SUMMARY
Patterning of vertebrate melanophores is essential for mate selection and protection from UV-induced damage. Patterning can be influenced by circulating long range factors such as hormones, but it is unclear how their activity is controlled in recipient cells to prevent excesses in cell number and migration. The zebrafish wanderlust mutant harbors a mutation in the sheddase bace2, and exhibits hyperdendritic and hyperproliferative melanophores that localize to aberrant sites. We performed a chemical screen to identify suppressors of the wanderlust phenotype, and found that inhibition of insulin/PI3Kγ/mTOR signaling rescues the defect. In normal physiology, Bace2 cleaves the insulin receptor, whereas its loss results in hyperactive insulin/PI3K/mTOR signaling. Insulin B, an isoform enriched in the head, drives the melanophore defect. These results suggest that insulin signaling is negatively regulated by melanophore-specific expression of a sheddase, highlighting how long distance factors can be regulated in a cell-type specific manner.
eTOC Blurb
Zhang et al. demonstrate that insulin signaling affects pigment patterning in zebrafish. They show that the local response to insulin signaling is regulated by Bace2, a melanocyte-enriched sheddase that cleaves the insulin receptor.
INTRODUCTION
Vertebrate pigmentation is an essential component of mate selection, camouflage and protection against UV damage (Price et al., 2008; Yamaguchi et al., 2007). Pigment cells patterning during development involves a precise spatiotemporal control of cell number, differentiation and localization. This patterning requires close coordination of both short-range and long-range extracellular factors with cell-intrinsic melanophore gene programs.
The major pigment cell type in humans is the black melanocyte, which corresponds to the black melanophore found in teleosts such as zebrafish. Across vertebrates, a cell-intrinsic transcriptional hierarchy orchestrates specification and differentiation (Mort et al., 2015). Melanocytes are derived from the neural crest, a population of sox10+ pluripotent cells that eventually give rise to lineage-restricted mitf+ melanoblasts (Ernfors, 2010; Sommer, 2011). mitf itself activates pigment genes such as dct, tyr and tyrp1 and the melanoblasts then differentiate into melanocytes/melanophores. These cells are characterized by black melanin production and formation of cell dendrites, which in mammals transfers melanin packets into surrounding keratinocytes (Goding, 2007; Lin and Fisher, 2007).
Local environmental factors crosstalk with these intrinsic melanophore gene programs to instruct their development. One example of this interaction is short range communication between endothelins (EDN1, EDN3) produced by keratinocytes after UV exposure (Garcia et al., 2008; Hara et al., 1995; Imokawa et al., 1992), which then act on nearby melanophores with endothelin-B receptor EDNRB on the membrane (Baynash et al., 1994; Parichy et al., 2000). Keratinocytes can also secrete other short range factors such as alpha-MSH which act on melanocortin receptors (MC1R) on the melanophores (Abdel-Malek et al., 1995; Chakraborty et al., 1991). These short-range factors act as strong positive regulators of melanophore proliferation and differentiation, and their effects are ultimately abrogated when the UV signal inducing their expression is reduced. Interactions between neighboring pigment cells also regulates their survival and localization(Hamada et al., 2014; Mahalwar et al., 2016). Adjacent xanthophores can exclude melanophores during stripe consolidation by extending specialized cell projections called airinemes which interact with microenvironmental macrophages (Eom and Parichy, 2017; Eom et al., 2015; Nakamasu et al., 2009).
Longer range factors such as endocrine hormones can also controls pigment patterning. Growth hormone/IGF can induce melanophore proliferation and differentiation (Edmondson et al., 1999; Tavakkol et al., 1992) and melanin concentrating hormone released from the pituitary modulates melanosome trafficking to facilitate camouflage (Logan et al., 2006; Richardson et al., 2008). Thyroid hormones are critical to promote xanthophore differentiation while attenuating melanophore proliferation and survival (McMenamin et al., 2014). Because these hormones can pass through the bloodstream, it remains unclear how these more “general” factors are regulated within the melanophore lineage in a spatiotemporal manner, and how the melanophore turns off this signal. Such negative regulation is essential to proper patterning, as excess melanophores could dramatically alter the appearance of the animal and render them susceptible to neoplastic growth. One hypothesis for this negative regulation is that these extracellular factors depend on lineage-specific gene programs within the melanophore to regulate their activity.
In this study, we utilized the zebrafish to study the role of distant factors in melanophore patterning. The zebrafish is an ideal system to address this question since it allows for facile genetic manipulation and chemical genetic strategies in a whole-organism context. Using this system, we characterized a mutant called wanderlust that has a disruption of normal melanophore patterning, with an excess number of hyperdendritic, hypermigratory melanophores. This mutant harbors an inactivating mutation in the sheddase Bace2, a melanophore-enriched gene which has been linked to abnormal migration but through an unknown mechanism (van Bebber et al., 2013). Here, we show that Bace2 acts to negatively regulate insulin/PI3K signaling within the melanophores. This occurs due to the ability of Bace2 to cleave and degrade the insulin receptor specifically on melanophores. When Bace2 is lost, this leads to unregulated melanophore proliferation and migration, suggesting a mechanism which connects distant hormonal signaling to melanophore-specific gene programs.
RESULTS
The zebrafish wanderlust mutant has hyperdendritic melanophores due to a loss of Bace2
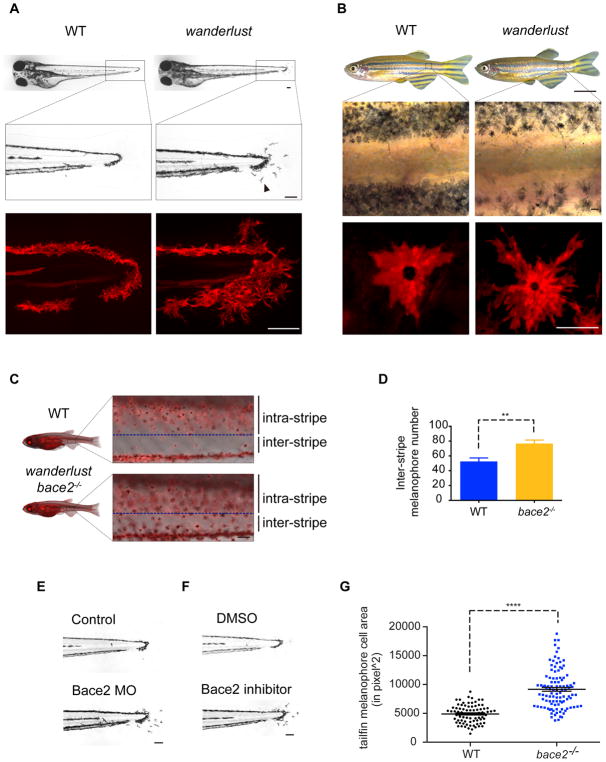
In order to identify processes involved in pigment patterning, we characterized a zebrafish mutant called wanderlust that has a nonsense mutation in the sheddase bace2 (van Bebber et al., 2013) (allele: bace2hu3332). This leads to an early stop codon in exon 1 (Figure S1A). During normal embryonic development, mitf+ melanophore precursors emerge from the neural crest from 16–24 hours post fertilization (hpf) and then migrate along the dorsolateral migration pathway, where they move first caudally and then ventrally along the somite boundaries (Kelsh et al., 2009). At the point of maximal migration, approximately 48hpf, melanophores are highly dendritic, but by 72hpf, after they have mostly ceased migration, the melanophores become compact and retract their dendrites (Cooper and Raible, 2009). In contrast, the wanderlust mutant has extensively hyperdendritic melanophores throughout development. These melanophores appear in atypical locations and abnormally migrate into the caudal tail fin (Figure 1A). The hyperdendritic phenotype is maintained throughout embryogenesis and into adulthood (Figure 1B). Consistent with this, during metamorphosis we find an increased number of ectopic melanophores in the inter-stripe region in the wanderlust mutant (Figure 1C–D). Transient knockdown of Bace2 using morpholinos (MO) in wild type (WT) embryos recapitulates the wanderlust embryo phenotype (Figure 1E), as does pharmacologic inhibition of Bace2 with the compound PF-06663195 (Figure 1F). Measurement of the tailfin melanophores shows that wanderlust melanophores occupy a larger cell area compared to WT embryos (Figure 1G). Collectively this demonstrates that it is the loss of Bace2 function that leads to the defect in melanophore localization and morphology.
Figure 1. The zebrafish wanderlust mutant has hyperdendritic melanophores due to a loss of Bace2.
(A) Brightfield imaging shows that the wanderlust melanophores are hyperdendritic compared to WT fish (arrowhead) in the tail fin at 72hpf. Labeling of the melanophore cell membranes (bottom panel) using the Tg(tyrp1b: membrane-mCherry) line demonstrates that this is due to a change in cell morphology rather than a redistribution of melanin.
(B) These hyperdendritic melanophores are maintained into adulthood, and yield irregular stripe boundaries (arrowhead).
(C–D) wanderlust mutants develop melanophores outside of the stripe during metamorphosis at 24 days post fertilization (dpf) (arrowhead). Inter-stripe melanophores number are quantified in (D); fish number: n(WT)=5, n(wanderlust)=5, two-tailed t test, **P<0.01.
(E–F) Bace2 loss of function using morpholino knockdown (E) or pharmacological inhibition (F) phenocopies the wanderlust mutant. WT embryos are treated with Bace2 inhibitor (PF-06663195, 100μM) from 48–72hpf.
(G) bace2−/− melanophores have larger cell area in the tail fin at 72hpf. Melanophore cell area is defined as the surface area covered by melanophores in the tailfin. Data are from five independent experiments, with total fish numbers: n(WT)=80, n(bace2−/−)=105, two-tailed t test, ****P<0.0001. All bar graphs are presented as mean ± s.e.m. Scale bars, 100μM (A, B middle and bottom panel, C, E, F), 0.5cm (B top panel). See also Figure S1.
Bace2 acts during melanophore differentiation
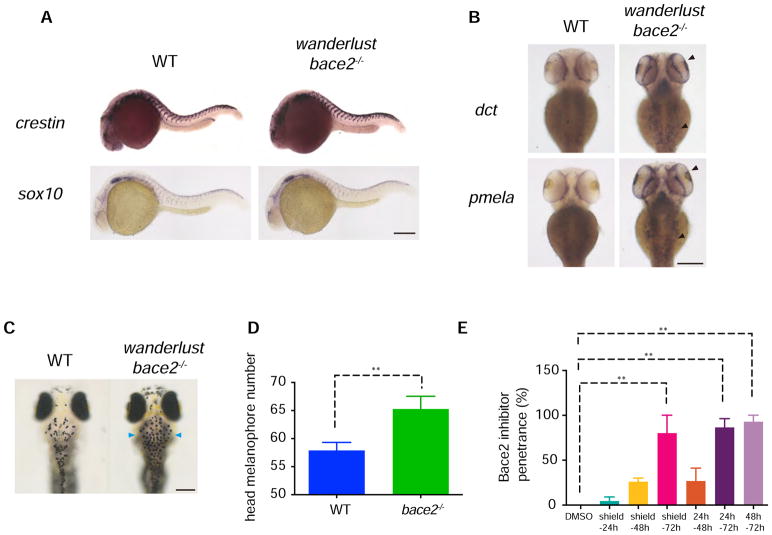
We used a panel of neural crest and melanophore markers to determine when during development Bace2 is required using whole mount in situ hybridization (ISH). Whereas expression of the neural crest markers crestin and sox10 at 24hpf was normal in the wanderlust mutant (Figure 2A and Figure S1B), by 72 hpf we noted an increase in pigmentation genes such as dct, pmela, tyr and tyrp1b in the mutant (Figure 2B and Figure S1C), especially noticeable in the head region. This increase in pigmentation gene expression corresponds to an increase in visibly pigmented melanophores in the head region (Figure 2C–D). To determine whether these excess head melanophores were due to proliferation, we utilized a previously developed assay (Hultman and Johnson, 2010; Lee et al., 2010; Lévesque et al., 2013; McMenamin et al., 2014) that uses existing melanin as a lineage tracer. In this assay, the animals are treated with the small molecule PTU, which prevents de novo melanin synthesis but does not affect existing melanin, so the appearance of new cells with melanin is indicative of division of a previously pigmented cell (Figure S2A–C). Using this assay, we find that PTU treatment did not prevent the increase in pigmented cell number, suggesting that Bace2 loss results in excess cell division of melanized cells (Figure S2D–E). To more precisely define the timing in which Bace2 acts, we treated WT embryos with Bace2 inhibitor PF-06663195 at various time windows from shield stage to 72hpf, and found that application from 48–72hpf is sufficient to give rise to the wanderlust phenotype (Figure 2E), coinciding with the period when the melanophores are actively pigmenting and differentiating.
Figure 2. Bace2 acts during melanophore differentiation.
(A) At 24hpf, ISH shows the bace2−/− mutants have unaffected expression for neural crest markers crestin and sox10.
(B) But at 72hpf, ISH shows mRNA for the pigmentation genes dct and pmela are elevated in bace2−/− mutants, especially in the head (arrowhead).
(C–D) Consistently, bace2−/− mutants have increased numbers of pigmented melanophores in the head (arrowhead). Fish number n(WT)=55, n(bace2−/−)=30, two-tailed t test, **P<0.01.
