Abstract
RNA transcript expression estimates are a promising method to study the mechanisms and classification of renal allograft rejections. Here we use the Nanostring platform to profile RNA expression in renal allografts in a non-human primate (NHP), the Cynomolgus monkey. We analyzed protocol and indication 278 archival renal allograft samples, both protocol and indication from 76 animals with diagnoses of chronic antibody (CAMR), acute cellular rejection (TCMR) and MIXED (both CAMR and TCMR), plus normals and samples with no pathological rejection using a Cynomolgus specific probe set of 67 genes. Analysis identified RNA expression heterogeneity of endothelial and NK genes within CAMR and TCMR, including the stages of CAMR. Three factors were partitioned into additional groups. One group with the longest allograft survival time is pure CAMR without NK or CD3. Three mixed groups show variation in NK and CD3. TCMR was split into two groups with variation in NK genes. Additional validation of the complete gene set correlated many of the genes with diagnoses of CAMR, MIXED, and TCMR rejections and with Banff histologic criteria defined in human subjects. These NHP data demonstrate the utility of RNA expression profiling to identify additional heterogeneity of endothelial and NK RNA gene expressions.
INTRODUCTION
RNA expression is now commonly used in transplantation to identify important inflammatory patterns in rejection. A large body of transcriptome-wide microarray data now exists to define the molecular phenotype of renal allograft rejection (1–8). Although exciting, results are variable as not all investigators identify uniform gene expression patterns of RNA expression (1, 6, 9–13).
The nCounter Analysis System (NanoString Technologies, Seattle, WA, USA) is a novel RNA gene expression platform, both highly multiplexed and flexible, and works reliably with RNA isolates derived from formalin fixed paraffin embedded (FFPE) tissue (Geiss 2008). More sensitive than microarrays and similar in sensitivity to real-time PCR (14, 15), this platform efficiently uses small archival FFPE tissue samples permitting correlation of RNA expression with covariates. In addition, for human subjects, this new technology might potentially permit its use on archival tissue to correlate retrospective outcome measures like allograft function, allograft survival, comorbidities and therapy (16). We have previously demonstrated the feasibility of the NanoString system with routine FFPE transplantation pathology samples in renal and cardiac allograft biopsies (17–19).
Cynomolgus renal allograft models using bone marrow transplantation (BMT, mixed chimerism), used extensively for the evaluation of novel transplantation protocols (20–24), employ no immunosuppression one month post induction, thereby allowing such renal allografts to represent the natural history of untreated allograft rejection post induction. These animals have protocol and indication kidney specimens, for which residual archived FFPE tissue remains.
The goal of the current study is to use FFPE Cynomolgus renal allograft specimens to identify additional variation in endothelial and NK gene expression with chronic antibody (CAMR) and T cell mediated (TCMR) rejections.
METHODS
Gene Set
The gene set includes 67 oligonucleotides previously described probes specific to the Macaca fascicularis transcriptome and was manufactured by Integrated DNA Technologies (Coralville, IA, USA) (17). The gene set includes a previously described gene-set comprised of endothelial, NK cell, and inflammation-related genes (17), plus additional transplantation immunology-associated genes, and 4 housekeeping genes. Only one B cell gene is present, MS4A1, (CD20). No genes for alternate macrophage activation are present. This gene set is derived from informative genes in human renal allograft rejection (25) (Table S1).
Gene expression analysis and RNA Isolation
Three consecutive 20-μm curls cut from each FFPE block were immediately transferred to sterile microcentrifuge tubes and stored at room temperature. Microtome blades were then replaced and equipment sterilization with RNase AWAY (Life Technologies, Carlsbad, CA, USA) between blocks. Curls Xylene deparaffinization and RNA extraction were performed with the RecoverAll™ Total Nucleic Acid Isolation Kit for FFPE (Life Technologies, Carlsbad, CA, USA). RNA concentration and purity were measured with a Nano-Drop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Gene expression was then quantified using the nCounter Elements assay (NanoStringTechnologies) with the FFPE tissue derived RNA isolates (17, 19). Quality control assessment and normalization were performed with nSolver Analysis Software version 3.0 (NanoString Technologies). The manufacturer recommended default parameters for quality control flagging were used. Each sample was first normalized to the geometric mean of the positive controls (with default flagging of normalization factors <0.3 and >3), followed by normalization to the geometric mean of the house keeping genes (with default flagging of normalization factors <0.1 and >10).
Animals
76 animals underwent one of two protocols, in which induction medications were used with bone marrow transplantation (BMT) at day zero (standard protocol) or delayed at about four months (delayed protocol). Induction cyclosporine was stopped at one month post BMT (20–24). Control animals did not receive BMT. The data set includes 76 animals with one to 11 biopsies over a time period of zero to 5983 days post transplantation (n = 278). 17 normal biopsies were from perioperative normal native kidneys from 17 allografted recipients were also included. Samples included protocol and indication biopsies, plus autopsy nephrectomies at euthanasia. Specimens were not included from animals with viral infections (BK or CMV), nephrolithiasis, or post-transplant lymphoproliferative disease. This animal model for the study of renal allograft rejection has many advantages compared to humans. The donors and recipients are healthy, and the graft is implanted within 30 minutes, eliminating the co-morbidities of deceased donors and variable peri-implantation time. In addition, the animals lack recurring native renal disease and human co-morbidities such as cardiac disease and diabetes. Chronic immunosuppression is not used eliminating the co-morbidities of drug toxicities. All surgical procedures and postoperative care were carried out in accordance with NIH guidelines and approved by the MGH subcommittee on Animal Research.
Alloantibodies
Alloantibodies were identified by indirect flow cytometry on donor T and B cells and scored as nominal data as either positive or negative (23). Anti-donor specific alloantibodies were assayed by flow cytometric analysis in post-treatment and all pre-treatment samples. Donor peripheral blood mononuclear cells (PBMC) were isolated from heparinized blood by gradient centrifugation over freshly prepared 60% Percoll (Pharmacia Biotech, Uppsala, Sweden). Contaminating red blood cells were removed by standard water-shock treatment. PBMCs were incubated with recipient sera for 30 min at 4◦C. After washing, FITC conjugated mouse anti-human IgG mAb was added and incubated for 30 min at 4◦C, then washed twice. PBMC were further incubated with PE conjugated anti-CD20 mAb (Becton-Dickinson, Mountain View, CA) for 30 min at 4◦C. After washing, PBMC were fixed with 2% paraformaldehyde. Cells were then with FACScan (Becton Dickinson, Mountain View, CA). A positive reaction was defined as a mean channel of fluorescence (MCF) of >10 for T cells and >50 for B cells. These thresholds are greater than two standard deviations above the mean channel level of donor cells stained with pre-treatment serum (4.8 ± 1.7 and 23.3 ± 12.5 for T and B cells, respectively). Anti-T cell and anti-B cell alloantibodies cross reactive on human HLA class I and class II antigens, respectively. Class I or class II alloantibodies were not distinguished.
