Abstract
Beige adipocytes have been recently shown to regulate energy dissipation when activated, and help organisms defend against hypothermia and obesity. Prior reports indicate beige-like adipocytes exist in adult humans and may present novel opportunities to curb the global epidemic in obesity and metabolic illnesses. In an effort to identify unique features of activated beige adipocytes, we uncovered that the cholinergic receptor nicotinic alpha 2 subunit (Chrna2) is induced in subcutaneous fat during the activation of these cells, and that acetylcholine-producing immune cells within this tissue regulate this signaling pathway via paracrine mechanisms. CHRNA2 functions selectively in uncoupling protein 1 (Ucp1)+ beige adipocytes, increasing thermogenesis through a cAMP and PKA pathway. Furthermore, this signaling via CHRNA2 is conserved and present in human subcutaneous adipocytes. Inactivation of Chrna2 in mice compromises the cold-induced thermogenic response selectively in subcutaneous fat and exacerbates high-fat diet-induced obesity and associated metabolic disorders, indicating that even partial loss of beige fat regulation in vivo leads to detrimental consequences. Our results reveal a beige-selective immune-adipose interaction mediated through CHRNA2 and identify a novel function of nicotinic acetylcholine receptors (nAChRs) in energy metabolism. These findings may lead to identification of therapeutic targets to counteract human obesity.
Keywords: Obesity, beige fat, brown fat, CHRNA2, acetylcholine
INTRODUCTION
The serious metabolic complications closely associated with obesity emphasize the urgency to develop counteracting strategies. It has recently been reported that beige-like thermogenic adipocytes are present in human adults1–4 and that the activation of these thermogenic fat cells increases energy expenditure and improves metabolic health in humans5–9. Whereas the regulation of white and classical brown fat cells has been investigated for decades, many aspects of the beige adipocyte function are still yet to be elucidated.
Previous studies of the regulation of thermogenic fat cells in vivo have mainly been focused on signaling through the β-adrenergic pathway10. Here we demonstrate that CHRNA2, a subunit of the nicotinic acetylcholine receptor family, is upregulated during beiging and specifically functions in beige fat cells from subcutaneous adipose depots. The nAChRs belong to a large superfamily of ligand-gated ion channels that are expressed throughout both the central and the peripheral nervous systems, as well as in non-neuronal cell populations11,12. At an individual-cell level resolution, we observed that CHRNA2-mediated signaling specifically occurs in Ucp1-expressing beige fat cells within subcutaneous culture, but not in either white or brown fat cells. Calcium imaging assays further revealed that primary fat cells from the human subcutaneous depot but not those from the perirenal depot respond to CHRNA2 agonist stimulation, suggesting that this beige-selective response is conserved in humans. We identified acetylcholine-producing immune cells within the subcutaneous fat depot that communicate with beige fat cells via CHRNA2 through paracrine signaling. Additionally, Chrna2 KO mice have a compromised response to cold specifically in beige fat and impaired metabolic homeostasis upon dietary challenges. Our results identify CHRNA2 as a functional beige-selective marker and suggest that this immune-adipose interaction through acetylcholine and CHRNA2 may lead to novel druggable targets to treat human obesity and the metabolic syndrome.
RESULTS
Chrna2 is induced in subcutaneous adipocytes during beiging
Rosiglitazone (Rosi), a thiazolidinedione (TZD) that acts as a PPARγ agonist, has been shown to induce the activation of “browning” in vitro and in vivo13–15. Thus, in order to investigate which signaling pathways activate beige adipocytes, we performed a microarray analysis with RNA samples from fully differentiated primary preadipocytes isolated from the murine inguinal subcutaneous fat depot (one where beige adipocytes are most abundant) followed by treatment in vitro with Rosi or a vehicle control. As expected, the thermogenic marker Ucp1 was induced in the Rosi-treated samples. It is of note that Chrna2, one of the subunits of nAChRs was greatly induced with Rosi treatment in these fat cultures (Fig. 1a). The Chrna2 induction was confirmed by quantitative PCR (qPCR) performed on primary inguinal fat cells from multiple strains of inbred mice (Fig. 1a and Supplementary Fig. 1a). Further analyses revealed that Chrna2 expresses at significant levels in subcutaneous adipocytes and among all nAChR subunits, it is the only one whose expression level was regulated during Rosi-induced beiging (Fig. 1b and Supplementary Fig. 1b,c).
Figure 1.
