Abstract
A highly efficient streamlined chemoenzymatic strategy for total synthesis of four prioritized ganglioside cancer antigens GD2, GD3, fucosyl GM1, and GM3 from commercially available lactose and phytosphingosine is demonstrated. Lactosyl sphingosine (LacβSph) was chemically synthesized (in 13-gram scale), subjected to sequential one-pot multienzyme (OPME) glycosylation reactions with facile C18-cartridge purification, followed by an improved acylation condition to form target gangliosides, including fucosyl GM1 which has never been synthesized before.
Table of Contents
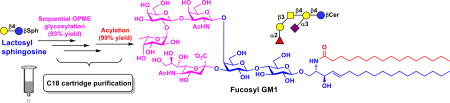
Highly efficient streamlined total synthesis of complex prioritized gangliosides was achieved chemoenzymatically by sequential one-pot multienzyme (OPME) reactions with facile C18 cartridge purification schemes followed by high-yield acylation.
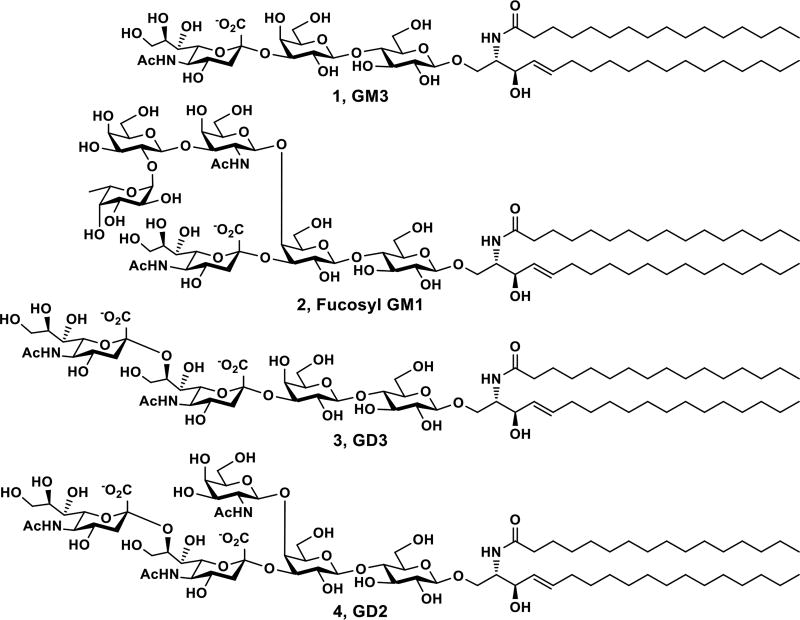
Gangliosides are sialic acid-containing glycosphingolipids presented broadly in vertebrate cells but are particularly abundant in the nervous cells.1, 2 They are involved in lipid raft formation,3 viral and bacterial infection,4, 5 immune regulation,6 and axon outgrowth.2 Exogenous GM1 has shown potential in treating central nervous system injuries and neurodegenerative diseases.7, 8 Aberrant expressing of some gangliosides is linked to cancer progression.9 In fact, among 75 cancer antigens prioritized in a 2009 National Cancer Institute pilot project report,10 four are gangliosides including GD2, GD3, fucosyl GM1, and GM3 (Fig. 1).
Fig. 1.
Structures of prioritized ganglioside cancer antigens GM3 (1), fucosyl GM1 (2), GD3 (3), and GD2 (4).
Due to their important functions, gangliosides are attractive but challenging synthetic targets.11, 12 For example, among four prioritized ganglioside cancer antigens, only GM3 has been synthesized chemically and chemoenzymatically.12–15 GD216 and GD317 have been synthesized by chemical methods only. Fucosyl GM1 has never been synthesized before and only its oligosaccharide portion has been chemically synthesized.18, 19 Chemical synthetic procedures for gangliosides encounter notable challenges including hard-to-control regio- and stereo-selectivities in sialylation, and low yields for glycosylating the oligosaccharide with the lipid.11 Chemoenzymatic synthesis of gangliosides has been reported only for GM3.12, 20, 21 Therefore, efficient synthetic approaches for diverse gangliosides are lacking but are urgently needed.
