Abstract
Malformations of the intrahepatic biliary structure cause cholestasis, a liver pathology that corresponds to poor bile flow, which leads to inflammation, fibrosis, and cirrhosis. Although the specification of biliary epithelial cells (BECs) that line the bile ducts is fairly well understood, the molecular mechanisms underlying intrahepatic biliary morphogenesis remain largely unknown. Wnt/β-catenin signaling plays multiple roles in liver biology; however, its role in intrahepatic biliary morphogenesis remains unclear. Using pharmacological and genetic tools that allow one to manipulate Wnt/β-catenin signaling, we show here that in zebrafish, both the suppression and overactivation of Wnt/β-catenin signaling impaired intrahepatic biliary morphogenesis. Hepatocytes, but not BECs, exhibited Wnt/β-catenin activity, and the global suppression of Wnt/β-catenin signaling reduced Notch activity in BECs. Hepatocyte-specific suppression of Wnt/β-catenin signaling also reduced Notch activity in BECs, indicating a cell non-autonomous role for Wnt/β-catenin signaling in regulating hepatic Notch activity. Reducing Notch activity to the same level as that observed in Wnt-suppressed livers also impaired biliary morphogenesis. Intriguingly, the expression of the Notch ligand genes, jag1b and jag2b, in hepatocytes was reduced in Wnt-suppressed livers and enhanced in Wnt-overactivated livers, revealing their regulation by Wnt/β-catenin signaling. Importantly, restoring Notch activity rescued the biliary defects observed in Wnt-suppressed livers. Conclusion: Wnt/β-catenin signaling cell non-autonomously controls Notch activity in BECs by regulating the expression of Notch ligand genes in hepatocytes, thereby regulating biliary morphogenesis.
Keywords: biliary morphogenesis, biliary epithelial cells, liver development, bile canaliculi, Notch signaling
Introduction
The intrahepatic biliary network is a complex three-dimensional network of conduits lined by biliary epithelial cells (BECs) and plays a crucial role in transporting bile from hepatocytes to the gallbladder. Hence, malformations of the network can cause bile leakage and lead to inflammation, fibrosis, and cirrhosis, as observed in patients with cholestatic liver diseases. Identification of mutations in JAG1, a Notch ligand gene, in most Alagille syndrome patients (1) provoked extensive studies about the role of Notch signaling in intrahepatic biliary development (2). During mammalian biliary development, bi-potent liver progenitor cells (i.e., hepatoblasts) can differentiate into either hepatocytes or BECs, the latter forming ductal plates, undergoing extensive morphogenesis, including tubulogenesis, and eventually establishing the intrahepatic biliary network (3, 4).
As a key regulator of biliary development, Notch signaling regulates early BEC induction and later morphogenesis processes (2). Hepatoblast-specific deletion of mouse Notch2 completely blocks BEC induction until the perinatal stage (5). Ectopic expression of the intracellular domain of NOTCH1 or NOTCH2 in mouse hepatoblasts initially induces excessive BECs and later, ectopic biliary tubules (6–8). In another hepatoblast-specific Notch2 deletion setting in mice, BEC induction is unaffected, but subsequent morphogenesis is impaired, thereby resulting in the impaired development of biliary tubules (9). The mouse knockout of the key Notch target gene, Hes1, also displays the same morphogenesis defect (10). Moreover, Notch signaling regulates BEC proliferation and survival (6, 11), which can contribute to biliary morphogenesis. In mice, Notch signaling regulates the formation of peripheral intrahepatic bile ducts in a dose-dependent manner: higher Notch activity leads to denser intrahepatic bile ducts (11). Besides Notch signaling, which is implicated in both biliary induction and morphogenesis, TGFβ signaling also regulates biliary induction (12). However, additional factors regulating biliary morphogenesis remain largely unknown.
Wnt/β-catenin signaling regulates multiple processes of liver development, including hepatoblast specification and proliferation, hepatocyte maturation and proliferation, and liver zonation (13). It is also implicated in biliary development; however, data supporting the role of Wnt/β-catenin signaling in this process are inconsistent. In liver explant culture studies, WNT3A promotes the differentiation of hepatoblasts into BECs (14); β-catenin stabilization with Apc deletion in mouse hepatoblasts induces their premature differentiation into BECs (15), supporting a positive role for Wnt/β-catenin signaling in biliary induction. In contrast, β-catenin deletion in mouse hepatoblasts does not affect their differentiation into BECs (16), suggesting that Wnt/β-catenin signaling is dispensable for biliary induction. This discrepancy makes the role of Wnt/β-catenin signaling in biliary development unclear. Although Wnt5a knockout mice exhibit excessive BECs, indicating a repressive role for Wnt5a in biliary induction, this effect is mediated by calcium/calmodulin-dependent protein kinase II, not by β-catenin (17).
We sought to determine the role of Wnt/β-catenin signaling in intrahepatic biliary morphogenesis. Using zebrafish as an animal model, we show here that Wnt/β-catenin signaling controls intrahepatic biliary network formation by regulating Notch signaling cell non-autonomously.
Experimental procedures
Zebrafish lines
Experiments were performed with approval of the Institutional Animal Care and Use Committee at the University of Pittsburgh. We used the following mutants and transgenic lines: lef1zd11, tcf7nkhg21c, jag1bgd2Gt, Tg(Tp1:GFP)um14, Tg(Tp1:VenusPEST)s940, Tg(Tp1:H2B-mCherry)s939, Tg(Tp1:mCherry)pt611, Tg(Tp1:mCherry-CAAX)s733, Tg(Tp1:GFP-utrophin)pt612, Tg(hs:Axin1)w35, Tg(hs:wnt2bb)pt603, Tg(hs:N3ICD)co17, Tg(hs:ca-β-catenin)w130, Tg(WRE:d2GFP)kyu1, and Tg(fabp10a:ca-β-catenin)s704. Their full names and references are listed in Supplementary Table 1.
Chemical treatment
1000x stock solutions of XAV939 (Selleckchem, Houston, TX), CHIR99021 (Selleckchem, Houston, TX), and LY411575 (Cayman Chemical, Ann Arbor, MI) were prepared in 100% DMSO and diluted to 10, 100, and 1 μM, respectively, with egg water. A 0.1% DMSO solution in egg water was used as a control.
PED6 and BODIPY C5 assays
PED6 assay was performed by treating larvae with 0.3 μg/ml PED6 (Life Technologies, Grand Island, NY) for 3 hours as previously described (18); BODIPY C5 assay was performed with 0.5 μM BODIPY C5 (Life Technologies, Grand Island, NY) for 2 hours as previously described (19). Images for PED6 and BODIPY C5 assays were obtained using epifluorescence and confocal microscopes, respectively. BODIPY C5-treated larvae were briefly rinsed, anesthetized with 0.016% tricaine/egg water, and then mounted in 1% low-melting agarose for confocal imaging.
Additional methods are available in Supporting Information.