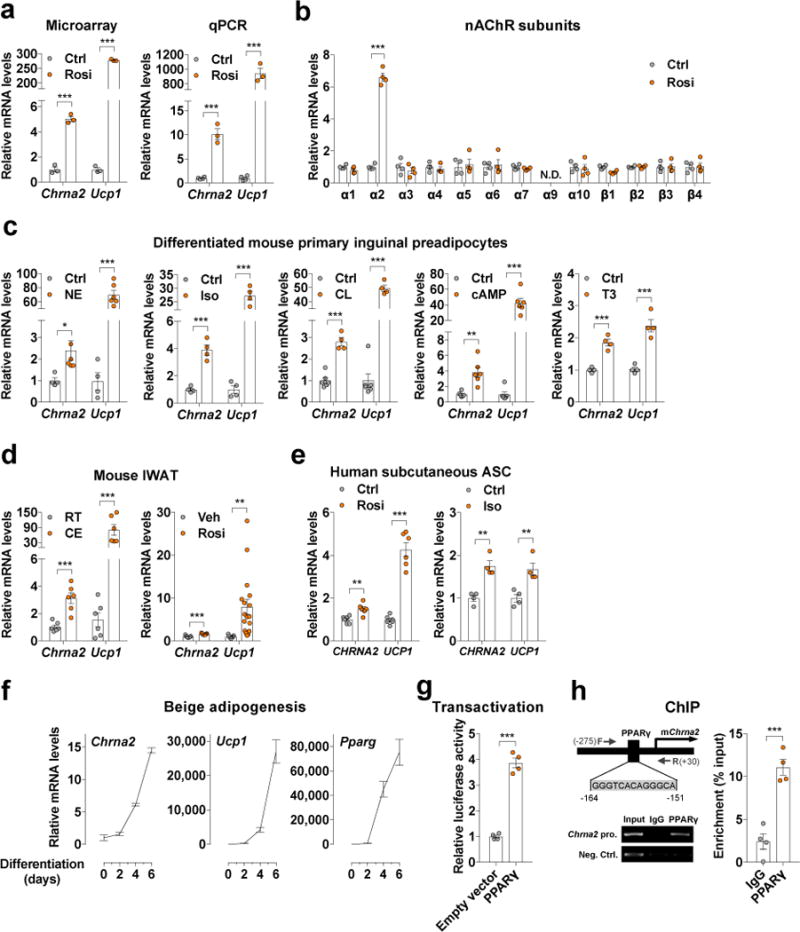
Chrna2 is induced in subcutaneous adipocytes of mice and humans during beiging. (a) Microarray and qPCR analyses of Chrna2 and Ucp1 mRNA expression in differentiated preadipocytes of wild type C57BL/6J (WT) mice with treatment of vehicle control (Ctrl) or 1 μM rosiglitazone (Rosi) for 4 d (n = 3 per group for microarray, n = 4 for Ctrl, n = 3 for Rosi in qPCR). (b) qPCR analyses of nicotinic acetylcholine receptor (nAChR) subunit mRNA levels in differentiated inguinal preadipocytes following 1 μM Rosi treatment for 2 d compared with control (n = 4 per group). (c) qPCR analyses of Chrna2 and Ucp1 mRNA expression in differentiated inguinal preadipocytes stimulated with vehicle (Ctrl), 0.2 μM norepinephrine (NE) for 2 d (n = 4 for Ctrl, 6 for NE), 10 μM isoproterenol (Iso) for 4 h (n = 4 per group), 0.1 μM CL-316,243 (CL) for 24 h (n = 6 for Ctrl, 4 for CL), 500 μM dibutyryl-cAMP (cAMP) for 6 h (n = 6 per group), or 1 μM triiodothyronine (T3) for 20 h (n = 4 per group). (d) qPCR analyses of Chrna2 and Ucp1 mRNA expression in inguinal adipose tissues of WT mice following cold exposure (CE) at 4°C for 2 d (n = 6 per group; room temperature, RT) (left) or daily oral gavage of vehicle (n = 9) or 20 mg/kg Rosi (n = 17) for 2 weeks (right). (e) qPCR analyses of CHRNA2 and UCP1 mRNA levels in differentiated human adipose stromal cells (ASC) from the subcutaneous depot exposed to vehicle, 1 μM Rosi for 4 d (n = 6 per group) or 10 μM Iso for 4 h (n = 4 per group). (f) qPCR analyses of Chrna2, Ucp1, and Pparg mRNA levels in the presence of 1 μM Rosi during WT inguinal preadipocyte differentiation (n = 3 per group). (g) Chrna2 transcriptional activity analysis using a murine Chrna2 promoter luciferase reporter construct with a PPARγ expressing vector or an empty vector as the control (n = 4 per group). (h) Representative gel image of chromatin immunoprecipitation (ChIP) analysis of PPARγ binding to the Chrna2 promoter under stimulation with 1 μM Rosi for 4 d in differentiated inguinal preadipocytes (left). Graph shows enrichment relative to input (n = 4 per group) (right). Data are presented as mean ± s.e.m. Data were analyzed by a two-tailed Student’s t-test (a–e,g,h). *P<0.05, **P<0.01, ***P<0.001. N.D., not detected.