Biosynthetically, gangliosides are formed in the Golgi from lactosylceramide (LacβCer) catalyzed by type II membrane protein glycosyltransferases.22 These processes are not readily duplicated in vitro as LacβCer is not soluble in water and the glycosyltransferases involved are immobilized on the Golgi membrane. Recently, we showed that α-Gal pentasaccharyl ceramide, a neutral glycosphingolipid, can be chemoenzymatically synthesized from water soluble lactosyl sphingosine (LacβSph) with simple C18-cartridge solid phase extraction (SPE) purification procedures followed by acylation.23 Herein, we develop high-yield streamlined chemoenzymatic processes for total synthesis of synthetically challenging complex gangliosides. A 13-gram-scale synthetic procedure is established for producing LacβSph from commercially available inexpensive lactose and phytosphingosine. Sequential one-pot multienzyme (OPME) chemoenzymatic synthetic methods, combined with facile C18-cartridge purification scheme, are successfully developed for the production of negatively charged complex gangliosides from LacβSph in a highly efficient manner. An improved acylation procedure is identified to convert glycosylsphingosines to target gangliosides in excellent 98–99% yields. The four prioritized ganglioside cancer antigens including GM3 (1), fucosyl GM1 (2), GD3 (3), and GD2 (4) (Figure 1) are successfully obtained in high yields.
To obtain target gangliosides, LacβSph (5)23 was prepared from lactose and phytosphingosine in a 13-gram scale to demonstrate the efficiency of the streamlined chemical synthetic procedure and to provide a sufficient amount of starting material for glycosphingolipid synthesis. The synthesis was achieved in 12 steps in an overall 40% yield (see ESI).
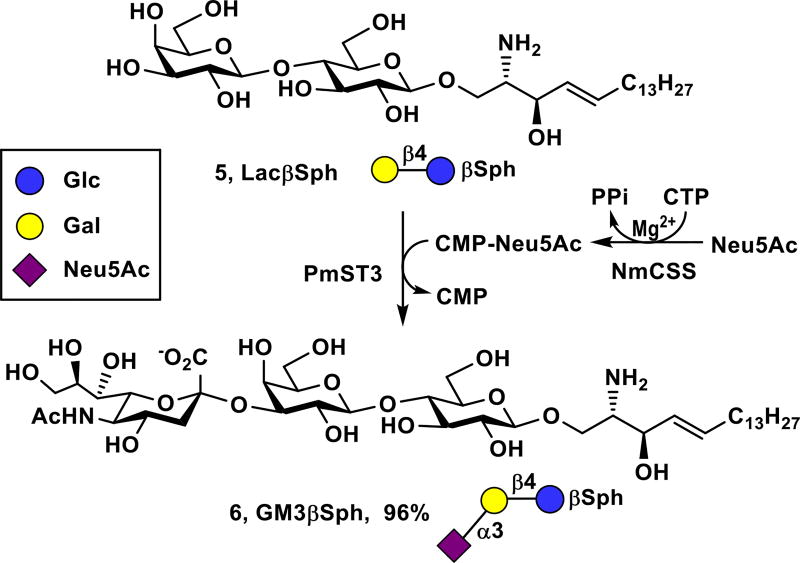
GM3 (1) is the simplest ganglioside and is a common precursor for more complex gangliosides. It is overexpressed by many types of tumors, including malignant melanoma, leukemia, pulmonary cancer, neuroectodermal tumor.24, 25 The corresponding GM3 sphingosine (GM3βSph, 6) lacking the fatty acyl chain was readily obtained from LacβSph (5) and monosaccharide N-acetylneuraminic acid (Neu5Ac) using a one-pot two-enzyme sialylation system containing Neisseria meningitidis CMP-sialic acid synthetase (NmCSS)26 and Pasteurella multocida α2–3-sialyltransferase 3 (PmST3) that can use both oligosaccharides and glycolipids as acceptor substrates27 (Scheme 1). PmST128 and its mutants29, 30 preferring oligosaccharides as acceptors are not suitable sialyltransferases for the reaction. A simple C18-cartridge SPE was used for purification by loading the sample to the C18-cartridge, washing the cartridge with water to completely remove excess Neu5Ac, cytidine 5’-monophosphate (CMP)-Neu5Ac, cytidine 5’-triphosphate (CTP), by-product CMP, and others, followed by eluting the cartridge with 60% acetonitrile (CH3CN) in water to obtain pure GM3βSph (6) in a gram-scale (1.44 g) with an excellent 96% yield. The remaining LacβSph was recovered by eluting the cartridge with pure CH3CN. The whole process took about 25 minutes in contrast to several hours using standard silica gel chromatography.