Results
Suppression of Wnt/β-catenin signaling impairs intrahepatic biliary network formation
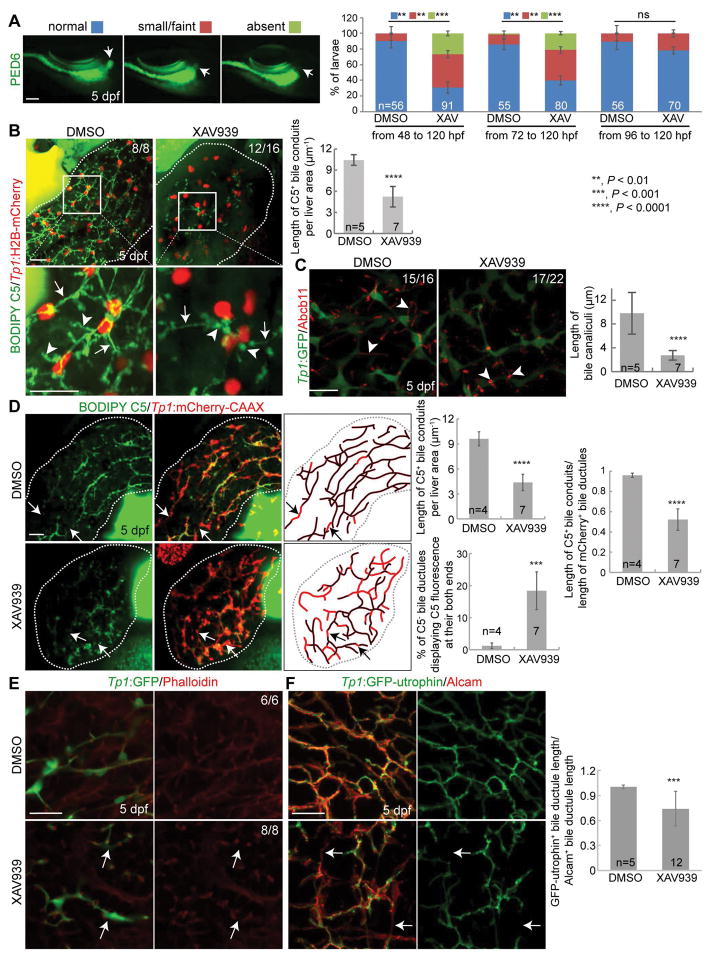
In zebrafish, hepatoblasts form around 22–24 hours post-fertilization (hpf) (20) and begin differentiating into BECs around 40–44 hpf (21, 22). By 5 days post-fertilization (dpf), BECs establish a fully functional, intricate intrahepatic biliary network (23). To visualize BECs in the liver, we utilized the Tg(Tp1:GFP) line, which expresses GFP under the control of the promoter containing the Notch-responsive element (22). Using this transgenic line, we and others previously reported the intrahepatic biliary morphogenesis process at the cellular level (21, 22). To determine if Wnt/β-catenin signaling regulates this morphogenesis process, we first suppressed Wnt/β-catenin signaling and examined the functionality of the biliary network. To suppress Wnt/β-catenin signaling, we used a Wnt inhibitor, XAV939, which stimulates β-catenin degradation by stabilizing Axin (24). To visualize biliary network functionality, we used a fluorescently labeled fatty acid reporter, PED6, which is metabolized into a component of bile in hepatocytes and accumulates in the gallbladder after biliary secretion (18). PED6 accumulation in the gallbladder was greatly reduced in larvae treated with XAV939 from 48 or 72, but not 96, hpf (Fig. 1A), suggesting the essential role of Wnt/β-catenin signaling in biliary morphogenesis prior to 96 hpf. To avoid impacting early liver development, as Wnt/β-catenin signaling also regulates hepatoblast specification and proliferation (20, 25, 26), we chose 72 hpf as the treatment start time for subsequent analyses.
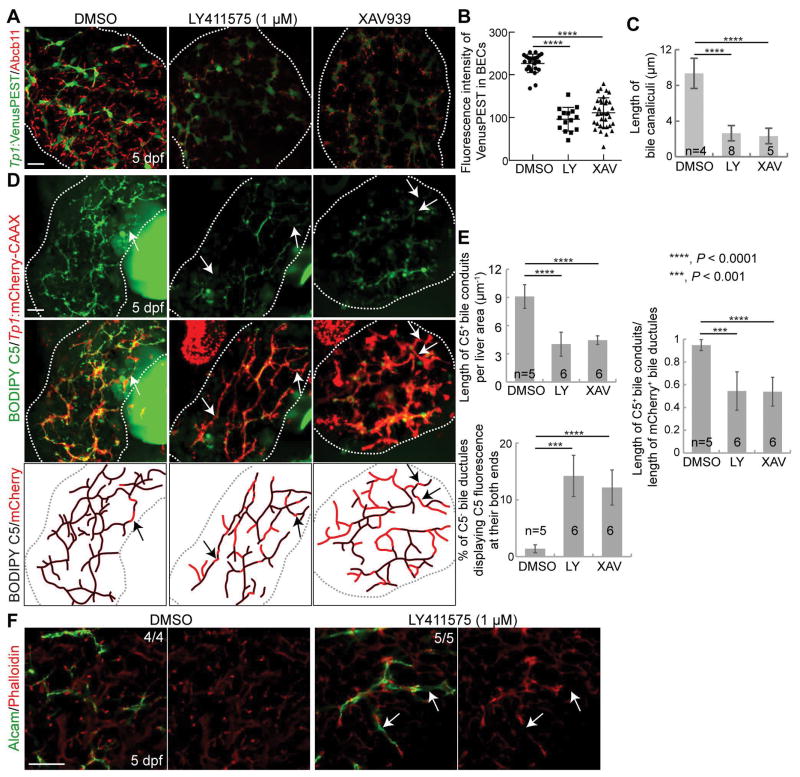
Figure 1. Suppression of Wnt/β-catenin signaling impairs intrahepatic biliary morphogenesis.
(A) Epifluorescence images showing PED6 accumulation in the gallbladder (arrows). Based on PED6 levels in the gallbladder, larvae were divided into three groups: normal, small/faint, and absent. Graph showing the percentage of larvae in each group. (B) Confocal projection images showing BODIPY C5 staining (green) and Tp1:H2B-mCherry expression (red; BEC nuclei) in the liver (dotted lines). Boxed regions are enlarged below. Arrows point to bile ductules; arrowheads to bile canaliculi. Quantification of the length of BODIPY C5+ bile conduits is shown. (C) Confocal projection images showing Abcb11 and Tp1:GFP expression in the liver. Arrowheads point to bile canaliculi. Numbers in the upper right corner indicate the proportion of larvae exhibiting the phenotype shown. Quantification of bile canalicular length is shown. For the quantification, 3–8 livers per condition and 10–20 bile canaliculi per liver were used. (D) Confocal projection images showing BODIPY C5 staining and Tp1:mCherry-CAAX expression in the liver, which reveal bile conduits and ductules, respectively. BODIPY C5+ bile conduits (black lines) and mCherry-CAAX+ bile ductules (red lines) are illustrated and their lengths are quantified in three ways: (1) the length of BODIPY C5+ bile conduits per liver area, (2) the length of BODIPY C5+ bile conduits divided by the length of mCherry-CAAX+ bile ductules, and (3) the percentage of BODIPY C5− bile ductules (arrows), calculated by dividing the length of BODIPY C5−/mCherry-CAAX+ ductules by the length of mCherry-CAAX+ ductules that exhibited BODIPY C5 fluorescence at both ends. (E) Single-optical section images showing Tp1:GFP and F-actin expression. (F) Confocal projection images showing Tp1:GFP-utrophin and Alcam expression. Quantification shows the length of GFP-utrophin+ bile ductules divided by the length of Alcam+ ductules. Arrows in D and E point to disconnected actin filaments. n indicates the number of larvae used for quantification. Scale bars: 100 (A), 20 (B–E) μm; error bars: ±SD.
