (Clontech), washed extensively with wash buffer (50 mM Tris-HCl pH 8.0, 500 mM NaCl, 5mM imidazole), and eluted with elution buffer (50mM Tris-HCl pH 8, 500 mM NaCl, 250 mM imidazole). Fractions containing protein were pooled and dialyzed overnight into buffer containing 50 mM Tris-HCl pH 8, 100 mM NaCl, 5% glycerol, 2 mM DTT, and RNase A (5mg). The protein was concentrated to ~5 mLs and loaded onto a 5 mL HiTrap MonoQ cartridge (GE Healthcare) equilibrated with wash buffer (50 mM Tris-HCl pH 8.0, 100 mM NaCl). The bound protein was washed with wash buffer and eluted with a linear gradient of high salt buffer (50 mM Tris pH 8.0, 1 M NaCl). The protein elutes between 30–40% high salt buffer. Fractions were collected and purified further with a 26/600 S200 gel filtration column (GE Healthcare) using a buffer containing 20 mM Tris-HCl pH 8.0 and 500 mM NaCl. Fractions containing purified protein were pooled, mixed with 5 mM DTT, and concentrated to 25 mg/ml. The final purified protein had an OD260/280 measurement greater than 1, indicating bound nucleic acid. The wild-type mCherry-A3H, mCherry-A3H-K52E, mCherry-A3H-E56A, and mCherry-A3H-E56A-K52E proteins were purified using the above protocol. The RNA binding defective mCherry-A3H mutant (W115A, R175E, R176E plus E56A to prevent bacterial toxicity) was purified by resuspending cell pellets in 50 mM Hepes pH 7.5, 1 M NaCl, 5 mM Imidazole, 100 μg/ml of RNase A (RNase A is not required for purification of hyperactive mutant but added to clarify cell lysates). The cells were lysed, clarified, and hyperactive protein was purified using Talon resin similar to wild-type mCherry-A3H, with the exception that the wash buffer contained 50 mM Hepes pH 7.5, 1 M NaCl, 5 mM Imidazole, and the elution buffer contained 50 mM Hepes pH 7.5, 500 mM NaCl, and 250 mM Imidazole. The protein was immediately purified further by a 26/600 S200 size exclusion column using the same buffer as wild-type mCherry-A3H proteins. The hyperactive A3H proteins are particularly prone to aggregation and should be prepared fresh. These proteins were used in studies shown in Figure 5 and Supplementary Figures S1–S3.
APOBEC3H Proteinase K, RNase A, and DNase I Treatment Experiments
A 4 μL aliquot of each mCherry-A3H protein solution (mCherry-A3Hwt, mCherry-A3H K52E, mCherry-A3H-E56A, mCherry-A3H-E56A-K52E; ~80ug total protein in buffers above) was diluted in 14 μL nuclease-free water and 2 μL of Proteinase K (Roche: 03115828001). The tubes were then incubated for 15 min each at 45°C, 55°C, 65°C, then 10 min at 95°C to inactivate Proteinase K. Then, 2 uL was taken from each tube and incubated with 1) 8 uL nuclease free water, 2) 1 uL DNase I in 7 uL Buffer RDD from Qiagen (79254), or 3) 1 mg/mL RNase A (Sigma: R5503) in 7 uL nuclease free water for 15 min at room temperature. Each sample was then mixed with 10 μL 2xDNA PAGE loading dye and half of the reaction was loaded on a 20% TBE-7M Urea gel and run at 10 W. The gel was stopped when the bromophenol blue band was about 99% of the way down the gel. The gel was stained with SYBR-Gold and imaged on a Typhoon FLA-7000 (GE Healthcare Life Sciences). 0.25 μL of the IDT oligo length standard (51-05-1501) was loaded to estimate size (approximately 25 ng/oligo). The results of these experiments are shown in Figure 5C.