Scheme 1.
Gram-scale (1.44 g) synthesis of GM3βSph (6) by one-pot multienzyme (OPME) sialylation of LacβSph (5).
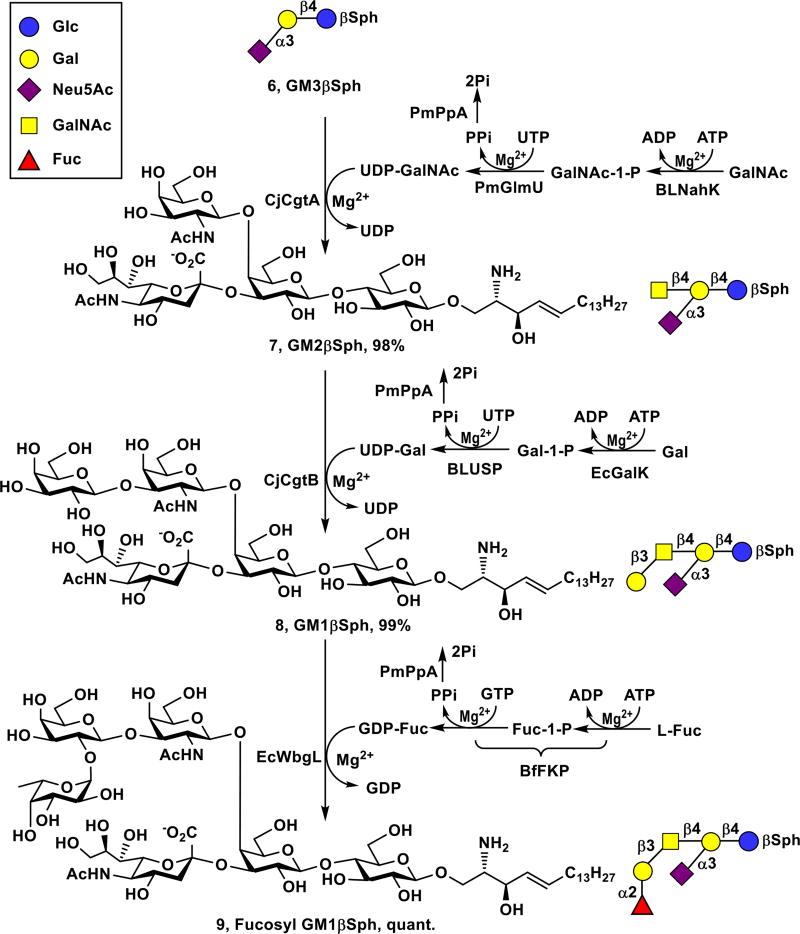
Fucosyl GM1 (2) has been found to be absent from normal tissues but be expressed on the cell surface of small-cell lung cancer (SCLC)31 which accounts for 10–15% of lung cancer cases and remains as one of the leading causes of death in the United States.32 Fucosyl GM1 is therefore an excellent candidate for cancer vaccine development and for the development of antibodies for detecting SCLC. The corresponding fucosyl GM1 sphingosine (Fuc-GM1βSph, 9) was obtained from GM3βSph (6) using three OPME reactions carried out in sequential (Scheme 2).
Scheme 2.
Sequential one-pot multienzyme (OPME) synthesis of Fuc-GM1βSph (9) from GM3βSph (6).