Pathology
Renal biopsies or autopsied nephrectomies (N =278) from 76 animals were scored and interpreted using Banff criteria (26), including Banff grades of rejection and Banff ordinal criteria scores. Banff rejection grades were computed in the Filemaker database with equations using Banff criteria. The stages of chronic antibody mediated rejection (23) are graded from zero to 4. Stage zero is without alloantibody or C4d and with a normal creatinine. Stage one is positive alloantibody without C4d and a normal creatinine. Stage two is positive alloantibody and C4d with a normal creatinine. Stage three is positive alloantibody and C4d, and cg>0 with normal or slightly elevated creatine. Stage 4 is positive alloantibody, C4d, and cg>1 (usually cg=3) with elevated and rising creatinine (> 1.5). Five broad diagnostic groups were created for analyses of gene validations: 1) normal native kidneys; 2) chronic antibody mediated rejection (CAMR), with no pathological evidence of TCMR (Banff less than borderline) but with alloantibodies, positive C4d, and various stages of chronic CAMR; 3) TCMR, Banff TCMR with grades from borderline to 3, and with no alloantibodies and no C4d; 4) MIXED rejections which showed various grades of TCMR and CAMR; 5) No evidence of pathological rejection (NPR) is defined as an allograft biopsy with no pathological evidence of either cellular (Banff less than borderline) or CAMR (without DSA and/or positive C4d, no transplant glomerulopathy (no CG) and a normal creatinine.
Statistical analyses
All analyses and graphical figures were performed using JMP Pro, version 13 software (SAS) with default parameters. Programs used were the Fit Model Platform Standard Least Squares (scaled estimates), K-means clustering, Mixed Model, Factor Analysis with Varimax rotation, Life Distribution, Proportional Hazard, and Validation. All p values were adjusted for false discovery (JMP Addin). Raw gene copy number data are non-parametric as are the log10 transformed raw data. Gene expression levels are highly interactive with collinearity. Initial multivariate regressions showed that sample type (biopsy vs autopsy), therapy type, and individual animals showed no significant effect on the RNA expression estimates. However, the time post-transplant affected both the magnitude of the scaled estimates and the p value of some gene estimates. Therefore, significant individual gene expressions and pathways were identified by the scaled estimates controlling for the time post-transplant as a fixed effect. Significant individual genes were quantified by the scaled estimates (Mean, Median = 0, Range -X to +X), controlling for the days post-transplant as a fixed effect.
RESULTS
Individual specimens were partitioned into five basic diagnostic groups for gene validation (NORMAL, NPR, TCMR, CAMR, AND MIXED). Table S2 shows the number in each group, the distributions of creatinines, and times post-transplant for animals.
Some endothelial genes were validated in a prior report (19). Validation of the complete gene set for Banff criteria is in the Supplementary Methods, Text, Data, and Tables S3 and S4.
Table 1 extends the prior validation (19) and shows significant estimates for the pathological diagnostic groups. The TCMR diagnostic group is dominated by T cell, and interferon inducible transcripts with little endothelial gene expression. These overall patterns are very similar to those previously reported in humans (11). No obvious partition of the expected RNA gene expression for Banff grades of TCMR was observed and is similar to prior reports showing the difficulty of partitioning some grades of cellular rejection by RNA gene profiling (27) (data not shown). This may also relate to the paucity of samples in some Banff groups without a component of alloantibody mediated injury. Alternatively, the Banff grades of TCMR may represent the accumulation of pathological injury over time and not distinct inflammatory gene expression pathways. Of the six NK genes (B3GAT1, FGFBP2, KLRB1, KLRF1, MYBL1, and SH2D1B), KLRB1, KLRF1, and SH2D1B best partitioned with TCMR.
TABLE 1.
SIGNIFICANT RNA EXPRESSSION IN ACUTE CELLULAR, CHRONIC ANTIBODY MEDIATED, AND MIXED REJECTIONS
| CHRONIC ANTIBODY REJECTION (CAMR) | MIXED REJECTION | ACUTE CELLULAR REJECTION (TCMR) | ||||||
|---|---|---|---|---|---|---|---|---|
| GENE | SE | FDRPV | GENE | SE | FDRPV | GENE | SE | FDRPV |
| VWF | 0.41 | 6.0E-10 | VWF | 0.41 | 3.3E-16 | CD3D | 0.49 | 5.1E-20 |
| CAV1 | 0.34 | 1.4E-10 | IL1RL1 | 0.40 | 1.4E-13 | PDCD1 | 0.48 | 6.7E-17 |
| DARC | 0.34 | 1.3E-07 | DARC | 0.33 | 1.1E-11 | CXCL11 | 0.44 | 4.3E-14 |
| MALL | 0.27 | 7.7E-05 | ITGAX | 0.24 | 1.6E-04 | CD8A | 0.44 | 6.8E-20 |
| GNLY | 0.23 | 3.9E-05 | CCL5 | 0.23 | 2.1E-05 | ITGAX | 0.42 | 1.3E-13 |
| PECAM1 | 0.20 | 6.8E-04 | CXCL11 | 0.22 | 9.5E-07 | ICOS | 0.39 | 1.9E-12 |
| FCGR3A | 0.20 | 7.9E-03 | CAV1 | 0.21 | 1.3E-04 | GZMB | 0.39 | 1.8E-11 |
| CXCL11 | 0.18 | 2.1E-02 | CD68 | 0.21 | 4.4E-08 | CCL5 | 0.36 | 5.4E-14 |
| CD68 | 0.16 | 6.2E-04 | PDCD1 | 0.19 | 6.7E-09 | IFNG | 0.32 | 4.4E-08 |
| PALMD | 0.15 | 4.7E-03 | CD3D | 0.19 | 4.0E-04 | TNFRSF18 | 0.31 | 5.7E-09 |
| ROBO4 | 0.15 | 2.0E-03 | GZMB | 0.18 | 1.8E-04 | CD69 | 0.29 | 1.5E-09 |