APOBEC3H Crystallization, X-Ray Data Collection and Structural Determination
Prior to crystallization, 10 mM DTT and trace amounts of human rhinovirus 3C protease was added to an aliquot of protein diluted to 20 mg/ml to remove the 6xHis tag. mCherry-A3H K52E was crystallized using sitting drop vapor diffusion in a solution containing 18% PEG 3350 (v/v) and 300 mM ammonium iodide. The crystal trays were incubated at 18ºC and protein crystals appeared overnight and grew to full size within 2–5 days. The crystals were preserved in cryprotectant solution containing 20% PEG3350, 300 mM ammonium iodide, and 20% glycerol. The mCherry-A3H-E56A-K52E protein was crystallized using similar methodology.
Initial x-ray diffraction data were collected at the Advanced Photon Source NE-CAT 24 ID-E beamline at the wavelength of 0.979 Å. The datasets were processed using HKL2000 (Otwinowski and Minor, 1997) and XDS (Kabsch, 2010), initially in the high-symmetry space group P6122 with good merging statistics. However, structure solution by molecular replacement was unsuccessful despite exhaustive attempts using mCherry and various A3 structures as search models. Even after reprocessing the data in a lower-symmetry space group P31, molecular replacement calculation with mCherry as the search model, which has 100% sequence homology with the mCherry tag used in crystallization and accounts for >50% of macromolecular content in the unit cell, did not yield any solution, implying less than ideal occupancy of mCherry in the crystal. Subsequently, high-multiplicity x-ray diffraction data were collected at the NE-CAT 24-ID-C beamline at the zinc K-edge wavelength of 1.282 Å. Single-wavelength anomalous dispersion (SAD) phasing using SHELX C/D/E (Sheldrick, 2010) and PHENIX (Adams et al., 2010) in space group P6122 produced an electron density map that clearly showed secondary structure elements of A3H and a segment of RNA double-helix, which has a deep and narrow major groove and a shallow and wide minor groove (Supplementary Figure S1). The A3H and RNA molecules were built into the experimental map using COOT (Emsley et al., 2010). The mFo-DFc difference map after initial refinement with phase-restraints against the SAD-derived phases revealed a β-barrel structure corresponding to partial mCherry. Including mCherry in the refinement lowered the Rfree by 1.0 % even though mCherry overlapped with its symmetry-related molecule. However, the Rfree stayed above 40%. Refinement in a lower symmetry space group P3112 resulted in significant improvement of Rfree and refining with a twin operator “-h,-k,l” increased the Rfree. Therefore we continued refinement in the space group P3112 without twinning. Subsequent iterative model building using COOT and refinement with PHENIX suite resulted in the final Rwork / Rfree of 35.3 / 36.2%. The slightly high R-factors are likely due to the uncertain location of mCherry. The model quality during refinement was checked constantly by combining the SAD-derived and model-based phases to minimize possible model bias. The asymmetric unit contains two A3H molecules, two RNA strands, and two copies of mCherry with occupancy refined to 0.5. However, we modeled two RNA double helices (mixture poly-A paired with poly-U) in the asymmetric unit, each with half-occupancy, to represent double-stranded RNA molecules comprising mixed populations of unknown complementary sequences and following crystallographic 2-fold symmetry as an ensemble. In spite of the positional ambiguity, mCherry contributes significantly to the total diffraction, as refinement with the mCherry removed led to ~5% increase in Rfree. To address overlaps between symmetry-related mCherry molecules, space group choices P31 or P1, with half of the mCherry molecules unmodeled, were considered. However, refinement in these space groups led to significantly poorer Rfree. Therefore, P3112 would be our top choice for space group, in which case mCherry does not have full occupancy. Fluorescent proteins related to mCherry have the tendency to have alternative packing modes in crystals, as reported earlier (Pletnev et al., 2009; Pletnev et al., 2014).
For mCherry-A3H-E56A-K52E, refinement was performed as described above, using the refined mCherry-A3H-K52E structure as the starting model. The summary of data collection and model refinement statistics is shown in Table 1. Images were produced using PYMOL (www.pymol.org). Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 6B0B and 6BBO. These structures are presented in Figure 5D and Supplementary Figures S1–S3.