(E) Treatment of WT embryos with 100μM Bace2 inhibitor (PF-0666195) from 48–72 hpf is sufficient to phenocopy bace2−/−. Data are from two independent experiments, with total fish numbers: n(DMSO)=21, n(shield-24h)=45, n(shield-48h)=56, n(shield-72h)=72, n(24h–48h)=58, n(24h–72h)=40, n(48h–72h)=49; One-way ANOVA followed by Holm-Sidak’s multiple comparisons test, **P<0.01. All bar graphs are presented as mean ± s.e.m. Scale bars, 200μM. See also Figure S1 and Figure S2.
Bace2 acts cell-intrinsically within the melanophore lineage
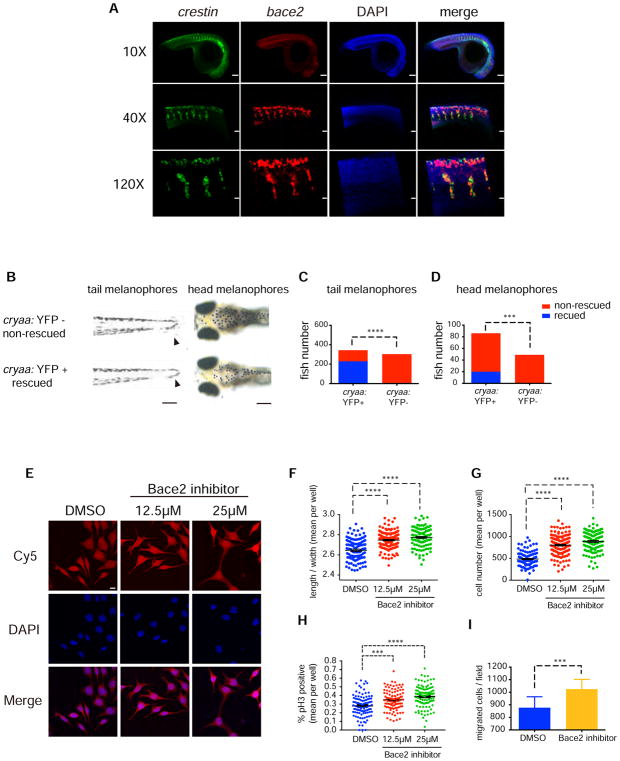
To test if Bace2 was acting cell-intrinsically within the melanophore lineage, we examined expression of bace2 by ISH and noted that it strongly overlaps with crestin, a pan neural crest marker that has been previously shown to mark all derivative lineages including melanophores (Figure 3A) (Kaufman et al., 2016; Luo et al., 2001; Rubinstein et al., 2000; White et al., 2011). To more directly test whether Bace2 was acting cell-intrinsically, we created a stable rescue transgenic line in which the dct promoter (Budi et al., 2011) drives wild-type Bace2. When crossed into the wanderlust mutant, we see a complete rescue of the melanophore defect in both the tail and head region, consistent with a cell intrinsic effect of Bace2 loss (Figure 3B–D).
Figure 3. Bace2 acts cell-intrinsically within the melanophore lineage.
(A) At 24hpf in WT embryos, double fluorescent ISH against the pan neural crest marker crestin (green) and bace2 (red) shows bace2 expression overlaps with crestin, suggesting bace2 is highly enriched in the neural crest lineage from which the melanophores are derived.
(B–D) Melanophore-specific transgenic rescue of the bace2−/− mutant. A stable transgenic line was created in which the dct promoter drives Bace2 in the melanophores (along with a cryaa: YFP transgene marker). Founders were identified by expression of the YFP marker in the eye. Founders were crossed into other uninjected bace2−/− adult to generate F1 embryos. F1 embryos were scored for dendritic tail melanophore rescue at 3dpF (B, arrowhead) or for rescue of the head melanophores at 5dpF. F1 were divided into four groups: rescued, non-rescued, YFP positive and YFP negative. Chi-square with Yate’s correction was performed to associate rescue phenotype with YFP positive eye (quantified in C and D). ***P<0.001, ****P<0.0001.
(E–G) The zebrafish melanoma cell line ZMEL1 was treated with 12.5μM or 25μM of the Bace2 inhibitor PF-06663195 for 72 hours, and resulted in hyperdendritic cells (arrowhead) similar to what was seen in vivo. A representative field of cells is shown in (E) and dendricity is quantified as an increase in cell length to width ratio (F), along with an increase in overall cell number (G). Data are from three independent experiments, one-way ANOVA followed by Holm-Sidak’s multiple comparisons test, ****P<0.0001.
(H) The increased cell number induced by Bace2 inhibitor is due to increased ZMEL1 cell proliferation, as measured by increased phospho-histone H3 (pH3) immunostaining. Data are from three independent experiments, one-way ANOVA followed by Holm-Sidak’s multiple comparisons test, ***P<0.001, ****P<0.0001.
(I) ZMEL1 treated with 25μM of the Bace2 inhibitor PF-06663195 for 48 hours have increased migration in Transwell assay. Data are from five independent experiments, two-tailed t test, ***P<0.001. All bar graphs are presented as mean ± s.e.m. Scale bars, 10 μM (A 120X, E), 30uM (A 40X), 100μM (A 10X, B left panel), 200μM (B right panel).
To further confirm this, we utilized the ZMEL1 cell line, which is a zebrafish-specific melanoma cell line expressing high levels of bace2 along with other key pigment genes such as mitfa, dct, and tyr (Heilmann et al., 2015; Kim et al., 2017). We treated the ZMEL1 cells with the Bace2 inhibitor PF-06663195, and found that inhibition of Bace2 in ZMEL1 cells caused a dose-dependent elongated/dendritic phenotype (Figure 3E–F) and a significant increase in cell number (Figure 3G). This increase in cell number is consistent with an increase in proliferation as measured by a significant increase in phospho-histone H3 (pH3) staining (Figure 3H) but not change in apoptosis as measured by caspase 3/7 activity (data not shown). This also lead to a strong increase in cell migration (Figure 3I), similar to what is seen in vivo during fish embryogenesis and metamorphosis. Taken together, these data indicate that Bace2 acts inside melanophores lineage and Bace2 deficiency leads to a hyperdendritic melanophore with increased proliferative and migratory capacity.
Pmela is a substrate for Bace2 but is not responsible for the wanderlust phenotype
Bace2 is a transmembrane aspartic protease that acts as a sheddase, typically cleaving extracellular fragments to be released into the local microenvironment. Numerous BACE2 substrates have been previously described, including PMEL in melanocytes (Rochin et al., 2013; Shimshek et al., 2016) and TMEM27 in pancreatic β cells (Esterházy et al., 2011; Stützer et al., 2013). In mammals, the cleavage of PMEL by BACE2 is required for proper melanosome formation. Similarly, we found that in the zebrafish, Bace2 is sufficient to cleave Pmela (Figure S3A) and loss of Bace2 leads to disrupted melanosomes as seen by electron microscopy (Figure S3B). However, genetic knockout of pmela (Figure S3C), or morpholino knockdown of pmela/pmelb (Burgoyne et al., 2015; Schonthaler et al., 2005) does not phenocopy the hyperdendritic/proliferative phenotype of the wanderlust mutant (Figure S3D–E). This suggests a Pmel-independent role for Bace2 in melanophore patterning and morphology.
Bace2 regulates melanophore dendricity via PI3K/mTOR signaling
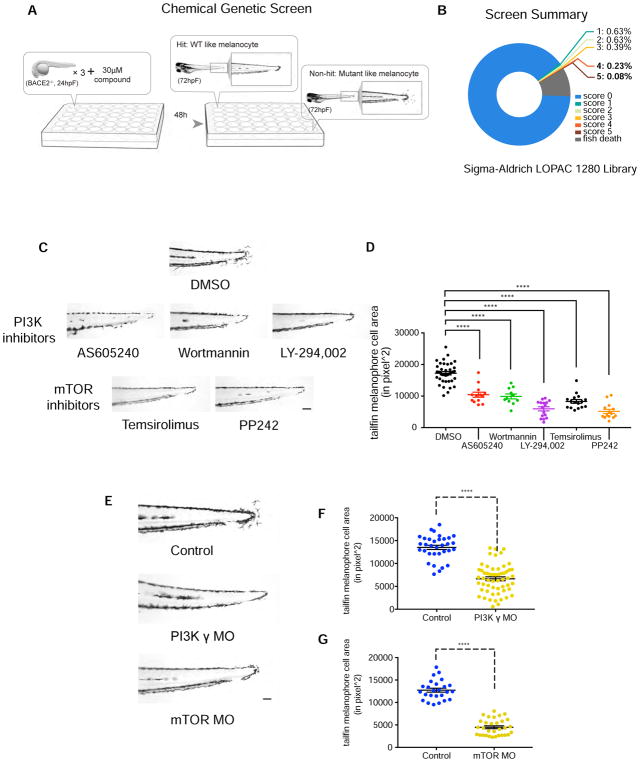
Because sheddases can affect a wide variety of substrates, we therefore wished to functionally look for factors that could rescue the melanophore defects in wanderlust. To do this, we performed a chemical genetic screen to identify suppressors of the wanderlust phenotype (Figure 4A). We screened 1280 chemicals using the Sigma LOPAC library to identify compounds that could rescue the hyperdendritic mutant phenotype. For the screen, we quantified the area of tailfin melanophores at 72hpf and identified chemicals which significant reduced this in the mutant and scored these on a scale from 0 (no rescue) to 5 (complete rescue) (see Figure S4A for examples). Similar to our experience with prior chemical screens (White et al., 2011), we found that most chemicals had either no effect (~90.3%) or were broadly toxic (~8.9%) (Figure 4B). We identified 4 chemical hits which strongly rescued the mutant phenotype, all of which converge on PI3K/mTOR signaling. Three PI3K inhibitors (AS605240, Wortmannin, LY-294,002) and one mTOR inhibitor (Temsirolimus) abrogated the hyperdendritic melanophores phenotype (Figure 4C–D). PP242, another mTOR inhibitor that is structurally distinct from Temsirolimus, phenocopied Temsirolimus. These compounds also rescued the structure of the hyperdendritic membrane structure, as seen using the tyrp1b:membrane-mCherry transgenic line (Figure S4B and Supplemental Movie 1–4). We determined if this rescue could be reversed by performing washout of the drugs at 72hpf, but found that the rescue persisted up to 108hpf (Figure S4C). To a much lesser extent, WT melanophore dendrites are responsive to inhibition of the pathway, especially during the time points when they are normally most dendritic (i.e. 48hpf) (Figure S4D–E).
Figure 4. Bace2 regulates melanophore dendricity via PI3K/mTOR signaling.
(A) Scheme for chemical suppressor screen of the bace2−/− mutant. 24hpf bace2−/− embryos were treated with each compound from the Sigma LOPAC 1280 library at 30μM for 48 hours, in order to identify chemicals which could rescue the melanophore defects.
(B) Compounds were scored with a range of 0 (non-rescued, mutant-like) to 5 (fully rescued, WT-like).
(C) Top hits from the screen (with score of 4 and 5) converge on PI3K/mTOR signaling pathway. PI3K inhibitors AS605240 (110nM), Wortmannin (230nM), LY-294,002 (15μM), and mTOR inhibitors Temsirolimus (30μM), PP242 (15μM) all fully rescue the bace2−/− hyperdendritic melanophores.
(D) Quantification of tailfin melanophore cell area at 72hpf with hits from the screen (n=each fish, one-way ANOVA followed by Holm-Sidak’s multiple comparisons test, ****P<0.0001).
(E–G) Morpholinos knockdown of the PI3K γ isoform and mTOR in bace2−/− mutants rescue the phenotype analogous to what is seen with compounds from the screen, and the resulting tailfin melanophore cell area is quantified in (F) and (G). The data are from three independent experiments. Uninjected bace2−/− siblings (Control) are scored in the same manner, two-tailed t test, ****P<0.0001. All bar graphs are presented as mean ± s.e.m. Scale bar, 100μM. See also Figure S3, Figure S4 and Figure S5.