To reveal bile canaliculi and intrahepatic bile conduits, in addition to the gallbladder, we utilized the fluorescently labeled lipid reporter, BODIPY C5 (27). In DMSO-treated control livers, intricate intrahepatic bile conduits were detected, whereas in XAV939-treated livers, the number of conduits was reduced (Fig. 1B, arrows). Bile canaliculi were thin and elongated in controls, whereas they were round and short in XAV939-treated livers (Fig. 1B, arrowheads), which was confirmed by the expression of Abcb11, a bile salt export pump present in the bile canaliculi of hepatocytes (28) (Fig. 1C, arrowheads). In addition to the pharmacological inhibition, we suppressed Wnt/β-catenin signaling using a genetic tool, Tg(hs:Axin1), which expresses Axin1 upon heat-shock (29). Axin1-overexpressing larvae also exhibited the same biliary defects as observed in XAV939-treated larvae: a poor connection of bile conduits (Fig. S1A, arrows) and stunted, round bile canaliculi (Fig. S1A and S1B, arrowheads).
Since bile canalicular defects can block the flow of bile from hepatocytes to bile ductules, bile conduits may not be fully revealed by BODIPY C5 fluorescence in the Wnt-compromised livers. To minimize the effect of non-functional bile canaliculi on revealing bile conduits by BODIPY C5 fluorescence, we examined BODIPY C5 fluorescence in the Tg(Tp1:mCherry-CAAX) line (30), which expresses a membrane-localized form of mCherry in BECs, allowing for visualization of bile ductules. We quantified the length of bile ductules that exhibited BODIPY C5 fluorescence at both ends, because the BODIPY C5 must pass through the ductules and reveal them if they are open. Quantification of these ductules revealed that ~18% of the length of the biliary network was not perfused by BODIPY C5 fluorescence in XAV939-treated livers compared to ~1.2% in control livers (Fig. 1D, arrows, % of C5− bile ductules displaying C5 fluorescence at their both ends), suggesting a defect in the formation of open ductules. Given the essential role of filamentous actin (F-actin) assembly in lumen (31) and bile conduit formation (32), we examined actin filaments in XAV939-treated livers. Although no difference in BEC morphology (i.e., Tp1:GFP expression) was observed between XAV939-treated and control livers, the F-actin filaments appeared disconnected in XAV939-treated livers (Fig. 1E, arrows). To observe F-actin specifically in the BECs, as F-actin is also present in other cell types in the liver, we generated a Tg(Tp1:GFP-utrophin) line, in which utrophin, fused to GFP, binds to F-actin (33). This transgenic line also revealed disconnected actin filaments in XAV939-treated livers but not in control livers; ~26% of Alcam+ bile ductules did not contain GFP-utrophin+ actin filaments (Fig. 1F, arrows). Altogether, these data suggest that Wnt/β-catenin signaling regulates intrahepatic biliary network formation.
Overactivation of Wnt/β-catenin signaling impairs intrahepatic biliary network formation
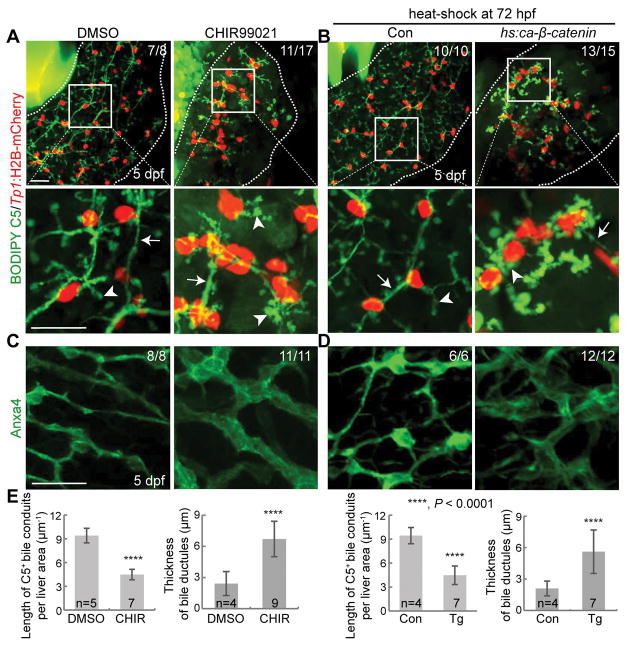
In addition to the suppression of Wnt/β-catenin signaling, we enhanced Wnt/β-catenin signaling using a GSK3 inhibitor, CHIR99021 (34). GSK3 inhibition leads to the accumulation of β-catenin and its subsequent nuclear translocation, thereby enhancing Wnt/β-catenin signaling. CHIR99021 treatment from 72 hpf impaired PED6 accumulation in the gallbladder (Fig. S2A) and resulted in a poor connection of bile conduits and stunted, round bile canaliculi (Fig. 2A). These phenotypes are similar to those observed in Wnt-suppressed livers (Fig. 1 and S1). However, CHIR99021-treated livers had much thicker bile ductules than controls, as clearly revealed by the expression pattern of another BEC marker Anxa4 (35) (Fig. 2C), whereas the thickness was comparable between XAV939-treated and control livers (Fig. 1C). Consistent with the pharmacological activation results, enhancing Wnt/β-catenin signaling with two different genetic tools, Tg(hs:ca-β-catenin) and Tg(hs:wnt2bb), which upon heat-shock, express constitutively active β-catenin and a Wnt ligand Wnt2bb, respectively, also resulted in a poor connection of bile conduits, stunted bile canaliculi, and thickened bile ductules (Fig. 2B, 2D, and S2B–E).
Figure 2. Overactivation of Wnt/β-catenin signaling impairs intrahepatic biliary morphogenesis.
(A, B) Confocal projection images showing BODIPY C5 staining and Tp1:H2B-mCherry expression in the liver (dotted lines) at 5 dpf. Boxed regions are enlarged below. Arrows point to bile ductules; arrowheads to bile canaliculi. (C, D) Confocal projection images showing Anxa4 expression in the liver. Larvae were treated with CHIR99021 from 72 hpf (A, C) or heat-shocked at 72 hpf (B, D). (E) Quantification of the length of BODIPY C5+ bile conduits and quantification of the thickness of bile ductules are shown. For the ductular thickness quantification, 10 bile ductules per liver were used. Scale bars: 20 μm; error bars: ±SD.