| KLRB1 | 0.15 | 1.9E-02 | CD4 | 0.18 | 1.5E-03 | CCR4 | 0.28 | 3.8E-08 |
| KLF4 | 0.14 | 3.0E-03 | TNFRSF18 | 0.17 | 1.5E-05 | FCGR3A | 0.28 | 3.9E-07 |
| CD74 | 0.12 | 1.7E-02 | CD74 | 0.15 | 1.8E-03 | GNLY | 0.26 | 5.4E-10 |
| FCGR3A | 0.14 | 3.8E-05 | CCR5 | 0.25 | 9.5E-10 | |||
| KLRB1 | 0.13 | 1.0E-02 | TBX21 | 0.25 | 2.5E-09 | |||
| GNLY | 0.13 | 3.6E-03 | PSMB10 | 0.23 | 1.9E-12 | |||
| ICOS | 0.13 | 1.2E-03 | IL1RL1 | 0.23 | 4.4E-06 | |||
| CD8A | 0.13 | 1.9E-02 | CD68 | 0.21 | 3.1E-09 | |||
| PSMB10 | 0.10 | 5.2E-03 | KLRB1 | 0.20 | 8.5E-06 | |||
| CD69 | 0.10 | 1.4E-03 | TGFB1 | 0.19 | 5.8E-04 | |||
| TBX21 | 0.10 | 2.8E-02 | CD74 | 0.19 | 2.1E-07 | |||
| TEK | −0.19 | 5.1E-04 | FAS | 0.18 | 1.8E-08 | |||
| SH2D1B | 0.18 | 9.8E-04 | ||||||
| KLRF1 | 0.15 | 2.5E-03 | ||||||
| CD4 | 0.15 | 1.7E-04 | ||||||
| FOXP3 | 0.15 | 4.0E-03 | ||||||
| MYBL1 | 0.12 | 2.7E-02 | ||||||
| IL6R | 0.10 | 3.1E-04 | ||||||
| IL4 | 0.09 | 3.5E-06 | ||||||
| BCL6 | 0.08 | 4.7E-03 | ||||||
| CAV1 | −0.09 | 1.2E-02 | ||||||
| TEK | −0.10 | 1.6E-02 | ||||||
| ROBO4 | −0.10 | 3.2E-03 | ||||||
| PALMD | −0.10 | 7.5E-03 | ||||||
| VWF | −0.17 | 2.8E-04 | ||||||
| PECAM1 | −0.17 | 7.9E-05 | ||||||
| MALL | −0.19 | 8.2E-05 | ||||||
The CAMR group (Table 1) is dominated by endothelial genes (VWF, CAV1, DARC, MALL, PECAM1, PALMD, ROBO4, KLF4, AND CD74), plus FCGR3A (CD16), CXCL11 and GNLY. Variation is present when CAMR is compared to MIXED rejections, in which fewer endothelial genes are highly expressed (VWF, DARC, CAV1, and CD74).
The MIXED group comprising both TCMR and CAMR includes both statistically significant endothelial, T cell and interferon inducible gene expressions. The MIXED group shows variations between the CAMR and TCMR groups. Most notably is the high expression of IL1RL1, a TH2 inducible receptor of CD33, which is absent in CAMR and much lower in TCMR. Some NK markers (SH2D1B and MYBL1) show less expression in the MIXED group as compared to the TCMR group.
Chronic alloantibody mediated rejection (CAMR) progresses through stages (22, 23). To identify endothelial gene expression in CAMR, Figure 1 shows the variation in endothelial genes in the stages of CAMR. The top panel (Figure 1A) shows the highest endothelial gene estimates for six genes with the most reliable estimates occurring when the specimen turns C4d positive (Stage 2). CDH5 is similar but slightly weaker (data not shown). IL1RL1, a receptor for CD33, is not known to be an endothelial gene is commonly identified in CAMR and may reflect an antibody induced pattern of gene expression. The middle panel (Figure 1B) shows CD74 with declining expression as CAMR progresses. KLF4, MALL, AND PALMD show intermediate levels of expression with MALL and PALMD declining with advancing stage. Most of the other endothelial genes show variable and weak expression, bottom panel (Figure 1C). Also, included in this latter group are RPS6, RPS6KB, and SELE (data not shown).
Figure 1.

Dynamic variation in Endothelial Expression in CAMR Stages
Because CAMR invariable includes some interstitial inflammation, we tested if endothelial gene expression varied with inflammation within CAMR stages. CAMR and MIXED rejection were analyzed within the stages of CAMR and MIXED rejection. Figure 2 illustrates this dynamic variation in endothelial RNA expression in CAMR stages with and without inflammation. The first pattern (CD34) shows positive estimates with CAMR stages 1 – 4 CAMR). The estimates fall if inflammation is present (MIXED rejection). CD74 is like CD34 (data not shown). The second pattern is that of IL1RL1, which is not known to be an endothelial gene but rises with CAMR stage and inflammation. The second pattern is also seen in VWF (DARC, CAV1, and PECAM1 are similar, data not shown), which is markedly elevated in both CAMR and in MIXED rejections. The endothelial gene expressions of these four seem less sensitive to inflammation as compared to CD34 and CD74. Three additional less informative patterns (data not shown) were observed. SELE and RPS6KB1 showed low estimates with little variation. KLF4 and MALL were higher in CAMR and PALMD was higher in Stage 3. Other endothelial genes (PLA1A, PLAT, RPS6, RHOJ, SOX7, TEK, AND THBD) showed inconsistent variation with CAMR stage and inflammation.
Figure 2.

Dynamic variation in endothelial Gene Expressions in CAMR Stages with (MIXED) and without inflammation (CAMR)
To explore the variation in endothelial and NK gene RNA expression (Table 1, Figures 1 & 2), three factors (28–30) derived solely from T cell and interferon inducible genes, endothelial and NK genes from non-tolerant animals (specimen number is 239) with Eigen values of 11, 7, and 4 were clustered into nine clusters by K-Mean clustering, Figure 3 and Table 2. Factor analysis was used to reduce the dataset into a smaller number of correlated variables (28–31). Table 2 contains the scaled estimates of the clustered groups, and Figure 3 illustrates by Hierarchical Clustering shows a heat map and a constellation plot for the nine clusters (A – H, plus normals).
Figure 3.
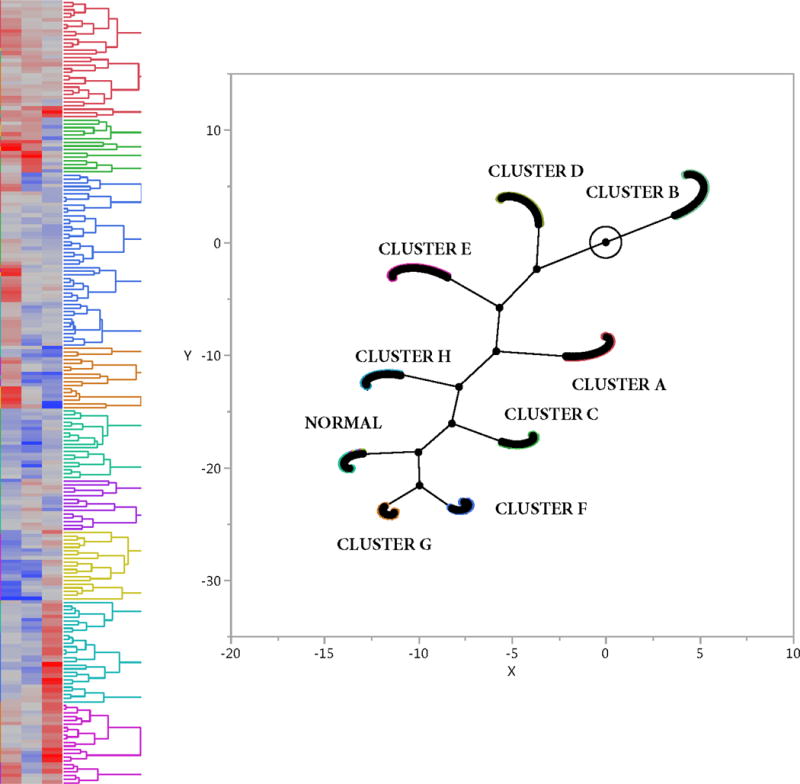
Heat Map and Constellation Plot from the Nine Clusters Derived from the Three Factors
TABLE 2.