A one-pot four-enzyme N-acetylgalactosamine (GalNAc)-activation and transfer system33 containing recombinant Bifidobacterium longum strain ATCC55813 N-acetylhexosamine-1-kinase (BLNahK),34 Pasteurella multocida N-acetylglucosamine uridylyltransferase (PmGlmU),35 Pasteurella multocida inorganic pyrophosphatase (PmPpA),36 and Campylobacter jejuni β1–4-N–acetylgalactosaminyltransferase (CjCgtA),33 was used to glycosylate GM3βSph (6) to form GM2βSph (7). Although GM3βSph (6) was a less favored acceptor for CjCgtA compared to GM3 oligosaccharide,33 adding a larger amount of CjCgtA was able to push the reaction to completion. GM2βSph (7, 120 mg, 98% yield) was readily purified by passing the reaction mixture through the C18 cartridge and eluting with 45% CH3CN in water.
GM1βSph (8) was then synthesized from GM2βSph (7) using a one-pot four-enzyme galactose-activation and transfer system containing Escherichia coli galactokinase (EcGalK),37 Bifidobacterium longum UDP-sugar pyrophosphorylase (BLUSP),38 PmPpA, and Campylobacter jejuni β1–3-galactosyltransferase (CjCgtB).39 Again, GM2βSph (7) was a less efficient acceptor for CjCgtB compared to GM2 oligosaccharide,33 but adding a larger amount of CjCgtB was able to complete the reaction without the complication of adding more than one galactose residue observed previously for galactosylation of GM2 oligosaccharide by CjCgtB.33 GM1βSph (8, 57 mg, 99% yield) was readily purified by passing the reaction mixture through the C18 cartridge and eluting with 40% CH3CN in water.
To synthesize Fuc-GM1βSph (9) from GM1βSph (8), a suitable α1–2-fucosyltransferase was needed. Despite its preference towards β1–3-linked galactoside acceptors, Thermosynechococcus elongates α1–2-fucosyltransferase (Te2FT)40 could not fucosylate GM1βSph (8). Escherichia coli O126 α1–2-fucosyltransferase (EcWbgL)41 that prefers β1–4-linked galactoside acceptors but is also active towards β1–3-linked galactosides was then cloned (see ESI) and found to be very efficient in using GM1βSph (8) as the acceptor. Fuc-GM1βSph (9, 55 mg) was synthesized in a quantitative yield from GM1βSph (8) using a one-pot three-enzyme fucose-activation and transfer system containing EcWbgL and guanosine 5'-diphosphate fucose (GDP-Fuc) biosynthetic enzymes including Bacteroides fragilis bifunctional L-fucokinase and guanidine 5'-diphosphate (GDP)-fucose pyrophosphorylase (BfFKP)42 and PmPpA (Scheme 2) followed by C18 cartridge-purification by eluting with 40% of CH3CN in water. Remarkable, three OPME glycosylation reactions carried out in sequential led to the formation of complex Fuc-GM1βSph (9) from GM3βSph (6) in an excellent 97% overall yield. The overall process for the formation of Fuc-GM1βSph (9) from chemically synthesized LacβSph (5) involved four sequential OPME reaction with a total yield of 93%.