We performed genetic knockdown experiments to determine the level at which this pathway caused the hyperdendritic phenotype. Amongst the small molecules we identified in our screen, the drug AS605240 was by far the most potent in rescuing the phenotype, which is a relatively selective inhibitor of the PI3K γ isoform. Consistent with this, we found that genetic knockdown of PI3K γ using a previously characterized morpholino (Li et al., 2015; Yoo et al., 2010), but not PI3K α, β or δ, rescued the melanophore dendricity (Figure 4E–F, Figure S5A). Morpholinos to mTOR itself resulted in near complete rescue of the phenotype (Figure 4E, G, Figure S5B–C). This is similar to what has been described for neuronal axon and dendrite patterning, in which PI3K signaling regulates downstream Rac/CDC42 signaling to drive dendrite neuronal morphology (Jiang et al., 2005; Markus et al., 2002; Shi et al., 2003). Our data suggests a specific role for PI3K/mTOR signaling in melanophore dendricity, which is exaggerated in the wanderlust mutant background.
bace2−/− mutants have increased PI3K/mTOR activity
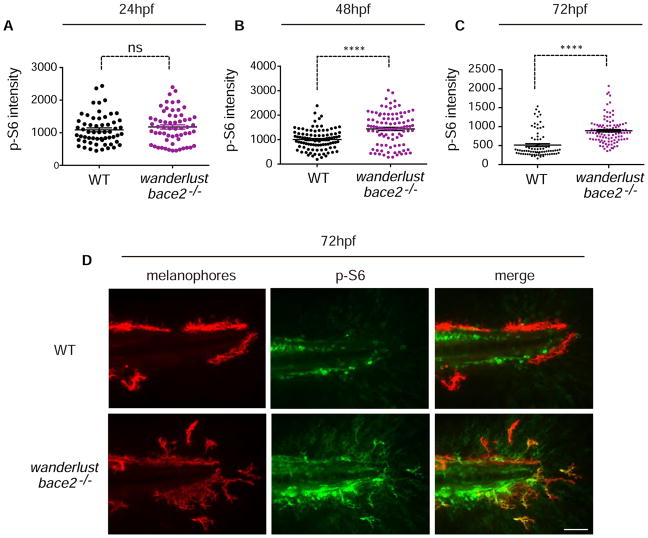
To directly assess whether the hypderdendritic melanophores in the wanderlust mutant had abnormalities of downstream PI3K/mTOR signaling, we performed immunostaining for phospho-S6 ribosomal protein (p-S6) in the WT versus mutant fish. We quantified the p-S6 signal intensity in the melanophores from 24–72hpf by examining co-staining with fluorophore-labelled melanophores. This revealed a significant increase in p-S6 staining in the mutant from 48–72hpf (Figure 5A–C), and showed specific p-S6 staining pattern in the dendrites themselves at 72hpf (Figure 5D). These data are indicative of increased output of the PI3K/mTOR pathway in the hyperdendritic melanophores of the wanderlust mutant.
Figure 5. bace2−/− mutants have increased PI3K/mTOR activity.
Immunostaining against phospho-S6 ribosomal protein(p-S6), a downstream readout of PI3K/mTOR activity, shows elevated p-S6 signal that overlaps with fluorophore-labelled melanophores at 48hpf (B) and 72hpf (C). A representative pictures of 72hpf is shown in (D) and overlapping signals are marked by arrowhead. The data are from three (A), five (B) and five (C) independent experiments, two-tailed t test, ****P<0.0001, ns=not significant. All bar graphs are presented as mean ± s.e.m. Scale bar, 50μM.
The insulin receptor is a substrate for Bace2 and regulates melanophore dendricity
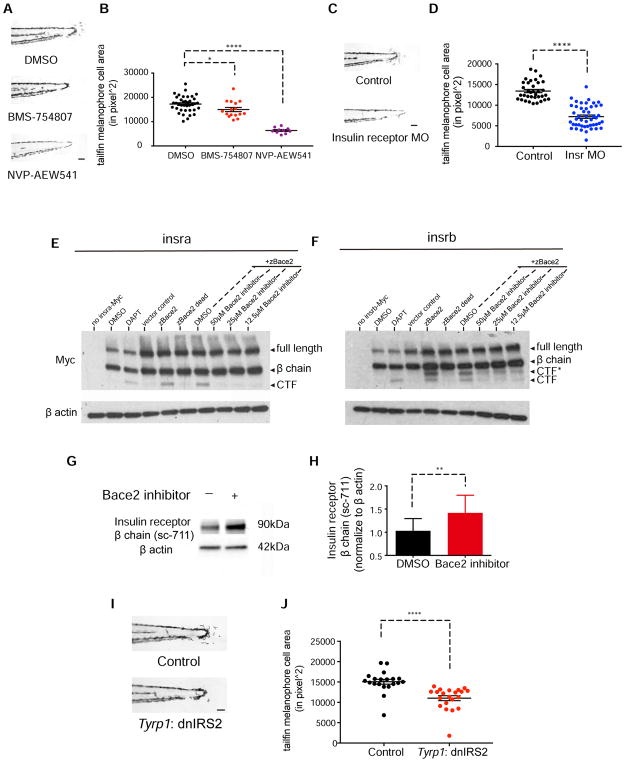
The PI3K/mTOR pathway can be activated by numerous growth factor signaling pathways. We turned our attention to the insulin pathway, a canonical regulator of PI3K, because human BACE2 is known to regulate trafficking of the insulin receptor in pancreatic β cells (Casas et al., 2010) and can cleave the closely related family member IGF2R (Stützer et al., 2013). This led us to hypothesize that insulin receptor signaling drives the PI3K pathway in the hyperdendritic melanophores. To test this, we treated wanderlust mutants with the insulin receptor inhibitors BMS-754807 and NVP-AEW541, and found that both inhibitors were able to rescue the mutant phenotype (Figure 6A–B). Due to a teleost genome duplication, zebrafish have two insulin receptors (Insra and Insrb) (Toyoshima et al., 2008) so we used morpholinos to knockdown both of these receptors. Whereas each receptor itself was able to mildly rescue the mutant phenotype, the combined knockdown was much more effective (Figure 6C–D, Figure S5D).
Figure 6. The insulin receptor is a substrate for Bace2 and regulates melanophore dendricity.
(A–B) Inhibition of the insulin receptor in bace2−/− embryos using the insulin receptor inhibitors BMS-754807 (7.5μM) and NVP-AEW541 (60μM) rescue the hyperdendritic melanophores in the mutant, quantified in (B). bace2−/− embryos are treated from 24–72hpf, and the data are from three independent experiments. One-way ANOVA followed by Holm-Sidak’s multiple comparisons test, *P<0.1, ****P<0.0001.
(C–D) Morpholino knockdown of insulin receptor (Insr) in bace2−/− embryos rescues the hyperdendritic melanophores, quantified in (D). Insra and insrb morpholinos were co-injected into bace2−/− embryos to deplete all insulin receptors. The data are from three independent experiments, two-tailed t test, ****P<0.0001.
(E–F) Bace2 cleaves the insulin receptor. Expression of zebrafish Insra-Myc (E) and Insrb-Myc (F) fusion protein in HEK 293T cells give rise to full length insulin receptor and the β chain. Inhibition of γ secretase using DAPT (2μM, 24 hours treatment) prevents the CTF from being cleaved, leading to CTF accumulation. Expression of zebrafish Bace2 (zBace2) on top of the Insra-Myc fusion protein leads to production of single CTF band (E). zBace2 addition leads to the production of two fragments from Insrb-Myc, one that migrates similarly to the CTF and additional larger fragment designated as CTF* (F). This is not seen with the enzymatic dead version of Bace2 (zBace2 dead) or after Bace2 inhibition with PF-06663195 (24 hours treatment). zBace2 dead was constructed by site-mutagenesis of two conserved catalytic sites aspartic acids into alanines (D98A and D292A).
(G–H) Bace2 inhibition with PF-06663195 (25μM, 24 hours treatment) in ZMEL1 cells, which normally express both Bace2 and the insulin receptor, leads to accumulation of endogenous insulin receptor β chain, quantified in (H). The data are from five independent experiments, two-tailed t test, **P<0.01.
(I–J) Melanophore-specific inhibition of insulin signaling in bace2−/− mutants rescues melanophores hyperdendricity in the tail fin. A mosaic F0 transgenic line was created in which the fugu tyrp1 promoter drives dominant negative insulin receptor substrate 2 (dnIRS2) fused to EGFP in the melanophores (along with a cmlc2: mCherry transgene marker). At 72hpf, tailfin melanophore cell area was quantified in uninjected control (Control) and transgene-positive injected embryos (J). The data are from three independent experiments, two-tailed t test, ****P<0.0001. All bar graphs are presented as mean ± s.e.m. Scale bar, 100μM. See also Figure S5 and Figure S6.
Insulin receptors are broadly expressed in multiple cell types (Garofalo and Rosen, 1988; Toyoshima et al., 2008; Uhlén et al., 2015), including melanophore lineage cells (Kim et al., 2017; Morvan et al., 2012). It is unclear how a general factor such as insulin can exert cell-type specific effects on cells such as melanophores. In mammals, the insulin receptor is produced as a precursor protein which is then proteolytically cleaved by the furin protease into α and β chains, which together form a functional insulin receptor (Bravo et al., 1994). Further cleavage of the β chain to a C-terminal fragment (CTF) has been variously ascribed to a lysosomal protease (Massague et al., 1981), a thiol protease (Knutson, 1991) or ADAM proteases (Kasuga et al., 2007). Given that Bace2 is a transmembrane β protease, we hypothesized that Bace2 is a melanophore-enriched protease that can cleave the insulin receptor β chain to dampen insulin/PI3K signaling. To test this, we expressed a fusion protein of the zebrafish insulin receptor and a C terminal c-Myc tag (Insr-Myc) along with zebrafish Bace2 in HEK293 cells (which do not express any fish proteins). Expression of the Insr-Myc fusion protein alone revealed the full-length insulin receptor (~180 kDa) and the β chain (~90 kDa) product of furin cleavage (Figure 6E–F). This fusion protein is robustly cleaved by concomitant expression of Bace2, as shown by an accumulation of the CTF for both Insra (Figure 6E) and Insrb (Figure 6F). This effect was not seen with the catalytically-dead Bace2, and can also be abolished when cells are treated with Bace2 inhibitor PF-06663195. This demonstrates that Bace2 is sufficient to induce insulin receptor cleavage (Figure S6A).
Since Bace2 can induce β chain cleavage, this raised the possibility that loss of Bace2 in melanophores could serve to stabilize the β chain. To test this, we identified an insulin receptor antibody that recognized the endogenous zebrafish insulin receptor β chain (Figure S5D), and then measured its level after Bace2 inhibition in the ZMEL1 cells. After 24 hours, we found a 37% increase in β chain of the insulin receptor in the Bace2 inhibited cells (Figure 6G–H), suggesting that Bace2 normally acts to limit insulin receptor levels on melanophores.
To directly test whether insulin receptor signaling itself is responsible for the wanderlust phenotype, we created a transgenic construct in which the fugu tyrp1 (Zou et al., 2006) promoter drives a dominant negative insulin receptor substrate 2 (dnIRS2) to inhibit insulin signaling only in the melanophores. We then quantified tail melanophore dendricity as we did in the screen, and found a significant decrease in the transgenic animals compared to controls (Figure 6I–J). We also found that CRISPR knockout of insra/insrb in ZMEL1 cells led to a significant decrease in the elongated/dendritic phenotype and cell number (Figure S6B–F). Taken together, these data are consistent with abnormal insulin receptor signaling induced by loss of bace2.
Insulin b is the upstream ligand that drives melanophore dendricity in wanderlust
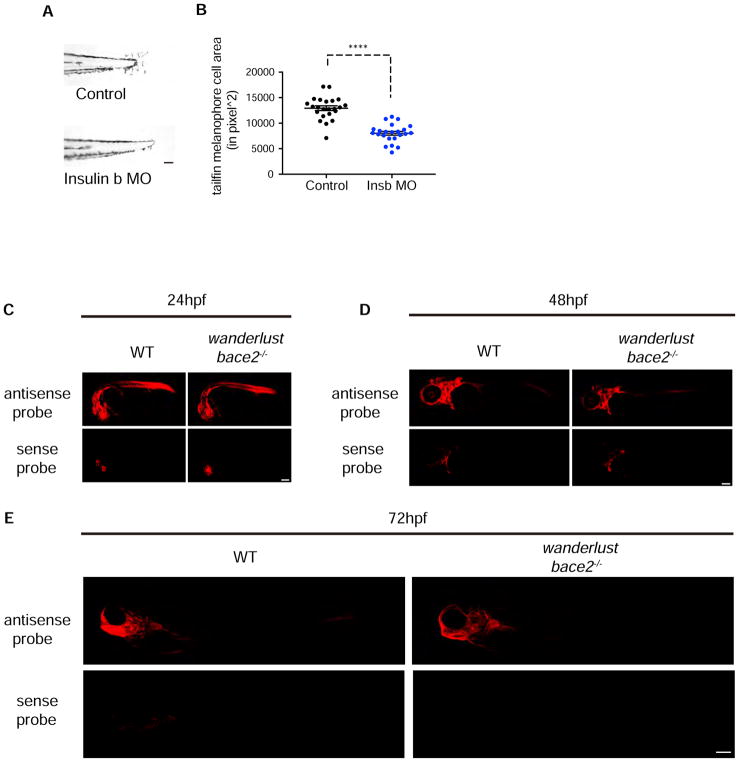
Finally, we wished to determine the source of the insulin ligand driving the wanderlust phenotype. Similar to the receptors, zebrafish have two insulin ligands - Insa and Insb (Papasani et al., 2006), and we knocked these down using morpholinos. Knockdown of Insa had little to no effect, whereas knockdown of Insb had a significant rescue of the wanderlust phenotype (Figure 7A–B, Figure S5E).
Figure 7. Insulin b is the upstream ligand that drives melanophore dendricity in wanderlust.
(A–B) Morpholino knockdown of the ligand insulin B (insb) in bace2−/− embryos rescues the hyperdendritic melanophores, quantified in (B). The data are from three independent experiments, two-tailed t test, ****P<0.0001.
(C–E) Fluorescent ISH shows zebrafish insb ligand mRNA is initially expressed ubiquitously at 24hpf and then restricted to the head/brain region (arrowhead) in bace2−/− embryos at 48–72hpf, sense probe controls for background signal. All bar graphs are presented as mean ± s.e.m. Scale bar, 100μM (A), 150μM (C, D), 200 μM (E). See also Figure S5 and S7.