| CLUSTER C | CLUSTER D | CLUSTER E | CLUSTER F | CLUSTER G | CLUSTER H | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GENE | SE | FDRPV | GENE | SE | FDRPV | GENE | SE | FDRPV | GENE | SE | FDRPV | GENE | SE | FDRPV | GENE | SE | FDRPV |
| VWF | 0.35 | 2.84E-05 | VWF | 0.47 | 6.28E-12 | VWF | 0.41 | 2.96E-09 | FCGR3A | 0.56 | 2.52E-09 | CD8A | 0.61 | 7.75E-11 | FCGR3A | 0.61 | 2.92E-14 |
| CAV1 | 0.27 | 3.85E-05 | CAV1 | 0.33 | 3.04E-10 | IL1RL1 | 0.38 | 5.72E-07 | ICOS | 0.54 | 8.79E-08 | B3GAT1 | 0.56 | 5.03E-06 | PDCD1 | 0.50 | 6.41E-09 |
| MALL | 0.27 | 2.66E-03 | FCGR3A | 0.33 | 6.86E-07 | DARC | 0.25 | 9.33E-05 | ITGAX | 0.50 | 3.05E-06 | PDCD1 | 0.55 | 7.59E-07 | ICOS | 0.41 | 1.16E-06 |
| DARC | 0.21 | 8.88E-03 | SOX7 | 0.31 | 4.06E-06 | CAV1 | 0.20 | 1.89E-04 | IL1RL1 | 0.45 | 1.57E-05 | CD3D | 0.53 | 1.62E-07 | ITGAX | 0.40 | 8.67E-06 |
| PECAM1 | 0.19 | 8.83E-03 | ROBO4 | 0.31 | 2.79E-13 | CD3D | 0.15 | 3.06E-02 | CD3D | 0.40 | 1.71E-05 | ICOS | 0.51 | 2.42E-06 | CD3D | 0.34 | 1.02E-05 |
| CDH5 | 0.18 | 4.75E-03 | DARC | 0.31 | 1.24E-06 | PECAM1 | 0.09 | 1.41E-01 | PDCD1 | 0.37 | 3.00E-04 | PSMB10 | 0.46 | 1.07E-11 | CD8A | 0.34 | 3.09E-06 |
| SOX7 | 0.17 | 4.69E-02 | MALL | 0.30 | 2.87E-05 | SOX7 | 0.07 | 3.53E-01 | CD74 | 0.36 | 8.87E-08 | ITGAX | 0.42 | 2.13E-04 | PLA1A | 0.29 | 1.79E-04 |
| PALMD | 0.15 | 1.17E-02 | CDH5 | 0.27 | 1.01E-07 | PALMD | 0.06 | 2.72E-01 | VWF | 0.33 | 5.55E-04 | CD68 | 0.42 | 8.82E-09 | CD74 | 0.28 | 9.75E-07 |
| TEK | 0.13 | 5.35E-02 | PECAM1 | 0.27 | 4.25E-06 | MALL | 0.06 | 4.69E-01 | SH2D1B | 0.30 | 1.06E-02 | FGFBP2 | 0.29 | 5.01E-02 | SH2D1B | 0.23 | 1.73E-02 |
| ROBO4 | 0.11 | 4.14E-02 | KLF4 | 0.26 | 6.42E-09 | RHOJ | 0.02 | 7.43E-01 | CAV1 | 0.28 | 1.50E-04 | KLRF1 | 0.21 | 7.08E-02 | PSMB10 | 0.19 | 2.10E-04 |
| PLAT | 0.10 | 3.20E-02 | PALMD | 0.25 | 3.25E-07 | PLAT | 0.00 | 9.26E-01 | CD8A | 0.28 | 1.30E-03 | SH2D1B | 0.21 | 9.46E-02 | CD68 | 0.16 | 4.31E-03 |
| PLA1A | 0.06 | 4.79E-01 | ITGAX | 0.21 | 5.70E-03 | CDH5 | −0.02 | 7.23E-01 | CD68 | 0.27 | 6.23E-05 | MYBL1 | 0.20 | 1.14E-01 | IL1RL1 | 0.15 | 8.72E-02 |
| KLF4 | 0.05 | 3.66E-01 | RHOJ | 0.17 | 9.30E-03 | THBD | −0.03 | 5.96E-01 | DARC | 0.26 | 3.96E-03 | KLRB1 | 0.20 | 5.59E-02 | KLRB1 | 0.13 | 1.04E-01 |
| KLRB1 | 0.00 | 9.68E-01 | PSMB10 | 0.16 | 2.39E-04 | CD68 | −0.04 | 3.89E-01 | PLA1A | 0.20 | 3.12E-02 | IL1RL1 | 0.19 | 1.07E-01 | KLRF1 | 0.09 | 3.39E-01 |
| THBD | −0.02 | 7.29E-01 | KLRF1 | 0.14 | 6.85E-02 | ROBO4 | −0.05 | 3.00E-01 | KLRB1 | 0.19 | 4.10E-02 | CD74 | 0.14 | 6.20E-02 | MYBL1 | 0.03 | 8.11E-01 |
| CD74 | −0.02 | 7.45E-01 | TEK | 0.14 | 1.33E-02 | MYBL1 | −0.05 | 5.54E-01 | PSMB10 | 0.19 | 2.21E-03 | RHOJ | 0.06 | 5.56E-01 | THBD | 0.00 | 9.96E-01 |
| RHOJ | −0.02 | 8.00E-01 | CD68 | 0.12 | 1.25E-02 | KLRB1 | −0.07 | 3.49E-01 | PLAT | 0.12 | 1.95E-02 | FCGR3A | 0.05 | 6.78E-01 | TEK | −0.02 | 8.20E-01 |
| B3GAT1 | −0.04 | 7.52E-01 | PDCD1 | 0.12 | 1.04E-01 | PLA1A | −0.07 | 2.89E-01 | KLRF1 | 0.08 | 4.68E-01 | THBD | 0.01 | 8.61E-01 | PLAT | −0.05 | 2.14E-01 |
| CD68 | −0.08 | 1.89E-01 | CD8A | 0.11 | 8.58E-02 | CD74 | −0.08 | 1.15E-01 | KLF4 | 0.06 | 3.84E-01 | DARC | −0.03 | 8.11E-01 | CDH5 | −0.06 | 3.18E-01 |
| MYBL1 | −0.11 | 2.77E-01 | THBD | 0.10 | 2.30E-02 | KLF4 | −0.09 | 6.35E-02 | CDH5 | 0.05 | 5.61E-01 | KLF4 | −0.09 | 2.26E-01 | KLF4 | −0.09 | 9.11E-02 |
| FCGR3A | −0.12 | 1.56E-01 | CD74 | 0.10 | 4.56E-02 | TEK | −0.14 | 1.57E-02 | PECAM1 | 0.04 | 7.00E-01 | PALMD | −0.16 | 3.96E-02 | CAV1 | −0.13 | 3.45E-02 |
| KLRF1 | −0.15 | 1.29E-01 | KLRB1 | 0.09 | 1.70E-01 | KLRF1 | −0.14 | 7.32E-02 | THBD | −0.01 | 8.62E-01 | PLAT | −0.19 | 5.05E-04 | ROBO4 | −0.18 | 4.54E-04 |
| FGFBP2 | −0.21 | 8.72E-02 | PLAT | 0.09 | 1.57E-02 | ITGAX | −0.14 | 6.65E-02 | MYBL1 | −0.03 | 8.54E-01 | SOX7 | −0.20 | 5.17E-02 | DARC | −0.22 | 4.76E-03 |