Wnt/β-catenin signaling in hepatocytes regulates intrahepatic network formation
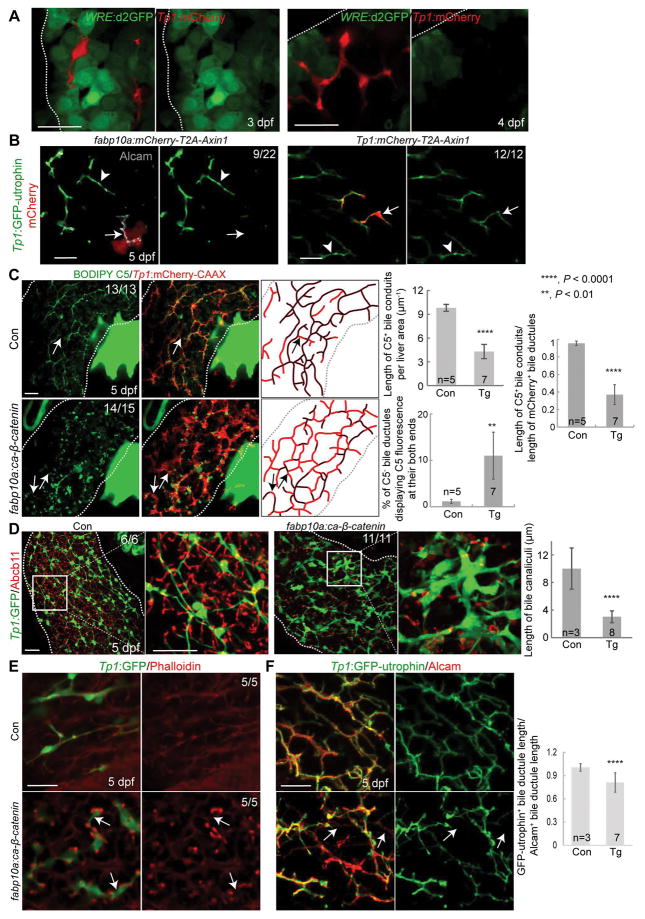
Using a Wnt/β-catenin signaling reporter line, Tg(WRE:d2GFP) (36), we next determined hepatic cell types that exhibit active Wnt/β-catenin signaling. WRE:d2GFP was expressed in hepatocytes, but not BECs, of the liver at 3 dpf (Fig. 3A). At 4 dpf, although WRE:d2GFP expression was still restricted to the hepatocytes, fewer hepatocytes exhibited Wnt activity and the d2GFP intensity was much weaker than at 3 dpf (Fig. 3A). This hepatocyte-specific Wnt/β-catenin activity suggests a cell non-autonomous effect of Wnt/β-catenin signaling on BECs. To test this non-autonomous effect, we injected the fabp10a:mCherry-T2A-Axin1 or Tp1:mCherry-T2A-Axin1 DNA construct to drive Axin1 expression and suppress Wnt/β-catenin signaling in either hepatocytes or BECs, respectively. All Axin1-overexpressing BECs had well-connected actin filaments; however, ~40% of bile ductules that contacted Axin1-overexpressing hepatocytes exhibited disconnected or absent actin filaments (Fig. 3B, arrows), confirming the cell non-autonomous role of Wnt/β-catenin signaling in biliary morphogenesis. Supporting this non-autonomous effect, hepatocyte-specific overactivation of Wnt/β-catenin signaling using the Tg(fabp10a:ca-β-catenin) line (37), which expresses constitutively active β-catenin under the hepatocyte-specific fabp10a promoter, resulted in a poor connection of bile conduits, stunted bile canaliculi, and thickened bile ductules (Fig. 3C and 3D), as observed upon the global overactivation of Wnt/β-catenin signaling (Fig. 2 and Fig. S2C–E). Moreover, Tg(fabp10a:ca-β-catenin) livers exhibited disconnected actin filaments in bile ductules (Fig. 3E and 3F, arrows).
Figure 3. Hepatocytes, but not BECs, exhibit Wnt activity.
(A) Single-optical section images showing WRE:d2GFP (green; Wnt activity) and Tp1:mCherry (red; BECs) expression in the liver (dotted lines). (B) Single-optical section images showing the expression of Tp1:GFP-utrophin (green), mCherry (red; Axin1-overexpressing cells), and Alcam (gray). Arrows point to BECs contacting Axin1-overexpressing hepatocytes or to BECs overexpressing Axin1; arrowheads to bile ductules distant from Axin1-overexpressing cells. Numbers indicate the proportion of clusters exhibiting the phenotype shown. (C) Confocal projection images showing BODIPY C5 staining and Tp1:mCherry-CAAX expression in the liver. BODIPY C5+ bile conduits (black lines) and mCherry-CAAX+ bile ductules (red lines) are illustrated and their lengths are quantified in three ways: (1) the length of BODIPY C5+ bile conduits per liver area, (2) the length of BODIPY C5+ bile conduits divided by the length of mCherry-CAAX+ bile ductules, and (3) the percentage of BODIPY C5− bile ductules (arrows). (D) Confocal projection images showing Abcb11 and Tp1:GFP expression in the liver (dotted lines). Boxed regions are enlarged below. Quantification of bile canalicular length is shown. (E) Single-optical section images showing Tp1:GFP and F-actin expression. (F) Confocal projection images showing Tp1:GFP-utrophin and Alcam expression. Quantification shows the length of GFP-utrophin+ bile ductules divided by that of Alcam+ ductules. Arrows in E and F point to disconnected actin filaments. Scale bars, 20 μm; error bars: ±SD.
Wnt/β-catenin signaling regulates Notch activity in BECs
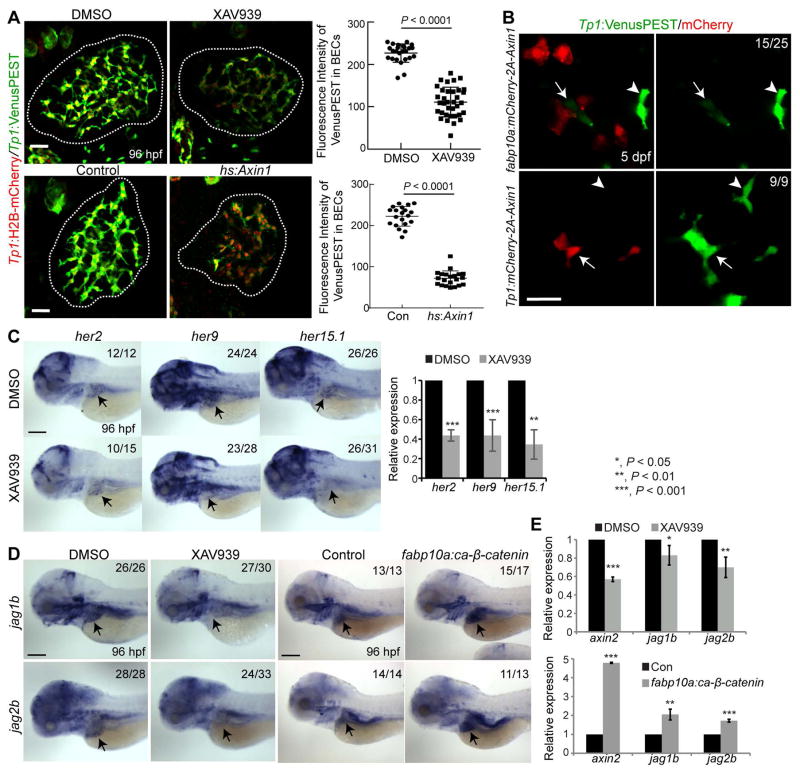
Given the role of Notch signaling in BEC induction and proliferation and biliary morphogenesis (2), we investigated whether Notch signaling was compromised in Wnt-suppressed livers. To reveal the strength of active Notch signaling, we used the Notch reporter line, Tg(Tp1:VenusPEST), which expresses Venus fluorescent proteins fused with a short-lived PEST domain under the promoter containing the Notch-responsive element (38). Tp1:VenusPEST expression was greatly reduced in XAV939-treated and Axin1-overexpressing livers compared with their controls (Fig. 4A), indicating that Wnt/β-catenin signaling positively regulates Notch activity in the developing liver. Tp1:H2B-mCherry expression also revealed such a difference, but the difference was not apparent as with VenusPEST, because H2B-mCherry is a very stable protein. In addition to the global suppression of Wnt/β-catenin signaling, the mosaic suppression of Wnt/β-catenin signaling in hepatocytes reduced Notch activity in BECs that contacted Axin1-overexpressing hepatocytes (Fig. 4B, arrows). However, Notch activity was not reduced in Axin1-overexpressing BECs (Fig. 4B), indicating the cell non-autonomous role of Wnt/β-catenin signaling in controlling hepatic Notch activity.