In humans, insulin is primarily produced in the pancreatic β-cells, whereas in teleosts and Drosophila, insulin peptides can be produced from other areas including the brain (Papasani et al., 2006). Whether humans produce insulin in the brain, or it is imported into the brain from the periphery, has been a source of some controversy although it is clear that insulin can have CNS-specific effects (Havrankova et al., 1979; Schwartz et al., 1992). To assess this in zebrafish, we performed whole-mount ISH for insa and insb ligand mRNA, and found that as predicted the insa transcript is exclusively produced in the pancreas (Figure S7A–B). In contrast, insb is initially expressed ubiquitously early in development but becomes gradually restricted to the head region and brain by 48–72hpf (Figure 7C–E). To functionally exclude a role for pancreatic insulin in the wanderlust dendricity phenotype, we used morpholinos against pdx1 (Kimmel et al., 2015) and hb9 (Harrison et al., 1999; Kimmel et al., 2011) to completely ablate the pancreas. Whereas this maneuver led to a complete loss of Insa in the pancreas (Figure S7C), it had no effect on the wanderlust dendricity phenotype (Figure S7D). In addition, we find no consistent change in the size of the pancreas in the wanderlust mutants (Figure S7E). These data indicate that it is the Insb ligand, which is enriched in the head, that drives the melanophore dendricity, highlighting how a long-range factor can influence pigment patterning.
Taken together, this data demonstrates that the sheddase Bace2 modulates melanophore morphology in a cell-type specific manner by regulating insulin receptor cleavage and accumulation, which alters sensitivity to distant ligands such as insb (see graphical abstract for model).
DISCUSSION
It is increasingly recognized that vertebrate coloration is due to continuous interactions between pigment producing cells and their neighboring cells (Nakamasu et al., 2009; Eom et al., 2015; Walderich et al., 2016). Although this has long been considered to consist primarily of short-range interactions (i.e. melanophore-iridophore, etc) (Frohnhöfer et al., 2013; Patterson and Parichy, 2013), recent data has highlighted a role for thyroid hormone in pigment patterning (McMenamin et al., 2014), and Aqp3a as a pore that can communicate with the external environment (Eskova et al., 2017). What has remained unclear is the manner in which long-range environmental factors interact with lineage-specific gene programs to regulate pigment pattern formation. Our data would suggest that a distant hormonal signal, insulin, can play a major role in this patterning by interacting with a melanophore-enriched gene, bace2.
A major question our study addresses is how circulating hormonal factors like insulin can ultimately yield cell-type specific phenotypes. One explanation for such specificity is that each cell type has a set of lineage-restricted genes that regulates the level and activity of the insulin receptor. In the present study, we identify Bace2 as one such candidate lineage factor in melanophores. Our data indicates a model in which the endogenous role of Bace2 in melanophores is to dampen insulin/PI3K signaling, and loss of Bace2 activity leads to hyperactivation of the pathway. This would be consistent with the phenotypes we observe, including hyperdendritic, proliferative and migratory melanophores. PI3K/mTOR signaling is well known to regulate dendrite formation in other cell types such as neurons (Jiang et al., 2005; Markus et al., 2002; Shi et al., 2003). Inactivation of PTEN, a negative regulator of PI3K, leads to highly migratory and metastatic melanoma cells (Dankort et al., 2009).
Sheddases such as Bace2 are membrane-bound proteases that cleave the extracellular domain of transmembrane proteins and shed the soluble ectodomain extracellularly. They can act as both positive or negative regulators of downstream signaling. Sheddases such as ADAM17 (also known as TACE) cleave and activate EGF ligands, and loss of ADAM phenocopies EGF ligand mutants (Luetteke et al., 1993; Peschon et al., 1998). Similarly, cleavage of the Notch receptor by ADAM10 and ADAM17 is required for release of the notch intracellular domain (NICD), where it can then move to the nucleus to influence downstream gene transcription (Brou et al., 2000; van Tetering et al., 2009). In contrast, sheddases can also acts as negative regulators of receptor activity by quenching free ligands using the ectodomain from cleaved receptors. For example, the soluble form of the Mer proto-oncogene tyrosine kinase (MerTK) can bind to its ligand to limit Akt signaling downstream of endogenous MerTK (Cai et al., 2016; Sather et al., 2007). A more straightforward way to dampen down receptor activity is to physically cleave the receptor proteins. ADAM10 inhibition leads to hepatocyte growth factor receptor (HGF-R/MET) accumulation and subsequent activation by its ligand HGF in liver metastasis (Kopitz et al., 2007; Miller et al., 2013; Schelter et al., 2011). Similarly, ADAM10 and ADAM17 can proteolytically cleave receptor tyrosine kinases such as AXL, and loss of ADAM cleavage leads to AXL accumulation on cell membrane and results in increase in mitogenic signaling activity (Miller et al., 2016; Orme et al., 2016). Our data suggests that Bace2 primarily acts as a negative regulator of insulin signaling by proteolytically cleaving the insulin receptor. Interestingly, it has been shown that downregulation of RTK cleavage can be hijacked by tumor cells to acquire drug resistance, suggesting that sheddases are also under dynamic feedback regulation (Miller et al., 2016).
In mammals, BACE2 has been shown to be enriched in pancreatic beta cells, where where it cleaves TMEM27 and leads to increased secretion of insulin (Esterházy et al., 2011). BACE2 is also known to be expressed in mammalian melanocytes, where it serves to cleave PMEL to achieve melanosome maturation (Rochin et al., 2013; Shimshek et al., 2016). In contrast, our studies highlight a Pmel independent role for Bace2 in pigmentation, since we find that loss of function of pmela/b does not resemble the zebrafish wanderlust mutant. Instead, our data shows that it is another substrate, the insulin receptor, which mediates the pigment phenotype. This highlights the functional plasticity of sheddases, which can have numerous different substrates to regulate cell-type specific effects. Whether Bace2 is important for zebrafish pancreas development remains to be determined, but is unlikely to play a role in pigment patterning since ablation of the pancreas with pdx1/hb9 morpholinos failed to rescue the wanderlust phenotype. Moreover, we did not see a strong signal of bace2 in pancreas during zebrafish embryogenesis by ISH, although this will need further study in the future. Insb is dynamically expressed. At 24hpf it is expressed fairly ubiquitously, but then becomes gradually restricted to the head region by 48–72hpf. We did not see a strong enrichment of insb in the pancreas as noted before (Papasani et al., 2006). Based on this, we also tried the same insb probe from (Papasani et al., 2006) but were not able to consistently see a greater signal in the antisense versus sense probe in the pancreas. Although we do not understand the nature of this discrepancy, we decided to primarily focus on the probe published by (Junker et al., 2014) for our studies since it correlated closely with their RNA-seq topography analysis and consistently gave us an excellent antisense:sense signal.
It is likely that insulin cooperates with other long-range endocrine-related factors such as thyroid hormone (McMenamin et al., 2014). Although not specifically addressed in our study, one interesting consideration is how hyperinsulinemia leads to hyperpigmentation in humans, a well described phenomenon that typically occurs as part of the disease acanthosis nigricans (Hermanns-Lê et al., 2004). This is a condition specifically linked to other endocrinopathies such as diabetes mellitus, hypothyroidism, and polycystic ovarian syndrome, and thus the melanocyte responsiveness to insulin may be part of this pathophysiology.
BACE inhibitors are being evaluated in clinical trials, primarily for the prevention or treatment of Alzheimer’s disease. This is due to the fact that BACE1 can cleave amyloid precursor protein, thought to be a major target in that disease. In contrast, BACE2 inhibitor may have utility for the treatment of diabetes, due to its capacity for increasing insulin secretion (Esterházy et al., 2011). Many of the molecules developed so far have some but not complete selectivity for either isoform. Our data would suggest that inhibition of BACE2, in the context of hyperinsulinemia, may have effects on pigmentation via the PI3K pathway, and may increase the migratory capacity of these hyperdendritic melanocytes. Whether this has clinical consequences, especially in regards to melanoma development, remains to be determined, but is an important area for future exploration.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
All materials, data and associated protocols will be available to readers promptly and without undue qualifications. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Richard White (whiter@mskcc.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish
Fish Husbandry
Fish stocks were reared under standard conditions at 28.5°C under 14:10 light:dark cycles. Healthy, immune-competent male and female adults of the AB strain were used for breeding (animals were typically ~8 months when bred). Animals were fed standard zebrafish diet consisting of brine shrimp followed by Zeigler pellets. For all mutant studies, we compared bace2−/− siblings to AB siblings derived from an initial incross of the AB and bace2−/− strain. Sex determination in embryos is not possible at the stages used in this study. Embryos were collected from natural mating and incubated in E3 buffer (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) at 28.5°C. The bace2−/− fish were obtained from the Wellcome Trust Sanger Institute.
Transgenic lines
One-cell-stage bace2−/− embryos were injected with the given DNA construct Tg(fugu dct:Z-bace2; crystallin:YFP) at 25 ng/μl with Tol2 mRNA at 20 ng/μl. Embryos were screened at 3 days post fertilization for the presence of yellow fluorescent protein (YFP) in the eye lens. YFP positive embryos were grown to adulthood and outcrossed so as to identify founders that gave germline transmission. To quantify melanophores rescue, founders were outcrossed to bace2−/− and the F1 were screened for melanophores rescue and the presence of YFP positive lenses.
One-cell-stage bace2−/− embryos were injected with the given DNA construct Tg(fugu tyrp1:dnIRS2-EGFP; cmlc2: mCherry) at 25 ng/μl with Tol2 mRNA at 20 ng/μl. Embryos were screened at 30hpf for the presence of mCherry in the heart. dnIRS2(Ye et al., 2016) is a previously published dominant negative insulin receptor substrate 2 that non-productively binds to the insulin receptor to inhibit insulin signaling. Tg(tyrp1b:membrane-mCherry) and Tg(mitfa:EGFP) fish strains were generous gifts from Dr. David Parichy and Dr. James Lister respectively. bace2−/− was crossed into these strains to generate bace2−/−; Tg(tyrp1b:membrane-mCherry) and bace2−/−; Tg(mitfa:EGFP).
Cell Lines
The ZMEL1 cell line(Heilmann et al., 2015) and HEK 293T cell line were maintained as previously described. All cells were cultured in DMEM with 10% FBS/penicillin/streptomycin/glutamine. The sex of the animal from which the ZMEL1 line was derived is unknown. HEK 293T cells are derived from a female fetus.
METHOD DETAILS
Genotyping of bace2−/− Fish
Tail clips from adult zebrafish were placed in microcentrifuge tubes or thermal cycler plates containing 50μl of 50 mM NaOH. Samples were boiled at 95°C for 30min, then cooled down with 5μl of 1M Tris-HCL (pH=8.0). A 1:10 dilution of the supernatant was used in PCR. Primer sequences for PCR are listed in Table S2. DNA was PCR amplified with Promega GoTag green mastermix and digested using Hpy188I enzyme (NEB catalog # R0617S). Limit Hpy188I digestion time to one hour to avoid non-specific digestion (star activity). bace2−/− confers a C to A mutation which abolishes the Hpy188I digestion site. WT fish have two bands at 450bp and 240bp, bace2−/− fish have one band at 690bp, bace2+/− fish have three bands at 240bp, 450bp and 690bp.
Chemical Screen
The chemical screen was performed similar to prior publications(Kaufman et al., 2009). We aimed to identify suppressors of the bace2−/− melanophores phenotype by treating bace2−/− embryos from 24hpf to 72hpf with 30μM chemicals from Sigma LOPAC 1280 library. Chemicals were prepared from 10mM stock and diluted into 30μM in 300μl E3 with 0.3% (v/v) DMSO. Embryos were staged at both shield stage and 24hpf prior to drug treatment. Three embryos per well in 48-well-plates were scored. Eight DMSO-only wells were included in each plate as negative control. Plates were foil wrapped to protect from light and incubated at 28.5°C from 24hpf to 72hpf. At 72hpf, embryos were anaesthetized with tricaine and were subject to direct observation under light microscope. Melanophores rescue was scored from 0–5, with 0 to be mutant-like melanophores (i.e. no rescue) and 5 to be completely rescued into WT-like melanophores. Chemicals with scores >3 were subject to a second round of validation, with different aliquots of the library. Positive hits were ordered from Sigma and were tested for minimal concentration required to rescue bace2−/− phenotype.
Embryo Drug Treatment
Embryos were treated with the following chemicals (with working concentration, treatment time and catalog number). All chemicals are dissolved in DMSO.
PF-06663195 (Bace2 inhibitor): 100μM, various treating time (Sigma, catalog # PZ0262).
AS605240: 110nM, 24hpf-72hpf (Sigma, catalog # A0233)
Wortmannin: 230nM, 24hpf-72hpf (Sigma, catalog # W1628)
LY-294,002: 15μM, 24hpf-72hpf (Sigma, catalog # L9908)
Temsirolimus: 30μM, 24hpf-72hpf (Sigma, catalog # PZ0020)
PP242: 15μM, 24hpf-72hpf (Abcam, catalog # ab141405)
BMS-754807: 7.5μM, 24hpf-72hpf (Sigma, catalog # BM0003)
NVP-AEW541: 60μM, 24hpf-72hpf (Selleck Chemicals, catalog # S1034)
PTU: 300μM, 3dpF-5dpF (Sigma, catalog # P7629).