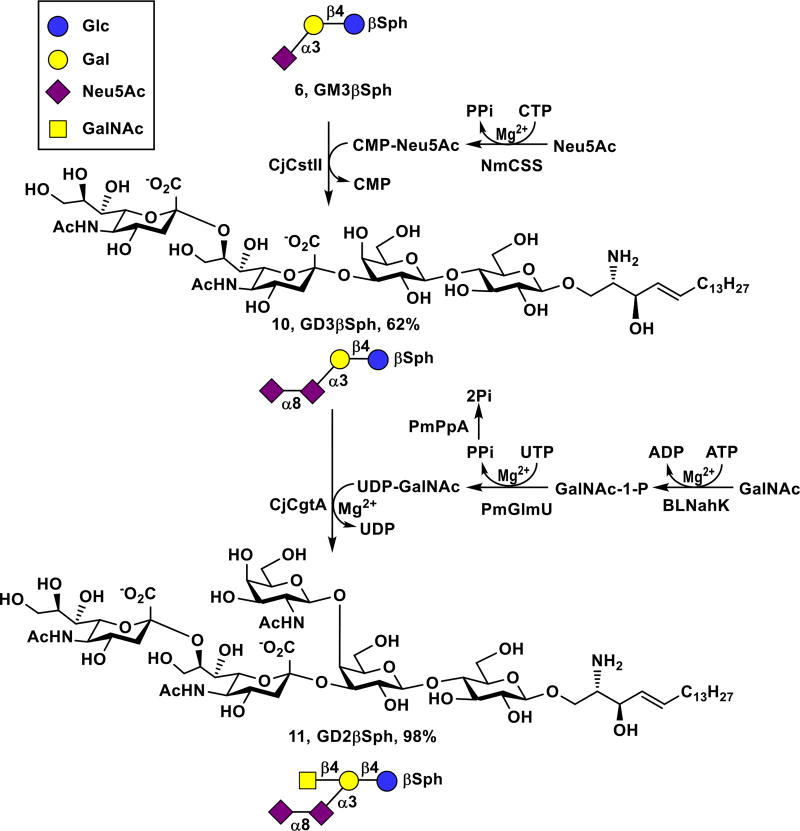
GD3 is a disialoganglioside overexpressed on melanomas, neuroectodermal tumors including neuroblastoma and glioma, as well as cancers of lung, breast, colon, prostate, and ovary.43, 44 Therefore, GD3 has received a considerable attention as a promising immunotherapeutic target for cancer therapy.9, 44, 45 The corresponding GD3βSph (10) was successfully synthesized from GM3βSph (6) and Neu5Ac using a one-pot two-enzyme sialylation reaction containing NmCSS and Campylobacter jejuni α2–3/8-sialyltransferase (CjCstII)46, 47 (Scheme 3). As CjCstII could continue to add an additional α2–8-linked Neu5Ac to GD3βSph (10), GT3βSph was produced as a minor product and was eluted from the C18-cartrige using 30% of CH3CN in water. The GD3βSph (10, 910 mg, 62% yield) was then readily obtained by eluting the cartridge with 35% of CH3CN in water.
Scheme 3.
Sequential one-pot multienzyme (OPME) synthesis of GD3βSph (10) and GD2βSph (11) from GM3βSph (6).
GD2 is another disialoganglioside cancer antigen expressed predominantly in the CD44hiCD24lo breast cancer stem cells isolated from human breast cancer cells.48 It has also been found in melanomas, gliomas, and neuroblastomas.49 Therapeutic effects of anti-GD2 monoclonal antibodies against neuroblastomas have been reported.50 GD2 (#12) has the highest priority compared to the other three prioritized ganglioside cancer antigens GM3 (#48), fucosyl GM1 (#41), and GD3 (#40).10 Similar to the synthesis of GM2βSph (7) from GM3βSph (6), GD2βSph (11) was readily synthesized from GD3βSph (10) using the one-pot multienzyme GalNAc-activation and glycosylation system containing BLNahK, PmGlmU, PmPpA, and CjCgtA (Scheme 3). As GD3βSph (10) was not an efficient substrate for CjCgtA comparing to GD2 oligosaccharide,33 a larger amount of CjCgtA was used to push the reaction to completion. GD2βSph (11, 34 mg) was easily purified by passing the reaction mixture through C18 cartridge and eluting with 35% of CH3CN in water with an excellent 98% yield.
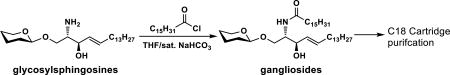
With the desired glycosylsphingosines in hand, the conditions for efficient acylation to form target glycosphingolipids were explored. Similar to that reported previously,23 palmitic acid in the presence of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride and hydroxybenzotriazole (EDC-HCl/HOBt) led to successful acylation of the amino group in glycosylsphingosines but the reaction could not reach completion. In comparison, reactions of glycosylsphingosines with palmitoyl chloride in a mixed solvent of tetrahydrofuran (THF) and saturated NaHCO3 solution (1:1, v/v) reached completion in 2 h. GM3 (1), GM2 (12), GM1 (13), fucosyl GM1 (2), GD3 (3), and GD2 (4) gangliosides were produced in 98–99% yields with facile C18 cartridge purification by eluting with 40–80% of CH3CN in water (Table 1).