Figure 4. Wnt/β-catenin signaling regulates Notch activity in BECs.
(A) Confocal projection images showing Tp1:VenusPEST and Tp1:H2B-mCherry expression in the liver at 96 hpf. Quantification of the fluorescence intensity of VenusPEST in BECs is shown. (B) Single-optical section images showing Tp1:VenusPEST (green) and mCherry (red; Axin1-overexpressing cells) expression. Arrows point to BECs contacting Axin1-overexpressing hepatocytes or to BECs expressing Axin1; arrowheads to BECs distant from Axin1-overexpressing cells. (C) WISH images showing her2, her9, and her15.1 expression. qPCR data showing the relative expression levels of these genes between control and XAV939-treated livers at 96 hpf. (D) WISH images showing jag1b and jag2b expression. Numbers indicate the proportion of larvae exhibiting the representative expression shown. Arrows point to the liver. Lateral view, anterior to the left. (E) qPCR data showing the relative expression levels of axin2, jag1b, and jag2b between control and XAV939-treated or ca-β-catenin-overexpressing livers at 96 hpf. axin2, a well-known target of Wnt/β-catenin signaling, was used as a positive control. Scale bars: 20 (A, B), 100 (C, D) μm; error bars: ±SD.
In addition to the Notch reporter lines, we examined the expression of Hes genes, well-known targets of Notch signaling. By whole-mount in situ hybridization (WISH) analysis, we first identified Hes genes that are restrictively expressed in BECs in the liver: her2, her9, and her15.1 (Fig. S3A; arrows). Their hepatic expression was significantly reduced in XAV939-treated larvae compared with controls (Fig. 4C), further supporting the positive role of Wnt/β-catenin signaling in controlling hepatic Notch signaling.
In colorectal cancer cells, Wnt/β-catenin signaling regulates Jag1 expression and thereby controls Notch activity (39). We investigated if such regulation also occurred in the developing liver. Jag1 is a key Notch ligand gene in the liver: mouse Jag1 mutants exhibit bile duct paucity (40) and human JAG1 mutation leads to Allagile syndrome (1). In zebrafish, although four jagged genes (jag1a, jag1b, jag2a, and jag2b) are expressed in the developing liver, jag1b and jag2b appear to be required for biliary formation (41). By WISH, we detected clear hepatic expression of all four jagged genes at 72 hpf (Fig. S3B; arrows). The overall staining in the entire liver suggests their expression in hepatocytes. Using a gene trap line that expresses GFP under the endogenous jag1b promoter (42), we confirmed jag1b expression in hepatocytes (Fig. S3C). Moreover, we observed the localized expression of jag1b around BECs at 60 and 72, but not 96, hpf (Fig. S3D). A subset of BECs at these earlier stages also expressed jag1b (Fig. S3D, arrows). Intriguingly, both jag1b and jag2b expression were reduced in XAV939-treated livers and enhanced in ca-β-catenin-overexpressing livers compared with their controls (Fig. 4D), which was further confirmed by quantitative RT-PCR (qPCR) (Fig. 4E). Using the jag1b:GFP line, we additionally revealed that the change of jag1b expression occurred in hepatocytes but not in BECs (Fig. S3E and S3F). Three of the four Notch receptor genes in zebrafish, notch1a, notch1b, and notch2, were broadly expressed in the liver, as previously reported (41), whereas notch3 was restrictively expressed in BECs (Fig. S4A). This BEC-restricted expression was confirmed by co-expression of notch3 with Tp1:GFP (Fig. S4B). Unlike jag1b and jag2b, notch3 expression was not significantly changed in Wnt-suppressed livers compared with controls (Fig. S4C).
Restoring Notch activity rescues intrahepatic biliary defects observed in XAV939-treated livers
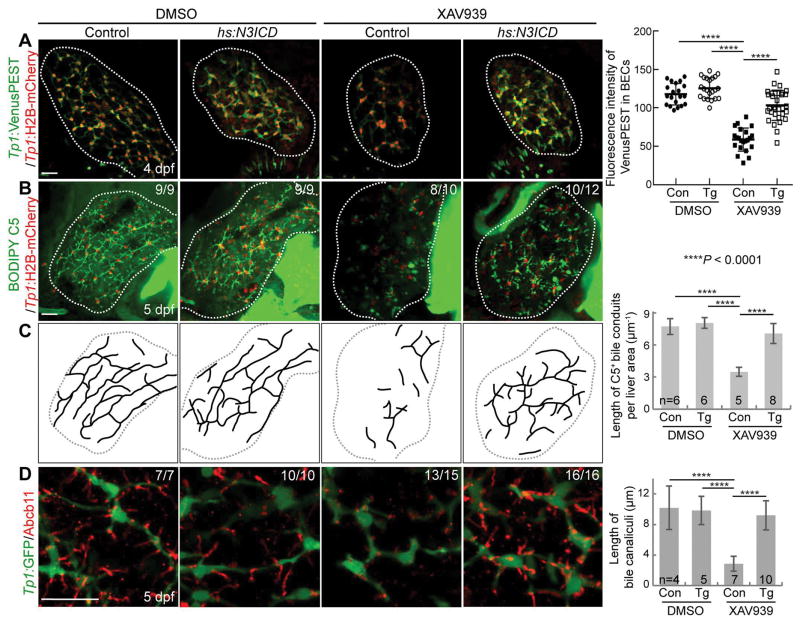
We next determined if the repressed Notch activity in XAV939-treated livers caused the intrahepatic biliary defects. If this were the case, reducing Notch activity to the same level as that detected in XAV939-treated livers should reproduce the biliary defects observed in XAV939-treated livers. First, we determined the dosage of a Notch inhibitor, LY411575, at which hepatic Notch activity is reduced to the level observed in XAV939-treated livers. Livers treated with 10 or 5 μM LY411575 exhibited almost no Notch activity (data not shown), whereas livers treated with 1 μM LY411575 exhibited a similar level of Notch activity to XAV939-treated livers (Fig. 5A and 5B). Importantly, 1 μM LY411575 treatment resulted in stunted bile canaliculi (Fig. 5C), a poor connection of bile conduits (Fig. 5D and 5E), and disconnected actin filaments (Fig. 5F, arrows), as XAV933 treatment did. To further prove that repressed Notch activity in XAV939-treated livers caused the intrahepatic biliary defects, we performed a rescue experiment using the Tg(hs:N3ICD) line that upon heat-shock, expresses the intracellular domain of Notch3 (43). Intriguingly, upon the expression of N3ICD via a single heat-shock at 72 hpf, the biliary defects observed in XAV939-treated livers were significantly rescued. Notch activity, which was assessed by Tp1:VenusPEST expression, was greatly increased at 4 dpf in N3ICD-overexpressing, XAV939-treated livers compared with only XAV939-treated livers (Fig. 6A), validating the efficacy of the Tg(hs:N3ICD) line. N3ICD overexpression significantly improved the connection of bile conduits (Fig. 6B and 6C) and the morphology of bile canaliculi (Fig. 6D) in XAV939-treated livers. These data clearly indicate that Wnt/β-catenin signaling controls intrahepatic biliary morphogenesis by regulating Notch signaling.