Morpholino Injections
All the morpholino oligonucleotides (MO) were synthesized by Gene ToolsTM LLC, resuspended in sterile water at a concentration of 1mM stock. MO stock was further diluted into working concentrations in sterile water and injected into zebrafish embryos at the one-cell stage by microinjection, at ~1.5nL/embryo. MO working concentration, sequence and reference information are listed in Table S1.
The efficacy of splicing morpholinos below have been confirmed using Reverse transcription PCR (RT-PCR), with primer information listed in Table S2.
The efficacy of mTOR splicing MO has been also confirmed using WB, with primary antibody: mTOR (7C10) Rabbit mAb (Cell Signaling Technology, catalog # 2983S). The efficacy of Insulin receptor ATG MO has been confirmed using WB, with primary antibody: insulin R β (C-19) (Santa Cruz Biotechnology, catalog # sc-711). The efficacy of Pdx1 and Hb9 ATG MO have been confirmed indirectly using immunostaining against Insulin protein, with primary antibody: insulin (Genetex, catalog # GTX128490).
Measurement of Tailfin Melanophore Cell Area
The extent of melanophore dendricity in the tailfin was calculated as the area of melanophores covering the tailfin mesenchyme. This was quantified by thresholding the mCherry intensity (in the context of Tyrp1: membrane-mCherry; bace2−/− for Figure 1G, 4D and 6B) or thresholding the bright field intensity (in the context of bace2−/− for Figure 4F/G, 6B, 6J and 7B) to highlight the pixels that represent melanophores in each image, and measuring the area they cover with FIJI software.
ZMEL1 phospho-Histone H3 (pH3) Immunostaining
ZMEL1 were seeded at 6000 cells/well in white wall, clear bottom 96-well plates (Fisher Scientific, catalog # 07-200-566) in DMEM/10% FBS/penicillin/streptomycin/glutamine. The cells were allowed to adhere overnight, and then media were changed to Bace2 inhibitor (PF-06663195) containing media at 12.5μM or 25μM with 0.025% (v/v) DMSO. Cells were grown for a total of 3 days after the addition of Bace2 inhibitor. At the end of the treatment, the cells were fixed with paraformaldehyde (4% in 1X PBS), permeabilized with 0.1% Triton-X-100 for 15 min at room temperature. Cells were rinsed with 1X PBS three times and then blocked with 1X PBS/5% goat serum/0.3% Triton X-100 for 1 hour at room temperature, and then stained with anti-pH3 primary antibody (1:1000, EMD Millipore, catalog # 05-806) diluted in antibody dilution buffer (1X PBS/1% BSA/0.3% Triton X-100) at 4°C overnight. Cells were rinsed with 1X PBS three times the next day and then stained with anti-mouse Alexa 647 (1:1000, CST, catalog # 4410) diluted in antibody dilution buffer for 1 hour at room temperature in the dark. Cells were rinsed three times with 1X PBS and stained with Hoechst 33342 (4μg/ml, Thermo Fisher Scientific, catalog # H3570) for 30 min at room temperature in the dark. Cells were rinsed three times with 1X PBS and imaged using a GE INCell 6000. 6 fields were captured per well using a Nikon 10X Plan Apo objective, 0.45NA. Images of the Hoechst signal was captured via the DAPI channel in order to identify the nucleus. The pH3 was captured via the far-red channel to identify the mitotic cells. The image analysis script was performed within the Perkin Elmer Columbus software. Percentage of mitotic cells were quantified by calculating pH3 positive cells as a fraction of the total number of cells in each well.
RT-PCR
Zebrafish embryos were homogenized in 1ml of Trizol (Life Technologies, catalog #15596 - 018) on ice using Bel-Art micro-tube homogenizer (Fisher Scientific, catalog # 03-421-215) for one minute, with a pool of 5 to 50 embryos at 3dpF. Then total RNA from zebrafish embryos were extracted using Direct-zol RNA miniprep kit (Zymo Research, catalog # R2050). cDNA library was constructed using SuperScript III First-Strand kit (Thermo Fisher Scientific, catalog # 18080-400). cDNA from both the control embryos and the morpholino injected embryos were used in downstream PCR reaction to confirm the efficacy of morpholino knockdown.
Cell Counting for Metamorphosis Zebrafish
24dpF age matched and size matched siblings were treated with epinephrine to contract pigment granules and also treated with 4mg/ml tricaine before imaging. Fish were tile scanned (22μm z step) using an upright Zeiss Discovery V16 equipped with a motorized stage in brightfield. Images were acquired with the Zeiss Zen software v1. Ten somites from dorsal and ventral sides of one fish (start the first somite in the beginning of dorsal fin, and count posteriorly) were defined as region of interest (ROI). Two artificial lines were drawn parallel to the vertebrate and aligned their one ends to the tail fin. Region inside of the two lines are defined as intra-stripe region, as WT zebrafish form melanophores mainly inside of this region; region outside of the two lines are defined as inter-stripe region. Inter-stripe melanophores reside within ROI were counted manually.
in situ Hybridization (ISH)
ISH were performed as described in(Thisse and Thisse, 2008). Embryos were fixed at 24hpf, 48hpf and 72hpf. Probes were diluted to a working solution of 0.8 ng/μl. Staining time varied between probes from couple hours at room temperature, or overnight at 4°C. Fluorescent ISH (FISH) was performed as described in(Welten et al., 2006). In brief, post methanol fixation embryos are treated with 3% H2O2 in methanol for 20 min at room temperature to inhibit endogenous peroxidase activity. Then FISH follows the same procedure with regular ISH except those modifications: performing all hybridization and wash steps at 55°C instead of 70°C, with 0.25ng/μl for both insb antisense and sense probe. Dilute anti-DIG-POD antibody (1:2000, Sigma-Aldrich, catalog # 11207733910) for anti-DIG-HRP detection in antibody buffer and pre-adsorb for 2 h at room temperature with slow agitation. Incubate embryos overnight in anti-DIG-POD antibody and next day wash 6 times for 15 minutes in PBST at room temperature. Dilute TSA Plus Cyanine 3 reagent (1:50, PerkinElmer, catalog # NEL744001KT) in the amplification buffer supplied and incubate embryos with diluted TSA/Cyanine 3 reagent for 30min at room temperature in the dark. Wash 8 times for 15 minutes in PBST and then image embryos. double FISH (dFISH) follows the same procedure with single probe FISH except for the following modifications. Hybridization at 65°C with bace2 riboprobe (3ng/μl) labelled with DIG (Sigma-Aldrich, catalog # 11277073910) and crestin riboprobe (2.92ng/μl) labelled with fluorescein (fluorescein-12-UTP, Sigma-Aldrich, catalog # 11685619910) in hybridization mix (HM+) with 5% dextran sulfate (Sigma-Aldrich, catalog # D8906-5G) to allow two targets to be visualized simultaneously. Embryos were then incubated overnight at 4°C with anti-DIG-AP antibody (1:5000, Sigma-Aldrich, catalog # 11093274910) and anti-fluorescein-HRP antibody (1:500, Sigma-Aldrich, catalog # 11426346910) followed by extensive washes in PBST. To detect HRP (crestin), embryos were incubated with TSA Plus fluorescein evaluation kit (1:50, PerkinElmer, catalog # NEL741E001KT) for 30 min in the dark in amplification buffer. Following the addition of 0.001% H2O2 signal was allowed to develop for 30 min. After two washes with PBST, Fast Red (Sigma, catalog # F4648) was used according to the manufacturer’s instructions to detect the AP (bace2). The embryos were then extensively washed 4×15min in PBST, and DAPI was used as a nuclear counterstain. Mount embryos in low melting agarose for imaging acquisition. Tile scan images were collected with a Leica TCS SP5 II (DMI6000 CFS; acquisition software Leica Application Suite AF v. 2.6.3.8173). Confocal images were acquired at 1,024 × 1,024 pixels (20.0-μm z step) using a 10X and 40X objectives, with zoom-in factor of 3 for 120X defection. The 3D reconstructions of the Insb, bace2 and crestin expression were performed using Imaris software.
Dct (Kelsh et al., 2000), mitfa (Lister et al., 1999), tyr (Mellgren and Johnson, 2004), sox10 (Dutton et al., 2001), crestin (Luo et al., 2001) and c-kit (Parichy et al., 1999) probes were generous gifts from Dr. David Parichy. Pmela (Schonthaler et al., 2005), ednrb1a (Parichy et al., 2000) and foxd3 (Odenthal and Nüsslein-Volhard, 1998) probes were generous gifts from Dr. James Lister. Tyrp1b probe DNA template was PCR amplified from MGC clone plasmid (Thermo Fisher Scientific, catalog # EDR1052-210947471), and in vitro transcribed using T3 RNA polymerase.
Bace2 probe DNA template was PCR amplified from plasmid pENTR-z-bace2, and in vitro transcribed using T3 RNA polymerase.
Insa probe was validated by (Thisse, B., Thisse, C. (2004) Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission (http://zfin.org).). Insa probe DNA template was PCR amplified from MGC clone plasmid (Dharmacon, catalog # MDR1734-202792960), and in vitro transcribed using T3 RNA polymerase.
Insb probe was validated by (Junker et al., 2014). Insb probe DNA template was amplified from zebrafish cDNA library and then TOPO TA cloned into pCR™ II-TOPO TA vector. The insb antisense probe was PCR amplified and then in vitro transcribed using T3 RNA polymerase. The insb sense probe (negative control) was digested with HindIII then in vitro transcribed using T7 RNA polymerase. Primer sequence is listed in Table S2. Probe sequence, source and reference information is listed in Table S3.
Embryos Protein Immunofluorescence
Embryos were kept in 300μM PTU to inhibit melanin synthesis. Embryos were fixed with 4% paraformaldehyde in PBS overnight at 4°C, then dehydrated in 100% methanol overnight at −20°C. Embryos were rehydrated with 50 % methanol/PBS then into PBST (PBS with 0.1% Tween-20) for 5min each, then permeabilized with 100% acetone at −20°C for 12 min (for 24hpf embryos) or 20 min (for 2–3dpf embryos). Embryos were blocked with blocking buffer (PBST+1%DMSO+4%BSA) for 1 hour at room temperature. Antibodies were diluted in blocking buffer, applied to embryos and incubated overnight at 4°C. Embryos were then washed with washing buffer (PBST+1%DMSO) twice every 15 minutes for 2 hours at room temperature. Embryos were Incubate with secondary antibody in blocking buffer for 2 hours at room temperature in dark. Embryos were then washed with washing buffer twice every 15 minutes for 1 hours at room temperature. For insulin staining: yolk was manually dissected off to visualize insulin staining in pancreas. For pS6 staining: pS6 signal co-localized with melanophores was measured to identify melanophore specific PI3K/mTOR activity. For 24hpf and 48hpf embryos, the transgenic mitfa: EGFP line was used to mark melanophores in EGFP. For 72hpf embryos, the transgenic tyrp1b: membrane-mCherry line was used to mark melanophores in mCherry. (This is because tyrp1b promoter is not on at 24hpf, and mitfa promoter is off at 72hpf). Embryos were mounted on agarose plate for imaging acquisition. Primary antibody: rabbit anti-zebrafish insulin (1:200, Genetex, catalog # GTX128490), rabbit anti-phospho-S6 Ribosomal Protein (Ser240/244) (D68F8) (1:800, CST, catalog # 5364), mouse anti-GFP (1:500, abcam, catalog # ab1218), chicken anti-mCherry (1:1000, abcam, catalog # ab205402). Secondary antibodies: Alexa-Fluor 488 anti-rabbit (1:200, CST, catalog # 4412), Alexa-Fluor 594 anti-rabbit (1:200, CST, catalog # 8889), Alexa-Fluor 594 anti-chicken (1:200, abcam, catalog # ab150176), Alexa-Fluor 488 anti-mouse (1:200, CST, catalog # 4408).