Table 1.
Gangliosides formation by acylation of glycosylsphingosines.
| |||
|---|---|---|---|
| Glycosylsphingosine | Ganglioside Product |
C18 cartridge elution condition |
Yield (%) |
| GM3βSph (6) | GM3 (1) | 80% CH3CN in H2O | 99 |
| GM2βSph (7) | GM2 (12) | 50% CH3CN in H2O | 98 |
| GM1βSph (8) | GM1 (13) | 50% CH3CN in H2O | 99 |
| Fuc-GM1βSph (9) | Fucosyl GM1 (2) | 50% CH3CN in H2O | 99 |
| GD3βSph (10) | GD3 (3) | 50% CH3CN in H2O | 99 |
| GD2βSph (11) | GD2 (4) | 40% CH3CN in H2O | 98 |
Conclusions
In conclusions, combined with easy acylation and facile C18 cartridge purification schemes, sequential one-pot multienzyme (OPME) glycosylation systems are highly efficient in synthesizing a diverse array of gangliosides. As many bacterial glycosyltransferases are involved in the synthesis of glycolipid repeating units for the production of lipopolysaccharides and capsular polysaccharides, they can use glycosylsphingosines as acceptors for glycosylation although larger amounts of the enzymes may be needed in some cases. The production of gangliosides by sequential OPME glycosylation of water soluble lactosyl sphingosine with C18 cartridge purification followed by high-yield acylation is a highly efficient streamlined process. It is reasonable to assume that by incorporating various glycosyltransferases, the OPME systems can be applied to the synthesis of other challenging complex glycosphingolipids.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH) Common Fund Glycoscience Program grant U01GM120419. Bruker Avance-800 NMR spectrometer was funded by NSF grant DBIO-722538.
Footnotes
Electronic Supplementary Information (ESI) available Experimental procedures, NMR and HRMS data, and NMR spectra of products. See DOI: 10.1039/x0xx00000x
Conflicts of interest
There are no conflicts to declare.
References
- 1.Furukawa K, Ohmi Y, Ohkawa Y, Tajima O, Furukawa K. Adv. Neurobiol. 2014;9:307–320. doi: 10.1007/978-1-4939-1154-7_14. [DOI] [PubMed] [Google Scholar]
- 2.Schnaar RL. J. Mol. Biol. 2016;428:3325–3336. doi: 10.1016/j.jmb.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lingwood D, Simons K. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 4.Jobling MG, Holmes RK. Infect. Immun. 2002;70:1260–1271. doi: 10.1128/IAI.70.3.1260-1271.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez MA, López S, Arias CF, Isa P. J. Virol. 2013;87:1115–1122. doi: 10.1128/JVI.01964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ledeen RW, Wu G. Trends Biochem. Sci. 2015;40:407–418. doi: 10.1016/j.tibs.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Mocchetti I. Cell. Mol. Life Sci. 2005;62:2283–2294. doi: 10.1007/s00018-005-5188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svennerholm L. Life Sci. 1994;55:2125–2134. doi: 10.1016/0024-3205(94)00393-9. [DOI] [PubMed] [Google Scholar]
- 9.Daniotti J, Vilcaes A, Torres Demichelis V, Ruggiero F, Rodriguez-Walker M. Front. Oncol. 2013;3:306. doi: 10.3389/fonc.2013.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, Mellman I, Prindiville SA, Viner JL, Weiner LM, Matrisian LM. Clin. Cancer Res. 2009;15:5323–5337. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiso M, Ishida H, Ando H, Imamura A. In: Glycoscience: Biology and Medicine. Taniguchi N, Endo T, Hart GW, Seeberger PH, Wong C-H, editors. Springer Japan, Tokyo: 2015. pp. 331–338. [DOI] [Google Scholar]