Fluorescent Microscopy Experiments
Approximately 10,000 293T or 8,000 HeLa cells were plated into a 96-well CellBIND microplate (Corning) and allowed to adhere overnight. Cells were transfected with 100 ng of the indicated untagged A3H construct. For localization controls, 100 ng of A3Bi-eGFP, A3G-eGFP, or eGFP expression vectors (above) were transfected. 48 hr post-transfection, cells were washed twice with PBS and fixed with 4% paraformaldehyde (20 min, RT). Cells were then washed twice with cold PBS and permeabilized with 0.5% Triton-X100 (10 min, 4°C). Cells were washed once with cold PBS, incubated in blocking buffer (4% BSA, 5% goat serum, in PBS) for 1 hr at RT, and then stained with 1:100 rabbit anti-A3H pAb (Novus Biologicals, NBP1-91682), or with a rabbit anti-A3B mAb (5210-87-13) at 1:500 that also cross-reacts with A3G (Leonard et al., 2015). Cells were washed with cold PBS and stained with 1:1000 goat anti-rabbit Alexa Flour 594 (ThermoFisher Scientific, A11037) for 1 hr at RT. After additional washing, cells were incubated in PBS containing 0.1% DAPI. Images were collected at 40x magnification using an EVOS FL Color microscope (ThermoFisher Scientific), and quantified using ImageJ software. Individual cells were scored and grouped into three categories (nuclear, whole cell, or cytoplasmic). Subcellular compartments were defined based on DAPI staining (nucleus), anti-A3G-eGFP staining (cytoplasm), and eGFP localization (whole cell) The nuclear:cytoplasmic A3 ratio was calculated by dividing the pixel density of the nuclear compartment (marked by DAPI) divided by the pixel density of cytoplasmic compartment (i.e., cytoplasmic signal is the whole cell A3 signal minus the nuclear A3 signal). The nuclear:cytoplasmic A3 ratio for each individual cell (n≥25 per condition) was graphed with Prism 6.0 (GraphPad Software). Finally, mean ratios +/− SEM were superimposed over each dot plot, and statistical significance was determined using a 2-tailed Student’s t-test.
A summary of experiments with untagged proteins including quantification is shown in Figure 6. A complementary series of experiments was done with N-terminally mCherry-tagged A3Hi constructs (constructions above) in 293T and HeLa cells, as above except live cells were imaged and mCherry fluorescence was detected using the RFP EVOS light cube (531/40 nm excitation; 593/40 nm emission). These additional experiments including quantification are presented in Supplementary Figure S4.
HIV-1 Packaging and Restriction Experiments
The HIV-1IIIB VifX26X27 provirus expression construct has been described (Albin et al., 2010; Haché et al., 2008). Analogous tandem stop codons were introduced into codons 26 and 27 of the full-length HIV-1 molecular clone pLAI.2 (Peden et al., 1991) (AIDS Reagent Program 2532) by subcloning the vif-vpr region into pJET1.2, performing site-directed mutagenesis (Stratagene), and shuttling the vif-vpr region back into the original vector using PshAI and SalI restriction sites.
HIV-1 packaging and restriction experiments were done by transfecting 50% confluent 293T cells (TransIt, Mirus) with 1 μg Vif-deficient provirus plasmid and an amount of each A3 expression construct or vector control indicated in the relevant figure legend. After 48 hr incubation, viral supernatants were cleared of cells by filtration (0.45 μm) and used to infect CEM-GFP cells to monitor infectivity via flow cytometry.
Cell and viral particle lysates were prepared for immunoblotting as follows. Cells were pelleted, washed, and then lysed with 2.5X Laemmli sample buffer. Virus-containing supernatants were filtered and pelleted via centrifugation through a 20% sucrose cushion, then lysed in 2.5X Laemmli sample buffer. Lysates were then subjected to SDS-PAGE followed by protein transfer to PVDF using a Bio-Rad Criterion system. Membranes were probed with a mouse anti-A3H mAb P1D8 (Refsland et al., 2014), a rabbit anti-A3H pAb (NBP1-91682, Novus), an anti-HIV-1 p24/CA mAb (AIDS Reagent Program 3537), and a mouse anti-tubulin mAb (Covance). Secondary antibodies were goat anti-rabbit IRdye 800CW (Li-COR 926-32211) and goat anti-mouse Alexa Fluor 680 (Molecular Probes A21057). Membranes were imaged using a Li-COR Odyssey instrument, and images were prepared for presentation using Image J.