| PSMB10 | −0.22 | 3.41E-05 | FGFBP2 | 0.08 | 4.36E-01 | SH2D1B | −0.16 | 5.98E-02 | SOX7 | −0.09 | 3.64E-01 | ROBO4 | −0.25 | 1.16E-04 | RHOJ | −0.22 | 3.76E-03 |
| SH2D1B | −0.24 | 1.73E-02 | CD3D | 0.07 | 3.39E-01 | B3GAT1 | −0.16 | 5.59E-02 | ROBO4 | −0.14 | 1.97E-02 | MALL | −0.27 | 1.46E-02 | B3GAT1 | −0.22 | 2.17E-02 |
| CD8A | −0.25 | 8.35E-04 | ICOS | 0.04 | 5.78E-01 | FGFBP2 | −0.16 | 1.06E-01 | TEK | −0.17 | 2.57E-02 | PECAM1 | −0.32 | 3.00E-04 | MALL | −0.23 | 7.75E-03 |
| CD3D | −0.31 | 1.18E-04 | MYBL1 | 0.04 | 6.39E-01 | PSMB10 | −0.24 | 1.17E-07 | RHOJ | −0.21 | 2.29E-02 | VWF | −0.33 | 1.30E-03 | PECAM1 | −0.24 | 5.77E-04 |
| IL1RL1 | −0.33 | 3.28E-04 | SH2D1B | −0.01 | 9.54E-01 | PDCD1 | −0.25 | 9.02E-04 | MALL | −0.21 | 3.59E-02 | CAV1 | −0.36 | 5.89E-06 | FGFBP2 | −0.24 | 3.85E-02 |
| ITGAX | −0.33 | 3.28E-04 | IL1RL1 | −0.03 | 7.18E-01 | CD8A | −0.27 | 1.48E-05 | PALMD | −0.22 | 1.34E-03 | PLA1A | −0.44 | 9.05E-06 | PALMD | −0.29 | 7.64E-07 |
| ICOS | −0.36 | 4.91E-05 | B3GAT1 | −0.03 | 7.18E-01 | ICOS | −0.27 | 1.58E-04 | B3GAT1 | −0.24 | 4.14E-02 | TEK | −0.50 | 1.95E-09 | VWF | −0.36 | 4.94E-06 |
| PDCD1 | −0.44 | 1.20E-06 | PLA1A | −0.11 | 1.13E-01 | FCGR3A | −0.33 | 9.72E-07 | FGFBP2 | −0.33 | 1.73E-02 | CDH5 | −0.52 | 4.02E-11 | SOX7 | −0.39 | 9.72E-07 |
The normal group shows mostly negative estimates. Cluster A (N=29), composed for low grade TCMR, Banff ACR1A and borderline with no DSA or C4d. Cluster B (N=39) was composed of No Pathological Rejection (Table S2) also showed low estimates. Pair wise estimates with adjusted False Discovery Rate p values showed that Clusters A and B had higher CD3 expression above the normal group. Data is not shown for the normal cluster and clusters A and B.
The most informative clusters, C – H, and their statistically significant estimates appear in Table 2. Cluster C (N=25), composed of just CAMR (Table S2), appears by RNA gene expression to be purely CAMR with only significantly elevated estimates endothelial gene expression and significantly lower estimates for NK, CD3D, and interferon induced gene expressions. Cluster D (N=40), composed of CAMR and mostly MIXED type rejections (Table S2) shows a more complex gene expression pattern with high endothelial gene expression, and some interferon induced genes (ITGAX, CXCL11, PSMB10), high FCGGR3A, likely an NK marker as CD68 is not identified, plus lower estimates for other NK cells (KLRF1, CD8A, KLRB1, FGFBP1, MYL1, and BEGAT1). CD3D is very low and insignificant so that the CD8A is likely NK related. The gene expression pattern in Cluster D suggests that much of the inflammation in this cluster is likely NK related. Cluster E (N=42), composed entirely of MIXED rejections (Table S2), shows high estimates for VWF, DARC, and CAV1 (alloantibody associated gene expressions), plus a high estimate for IL1RL1. Other endothelial gens show low or insignificant estimates. CD3D is weakly positive. FCGR3A (CD16) shows a high estimate and is likely NK related. Cluster F (N=21), composed of only MIXED rejections (Table S2) shows a MIXED gene expression pattern with interferon inducible genes, CD3D, high FCGR3A and positive estimate for some NK genes (SH2D1B, KLRB1, KLRF1) but not MYBL1, B3GAT1, or FGFBP2. CD68 is significantly elevated so that the high FCGR3A may not represent an NK marker. Clusters G and H (N=16 and 27), both composed of just TCMR (Table S2), both show high levels of TCMR associated RNA expression with low levels of endothelial expression. Clusters G and H show differential expression of B3GAT1 (CD57) and FCGR3A. B3GAT1 is high in cluster G and low in cluster H, whereas FCGR3A is low in cluster G and high in cluster H. Cluster G has higher estimates of CD3 and interferon inducible genes.