- 12.Liu Y, Wen L, Li L, Gadi MR, Guan W, Huang K, Xiao Z, Wei M, Ma C, Zhang Q, Yu H, Chen X, Wang PG, Fang J. Eur. J. Org. Chem. 2016;2016:4315–4320. doi: 10.1002/ejoc.201600950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito Y, Paulson JC. J. Am. Chem. Soc. 1993;115:1603–1605. [Google Scholar]
- 14.Duclos RI. Carbohydr. Res. 2000;328:489–507. doi: 10.1016/s0008-6215(00)00121-x. [DOI] [PubMed] [Google Scholar]
- 15.Murase T, Ishida H, Kiso M, Hasegawa A. Carbohydr. Res. 1989;188:71–80. doi: 10.1016/0008-6215(89)84060-1. [DOI] [PubMed] [Google Scholar]
- 16.Matsuzaki Y, Nunomura S, Ito Y, Sugimoto M, Nakahara Y, Ogawa T. Carbohydr. Res. 1993;242:C1–6. doi: 10.1016/0008-6215(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 17.Ishida H, Ohta Y, Tsukada Y, Kiso M, Hasegawa A. Carbohydr. Res. 1993;246:75–88. doi: 10.1016/0008-6215(93)84025-2. [DOI] [PubMed] [Google Scholar]
- 18.Allen JR, Danishefsky SJ. J. Am. Chem. Soc. 1999;121:10875–10882. [Google Scholar]
- 19.Mong TK-K, Lee H-K, Durón SG, Wong C-H. Proc. Natl. Acad. Sci. 2003;100:797–802. doi: 10.1073/pnas.0337590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaughan MD, Johnson K, DeFrees S, Tang X, Warren RA, Withers SG. J. Am. Chem. Soc. 2006;128:6300–6301. doi: 10.1021/ja058469n. [DOI] [PubMed] [Google Scholar]
- 21.Nishimura S-I, Yamada K. J. Am. Chem. Soc. 1997;119:10555–10556. [Google Scholar]
- 22.Groux-Degroote S, Guerardel Y, Delannoy P. Chembiochem. 2017;18:1146–1154. doi: 10.1002/cbic.201600705. [DOI] [PubMed] [Google Scholar]
- 23.Santra A, Li Y, Yu H, Slack TJ, Wang PG, Chen X. Chem. Commun. 2017;53:8280–8283. doi: 10.1039/c7cc04090c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan XL, Izumi T, Yamada H, Akiyoshi K, Suenobu S, Yokoyama S. Brain Develop. 2000;22:196–198. doi: 10.1016/s0387-7604(00)00091-7. [DOI] [PubMed] [Google Scholar]
- 25.Seyfried TN, Mukherjee P. J. Oncol. 2010;2010:961243. doi: 10.1155/2010/961243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu H, Yu H, Karpel R, Chen X. Bioorg. Med. Chem. 2004;12:6427–6435. doi: 10.1016/j.bmc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 27.Thon V, Li Y, Yu H, Lau K, Chen X. Appl. Microbiol. Biotechnol. 2012;94:977–985. doi: 10.1007/s00253-011-3676-6. [DOI] [PubMed] [Google Scholar]
- 28.Yu H, Chokhawala H, Karpel R, Yu H, Wu B, Zhang J, Zhang Y, Jia Q, Chen X. J. Am. Chem. Soc. 2005;127:17618–17619. doi: 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- 29.Sugiarto G, Lau K, Li Y, Khedri Z, Yu H, Le DT, Chen X. Mol. Biosyst. 2011;7:3021–3027. doi: 10.1039/c1mb05182b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugiarto G, Lau K, Qu J, Li Y, Lim S, Mu S, Ames JB, Fisher AJ, Chen X. ACS Chem. Biol. 2012;7:1232–1240. doi: 10.1021/cb300125k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickler MN, Ragupathi G, Liu NX, Musselli C, Martino DJ, Miller VA, Kris MG, Brezicka F-T, Livingston PO, Grant SC. Clin. Cancer Res. 1999;5:2773–2779. [PubMed] [Google Scholar]
- 32.Chu QSC, Markman B, Leighl N, Krug L, Rudin C, Lathers D, Basciano P, Fracasso PM, Kollia G, Phillips P, Kolaitis G, Williams D, Jackson J, Ready N. Ann. Oncol. 2016;27:1427PD–1427PD. [Google Scholar]