For Vif-null HIV-1IIIB dose response experiments, the mean viral infectivity +/− SD from 3 biologically independent experiments was determined and quantification of the levels of A3H packaging relative to WT was done for relevant RNA binding mutants. First, viral particle A3H and p24 levels and cellular A3H and tubulin levels were quantified. Second, the relative packaging efficiency (RPE) was calculated for each condition using the following formula: RPE: (VLP A3H/p24)/(cellular A3H/TUB). Lastly, normalization was done by dividing the RPE value for each RNA binding mutant by the RPE value for WT A3H, and statistical significance was determined from 3 biologically independent experiments by comparing mean RPE +/− SEM using a one-sided Student’s t-test. HIV-1 packaging and infectivity data are reported in Figure 7 and Supplementary Figures S5 and S6.
Quantification and Statistical Analysis
Key RifR mutation data were quantified by comparing mean +/− SEM of the median mutation frequency from 3 independent experiments and assessing statistical significance using the Student’s t-test. X-Ray data collection and refinement statistics for crystal structures are shown in Table 1. A3H localization and packaging experiments were quantified and assessed statistically as described above.
Data and Software Availability
The coordinates and structure factors for mCherry-K52E and mCherry-E56A-K52E are deposited in the Protein Data Bank with accession codes 6B0B and 6BBO, respectively. All original gel and microscopy images from Figures 1–7 are deposited in Mendeley (http://dx.doi.org/10.17632/6mdtnv4kjb.1).
Supplementary Material
Acknowledgments
We thank D. Ebrahimi for providing helpful comments and M. Jarvis and J. Zhang for technical assistance. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: pLAI.2 from K. Peden, courtesy of the MRC AIDS Directed Program, and anti-HIV-1 p24/CA mAb courtesy of B. Chesebro and K. Wehrly. This work was supported by grants from the US National Institutes of Health (NIGMS R01-GM118000 to RSH and HA, NIGMS R35-GM118047 to HA, NCI R21-CA206309 to RSH), the Prospect Creek Foundation (RSH), and the University of Minnesota (College of Biological Sciences, Academic Health Center, and Masonic Cancer Center to RSH). Partial salary support for RLK, CMR, and DS was provided by NIH training grants (T32-HL007062, T32-AI83196, and T90-DE022732, respectively). This work is based in part upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the US National Institutes of Health (NIGMS P41-GM103403). The Pilatus 6M detector on 24-ID-C beamline is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. RSH is the Margaret Harvey Schering Land Grant Chair for Cancer Research, a Distinguished University McKnight Professor, and an Investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
NMS and RSH conceived and designed the studies. KVL, MWL, and NMS created DNA constructs, apart from the Vif-null HIV-1 proviral construct (CMR), the wild-type A3Hi construct (RLK), and the mCherry localization vectors (DS). KVL, MWL, and MAC performed bacterial mutation experiments. MAC performed DNA deamination activity assays. NMS purified and crystallized A3H. SB and KS collected and phased diffraction data. KS determined the structure, and KS, NMS, HA, and RSH analyzed structural data. DS performed localization studies. CMR and JW performed HIV-1 restriction experiments. WLB produced antibodies and contributed to lab management. NMS and RSH drafted the manuscript, and all authors contributed to figure preparation and revisions.
COMPETING FINANCIAL INTERESTS
RSH is a co-founder, shareholder, and consultant of ApoGen Biotechnologies Inc. HA is a consultant for ApoGen Biotechnologies Inc. The other authors have no competing financial interests to declare.