Table 3 contains summaries for creatinines, days post BMT for the specimen, and the terminal survival times for the clustered groups C – H. The survival time for Cluster C is significantly longer as compared to Clusters D, E, and F, P <0.01. Cluster C is purely CAMR without evidence of significant NK or CD3 gene expressions, whereas Clusters D, E, and F are CAMR with evidence of NK, or NK/CD3 (MIXED rejections). These findings suggest that any type of inflammation in CAMR (MIXED) shortens allograft survival and that pure CAMR (no inflammation) is less pathological. The shorter allograft survivals in clusters G and H are predominantly TCMR (no C4d, DSA) with differential expression of B3GAT1 and FCGR3A. Cluster G shows stronger TCMR related estimates and shorter allograft survival than cluster H.
TABLE 3.
| GROUP | CHARACTERISTICS | MEDIAN CREATININE |
RANGE | MEDIAN DAYS TERMINAL |
RANGE |
|---|---|---|---|---|---|
| C | PURE CAMR: NO NK NO CD3 | 1.5 | 1.1–4.6 | 815 | 477–2023 |
| D | MIXED: CAMR + NK FCGR3A | 2.8 | 0.9–5.1 | 386 | 71–1379 |
| E | MIXED:CAMR + NK FCGR3A > CD3 | 3 | 1.0–8.0 | 470 | 75–838 |
| F | MIXED: CAMR + NK + CD3 | 2.8 | 0.8–8.7 | 378 | 110–930 |
| G | STRONG TCMR: SOME NK | 3 | 1.3–14 | 173 | 58–282 |
| H | WEAKER TCMR: SOME NK | 1.2 | 1.0–3.9 | 164 | 69–910 |
Terminal graft survival for Group C is significantly different from Groups D, E, or F: FDRPV < 0.01
Terminal graft survivals and creatinines for Groups G and H are signficantly different: FDRPV < 0.03
The MIXED rejection group comprised of both TCMR and CAMR is not classified as a separate diagnostic category within the Banff classification scheme (26). Nevertheless, some interstitial inflammation is commonly present in CAMR, though mostly low grade. In this dataset, there are by pathological criteria 42 pure CAMR samples with Banff ACR less than borderline but only 25 of 42 samples showed no inflammation by gene expression (cluster C). 17 of the 42 CAMR samples by pathological criteria alone (no TCMR) were re clustered into clusters D – F as MIXED rejections. There are 86 MIXED samples with Banff TCMR grades of borderline (n = 31, 36%), 1A (n= 20, 23%), 1B (n = 4, 4.5%), 2A (n = 17, 20%), 2B (n = 1, 1.1%), and grade 3 (n = 13, 15%).
64 animals with multiple specimens were compared for the patterns of clusters C – H. 53.3 percent were stable with the same clusters. 18 percent changed from CAMR to MIXED (clusters C or D to clusters E OR F). 28.6 percent changed from TCMR to MIXED (clusters G or H to clusters E or F). Clusters C, D, and E were mostly (>50%) CAMR stage 4, whereas Cluster F was mostly (>50%) CAMR stage 1.
Additional analyses of the biological relevance of Cluster C – H is complicated by the time post-transplant because samples occur at different times post-transplant. Gene expression from the same animal in an earlier sample may or may not be relevant or comparable to latter samples. To address this issue additional analyses were done and compared both per sample and per animal. Figure 4 shows Life Distribution Curves per cluster per sample (A) and per animal (B). In both Cluster C (CAMR without inflammation) shows the longest survival (although with eventual rejection) as compared to the other clusters. The relative risk by Proportional Hazard are lowest for Cluster C. These data suggest that CAMR without inflammation (Cluster C) has a longer survival time as compared to CAMR with inflammation (MIXED rejections) in Clusters D – F).
Figure 4.

Life Distribution Curves and Proportional Hazard Relative Risk Analysis of Cluster C – H. Life Distribution by Groups using terminal time vs Groups. Plotted as Probability of Survival vs Times Post Transplant, JMP. Risk Ratios by Cox Proportional Hazard, JMP
DISCUSSION
This report supports the utility of Nanostring RNA expression using a set of known informative genes to analyze retrospectively archival renal allograft specimens from Cynomolgus monkeys using the consistent quality and quantity of RNA derived even from small archival biopsies.
These findings demonstrate RNA expression profiling readily identifies both the informative RNA transcripts relevant to the diagnosis of renal allograft rejection and the marked heterogeneity of gene expression of endothelial and NK gene expression within the broad diagnostic groups of CAMR, TCMR, and MIXED rejection types, all of which may be split further into additional groups based solely on gene expression. Endothelial gene expression is not uniform and shows marked variation in CAMR and change over time with and without inflammation as the CAMR advances through its progressive stages. Some endothelial genes (VWF, CAV1, DARC and PECAM1) are reliable markers for alloantibody in the progressive stages of CAMR even with inflammation in MIXED rejections. The CAMR groups is dominated by endothelial genes, plus FCGR3A (CD16), likely not an NK marker as CD68, a macrophage marker is present. CXCL11 and GNLY are also present, possibly related to NK cells.
CD34 and CD74 both seem to decrease with either CAMR stage and/or with inflammation. The decrease in CD34 is likely associated with inflammation as by linear regression CD34 negatively correlates with CD3, CCR5 (a TH1 cytokine), TBX21 (tBET), and selectin (FDRPV < 0.05, data not shown). Endothelial protein expression is variably increased with single cytokine stimulation (TNF, IFNG, IL2) but some including CD34 shown both synergistic and antagonistic variation with multiple cytokine stimulations (32). PECAM protein expression is increased with both single and multiple cytokine stimulations (32). KLRF, MALL, and PALMD show intermediate estimates. Many of the endothelial genes (lower panel Figure 1) have low estimates and lack utility identifying CAMR and MIXED rejections. NK and CD3 gene expressions also show variation. These data are like those previously reported showing the variation the NK and T cells transcripts in antibody and cellular rejections (33, 34).