Creation of pmela CRISPR Mutant
Creation of pmela CRISPR knockout mutant was adapted from(Gagnon et al., 2014). Guide RNA (gRNA) was designed against the Pmela coding sequence, and synthesized via in vitro transcription. The gRNA was incubated with purified recombinant Cas9-3NLS protein (IDT, catalog # 1074181), and these were coinjected (300ng/μl gRNA+1μg/μl protein) into single cell zebrafish embryos, with 1.5nL injection volume per embryo. F0 embryos displayed mosaic knockout phenotype similar to fading vision mutant(Schonthaler et al., 2005), and were raised into adulthood. F0 founders were incrossed to yield trans-heterozygous F1 fish. Pmela gRNA sequence is listed in the Key Resource Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-zebrafish insulin | Genetex | cat # GTX128490 |
| rabbit anti-mTOR (7C10) | Cell Signaling Technology | cat # 2983S |
| rabbit anti-insulin R β (C-19) | Santa Cruz Biotechnology | cat # sc-711 |
| mouse anti-c-MYC | Santa Cruz Biotechnology | cat # sc-40 |
| mouse anti-FLAG | Sigma-Aldrich | cat # F3165 |
| rabbit anti-mouse IgG H&L (HRP) | Abcam | cat # ab97046 |
| goat anti-rabbit IgG H&L (HRP) | Abcam | cat # ab97051 |
| mouse anti-phospho-Histone H3 (Ser10) | EMD Millipore | cat # 05-806 |
| rabbit anti-phospho-S6 Ribosomal Protein (Ser240/244) | Cell Signaling Technology | cat # 5364 |
| mouse anti-GFP | Abcam | cat # ab1218 |
| chicken anti-mCherry | Abcam | cat # ab205402 |
| sheep anti-DIG-POD | Sigma-Aldrich | cat # 11207733910 |
| sheep anti-DIG-AP | Sigma-Aldrich | cat # 11093274910 |
| sheep anti-fluorescein-POD | Sigma-Aldrich | cat # 11426346910 |
| Alexa-Fluor 647 anti-mouse | Cell Signaling Technology | cat # 4410 |
| Alexa-Fluor 488 anti-rabbit | Cell Signaling Technology | cat # 4412S |
| Alexa-Fluor 594 anti-rabbit | Cell Signaling Technology | cat # 8889 |
| Alexa-Fluor 594 anti-chicken | Abcam | cat # ab150176 |
| Alexa-Fluor 488 anti-mouse | Cell Signaling Technology | cat # 4408 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| PF-06663195 (Bace2 inhibitor) | Sigma-Aldrich | cat # PZ0262 |
| AS605240 | Sigma-Aldrich | cat # A0233 |
| Wortmannin | Sigma-Aldrich | cat # W1628 |
| LY-294,002 | Sigma-Aldrich | cat # L9908 |
| PP242 | Abcam | cat # ab141405 |
| BMS-754807 | Sigma-Aldrich | cat # BM0003 |
| NVP-AEW541 | Selleck Chemicals | cat # S1034 |
| PTU | Sigma-Aldrich | cat # P7629 |
| Temsirolimus | Sigma-Aldrich | cat # PZ0020 |
| DAPT | Sigma-Aldrich | cat # D5942-5MG |
| T3 RNA Polymerase | Promega | cat # P2083 |
| T7 RNA Polymerase | Promega | cat # P2075 |
| DIG RNA Labeling Mix | Sigma-Aldrich | cat # 11277073910 |
| Fluorescein RNA Labeling Mix | Sigma-Aldrich | cat # 11685619910 |
| Fast Red TR/Naphthol AS-MX Tablets | Sigma-Aldrich | cat # F4648-50SET |
| Hpy188I restriction endonuclease | NEB | cat # R0617S |
| HCS CellMask Deep Red Stain | Thermo Fisher Scientific | cat # H32721 |
| Surveyor nuclease | IDT | cat # 706020 |
| TracrRNA ATTO™ 550 | IDT | cat # 1075928 |
| Alt-R® Cas9 Electroporation Enhancer | IDT | cat # 1075916 |
| recombinant Cas9-3NLS protein | IDT | cat # 1074181 |
| Critical Commercial Assays | ||
| TSA/Cyanine 3 reagent | PerkinElmer | cat # NEL744001KT |
| TSA Plus fluorescein evaluation kit | PerkinElmer | cat # NEL741E001KT |
| NEON transfection system | Thermo Fisher Scientific | cat # MPK10096 |
| Experimental Models: Cell Lines | ||
| ZMEL1 | S. Heilmann et al. 2015. Cancer Research | |
| HEK 293T | ATCC | cat # CRL-3216 |
| Experimental Models: Organisms/Strains | ||
| bace2−/− | Wellcome Trust Sanger Institute | Allele name: hu3332 |
| bace2−/−; Tg(fugu dct:Z-bace2; crystalline:YFP) | This paper | |
| bace2+/+; Tg(tyrp1b:membrane-mCherry) | Eom et al. 2015. eLife | |
| bace2−/−; Tg(tyrp1b:membrane-mCherry) | This paper | |
| bace2+/+; Tg(mitfa:EGFP) | Curran K et al. 2009 Dev Biol. | |
| bace2−/−; Tg(mitfa:EGFP) | This paper | |
| bace2−/−; Tg(fugu tyrp1:dnIRS2-GFP; cmlc2:mCherry) | This paper | |
| Oligonucleotides | ||
| Pmela gRNA | IDT | ATTTAATACGACTCACTATAGAATGAACTTGTCTTTGCCGGTTTAAGAGCTATGCTGG |
| Insra Alt-R gRNA | IDT | TTGGTCATCAACATCCGCGG |
| Insrb Alt-R gRNA | IDT | CGTGGAGAAACGCTGGACGG |
| Non-target Alt-R gRNA | IDT | AACCTACGGGCTACGATACG |
| See Table S1 for morpholinos working concentration, sequence and reference | ||
| See Table S2 for primer sequence | ||
| See Table S3 for in situ hybridization (ISH) probes, source and reference | ||
| Recombinant DNA | ||
| Fugu dct: Z-bace2; crystallin:YFP | This paper | |
| CMV: Z-bace2-2a-GFP-394 | This paper | |
| CMV: Z-bace2 enzymatic dead-2a-GFP-394 | This paper | |
| CMV: Z-insra-Myc-394 | This paper | |
| CMV: Z-insrb-Myc-394 | This paper | |
| CMV: tdtomato-394 | This paper | |
| CMV: Z-pmela-FLAG | This paper | |
| Fugu tyrp1:dnIRS2-EGFP; cmlc2: mCherry | This paper | |
| Software and Algorithms | ||
| Columbus software | Perkin Elmer | http://www.perkinelmer.com/product/image-data-storage-and-analysis-system-columbus |
| CHOPCHOP | http://chopchop.cbu.uib.no/# | |
Insa Positive Cell Area Measurement
Insa ISH pictures were used to measure the insa positive cell area. Insa positive cell area is quantified by thresholding the bright field intensity to highlight the pixels that represent insa in each image, and measuring the area they cover with FIJI software.
High Content Morphology Analysis of ZMEL1
High-content image analysis was used to measure the effects of pharmacological inhibition of Bace2 on ZMEL1 cell morphology. ZMEL1 were seeded at 4000 cells/well in black wall, clear bottom 96-well plates (PerkinElmer, catalog #6005182) in DMEM/10% FBS/penicillin/streptomycin/glutamine. The cells were allowed to adhere overnight, and then media were changed to Bace2 inhibitor (PF-06663195) containing media at 12.5μM or 25μM with 0.025% (v/v) DMSO. Cells were grown for a total of 3 days after the addition of Bace2 inhibitor. At the end of the treatment, the cells were fixed with paraformaldehyde (4% in 1X PBS), permeabilized with 0.1% Triton-X-100 and stained using Hoechst 33342 (4μg/ml, Thermo Fisher Scientific, catalog # H3570) and HCS CellMask Deep Red Stain (1.7μg/ml, Thermo Fisher Scientific, catalog # H32721). Using a GE INCell 6000, 6 fields were captured per well using a Nikon 10X Plan Apo objective, 0.45NA. Images of the Hoechst signal was captured via the DAPI channel in order to identify the nucleus. The HCS Cellmask Deep Red was captured via the far-red channel to identify the cell body.
The image analysis script was performed within the Perkin Elmer Columbus software. Briefly the Hoechst stained nuclei was used to initially segment objects and act as a seed region to segment the cells stained with HCS CellMask Deep Red. Cell morphology is the primary readout of this high-content assay and the primary cell morphology parameters used are length to width ratio. Cell length is defined as the maximum distance (in microns) between two points on the cell mask. Cell width is defined as the diameter of the largest circle that will fit within the cell or nuclear mask.
Knockout insr in ZMEL1
Alt-R CRISPR-Cas9 gRNAs against zebrafish insra and insrb were designed using CHOPCHOP website(Labun et al., 2016) and synthesized by IDT. The RNP complex containing gRNAs (11μM of each insra and insrb gRNA mixture), tracrRNA ATTO™ 550 (22μM, IDT, catalog # 1075928) guide complex and an S. pyogenes recombinant Cas9 endonuclease (18.3μM, IDT, catalog # 1074181) were prepared at room temperature following manufacturer’s instruction. A non-target gRNA was included as negative control (22μM). The final complex (10μl RNP and 20μl of electroporation enhancer at 10.8μM) was nucleofected into 90μl cell suspension containing 8 million ZMEL1 cells using NEON transfection system (100μl kit, Thermo fisher Scientific, catalog # MPK10096, pulse voltage: 1400, pulse width: 20 and pulse number: 2) and plated back to fibronectin-coated flask. The tracrRNA ATTO™ 550 labelled ZMEL1 cells with RFP and was used to FACS sort RFP+ cells 3 days post nucleofection. Genomic DNA and protein lysates were extracted from sorted cells to assay mutations using Surveyor assay and protein using Western Blot respectively. For surveyor assay, insra and insrb genomic sequence adjacent to gRNA PAM sites were PCR amplified and then digested with surveyor nuclease (IDT, catalog # 706020). gRNA sequences are listed in the Key Resource Table. Primers for Surveyor PCR are listed in Table S2.
Plasmid Construction and Cell Line Expression
The following plasmids were constructed using the Gateway Tol2kit.
Fugu dct: Z-bace2 (zebrafish bace2); cryaa:YFP
Fugu tyrp1:dnIRS2-EGFP; cmlc2: mCherry
CMV: Z-bace2-2a-GFP-394
CMV: Z-bace2 enzymatic dead-2a-GFP-394
CMV: Z-insra-Myc-394
CMV: Z-insrb-Myc-394
CMV: tdtomato-394 (used as vector control)
Zebrafish Bace2 (Z-bace2) cDNA was amplified from WT embryos’ cDNA library and cloned into pENTR/D-TOPO vector.
Zebrafish Bace2 enzymatic dead cDNA was constructed by site-mutagenesis of two conserved protease sites (aspartic acids) into alanines (D98A and D292A) respectively.
Fugu tyrp1 promoter was a generous gift from (Zou et al., 2006).
Zebrafish dnIRS2-GFP was a generous gift from (Ye et al., 2016) and then further cloned into pENTR/D-TOPO vector.
Zebrafish insra and insrb cDNA was amplified from WT embryos’ cDNA library and cloned into pENTR/D-TOPO vector.
CMV: Z-pmela-FLAG was constructed by In-fusion cloning to clone Z-pmela cDNA into pCS2FLAG backbone (Addgene, catalog #16331). The FLAG tag was C-terminal fused to Z-pmela cDNA. Z-pmela cDNA was amplified from MGC clone plasmid (Open Biosystems, catalog # MDR1734-202779066). All primers are listed in Table S2. Lipofectamine 2000 (Life Technologies, catalog # 11668019) was used to overexpress plasmids in HEK 293T cells.
Western Blot
Cell lysates were collected using RIPA buffer (Thermo Fisher Scientific, catalog # 89901), with addition of 1X protease inhibitor (Thermo Fisher Scientific, catalog # 78430), 1X phosphatase Inhibitor (Fisher Scientific, catalog #78441) and 5mM EDTA. Proteins concentrations were measured with Bradford (Sigma-Aldrich, catalog # B6916) according to kit direction. Protein were mixed with 6X SDS loading dye (Boston BioProducts, catalog # BP-111R) and boiled at 95°C for 10min. Proteins were run on pre-made mini protean TGX gels (Bio-Rad). Gels were transferred using Turbo™ Mini Nitrocellulose Transfer Pack (Bio-Rad, catalog # 1704158). Membranes were blocked in 5% milk for 1 hour at room temperature prior to addition of primary antibody overnight at 4°C. Membranes were washed with 1X TBST (0.1% Tween 20) for 3×10min, then incubated with secondary antibody for 1 hour at room temperature. Membranes were washed with 1X TBST for 3×10min. Signals were developed using ECL prime (GE Healthcare, catalog # RPN2236). Primary antibodies used for Western blot are: mTOR (7C10) mAb (1:1000, CST, catalog # 2983S), insulin R β (C-19) (1:200, Santa Cruz Biotechnology, catalog # sc-711), c-MYC (1:1000, Santa Cruz Biotechnology, catalog # sc-40), FLAG (1:500, Sigma-Aldrich, catalog # F3165). Secondary antibodies were: rabbit anti-mouse IgG H&L (HRP) (1:10,000, Abcam, catalog # ab97046), goat anti-rabbit IgG H&L (HRP) (1:10,000, Abcam, catalog # ab97051).
Transwell Assay
1×105 ZMEL1 cells were seeded into 3.0μM transwell inserts (Corning catalog # 353492) in 500μl of culture media, in the presence or absence of 25μM Bace2 inhibitor PF-06663195 with 0.025% (v/v) DMSO. The receiving plates (Corning catalog # 353504) were filled with 500μl culture media, also in the presence or absence of 25μM Bace2 inhibitor. ZMEL1 were allowed to migrate for 48 hours, then sterile cotton swabs were used to gently clean the inside of each membrane 3 times. Hoechst 33342 were added to final concentration of 4μg/ml to facilitate cell counting. Image five different 20X fields per well and quantify cell numbers by image analysis software FIJI.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical comparisons were performed with the aid of GraphPad PRISM 7 and the statistical details including sample size can be found in the figure legends. A p value of >0.05 is not considered statistically significant. * indicates p<0.05, ** indicates p<0.01, *** indicates p<0.001 and **** indicates p<0.0001.
Supplementary Material
Confocal image of tailfin melanophores in DMSO treated bace2−/−; Tg(tyrp1b: membrane-mCherry) fish to visualize melanophore membrane structure. The 3D reconstruction movie was assembled using Imaris software.
Confocal image of tailfin melanophores in response to the PI3K inhibitor AS605240 (110nM) in bace2−/−; Tg(tyrp1b: membrane-mCherry) fish to visualize melanophore membrane structure. The 3D reconstruction movie was assembled using Imaris software.