BIBLIOGRAPHY
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin JS, Hache G, Hultquist JF, Brown WL, Harris RS. Long-term restriction by APOBEC3F selects human immunodeficiency virus type 1 variants with restored Vif function. J Virol. 2010;84:10209–10219. doi: 10.1128/JVI.00632-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apolonia L, Schulz R, Curk T, Rocha P, Swanson CM, Schaller T, Ule J, Malim MH. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog. 2015;11:e1004609. doi: 10.1371/journal.ppat.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ara A, Love RP, Chelico L. Different mutagenic potential of HIV-1 restriction factors APOBEC3G and APOBEC3F is determined by distinct single-stranded DNA scanning mechanisms. PLoS Pathog. 2014;10:e1004024. doi: 10.1371/journal.ppat.1004024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Cullen BR. Single-stranded RNA facilitates nucleocapsid: APOBEC3G complex formation. RNA. 2008;14:1228–1236. doi: 10.1261/rna.964708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzidi MS, Caval V, Suspene R, Hallez C, Pineau P, Wain-Hobson S, Vartanian JP. APOBEC3DE antagonizes hepatitis B virus restriction factors APOBEC3F and APOBEC3G. J Mol Biol. 2016;428:3514–3528. doi: 10.1016/j.jmb.2016.05.022. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP, Allers C, Landau NR. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387:313–321. doi: 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, Leonard B, Shandilya SM, Bohn MF, Schiffer CA, et al. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J Biol Chem. 2012;287:34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S, Guo F, Niu M, Saadatmand J, Deflassieux J, Kleiman L. The interaction between HIV-1 Gag and APOBEC3G. J Biol Chem. 2004;279:33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′ --> 5′ on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, Greene WC. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Durandy A, Taubenheim N, Peron S, Fischer A. Pathophysiology of B-cell intrinsic immunoglobulin class switch recombination deficiencies. Adv Immunol. 2007;94:275–306. doi: 10.1016/S0065-2776(06)94009-7. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Goubran MH, Follack TB, Chelico L. Deamination-independent restriction of LINE-1 retrotransposition by APOBEC3H. Sci Rep. 2017;7:10881. doi: 10.1038/s41598-017-11344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossat N, Tam PP. Re-editing the paradigm of cytidine (C) to uridine (U) RNA editing. RNA Biology. 2014;11:1233–1237. doi: 10.1080/15476286.2014.996054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois-Montbrun S, Holmes RK, Swanson CM, Fernandez-Ocana M, Byers HL, Ward MA, Malim MH. Comparison of cellular ribonucleoprotein complexes associated with the APOBEC3F and APOBEC3G antiviral proteins. J Virol. 2008;82:5636–5642. doi: 10.1128/JVI.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, Malim MH. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Chen Q, Xiao X, Ito F, Wolfe A, Chen XS. Biochemical characterization of APOBEC3H variants: implications for their HIV-1 restriction activity and mC modification. J Mol Biol. 2016;428:4626–4638. doi: 10.1016/j.jmb.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haché G, Shindo K, Albin JS, Harris RS. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr Biol. 2008;18:819–824. doi: 10.1016/j.cub.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari A, Ooms M, Mulder LC, Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol. 2009;83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Dudley JP. APOBECs and virus restriction. Virology. 2015;479–480C:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol. 2011;85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Burdick R, Shigemi M, Plisov S, Hu WS, Pathak VK. Mov10 and APOBEC3G localization to processing bodies is not required for virion incorporation and antiviral activity. J Virol. 2013;87:11047–11062. doi: 10.1128/JVI.02070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr, Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Kao S, Miyagi E, Takeuchi H, Goila-Gaur R, Opi S, Gipson CL, Parslow TG, Ly H, Strebel K. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J Virol. 2005;79:5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock J, Blum HE. Hypermutation of hepatitis B virus genomes by APOBEC3G, APOBEC3C and APOBEC3H. J Gen Virol. 2008;89:1184–1191. doi: 10.1099/vir.0.83507-0. [DOI] [PubMed] [Google Scholar]