A notable finding, after the three factors partitioned CAMR and MIXED rejection into four clusters is that kidney allografts with pure CAMR group (cluster C) without both pathological (i = 0) or RNA expression evidence of inflammation show a much-prolonged survival time as compared to CAMR with evidence of NK and/or CD3 RNA expression (MIXED rejections, clusters D – F). Cluster C has a longer survival time as compared to MIXED rejections with NK and/or NK/CD3 inflammation (clusters D – F) suggesting that the inflammation in CAMR (MIXED rejections) causes shorter allograft survival as compared to pure CAMR (Cluster C), which lacks inflammation by RNA expression or by pathological review. Pure CAMR (no inflammation by RNA expression), therefore, seems less pathological with longer allograft survival suggesting that inflammation in CAMR (MIXED rejection) adds additional risk to allograft survival, in addition to DSA, C4d, and transplant glomerulopathy. One MIXED type, cluster D contains NK, FCGR3A (CD16). MIXED cluster (E) shows NK and weaker CD3. Cluster F, another MIXED type, contains both multiple NK and CD3. These data confirm the complexity of gene expression in CAMR and MIXED rejections (35). Because there is no survival variation among clusters D – F (MIXED rejections) with variations in NK/CD3 gene expressions, the biological significance of such NK/CD3 variation is unclear and may not be biologically important. Such gene expression variation within clusters D – F may be due to case mix or just variation in expression patterns (rank). 40 percent of CAMR defined only by Banff criteria (no TCMR) contained inflammation by RNA expression suggesting that RNA expression is more sensitive, in some samples, than pathological review alone. The TCMR clustered NK expression differently with one (G) with high B3GAT1 (CD57) and one (H) with high FCGR3A with cluster G showing higher TCMR related expression. Because many of the samples in TCMR clusters G and H are from the same animals, the variation likely just represents variation in the progressing TCMR.
Although this Cynomolgus model is useful to analyze renal allograft rejection, extrapolation to human subjects is challenging due to human covariates including different therapies, chronic immunosuppression, and medical co-morbidities. Such human covariates may alter the patterns of gene expression in human kidney allografts as compared to this Cynomolgus model. Nevertheless, as the derived gene set is from human studies, and the gene set validates well against various rejections patterns, also derived from human studies and Banff criteria, also derived from human studies, data in this report are likely informative for human studies.
Overall, these findings identify additional heterogeneity within CAMR, MIXED, and TCMR rejections, which may be partitioned into additional diagnostic groups by RNA expression and provide additional diagnostic information unavailable solely with just pathological review.
Supplementary Material
Table S1: Gene Probe Set
Table S2: Distributions of creatinines and times post-transplant for the five diagnostic groups.
Tables S3 and S4: Significant scaled estimates within Banff Criteria, sorted highest to lowest by estimates. Banff criteria, C4d, and DSA are coded as nominal.
Acknowledgments
This work was supported by funding from Astellas Pharma Canada, Inc., Canadian Foundation for Innovation, and National Institutes of Health (NIH PO1 HL18646).
ABBREVIATIONS
- CAMR
chronic antibody-mediated rejection
- BMT
bone marrow transplant
- cg
transplant glomerulopathy
- DSA
donor specific antibody
- FDPV
False discovery p value, adjusted p value
- FFPE
formalin-fixed paraffin-embedded
- MIXED
both TCMR and CAMR
- NHP
nonhuman primate
- Normal
native perioperative kidney biopsies
- NPR
no pathological rejection
- SE
scaled estimate
- TCMR
T-cell mediated rejection
Footnotes
DR BENJAMIN ALEXANDER ADAM (Orcid ID : 0000-0003-1908-1739)
DISCLOSURE
The authors of this manuscript have conflicts of interest to disclose as described by the American Journal of Transplantation. M.M. received consultancy honoraria from Astellas Pharma Canada, Inc. The other authors have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supporting Information
Additional Supporting Information may be found online in the supporting information tab for this article.
References
- 1.Akalin E, Hendrix RC, Polavarapu RG, Pearson TC, Neylan JF, Larsen CP, et al. Gene expression analysis in human renal allograft biopsy samples using high-density oligoarray technology. Transplantation. 2001;72(5):948–953. doi: 10.1097/00007890-200109150-00034. [DOI] [PubMed] [Google Scholar]
- 2.Halloran PF, de Freitas DG, Einecke G, Famulski KS, Hidalgo LG, Menge LM, et al. An integrated view of molecular changes, histopathology and outcomes in kidney transplants. Am J Transplant. 2010;10(10):2223–2230. doi: 10.1111/j.1600-6143.2010.03268.x. [DOI] [PubMed] [Google Scholar]
- 3.Halloran PF, de Freitas DG, Einecke G, Famulski KS, Hidalgo LG, Mengel M, et al. The molecular phenotype of kidney transplants. Am J Transplant. 2010;10(10):2215–2222. doi: 10.1111/j.1600-6143.2010.03267.x. [DOI] [PubMed] [Google Scholar]
- 4.Halloran PF, Einecke G. Microarrays and transcriptome analysis in renal transplantation. Nat Clin Pract Nephrol. 2006;2(1):2–3. doi: 10.1038/ncpneph0066. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann SC, Hale DA, Kleiner DE, Mannon RB, Kampen RL, Jacobson LM, et al. Functionally significant renal allograft rejection is defined by transcriptional criteria. Am J Transplant. 2005;5(3):573–581. doi: 10.1111/j.1600-6143.2005.00719.x. [DOI] [PubMed] [Google Scholar]
- 6.Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125–138. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 7.Scherer A, Gwinner W, Mengel M, Kirsch T, Raulf F, Szustakowski JD, et al. Transcriptome changes in renal allograft protocol biopsies at 3 months precede the onset of interstitial fibrosis/tubular atrophy (IF/TA) at 6 months. Nephrol Dial Transplant. 2009;24(8):2567–2575. doi: 10.1093/ndt/gfp183. [DOI] [PubMed] [Google Scholar]
- 8.Stegall MD, Park WD, Kim D, Kremers W. Gene expression during acute allograft rejection: novel statistical analysis of microarray data. Am J Transplant. 2002;2(10):913–925. doi: 10.1034/j.1600-6143.2002.21007.x. [DOI] [PubMed] [Google Scholar]
- 9.Flechner SM, Kurian SM, Head SR, Sharp SM, Whisenant TC, Zhang J, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4(9):1475–1489. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khatri P, Roedder S, Kimura N, De Vusser K, Morgan AA, Gong Y, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med. 2013;210(11):2205–2221. doi: 10.1084/jem.20122709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeve J, Sellares J, Mengel M, Sis B, Skene A, Hidalgo L, et al. Molecular diagnosis of T cell-mediated rejection in human kidney transplant biopsies. Am J Transplant. 2013;13(3):645–655. doi: 10.1111/ajt.12079. [DOI] [PubMed] [Google Scholar]