Confocal image of tailfin melanophores in response to the mTOR inhibitor PP242 (15μM) in bace2−/−; Tg(tyrp1b: membrane-mCherry) fish to visualize melanophore membrane structure. The 3D reconstruction movie was assembled using Imaris software.
Confocal image of tailfin melanophores in response to the insulin receptor antagonist NVP-AEW541 (60μM) in bace2−/−; Tg(tyrp1b: membrane-mCherry) fish to visualize melanophore membrane structure. The 3D reconstruction movie was assembled using Imaris software.
Highlights.
bace2 mutation causes defects in melanocyte patterning and morphology
bace2 mutant phenotype can be rescued by inhibitors of insulin/PI3K/mTOR signaling
Bace2 normally cleaves the insulin receptor, making mutants hypersensitive to insulin
Bace2 is a melanocyte enriched sheddase that limits response to distant insulin signaling
Acknowledgments
We thank Pfizer, Inc for the Bace2 inhibitor PF-06663195. We thank Wenjing Wu for assistance with artwork. This work was supported by the NIH Director’s New Innovator Award (DP2CA186572), Mentored Clinical Scientist Research Career Development Award (K08AR055368), the Melanoma Research Alliance, The Pershing Square Sohn Foundation, The Alan and Sandra Gerry Metastasis Research Initiative at the Memorial Sloan Kettering Cancer Center, The Harry J. Lloyd Foundation and Consano (all to R.M.W.). Y.M.Z is funded by the NCI Predoctoral to Postdoctoral Fellow Transition Award (F99/K00) under the award number 1F99CA212436-01. J.J.B-C. is funded by an NCI K22CA196750 grant and the TCI Young Scientist Cancer Research Award JJR Fund (P30 CA196521). J.D.M. is the recipient of a postdoctoral fellowship from the ARC Foundation. S.J.C is supported by the Kirschstein-NRSA predoctoral fellowship (F31CA196305), the Joanna M. Nicolay Melanoma Foundation Research Scholar Award 2014 and the Robert B. Catell Fellowship. N.R.C. is supported by the Kirschstein-NRSA predoctoral fellowship (F30) under award number F30CA220954, by a research grant from the Melanoma Research Foundation, and by a Medical Scientist Training Program grant under award number T32GM007739.
Footnotes
AUTHORS CONTRIBUTIONS
R.M.W. and Y.M.Z. conceived the study and wrote the manuscript. Y.M.Z. designed, performed, and interpreted all experiments except as noted. M.Z. conceived the study, performed high resolution imaging, Bace inhibitor studies, ISH for pigment genes, and generated the bace2−/−; Tg(tyrp1b: membrane-mCherry) and bace2−/−; Tg(mitfa: EGFP) lines, and performed electron microscopy. T.G. and T.T. assisted with the chemical screen. S.J.C. assisted with molecular cloning. R.G., M.F. and N.C. assisted with the ZMEL1 morphology analysis. C.M., J.M. and J.J.B-C. assisted with cell culture assays.
DECLARATION of INTEREST
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Malek Z, Swope VB, Suzuki I, Akcali C, Harriger MD, Boyce ST, Urabe K, Hearing VJ. Mitogenic and melanogenic stimulation of normal human melanocytes by melanotropic peptides. Proc Natl Acad Sci USA. 1995;92:1789–1793. doi: 10.1073/pnas.92.5.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baynash AG, Hosoda K, Giaid A, Richardson JA, Emoto N, Hammer RE, Yanagisawa M. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277–1285. doi: 10.1016/0092-8674(94)90018-3. [DOI] [PubMed] [Google Scholar]
- van Bebber F, Hruscha A, Willem M, Schmid B, Haass C. Loss of Bace2 in zebrafish affects melanocyte migration and is distinct from Bace1 knock out phenotypes. J Neurochem. 2013;127:471–481. doi: 10.1111/jnc.12198. [DOI] [PubMed] [Google Scholar]
- Bravo DA, Gleason JB, Sanchez RI, Roth RA, Fuller RS. Accurate and efficient cleavage of the human insulin proreceptor by the human proprotein-processing protease furin. Characterization and kinetic parameters using the purified, secreted soluble protease expressed by a recombinant baculovirus. J Biol Chem. 1994;269:25830–25837. [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israël A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Budi EH, Patterson LB, Parichy DM. Post-embryonic nerve-associated precursors to adult pigment cells: genetic requirements and dynamics of morphogenesis and differentiation. PLoS Genet. 2011;7:e1002044. doi: 10.1371/journal.pgen.1002044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne T, O’Connor MN, Seabra MC, Cutler DF, Futter CE. Regulation of melanosome number, shape and movement in the zebrafish retinal pigment epithelium by OA1 and PMEL. J Cell Sci. 2015;128:1400–1407. doi: 10.1242/jcs.164400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B, Thorp EB, Doran AC, Subramanian M, Sansbury BE, Lin CS, Spite M, Fredman G, Tabas I. MerTK cleavage limits proresolving mediator biosynthesis and exacerbates tissue inflammation. Proc Natl Acad Sci USA. 2016;113:6526–6531. doi: 10.1073/pnas.1524292113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas S, Casini P, Piquer S, Altirriba J, Soty M, Cadavez L, Gomis R, Novials A. BACE2 plays a role in the insulin receptor trafficking in pancreatic β-cells. Am J Physiol Endocrinol Metab. 2010;299:E1087–95. doi: 10.1152/ajpendo.00420.2010. [DOI] [PubMed] [Google Scholar]
- Chakraborty AK, Orlow SJ, Bolognia JL, Pawelek JM. Structural/functional relationships between internal and external MSH receptors: modulation of expression in Cloudman melanoma cells by UVB radiation. J Cell Physiol. 1991;147:1–6. doi: 10.1002/jcp.1041470102. [DOI] [PubMed] [Google Scholar]
- Cooper CD, Raible DW. Mechanisms for reaching the differentiated state: Insights from neural crest-derived melanocytes. Semin Cell Dev Biol. 2009;20:105–110. doi: 10.1016/j.semcdb.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, You MJ, DePinho RA, McMahon M, Bosenberg M. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- Edmondson SR, Russo VC, McFarlane AC, Wraight CJ, Werther GA. Interactions between growth hormone, insulin-like growth factor I, and basic fibroblast growth factor in melanocyte growth. J Clin Endocrinol Metab. 1999;84:1638–1644. doi: 10.1210/jcem.84.5.5692. [DOI] [PubMed] [Google Scholar]
- Eom DS, Parichy DM. A macrophage relay for long-distance signaling during postembryonic tissue remodeling. Science. 2017;355:1317–1320. doi: 10.1126/science.aal2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom DS, Bain EJ, Patterson LB, Grout ME, Parichy DM. Long-distance communication by specialized cellular projections during pigment pattern development and evolution. Elife. 2015:4. doi: 10.7554/eLife.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernfors P. Cellular origin and developmental mechanisms during the formation of skin melanocytes. Exp Cell Res. 2010;316:1397–1407. doi: 10.1016/j.yexcr.2010.02.042. [DOI] [PubMed] [Google Scholar]
- Eskova A, Chauvigné F, Maischein HM, Ammelburg M, Cerdà J, Nüsslein-Volhard C, Irion U. Gain-of-function mutations in Aqp3a influence zebrafish pigment pattern formation through the tissue environment. Development. 2017;144:2059–2069. doi: 10.1242/dev.143495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterházy D, Stützer I, Wang H, Rechsteiner MP, Beauchamp J, Döbeli H, Hilpert H, Matile H, Prummer M, Schmidt A, et al. Bace2 is a β cell-enriched protease that regulates pancreatic β cell function and mass. Cell Metab. 2011;14:365–377. doi: 10.1016/j.cmet.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Frohnhöfer HG, Krauss J, Maischein HM, Nüsslein-Volhard C. Iridophores and their interactions with other chromatophores are required for stripe formation in zebrafish. Development. 2013;140:2997–3007. doi: 10.1242/dev.096719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia RJ, Ittah A, Mirabal S, Figueroa J, Lopez L, Glick AB, Kos L. Endothelin 3 induces skin pigmentation in a keratin-driven inducible mouse model. J Invest Dermatol. 2008;128:131–142. doi: 10.1038/sj.jid.5700948. [DOI] [PubMed] [Google Scholar]
- Garofalo RS, Rosen OM. Tissue localization of Drosophila melanogaster insulin receptor transcripts during development. Mol Cell Biol. 1988;8:1638–1647. doi: 10.1128/mcb.8.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goding CR. Melanocytes: the new black. Int J Biochem Cell Biol. 2007;39:275–279. doi: 10.1016/j.biocel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Hamada H, Watanabe M, Lau HE, Nishida T, Hasegawa T, Parichy DM, Kondo S. Involvement of Delta/Notch signaling in zebrafish adult pigment stripe patterning. Development. 2014;141:318–324. doi: 10.1242/dev.099804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Yaar M, Gilchrest BA. Endothelin-1 of keratinocyte origin is a mediator of melanocyte dendricity. J Invest Dermatol. 1995;105:744–748. doi: 10.1111/1523-1747.ep12325522. [DOI] [PubMed] [Google Scholar]
- Harrison KA, Thaler J, Pfaff SL, Gu H, Kehrl JH. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat Genet. 1999;23:71–75. doi: 10.1038/12674. [DOI] [PubMed] [Google Scholar]
- Havrankova J, Roth J, Brownstein MJ. Concentrations of insulin and insulin receptors in the brain are independent of peripheral insulin levels. Studies of obese and streptozotocin-treated rodents. J Clin Invest. 1979;64:636–642. doi: 10.1172/JCI109504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann S, Ratnakumar K, Langdon E, Kansler E, Kim I, Campbell NR, Perry E, McMahon A, Kaufman C, van Rooijen E, et al. A quantitative system for studying metastasis using transparent zebrafish. Cancer Res. 2015;75:4272–4282. doi: 10.1158/0008-5472.CAN-14-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanns-Lê T, Scheen A, Piérard GE. Acanthosis nigricans associated with insulin resistance: pathophysiology and management. Am J Clin Dermatol. 2004;5:199–203. doi: 10.2165/00128071-200405030-00008. [DOI] [PubMed] [Google Scholar]
- Hultman KA, Johnson SL. Differential contribution of direct-developing and stem cell-derived melanocytes to the zebrafish larval pigment pattern. Dev Biol. 2010;337:425–431. doi: 10.1016/j.ydbio.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imokawa G, Yada Y, Miyagishi M. Endothelins secreted from human keratinocytes are intrinsic mitogens for human melanocytes. J Biol Chem. 1992;267:24675–24680. [PubMed] [Google Scholar]
- Jiang H, Guo W, Liang X, Rao Y. Both the establishment and the maintenance of neuronal polarity require active mechanisms: critical roles of GSK-3beta and its upstream regulators. Cell. 2005;120:123–135. doi: 10.1016/j.cell.2004.12.033. [DOI] [PubMed] [Google Scholar]
- Junker JP, Noël ES, Guryev V, Peterson KA, Shah G, Huisken J, McMahon AP, Berezikov E, Bakkers J, van Oudenaarden A. Genome-wide RNA Tomography in the zebrafish embryo. Cell. 2014;159:662–675. doi: 10.1016/j.cell.2014.09.038. [DOI] [PubMed] [Google Scholar]
- Kasuga K, Kaneko H, Nishizawa M, Onodera O, Ikeuchi T. Generation of intracellular domain of insulin receptor tyrosine kinase by gamma-secretase. Biochem Biophys Res Commun. 2007;360:90–96. doi: 10.1016/j.bbrc.2007.06.022. [DOI] [PubMed] [Google Scholar]
- Kaufman CK, White RM, Zon L. Chemical genetic screening in the zebrafish embryo. Nat Protoc. 2009;4:1422–1432. doi: 10.1038/nprot.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman CK, Mosimann C, Fan ZP, Yang S, Thomas AJ, Ablain J, Tan JL, Fogley RD, van Rooijen E, Hagedorn EJ, et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science. 2016;351:aad2197. doi: 10.1126/science.aad2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsh RN, Schmid B, Eisen JS. Genetic analysis of melanophore development in zebrafish embryos. Dev Biol. 2000;225:277–293. doi: 10.1006/dbio.2000.9840. [DOI] [PubMed] [Google Scholar]
- Kelsh RN, Harris ML, Colanesi S, Erickson CA. Stripes and belly-spots -- a review of pigment cell morphogenesis in vertebrates. Semin Cell Dev Biol. 2009;20:90–104. doi: 10.1016/j.semcdb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IS, Heilmann S, Kansler ER, Zhang Y, Zimmer M, Ratnakumar K, Bowman RL, Simon-Vermot T, Fennell M, Garippa R, et al. Microenvironment-derived factors driving metastatic plasticity in melanoma. Nat Commun. 2017;8:14343. doi: 10.1038/ncomms14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel RA, Onder L, Wilfinger A, Ellertsdottir E, Meyer D. Requirement for Pdx1 in specification of latent endocrine progenitors in zebrafish. BMC Biol. 2011;9:75. doi: 10.1186/1741-7007-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel RA, Dobler S, Schmitner N, Walsen T, Freudenblum J, Meyer D. Diabetic pdx1-mutant zebrafish show conserved responses to nutrient overload and anti-glycemic treatment. Sci Rep. 2015;5:14241. doi: 10.1038/srep14241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson VP. Proteolytic processing of the insulin receptor beta subunit is associated with insulin-induced receptor down-regulation. J Biol Chem. 1991;266:15656–15662. [PubMed] [Google Scholar]