- Koito A, Ikeda T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbiol. 2013;4:28. doi: 10.3389/fmicb.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouno T, Silvas TV, Hilbert BJ, Shandilya SMD, Bohn MF, Kelch BA, Royer WE, Somasundaran M, Kurt Yilmaz N, Matsuo H, et al. Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity. Nat Commun. 2017;8:15024. doi: 10.1038/ncomms15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J Biol Chem. 2006;281:29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- LaRue RS, Jónsson SR, Silverstein KAT, Lajoie M, Bertrand D, El-Mabrouk N, Hötzel I, Andrésdóttir V, Smith TPL, Harris RS. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard B, McCann JL, Starrett GJ, Kosyakovsky L, Luengas EM, Molan AM, Burns MB, McDougle RM, Parker PJ, Brown WL, et al. The PKC/NF-kappaB signaling pathway induces APOBEC3B expression in multiple human cancers. Cancer Res. 2015;75:4538–4547. doi: 10.1158/0008-5472.CAN-15-2171-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen Y, Li M, Carpenter MA, McDougle RM, Luengas EM, Macdonald PJ, Harris RS, Mueller JD. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J Mol Biol. 2014;426:1296–1307. doi: 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Emerman M. Polymorphism in human APOBEC3H affects a phenotype dominant for subcellular localization and antiviral activity. J Virol. 2011;85:8197–8207. doi: 10.1128/JVI.00624-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Bieniasz PD. HIV restriction factors and mechanisms of evasion. Cold Spring Harbor Perspectives in Medicine. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall WM, Smith HC. Direct evidence that RNA inhibits APOBEC3G ssDNA cytidine deaminase activity. Biochem Biophys Res Commun. 2011;412:612–617. doi: 10.1016/j.bbrc.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra M, Singer D, Mano Y, Hritz J, Nam G, Gorelick RJ, Byeon IJ, Gronenborn AM, Iwatani Y, Levin JG. Sequence and structural determinants of human APOBEC3H deaminase and anti-HIV-1 activities. Retrovirology. 2015;12:3. doi: 10.1186/s12977-014-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi E, Opi S, Takeuchi H, Khan M, Goila-Gaur R, Kao S, Strebel K. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J Virol. 2007;81:13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münk C, Beck T, Zielonka J, Hotz-Wagenblatt A, Chareza S, Battenberg M, Thielebein J, Cichutek K, Bravo IG, O’Brien SJ, et al. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 2008;9:R48. doi: 10.1186/gb-2008-9-3-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Misawa N, Juarez-Fernandez G, Moriwaki M, Nakaoka S, Funo T, Yamada E, Soper A, Yoshikawa R, Ebrahimi D, et al. HIV-1 competition experiments in humanized mice show that APOBEC3H imposes selective pressure and promotes virus adaptation. PLoS Pathog. 2017;13:e1006348. doi: 10.1371/journal.ppat.1006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak U, Matthews AJ, Zheng S, Chaudhuri J. The splicing regulator PTBP2 interacts with the cytidine deaminase AID and promotes binding of AID to switch-region DNA. Nat Immunol. 2011;12:160–166. doi: 10.1038/ni.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms M, Brayton B, Letko M, Maio SM, Pilcher CD, Hecht FM, Barbour JD, Simon V. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe. 2013;14:411–421. doi: 10.1016/j.chom.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Ooms M, Krikoni A, Kress AK, Simon V, Munk C. APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1. J Virol. 2012;86:6097–6108. doi: 10.1128/JVI.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- Phalora PK, Sherer NM, Wolinsky SM, Swanson CM, Malim MH. HIV-1 replication and APOBEC3 antiviral activity are not regulated by P bodies. J Virol. 2012;86:11712–11724. doi: 10.1128/JVI.00595-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletnev S, Morozova KS, Verkhusha VV, Dauter Z. Rotational order-disorder structure of fluorescent protein FP480. Acta Crystallogr D Biol Crystallogr. 