Figure 5. Reducing Notch activity results in intrahepatic biliary defects, as observed in XAV939-treated livers.
(A–C) Confocal projection images showing Abcb11 and Tp1:VenusPEST expression in the liver. Quantification of the fluorescence intensity of VenusPEST in BECs (B) and of bile canalicular length (C) is shown. Larvae were treated with LY411575 or XAV939 from 72 hpf to 5 dpf. (D) Confocal projection images showing BODIPY C5 staining and Tp1:mCherry-CAAX expression in the liver. BODIPY C5+ bile conduits (black lines) and mCherry-CAAX+ bile ductules (red lines) are illustrated. (E) Quantification of the data shown in (D) in three ways: (1) the length of BODIPY C5+ bile conduits per liver area, (2) the length of BODIPY C5+ bile conduits divided by the length of mCherry-CAAX+ bile ductules, and (3) the percentage of BODIPY C5− bile ductules (arrows). (F) Quantification of the length of bile conduits drawn in (E). Scale bars: 20 μm; error bars: ±SD.
Figure 6. Restoring Notch activity rescues the intrahepatic biliary defects observed in XAV939-treated livers.
(A) Confocal projection images showing Tp1:VenusPEST and Tp1:H2B-mCherry expression in the liver. Quantification of the fluorescence intensity of VenusPEST in BECs is shown. (B) Confocal projection images showing Tp1:H2B-mCherry expression and BODIPY C5 staining in the liver. (C) Illustration showing bile conduits revealed by BODIPY C5 staining shown in B and its quantification. (D) Confocal projection images showing Abcb11 (red; bile canaliculi) and Tp1:GFP (green; BECs) expression in the liver. Quantification of bile canalicular length is shown. Scale bars: 20 μm; error bars: ±SD.
Discussion
In this study, we provide several lines of evidence supporting the role of Wnt/β-catenin signaling in intrahepatic biliary morphogenesis. First, suppressing or enhancing Wnt/β-catenin signaling with pharmacological or genetic tools impaired intrahepatic biliary morphogenesis. Second, the suppression of Wnt/β-catenin signaling greatly reduced Notch activity in BECs. Third, Wnt/β-catenin signaling in hepatocytes regulated the expression of the Notch ligand genes, jag1b and jag2b, thereby controlling Notch activity. Last, restoring Notch activity rescued intrahepatic biliary defects observed in Wnt-suppressed livers. Altogether, these data demonstrate a novel role for Wnt/β-catenin signaling in intrahepatic biliary network formation as the upstream regulator of Notch signaling.
Notch signaling is a key regulator of biliary development: it regulates biliary specification and intrahepatic biliary morphogenesis (2). Despite these key roles, the upstream regulator of Notch signaling in the liver remains unknown. Our finding that Wnt/β-catenin signaling regulates Notch activity by promoting the expression of Notch ligand genes provides the first example of such an upstream regulator in the developing liver. Wnt/β-catenin signaling has been known to regulate Notch activity in other tissues. Wnt/β-catenin signaling directly regulates Jag1 expression, thereby inducing Notch signaling in colorectal cancer cells (39); it also regulates the expression of Dll1, another Notch ligand gene, during somitogenesis, thereby inducing Notch activity in the pre-somitic mesoderm (44). In addition, activated β-catenin stimulates Jag1 expression in osteoblasts, thereby inducing Notch activity in hematopoietic stem cell progenitors, which can lead to leukemia (45). Likewise, we found that in the developing zebrafish liver, Wnt/β-catenin signaling in hepatocytes regulates jag1b and jag2b expression, thereby controlling Notch activity in BECs.
Although suppressing and overactivating Wnt/β-catenin signaling both impaired biliary morphogenesis, the biliary defects observed in Wnt-overactivated livers did not appear to be due to abnormal Notch activity, because Notch activity did not appear to be affected in Wnt-overactivated livers (Fig. S5). Moreover, overactivation of Notch signaling via N3ICD expression did not impair biliary morphogenesis (Fig. 6B), suggesting that increased Notch activity does not compromise biliary morphogenesis. N3ICD expression also failed to rescue biliary defects in ca-β-catenin-expressing livers (Fig. S6) unlike in XAV939-treated livers (Fig. 6), suggesting that Notch activity was not reduced in Wnt-overactivated livers. Although Wnt overactivation increased hepatic expression of jag1b and jag2b, it appears to affect biliary morphogenesis via a mediator other than Notch signaling.
During zebrafish biliary morphogenesis, Wnt and Notch activities were observed only in hepatocytes and BECs, respectively. However, global suppression of Notch signaling affected not only bile conduits but also the bile canaliculi of hepatocytes. In addition, global activation of Notch signaling rescued the defects in the morphology of bile canaliculi in Wnt-suppressed livers. These data raise the possibility that the formation of bile canaliculi and bile ductules are interdependent. Supporting this possibility, blocking the formation of bile canaliculi impairs intrahepatic biliary network formation in mice (3, 46). This interdependence makes it very difficult to determine whether the suppression of Wnt/β-catenin signaling primarily affects bile canaliculi or bile ductules.
Intrahepatic biliary morphogenesis continues between 4 and 5 dpf: bile ductules further elongate and become thinner (21, 22, 32). However, suppressing Wnt/β-catenin signaling during this period did not impair intrahepatic biliary morphogenesis or reduce Notch activity (Fig. S7). In contrast, the complete inhibition of Notch signaling during this period impairs biliary morphogenesis (22), indicating the continuous role of Notch signaling in biliary morphogenesis between 4 and 5 dpf. Given that hepatic Wnt activity is very weak at 96 hpf and only a small subset of hepatocytes exhibit the weak Wnt activity (Fig. 3A), Wnt/β-catenin signaling does not appear to regulate hepatic Notch activity from 4 dpf. It will be interesting to identify another upstream regulator of Notch signaling in the developing liver.
We sought to identify key component genes of the Wnt/β-catenin and Notch signaling pathways that are essential for intrahepatic biliary morphogenesis. For Wnt/β-catenin signaling, we examined biliary functions and morphology in lef1 and tcf7 single and double mutants, given their relatively high expression in the developing liver compared with three other Tcf/Lef family member genes, tcf7l1a, tcf7l1b, and tcf7l2 (Fig. S8C). However, there was no biliary defect in these mutants (data not shown), suggesting that the three other genes may compensate for the absence of lef1 and tcf7 in the developing liver. We also examined the expression of Wnt receptor fzd and co-receptor lrp genes, and found that six fzd genes and both lrp5 and lrp6 were expressed in the developing liver (Fig. S8A and B; arrows). For Notch signaling, we examined biliary functions and morphology in jag1b mutants, given its positive regulation by Wnt/β-catenin signaling (Fig. 4E and S3D). However, there was no biliary defect in these mutants (data not shown), suggesting the involvement of other Notch ligand genes, such as jag1a, jag2a, and jag2b, in biliary morphogenesis. Given published morpholino-mediated jagged knockdown studies (41) and the positive regulation of jag2b in the developing liver by Wnt/β-catenin signaling (Fig. 4E), jag2b together with jag1b may regulate intrahepatic biliary morphogenesis in zebrafish. Although our mutant analyses failed to reveal a key component of the Wnt/β-catenin or Notch signaling pathway that is essential for biliary morphogenesis, these negative data suggest high redundancy among the Tcf/Lef family member and Notch ligand genes.