- Kopitz C, Gerg M, Bandapalli OR, Ister D, Pennington CJ, Hauser S, Flechsig C, Krell H-W, Antolovic D, Brew K, et al. Tissue inhibitor of metalloproteinases-1 promotes liver metastasis by induction of hepatocyte growth factor signaling. Cancer Res. 2007;67:8615–8623. doi: 10.1158/0008-5472.CAN-07-0232. [DOI] [PubMed] [Google Scholar]
- Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–6. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Nachtrab G, Klinsawat PW, Hami D, Poss KD. Ras controls melanocyte expansion during zebrafish fin stripe regeneration. Dis Model Mech. 2010;3:496–503. doi: 10.1242/dmm.004515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque M, Feng Y, Jones RA, Martin P. Inflammation drives wound hyperpigmentation in zebrafish by recruiting pigment cells to sites of tissue damage. Dis Model Mech. 2013;6:508–515. doi: 10.1242/dmm.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Lahvic JL, Binder V, Pugach EK, Riley EB, Tamplin OJ, Panigrahy D, Bowman TV, Barrett FG, Heffner GC, et al. Epoxyeicosatrienoic acids enhance embryonic haematopoiesis and adult marrow engraftment. Nature. 2015;523:468–471. doi: 10.1038/nature14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- Logan DW, Burn SF, Jackson IJ. Regulation of pigmentation in zebrafish melanophores. Pigment Cell Res. 2006;19:206–213. doi: 10.1111/j.1600-0749.2006.00307.x. [DOI] [PubMed] [Google Scholar]
- Luetteke NC, Qiu TH, Peiffer RL, Oliver P, Smithies O, Lee DC. TGF alpha deficiency results in hair follicle and eye abnormalities in targeted and waved-1 mice. Cell. 1993;73:263–278. doi: 10.1016/0092-8674(93)90228-i. [DOI] [PubMed] [Google Scholar]
- Luo R, An M, Arduini BL, Henion PD. Specific pan-neural crest expression of zebrafish Crestin throughout embryonic development. Dev Dyn. 2001;220:169–174. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1097>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Mahalwar P, Singh AP, Fadeev A, Nüsslein-Volhard C, Irion U. Heterotypic interactions regulate cell shape and density during color pattern formation in zebrafish. Biol Open. 2016;5:1680–1690. doi: 10.1242/bio.022251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus A, Zhong J, Snider WD. Raf and akt mediate distinct aspects of sensory axon growth. Neuron. 2002;35:65–76. doi: 10.1016/s0896-6273(02)00752-3. [DOI] [PubMed] [Google Scholar]
- Massague J, Pilch PF, Czech MP. A unique proteolytic cleavage site on the beta subunit of the insulin receptor. J Biol Chem. 1981;256:3182–3190. [PubMed] [Google Scholar]
- McMenamin SK, Bain EJ, McCann AE, Patterson LB, Eom DS, Waller ZP, Hamill JC, Kuhlman JA, Eisen JS, Parichy DM. Thyroid hormone-dependent adult pigment cell lineage and pattern in zebrafish. Science. 2014;345:1358–1361. doi: 10.1126/science.1256251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellgren EM, Johnson SL. A requirement for kit in embryonic zebrafish melanocyte differentiation is revealed by melanoblast delay. Dev Genes Evol. 2004;214:493–502. doi: 10.1007/s00427-004-0428-y. [DOI] [PubMed] [Google Scholar]
- Miller MA, Meyer AS, Beste MT, Lasisi Z, Reddy S, Jeng KW, Chen CH, Han J, Isaacson K, Griffith LG, et al. ADAM-10 and -17 regulate endometriotic cell migration via concerted ligand and receptor shedding feedback on kinase signaling. Proc Natl Acad Sci USA. 2013;110:E2074–83. doi: 10.1073/pnas.1222387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Oudin MJ, Sullivan RJ, Wang SJ, Meyer AS, Im H, Frederick DT, Tadros J, Griffith LG, Lee H, et al. Reduced Proteolytic Shedding of Receptor Tyrosine Kinases Is a Post-Translational Mechanism of Kinase Inhibitor Resistance. Cancer Discov. 2016;6:382–399. doi: 10.1158/2159-8290.CD-15-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort RL, Jackson IJ, Patton EE. The melanocyte lineage in development and disease. Development. 2015;142:1387. doi: 10.1242/dev.123729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morvan D, Steyaert JM, Schwartz L, Israel M, Demidem A. Normal human melanocytes exposed to chronic insulin and glucose supplementation undergo oncogenic changes and methyl group metabolism cellular redistribution. Am J Physiol Endocrinol Metab. 2012;302:E1407–18. doi: 10.1152/ajpendo.00594.2011. [DOI] [PubMed] [Google Scholar]
- Nakamasu A, Takahashi G, Kanbe A, Kondo S. Interactions between zebrafish pigment cells responsible for the generation of Turing patterns. Proc Natl Acad Sci USA. 2009;106:8429–8434. doi: 10.1073/pnas.0808622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenthal J, Nüsslein-Volhard C. fork head domain genes in zebrafish. Dev Genes Evol. 1998;208:245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- Orme JJ, Du Y, Vanarsa K, Mayeux J, Li L, Mutwally A, Arriens C, Min S, Hutcheson J, Davis LS, et al. Heightened cleavage of Axl receptor tyrosine kinase by ADAM metalloproteases may contribute to disease pathogenesis in SLE. Clin Immunol. 2016;169:58–68. doi: 10.1016/j.clim.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papasani MR, Robison BD, Hardy RW, Hill RA. Early developmental expression of two insulins in zebrafish (Danio rerio) Physiol Genomics. 2006;27:79–85. doi: 10.1152/physiolgenomics.00012.2006. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Rawls JF, Pratt SJ, Whitfield TT, Johnson SL. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development. 1999;126:3425–3436. doi: 10.1242/dev.126.15.3425. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Mellgren EM, Rawls JF, Lopes SS, Kelsh RN, Johnson SL. Mutational analysis of endothelin receptor b1 (rose) during neural crest and pigment pattern development in the zebrafish Danio rerio. Dev Biol. 2000;227:294–306. doi: 10.1006/dbio.2000.9899. [DOI] [PubMed] [Google Scholar]
- Patterson LB, Parichy DM. Interactions with iridophores and the tissue environment required for patterning melanophores and xanthophores during zebrafish adult pigment stripe formation. PLoS Genet. 2013;9:e1003561. doi: 10.1371/journal.pgen.1003561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- Price AC, Weadick CJ, Shim J, Rodd FH. Pigments, patterns, and fish behavior. Zebrafish. 2008;5:297–307. doi: 10.1089/zeb.2008.0551. [DOI] [PubMed] [Google Scholar]
- Richardson J, Lundegaard PR, Reynolds NL, Dorin JR, Porteous DJ, Jackson IJ, Patton EE. mc1r Pathway regulation of zebrafish melanosome dispersion. Zebrafish. 2008;5:289–295. doi: 10.1089/zeb.2008.0541. [DOI] [PubMed] [Google Scholar]
- Rochin L, Hurbain I, Serneels L, Fort C, Watt B, Leblanc P, Marks MS, De Strooper B, Raposo G, van Niel G. BACE2 processes PMEL to form the melanosome amyloid matrix in pigment cells. Proc Natl Acad Sci USA. 2013;110:10658–10663. doi: 10.1073/pnas.1220748110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein AL, Lee D, Luo R, Henion PD, Halpern ME. Genes dependent on zebrafish cyclops function identified by AFLP differential gene expression screen. Genesis. 2000;26:86–97. doi: 10.1002/(sici)1526-968x(200001)26:1<86::aid-gene11>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Sather S, Kenyon KD, Lefkowitz JB, Liang X, Varnum BC, Henson PM, Graham DK. A soluble form of the Mer receptor tyrosine kinase inhibits macrophage clearance of apoptotic cells and platelet aggregation. Blood. 2007;109:1026–1033. doi: 10.1182/blood-2006-05-021634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelter F, Grandl M, Seubert B, Schaten S, Hauser S, Gerg M, Boccaccio C, Comoglio P, Krüger A. Tumor cell-derived Timp-1 is necessary for maintaining metastasis-promoting Met-signaling via inhibition of Adam-10. Clin Exp Metastasis. 2011;28:793–802. doi: 10.1007/s10585-011-9410-z. [DOI] [PubMed] [Google Scholar]
- Schonthaler HB, Lampert JM, von Lintig J, Schwarz H, Geisler R, Neuhauss SCF. A mutation in the silver gene leads to defects in melanosome biogenesis and alterations in the visual system in the zebrafish mutant fading vision. Dev Biol. 2005;284:421–436. doi: 10.1016/j.ydbio.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Schwartz MW, Figlewicz DP, Baskin DG, Woods SC, Porte D. Insulin in the brain: a hormonal regulator of energy balance. Endocr Rev. 1992;13:387–414. doi: 10.1210/edrv-13-3-387. [DOI] [PubMed] [Google Scholar]
- Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- Shimshek DR, Jacobson LH, Kolly C, Zamurovic N, Balavenkatraman KK, Morawiec L, Kreutzer R, Schelle J, Jucker M, Bertschi B, et al. Pharmacological BACE1 and BACE2 inhibition induces hair depigmentation by inhibiting PMEL17 processing in mice. Sci Rep. 2016;6:21917. doi: 10.1038/srep21917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer L. Generation of melanocytes from neural crest cells. Pigment Cell Melanoma Res. 2011;24:411–421. doi: 10.1111/j.1755-148X.2011.00834.x. [DOI] [PubMed] [Google Scholar]
- Stützer I, Selevsek N, Esterházy D, Schmidt A, Aebersold R, Stoffel M. Systematic proteomic analysis identifies β-site amyloid precursor protein cleaving enzyme 2 and 1 (BACE2 and BACE1) substrates in pancreatic β-cells. J Biol Chem. 2013;288:10536–10547. doi: 10.1074/jbc.M112.444703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakkol A, Elder JT, Griffiths CE, Cooper KD, Talwar H, Fisher GJ, Keane KM, Foltin SK, Voorhees JJ. Expression of growth hormone receptor, insulin-like growth factor 1 (IGF-1) and IGF-1 receptor mRNA and proteins in human skin. J Invest Dermatol. 1992;99:343–349. doi: 10.1111/1523-1747.ep12616668. [DOI] [PubMed] [Google Scholar]
- van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem. 2009;284:31018–31027. doi: 10.1074/jbc.M109.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Toyoshima Y, Monson C, Duan C, Wu Y, Gao C, Yakar S, Sadler KC, LeRoith D. The role of insulin receptor signaling in zebrafish embryogenesis. Endocrinology. 2008;149:5996–6005. doi: 10.1210/en.2008-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Walderich B, Singh AP, Mahalwar P, Nüsslein-Volhard C. Homotypic cell competition regulates proliferation and tiling of zebrafish pigment cells during colour pattern formation. Nat Commun. 2016;7:11462. doi: 10.1038/ncomms11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welten MCM, de Haan SB, van den Boogert N, Noordermeer JN, Lamers GEM, Spaink HP, Meijer AH, Verbeek FJ. ZebraFISH: fluorescent in situ hybridization protocol and three-dimensional imaging of gene expression patterns. Zebrafish. 2006;3:465–476. doi: 10.1089/zeb.2006.3.465. [DOI] [PubMed] [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Brenner M, Hearing VJ. The regulation of skin pigmentation. J Biol Chem. 2007;282:27557–27561. doi: 10.1074/jbc.R700026200. [DOI] [PubMed] [Google Scholar]
- Ye L, Robertson MA, Mastracci TL, Anderson RM. An insulin signaling feedback loop regulates pancreas progenitor cell differentiation during islet development and regeneration. Dev Biol. 2016;409:354–369. doi: 10.1016/j.ydbio.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM, Huttenlocher A. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev Cell. 2010;18:226–236. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Beermann F, Wang J, Kawakami K, Wei X. The Fugu tyrp1 promoter directs specific GFP expression in zebrafish: tools to study the RPE and the neural crest-derived melanophores. Pigment Cell Res. 2006;19:615–627. doi: 10.1111/j.1600-0749.2006.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confocal image of tailfin melanophores in DMSO treated bace2−/−; Tg(tyrp1b: membrane-mCherry) fish to visualize melanophore membrane structure. The 3D reconstruction movie was assembled using Imaris software.
Confocal image of tailfin melanophores in response to the PI3K inhibitor AS605240 (110nM) in bace2−/−; Tg(tyrp1b: membrane-mCherry) fish to visualize melanophore membrane structure. The 3D reconstruction movie was assembled using Imaris software.
Confocal image of tailfin melanophores in response to the mTOR inhibitor PP242 (15μM) in bace2−/−; Tg(tyrp1b: membrane-mCherry) fish to visualize melanophore membrane structure. The 3D reconstruction movie was assembled using Imaris software.
Confocal image of tailfin melanophores in response to the insulin receptor antagonist NVP-AEW541 (60μM) in bace2−/−; Tg(tyrp1b: membrane-mCherry) fish to visualize melanophore membrane structure. The 3D reconstruction movie was assembled using Imaris software.