- 12.Saint-Mezard P, Berthier CC, Zhang H, Hertig A, Kaiser S, Schumacher M, et al. Analysis of independent microarray datasets of renal biopsies identifies a robust transcript signature of acute allograft rejection. Transpl Int. 2009;22(3):293–302. doi: 10.1111/j.1432-2277.2008.00790.x. [DOI] [PubMed] [Google Scholar]
- 13.Sellares J, de Freitas DG, Mengel M, Reeve J, Einecke G, Sis B, et al. Understanding the causes of kidney transplant failure: the dominant role of antibody-mediated rejection and nonadherence. Am J Transplant. 2012;12(2):388–399. doi: 10.1111/j.1600-6143.2011.03840.x. [DOI] [PubMed] [Google Scholar]
- 14.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 15.Reis PP, Waldron L, Goswami RS, Xu W, Xuan Y, Perez-Ordonez B, et al. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol. 2011;11:46. doi: 10.1186/1472-6750-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith KA, Hayward RA. Performance measurement in chronic kidney disease. J Am Soc Nephrol. 2011;22(2):225–234. doi: 10.1681/ASN.2010111152. [DOI] [PubMed] [Google Scholar]
- 17.Adam B, Afzali B, Dominy KM, Chapman E, Gill R, Hidalgo LG, et al. Multiplexed color-coded probe-based gene expression assessment for clinical molecular diagnostics in formalin-fixed paraffin-embedded human renal allograft tissue. Clin Transplant. 2016;30(3):295–305. doi: 10.1111/ctr.12689. [DOI] [PubMed] [Google Scholar]
- 18.Afzali B, Chapman E, Racape M, Adam B, Bruneval P, Gil F, et al. Molecular Assessment of Microcirculation Injury in Formalin-Fixed Human Cardiac Allograft Biopsies With Antibody-Mediated Rejection. Am J Transplant. 2016 doi: 10.1111/ajt.13956. [DOI] [PubMed] [Google Scholar]
- 19.Adam BA, Smith RN, Rosales IA, Matsunami M, Afzali B, Oura T, et al. Chronic Antibody-Mediated Rejection in Nonhuman Primate Renal Allografts: Validation of Human Histological and Molecular Phenotypes. Am J Transplant. 2017 doi: 10.1111/ajt.14327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cosimi AB, Burton RC, Kung PC, Colvin RB, Goldstein G, Lifter J, et al. Evaluation in primate renal allograft recipients of monoclonal antibody to human T-cell subclasses. Transplant Proc. 1981;13(1 Pt 1):499–503. [PubMed] [Google Scholar]
- 21.Kawai T, Sogawa H, Boskovic S, Abrahamian G, Smith RN, Wee SL, et al. CD154 blockade for induction of mixed chimerism and prolonged renal allograft survival in nonhuman primates. Am J Transplant. 2004;4(9):1391–1398. doi: 10.1111/j.1600-6143.2004.00523.x. [DOI] [PubMed] [Google Scholar]
- 22.Smith RN, Kawai T, Boskovic S, Nadazdin O, Sachs DH, Cosimi AB, et al. Chronic antibody mediated rejection of renal allografts: pathological, serological and immunologic features in nonhuman primates. Am J Transplant. 2006;6(8):1790–1798. doi: 10.1111/j.1600-6143.2006.01351.x. [DOI] [PubMed] [Google Scholar]
- 23.Smith RN, Kawai T, Boskovic S, Nadazdin O, Sachs DH, Cosimi AB, et al. Four stages and lack of stable accommodation in chronic alloantibody-mediated renal allograft rejection in Cynomolgus monkeys. Am J Transplant. 2008;8(8):1662–1667. doi: 10.1111/j.1600-6143.2008.02303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada Y, Boskovic S, Aoyama A, Murakami T, Putheti P, Smith RN, et al. Overcoming memory T-cell responses for induction of delayed tolerance in nonhuman primates. Am J Transplant. 2012;12(2):330–340. doi: 10.1111/j.1600-6143.2011.03795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mueller TF, Einecke G, Reeve J, Sis B, Mengel M, Jhangri GS, et al. Microarray analysis of rejection in human kidney transplants using pathogenesis-based transcript sets. Am J Transplant. 2007;7(12):2712–2722. doi: 10.1111/j.1600-6143.2007.02005.x. [DOI] [PubMed] [Google Scholar]
- 26.Loupy A, Haas M, Solez K, Racusen L, Glotz D, Seron D, et al. The Banff 2015 Kidney Meeting Report: Current Challenges in Rejection Classification and Prospects for Adopting Molecular Pathology. Am J Transplant. 2017;17(1):28–41. doi: 10.1111/ajt.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mengel M. What is the significance of subclinical inflammation in human renal allografts? It depends! Transplantation. 2012;93(1):22–23. doi: 10.1097/TP.0b013e31823bb6ae. [DOI] [PubMed] [Google Scholar]
- 28.Anand Brown A, Ding Z, Vinuela A, Glass D, Parts L, Spector T, et al. Pathway-based factor analysis of gene expression data produces highly heritable phenotypes that associate with age. G3 (Bethesda) 2015;5(5):839–847. doi: 10.1534/g3.114.011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kustra R, Shioda R, Zhu M. A factor analysis model for functional genomics. BMC Bioinformatics. 2006;7:216. doi: 10.1186/1471-2105-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc. 2012;7(3):500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parts L, Stegle O, Winn J, Durbin R. Joint genetic analysis of gene expression data with inferred cellular phenotypes. PLoS Genet. 2011;7(1):e1001276. doi: 10.1371/journal.pgen.1001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raab M, Daxecker H, Markovic S, Karimi A, Griesmacher A, Mueller MM. Variation of adhesion molecule expression on human umbilical vein endothelial cells upon multiple cytokine application. Clin Chim Acta. 2002;321(1–2):11–16. doi: 10.1016/s0009-8981(02)00048-7. [DOI] [PubMed] [Google Scholar]
- 33.Hidalgo LG, Einecke G, Allanach K, Halloran PF. The transcriptome of human cytotoxic T cells: similarities and disparities among allostimulated CD4(+) CTL, CD8(+) CTL and NK cells. Am J Transplant. 2008;8(3):627–636. doi: 10.1111/j.1600-6143.2007.02128.x. [DOI] [PubMed] [Google Scholar]
- 34.Hidalgo LG, Sis B, Sellares J, Campbell PM, Mengel M, Einecke G, et al. NK cell transcripts and NK cells in kidney biopsies from patients with donor-specific antibodies: evidence for NK cell involvement in antibody-mediated rejection. Am J Transplant. 2010;10(8):1812–1822. doi: 10.1111/j.1600-6143.2010.03201.x. [DOI] [PubMed] [Google Scholar]
- 35.Halloran PF, Merino Lopez M, Barreto Pereira A. Identifying Subphenotypes of Antibody-Mediated Rejection in Kidney Transplants. Am J Transplant. 2016;16(3):908–920. doi: 10.1111/ajt.13551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Gene Probe Set
Table S2: Distributions of creatinines and times post-transplant for the five diagnostic groups.
Tables S3 and S4: Significant scaled estimates within Banff Criteria, sorted highest to lowest by estimates. Banff criteria, C4d, and DSA are coded as nominal.