In the zebrafish liver, most BECs directly contact surrounding hepatocytes (47), whereas in the mammalian liver, only BECs that reside in intrahepatic bile ductules directly contact hepatocytes. Although in the healthy mouse liver, BECs in bile ductules are a small portion of the total BECs, in the injured liver, bile ductules elongate via the proliferation of BECs, making longer and denser bile ductules (48, 49). These extended ductules appear to replace collapsed bile canaliculi between damaged hepatocytes, thereby restoring bile drain. This temporarily established intrahepatic biliary network is very similar to the intrahepatic biliary network in zebrafish because BECs in the network directly contact hepatocytes. In the healthy adult liver, Wnt activity is restricted to hepatocytes near the central veins (i.e., pericentral hepatocytes); however, upon liver injury, Wnt activity is induced in other hepatocytes (13). Intriguingly, in β-catenin conditional knockout mice which lack β-catenin in both hepatocytes and BECs, the number of activated BECs, also called oval cells, induced by 3,5-diethoxycarbonyl-1,4-dihydrocollidine treatment was dramatically reduced compared with wild-type controls (50). Since the reduced BEC number should cause shorter and sparser bile ductules, the mutant data suggest the involvement of Wnt/β-catenin signaling in intrahepatic biliary morphogenesis during liver injury. In the future, it will be interesting to test whether the Wnt/β-catenin and Notch crosstalk observed during zebrafish liver development also regulates intrahepatic biliary morphogenesis following liver injury.
Supplementary Material
Acknowledgments
Financial support: The work was supported in part by grants from the NIH to D.S. (DK101426, DK109365), M.K. (DK105714), K.J.E. (K08CA172288), and D.Y.R.S. (DK60322) and from the Packard foundation to D.Y.R.S.
We thank Bruce Appel for Tg(hs:N3ICD), Jian Zhang for jag1b, Richard Dorsky for lef1, and Koichi Kawakami for tcf7 fish. We also thank Jinrong Peng for anti-Bhmt antibodies, Beth Roman for in situ probes, and Neil Hukriede and Michael Tsang for discussion.
List of abbreviations
- BECs
biliary epithelial cells
- hpf
hours post-fertilization
- dpf
days post-fertilization
- F-actin
filamentous actin
- WISH
whole-mount in situ hybridization
- qPCR
quantitative RT-PCR
References
Author names in bold designate shared co-first authorship.
- 1.Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 2.Geisler F, Strazzabosco M. Emerging roles of Notch signaling in liver disease. Hepatology. 2015;61:382–392. doi: 10.1002/hep.27268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanimizu N, Kaneko K, Itoh T, Ichinohe N, Ishii M, Mizuguchi T, Hirata K, et al. Intrahepatic bile ducts are developed through formation of homogeneous continuous luminal network and its dynamic rearrangement in mice. Hepatology. 2016;64:175–188. doi: 10.1002/hep.28521. [DOI] [PubMed] [Google Scholar]
- 4.Raynaud P, Carpentier R, Antoniou A, Lemaigre FP. Biliary differentiation and bile duct morphogenesis in development and disease. Int J Biochem Cell Biol. 2011;43:245–256. doi: 10.1016/j.biocel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 5.Falix FA, Weeda VB, Labruyere WT, Poncy A, de Waart DR, Hakvoort TB, Lemaigre F, et al. Hepatic Notch2 deficiency leads to bile duct agenesis perinatally and secondary bile duct formation after weaning. Dev Biol. 2014;396:201–213. doi: 10.1016/j.ydbio.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Tchorz JS, Kinter J, Muller M, Tornillo L, Heim MH, Bettler B. Notch2 signaling promotes biliary epithelial cell fate specification and tubulogenesis during bile duct development in mice. Hepatology. 2009;50:871–879. doi: 10.1002/hep.23048. [DOI] [PubMed] [Google Scholar]
- 7.Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136:1727–1739. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeliazkova P, Jors S, Lee M, Zimber-Strobl U, Ferrer J, Schmid RM, Siveke JT, et al. Canonical Notch2 signaling determines biliary cell fates of embryonic hepatoblasts and adult hepatocytes independent of Hes1. Hepatology. 2013;57:2469–2479. doi: 10.1002/hep.26254. [DOI] [PubMed] [Google Scholar]
- 9.Geisler F, Nagl F, Mazur PK, Lee M, Zimber-Strobl U, Strobl LJ, Radtke F, et al. Liver-specific inactivation of Notch2, but not Notch1, compromises intrahepatic bile duct development in mice. Hepatology. 2008;48:607–616. doi: 10.1002/hep.22381. [DOI] [PubMed] [Google Scholar]
- 10.Kodama Y, Hijikata M, Kageyama R, Shimotohno K, Chiba T. The role of notch signaling in the development of intrahepatic bile ducts. Gastroenterology. 2004;127:1775–1786. doi: 10.1053/j.gastro.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Sparks EE, Huppert KA, Brown MA, Washington MK, Huppert SS. Notch signaling regulates formation of the three-dimensional architecture of intrahepatic bile ducts in mice. Hepatology. 2010;51:1391–1400. doi: 10.1002/hep.23431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, et al. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 2005;19:1849–1854. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monga SP. Role and regulation of beta-catenin signaling during physiological liver growth. Gene Expr. 2014;16:51–62. doi: 10.3727/105221614X13919976902138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussain SZ, Sneddon T, Tan X, Micsenyi A, Michalopoulos GK, Monga SPS. Wnt impacts growth and differentiation in ex vivo liver development. Experimental Cell Research. 2004;292:157–169. doi: 10.1016/j.yexcr.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Decaens T, Godard C, de Reynies A, Rickman DS, Tronche F, Couty JP, Perret C, et al. Stabilization of beta-catenin affects mouse embryonic liver growth and hepatoblast fate. Hepatology. 2008;47:247–258. doi: 10.1002/hep.21952. [DOI] [PubMed] [Google Scholar]
- 16.Cordi S, Godard C, Saandi T, Jacquemin P, Monga SP, Colnot S, Lemaigre FP. Role of beta-catenin in development of bile ducts. Differentiation. 2016;91:42–49. doi: 10.1016/j.diff.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiyohashi K, Kakinuma S, Kamiya A, Sakamoto N, Nitta S, Yamanaka H, Yoshino K, et al. Wnt5a signaling mediates biliary differentiation of fetal hepatic stem/progenitor cells in mice. Hepatology. 2013;57:2502–2513. doi: 10.1002/hep.26293. [DOI] [PubMed] [Google Scholar]
- 18.Farber SA, Pack M, Ho SY, Johnson ID, Wagner DS, Dosch R, Mullins MC, et al. Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science. 2001;292:1385–1388. doi: 10.1126/science.1060418. [DOI] [PubMed] [Google Scholar]