2009;65:906–912. doi: 10.1107/S0907444909020927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletnev S, Subach FV, Verkhusha VV, Dauter Z. The rotational order-disorder structure of the reversibly photoswitchable red fluorescent protein rsTagRFP. Acta Crystallogr D Biol Crystallogr. 2014;70:31–39. doi: 10.1107/S1399004713024644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, Wang L, Meng FL, Hwang JK, Alt FW, Wu H. AID recognizes structured DNA for class switch recombination. Mol Cell. 2017;67:361–373 e364. doi: 10.1016/j.molcel.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Hultquist JF, Luengas EM, Ikeda T, Shaban NM, Law EK, Brown WL, Reilly C, Emerman M, Harris RS. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 2014;10:e1004761. doi: 10.1371/journal.pgen.1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Nussenzweig MC. Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu Rev Pathol. 2013;8:79–103. doi: 10.1146/annurev-pathol-020712-164004. [DOI] [PubMed] [Google Scholar]
- Schafer A, Bogerd HP, Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Schumacher AJ, Haché G, MacDuff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sharma S, Patnaik SK, Taggart RT, Kannisto ED, Enriquez SM, Gollnick P, Baysal BE. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat Commun. 2015;6:6881. doi: 10.1038/ncomms7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K, Carpenter MA, Banerjee S, Shaban NM, Kurahashi K, Salamango DJ, McCann JL, Starrett GJ, Duffy JV, Demir O, et al. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat Struct Mol Biol. 2017;24:131–139. doi: 10.1038/nsmb.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K, Carpenter MA, Kurahashi K, Harris RS, Aihara H. Crystal structure of the DNA deaminase APOBEC3B catalytic domain. J Biol Chem. 2015;290:28120–28130. doi: 10.1074/jbc.M115.679951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Lushnikov AJ, Li M, Harris RS, Lyubchenko YL. Interaction of APOBEC3A with DNA assessed by atomic force microscopy. PloS One. 2014;9:e99354. doi: 10.1371/journal.pone.0099354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Lushnikov AY, Li M, Lackey L, Harris RS, Lyubchenko YL. Atomic force microscopy studies provide direct evidence for dimerization of the HIV restriction factor APOBEC3G. J Biol Chem. 2011;286:3387–3395. doi: 10.1074/jbc.M110.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon V, Bloch N, Landau NR. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol. 2015;16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starrett GJ, Luengas EM, McCann JL, Ebrahimi D, Temiz NA, Love RP, Feng Y, Adolph MB, Chelico L, Law EK, et al. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat Commun. 2016;7:12918. doi: 10.1038/ncomms12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Matsuo H, Harris RS. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J Virol. 2008;82:9591–9599. doi: 10.1128/JVI.02471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarovskaia ES, Xu H, Mbisa JL, Barr R, Gorelick RJ, Ono A, Freed EO, Hu WS, Pathak VK. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J Biol Chem. 2004;279:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- Tan L, Sarkis PT, Wang T, Tian C, Yu XF. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. Faseb J. 2009;23:279–287. doi: 10.1096/fj.07-088781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- Thielen BK, Klein KC, Walker LW, Rieck M, Buckner JH, Tomblingson GW, Lingappa JR. T cells contain an RNase-insensitive inhibitor of APOBEC3G deaminase activity. PLoS Pathog. 2007;3:1320–1334. doi: 10.1371/journal.ppat.0030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Sarkis PT, Yu XF. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J Mol Biol. 2008;375:1098–1112. doi: 10.1016/j.jmb.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol. 2011;85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York A, Kutluay SB, Errando M, Bieniasz PD. The RNA binding specificity of human APOBEC3 proteins resembles that of HIV-1 nucleocapsid. PLoS Pathog. 2016;12:e1005833. doi: 10.1371/journal.ppat.1005833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen A, Du J, Zhou X, Xiong Y, Yu XF. Reduced APOBEC3H variant anti-viral activities are associated with altered RNA binding activities. PloS One. 2012;7:e38771. doi: 10.1371/journal.pone.0038771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.