- 33.Yu H, Li Y, Zeng J, Thon V, Nguyen DM, Ly T, Kuang HY, Ngo A, Chen X. J. Org. Chem. 2016;81:10809–10824. doi: 10.1021/acs.joc.6b01905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Yu H, Chen Y, Lau K, Cai L, Cao H, Tiwari VK, Qu J, Thon V, Wang PG, Chen X. Molecules. 2011;16:6396–6407. doi: 10.3390/molecules16086396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Thon V, Li Y, Yu H, Ding L, Lau K, Qu J, Hie L, Chen X. Chem. Commun. 2011;47:10815–10817. doi: 10.1039/c1cc14034e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lau K, Thon V, Yu H, Ding L, Chen Y, Muthana MM, Wong D, Huang R, Chen X. Chem. Commun. 2010;46:6066–6068. doi: 10.1039/c0cc01381a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X, Fang J, Zhang J, Liu Z, Shao J, Kowal P, Andreana P, Wang PG. J. Am. Chem. l Soc. 2001;123:2081–2082. doi: 10.1021/ja005738v. [DOI] [PubMed] [Google Scholar]
- 38.Muthana MM, Qu J, Li Y, Zhang L, Yu H, Ding L, Malekan H, Chen X. Chem. Commun. 2012;48:2728–2730. doi: 10.1039/c2cc17577k. [DOI] [PubMed] [Google Scholar]
- 39.Malekan H, Fung G, Thon V, Khedri Z, Yu H, Qu J, Li Y, Ding L, Lam KS, Chen X. Bioorg. Med. Chem. 2013;21:4778–4785. doi: 10.1016/j.bmc.2013.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao C, Wu Y, Yu H, Shah IM, Li Y, Zeng J, Liu B, Mills DA, Chen X. Chem. Commun. 2016;52:3899–3902. doi: 10.1039/c5cc10646j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engels L, Elling L. Glycobiology. 2014;24:170–178. doi: 10.1093/glycob/cwt096. [DOI] [PubMed] [Google Scholar]
- 42.Yi W, Liu X, Li Y, Li J, Xia C, Zhou G, Zhang W, Zhao W, Chen X, Wang PG. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4207–4212. doi: 10.1073/pnas.0812432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamamura K, Furukawa K, Hayashi T, Hattori T, Nakano J, Nakashima H, Okuda T, Mizutani H, Hattori H, Ueda M, Urano T, Lloyd KO, Furukawa K. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11041–11046. doi: 10.1073/pnas.0503658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo ASY, Ma Q, Liu DL, Junghans RP. Clin. Cancer Res. 2010;16:2769–2780. doi: 10.1158/1078-0432.CCR-10-0043. [DOI] [PubMed] [Google Scholar]
- 45.Torres Demichelis V, Vilcaes AA, Iglesias-Bartolomé R, Ruggiero FM, Daniotti JL. PLoS One. 2013;8(1):e55304. doi: 10.1371/journal.pone.0055304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng J, Yu H, Lau K, Huang S, Chokhawala HA, Li Y, Tiwari VK, Chen X. Glycobiology. 2008;18:686–697. doi: 10.1093/glycob/cwn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilbert M, Brisson J-R, Karwaski M-F, Michniewicz J, Cunningham A-M, Wu Y, Young NM, Wakarchuk WW. J. Biol. Chem. 2000;275:3896–3906. doi: 10.1074/jbc.275.6.3896. [DOI] [PubMed] [Google Scholar]
- 48.Battula VL, Shi Y, Evans KW, Wang R-Y, Spaeth EL, Jacamo RO, Guerra R, Sahin AA, Marini FC, Hortobagyi G, Mani SA, Andreeff M. J. Clin. Invest. 2012;122:2066–2078. doi: 10.1172/JCI59735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hakomori S-I. Cancer Res. 1996;56:5309–5318. [PubMed] [Google Scholar]
- 50.Kushner BH, Kramer K, Modak S, Cheung N-KV. J. Clin. Oncol. 2011;29:1168–1174. doi: 10.1200/JCO.2010.28.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.