- 19.Carten JD, Bradford MK, Farber SA. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Developmental Biology. 2011;360:276–285. doi: 10.1016/j.ydbio.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- 21.Ningappa M, So J, Glessner J, Ashokkumar C, Ranganathan S, Min J, Higgs BW, et al. The role of ARF6 in biliary atresia. PLoS One. 2015 doi: 10.1371/journal.pone.0138381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorent K, Moore JC, Siekmann AF, Lawson N, Pack M. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev Dyn. 2010;239:855–864. doi: 10.1002/dvdy.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkins BJ, Pack M. Zebrafish models of human liver development and disease. Compr Physiol. 2013;3:1213–1230. doi: 10.1002/cphy.c120021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang S-Ma, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud Ga, Charlat O, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 25.Shin D, Lee Y, Poss KD, Stainier DYR. Restriction of hepatic competence by Fgf signaling. Development. 2011;138:1339–1348. doi: 10.1242/dev.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goessling W, North TE, Lord AM, Ceol C, Lee S, Weidinger G, Bourque C, et al. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Developmental Biology. 2008;320:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- 27.Delous M, Yin C, Shin D, Ninov N, Debrito Carten J, Pan L, Ma TP, et al. Sox9b is a key regulator of pancreaticobiliary ductal system development. PLoS Genet. 2012;8:e1002754. doi: 10.1371/journal.pgen.1002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerloff T, Stieger B, Hagenbuch B, Madon J, Landmann L, Roth J, Hofmann AF, et al. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J Biol Chem. 1998;273:10046–10050. doi: 10.1074/jbc.273.16.10046. [DOI] [PubMed] [Google Scholar]
- 29.Kagermeier-Schenk B, Wehner D, Ozhan-Kizil G, Yamamoto H, Li J, Kirchner K, Hoffmann C, et al. Waif1/5T4 inhibits Wnt/beta-catenin signaling and activates noncanonical Wnt pathways by modifying LRP6 subcellular localization. Dev Cell. 2011;21:1129–1143. doi: 10.1016/j.devcel.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 30.Pestel J, Ramadass R, Gauvrit S, Helker C, Herzog W, Stainier DY. Real-time 3D visualization of cellular rearrangements during cardiac valve formation. Development. 2016;143:2217–2227. doi: 10.1242/dev.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hogan BL, Kolodziej PA. Organogenesis: molecular mechanisms of tubulogenesis. Nat Rev Genet. 2002;3:513–523. doi: 10.1038/nrg840. [DOI] [PubMed] [Google Scholar]
- 32.Dimri M, Bilogan C, Pierce LX, Naegele G, Vasanji A, Gibson I, McClendon A, et al. Three-dimensional structural analysis reveals a Cdk5-mediated kinase cascade regulating hepatic biliary network branching in zebrafish. Development. 2017;144:2595–2605. doi: 10.1242/dev.147397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burkel BM, von Dassow G, Bement WM. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil Cytoskeleton. 2007;64:822–832. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ring DB, Johnson KW, Henriksen EJ, Nuss JM, Goff D, Kinnick TR, Ma ST, et al. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes. 2003;52:588–595. doi: 10.2337/diabetes.52.3.588. [DOI] [PubMed] [Google Scholar]
- 35.Zhang D, Golubkov VS, Han W, Correa RG, Zhou Y, Lee S, Strongin AY, et al. Identification of Annexin A4 as a hepatopancreas factor involved in liver cell survival. Dev Biol. 2014;395:96–110. doi: 10.1016/j.ydbio.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu N, Kawakami K, Ishitani T. Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/beta-catenin signaling-reporter transgenic zebrafish. Developmental Biology. 2012;370:71–85. doi: 10.1016/j.ydbio.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 37.Evason KJ, Francisco MT, Juric V, Balakrishnan S, del Lopez Pazmino MP, Gordan JD, Kakar S, et al. Identification of Chemical Inhibitors of beta-Catenin-Driven Liver Tumorigenesis in Zebrafish. PLoS Genet. 2015;11:e1005305. doi: 10.1371/journal.pgen.1005305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ninov N, Borius M, Stainier DYR. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development. 2012;139:1557–1567. doi: 10.1242/dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodilla V, Villanueva A, Obrador-Hevia A, Robert-Moreno A, Fernandez-Majada V, Grilli A, Lopez-Bigas N, et al. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc Natl Acad Sci U S A. 2009;106:6315–6320. doi: 10.1073/pnas.0813221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofmann JJ, Zovein AC, Koh H, Radtke F, Weinmaster G, Iruela-Arispe ML. Jagged1 in the portal vein mesenchyme regulates intrahepatic bile duct development: insights into Alagille syndrome. Development. 2010;137:4061–4072. doi: 10.1242/dev.052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorent K, Yeo SY, Oda T, Chandrasekharappa S, Chitnis A, Matthews RP, Pack M. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development. 2004;131:5753–5766. doi: 10.1242/dev.01411. [DOI] [PubMed] [Google Scholar]
- 42.Ma WR, Zhang J. Jag1b is essential for patterning inner ear sensory cristae by regulating anterior morphogenetic tissue separation and preventing posterior cell death. Development. 2015;142:763–773. doi: 10.1242/dev.113662. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Pan L, Moens CB, Appel B. Notch3 establishes brain vascular integrity by regulating pericyte number. Development. 2014;141:307–317. doi: 10.1242/dev.096107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hofmann M, Schuster-Gossler K, Watabe-Rudolph M, Aulehla A, Herrmann BG, Gossler A. WNT signaling, in synergy with T/TBX6, controls Notch signaling by regulating Dll1 expression in the presomitic mesoderm of mouse embryos. Genes Dev. 2004;18:2712–2717. doi: 10.1101/gad.1248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kode A, Manavalan JS, Mosialou I, Bhagat G, Rathinam CV, Luo N, Khiabanian H, et al. Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature. 2014;506:240–244. doi: 10.1038/nature12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woods A, Heslegrave AJ, Muckett PJ, Levene AP, Clements M, Mobberley M, Ryder TA, et al. LKB1 is required for hepatic bile acid transport and canalicular membrane integrity in mice. Biochem J. 2011;434:49–60. doi: 10.1042/BJ20101721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao YL, Lin JX, Yang P, Chen QS, Chu XH, Gao C, Hu JH. Fine Structure, Enzyme Histochemistry, and Immunohistochemistry of Liver in Zebrafish. Anatomical Record-Advances in Integrative Anatomy and Evolutionary Biology. 2012;295:567–576. doi: 10.1002/ar.22416. [DOI] [PubMed] [Google Scholar]
- 48.Kaneko K, Kamimoto K, Miyajima A, Itoh T. Adaptive remodeling of the biliary architecture underlies liver homeostasis. Hepatology. 2015;61:2056–2066. doi: 10.1002/hep.27685. [DOI] [PubMed] [Google Scholar]
- 49.Kamimoto K, Kaneko K, Kok CYY, Okada H, Miyajima A, Itoh T. Heterogeneity and stochastic growth regulation of biliary epithelial cells dictate dynamic epithelial tissue remodeling. Elife. 2016:5. doi: 10.7554/eLife.15034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Apte U, Thompson MD, Cui SS, Liu B, Cieply B, Monga SPS. Wnt/beta-catenin signaling mediates oval cell response in rodents. Hepatology. 2008;47:288–295. doi: 10.1002/hep.21973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.