A spectrum of signaling pathways and molecules have been reported to activate adaptive thermogenesis in adipocytes10. Chrna2 expression was induced by many of these beiging stimuli, including norepinephrine, isoproterenol (Iso, a pan adrenergic agonist), CL-316,243 (CL, a β3-adrenergic specific agonist), dibutyryl-cAMP (cAMP, a cyclic nucleotide derivative that mimics endogenous cAMP), or triiodothyronine (T3, a thyroid hormone that has long been known to play a role in regulating thermogenesis10) (Fig. 1c). Furthermore, Chrna2 mRNA expression was induced during in vivo “beiging” in the inguinal fat after cold exposure (CE) or Rosi treatment (Fig. 1d and Supplementary Fig. 1d). Conversely, Chrna2 expression was significantly lower in inguinal fat of obese mice compared to that of lean mice (Supplementary Fig. 1e). It has been previously reported that differentiated human adipose precursors from subcutaneous fat respond to thermogenic stimulants16,17, suggesting that besides their presence in the supraclavicular region, thermogenic adipocytes may also exist in the human subcutaneous depot. Similar to their murine counterparts, induction of CHRNA2 expression was observed in differentiated primary adipose stromal cells (ASC) isolated from the human subcutaneous depot upon treatments with Rosi and Iso (Fig. 1e), indicating that this observed Chrna2 induction is relevant to human physiology.
Mechanistic studies revealed that Chrna2 expression was markedly induced during the adipogenesis of inguinal preadipocytes alongside with adipogenic and thermogenic markers, such as Pparg and Ucp1, under chronic Rosi treatment (Fig. 1f). A promoter fragment that contains 1.1kb of the 5′ flanking region of murine Chrna2 was activated (induced by 4-fold) by the expression of PPARγ (Fig. 1g). Chromatin ImmunoPrecipitation (ChIP) assays confirmed that PPARγ recruitment was enriched on the promoter region of Chrna2 between −164bp and −151bp to stimulate Chrna2 transcription in inguinal adipocytes (Fig. 1h). Together these data suggest that Chrna2 was regulated by PPARγ as a direct target during beige adipogenesis (Supplementary Fig. 2).
CHRNA2 signaling is beige adipocyte-selective
To directly examine whether CHRNA2-containing ion channels are functional in fat cells, particularly beige adipocytes as Chrna2 is inducible during the beiging process, we generated “beige-fat-reporter mice” by crossing Ucp1-CRE transgenic mice18 and Ai14 reporter mice19 in which Ucp1-expressing adipocytes are labeled with red fluorescent protein (RFP). (Fig. 2a). We validated the presence of RFP+ cells through analysis of Rfp (tdTomato) expression in differentiated primary inguinal preadipocytes as well as in the inguinal adipose tissue of Ucp1-CRE-RFP mice (Supplementary Fig. 3). Both acetylcholine (Ach) and nicotine (Nic), agonists of nAChR channels, induced an increase in intracellular calcium in the RFP-labeled beige fat cells (Fig. 2b and Supplementary Fig. 4b). This agonist-induced calcium uptake in RFP+ beige adipocytes was further increased in the presence of Rosi, consistent with the notion that CHRNA2 forms a functional ion channel in activated beige adipocytes. (Supplementary Fig. 4g). However, no response to either agonist was detected in RFP-negative white adipocytes (Fig. 2b and Supplementary Fig. 4a). We did not observe any response to acetylcholine or nicotine in the RFP+ brown fat cells isolated from the interscapular depot of the same mice (Fig. 2c and Supplementary Fig. 4c). This strongly suggests that the response to CHRNA2 agonists is beige fat-specific. No cell from the Chrna2 KO inguinal fat cultures responded to agonist stimulation, suggesting that the observed response is CHRNA2-dependent (Fig. 2d and Supplementary Fig. 4d, 5a). A similar response to nicotine and acetylcholine was observed via a calcium imaging assay on primary human subcutaneous fat cultures (Fig. 2e, f and Supplementary Fig. 4e), and could be blocked by CHRNA2 knockdown in these human subcutaneous adipose cultures (Supplementary Fig. 5b), indicating that beige-like thermogenic fat cells exist in human subcutaneous depots and can be functionally regulated through CHRNA2 signaling. We further tested CHRNA2 signaling in differentiated preadipocytes isolated from human perirenal fat, a depot that has been reported to contain thermogenic adipocytes more similar to murine classical brown fat20–22. Human perirenal adipocytes, while perfectly responsive to other stimuli (e.g. menthol23), showed no response to either acetylcholine or nicotine (Fig. 2g and Supplementary Fig. 4f, 5c). Since activation of CHRNA2 was detected within the human subcutaneous but not perirenal fat culture, our results provide functional proof for the distinction of these two types of thermogenic fat in adult humans.
Figure 2.
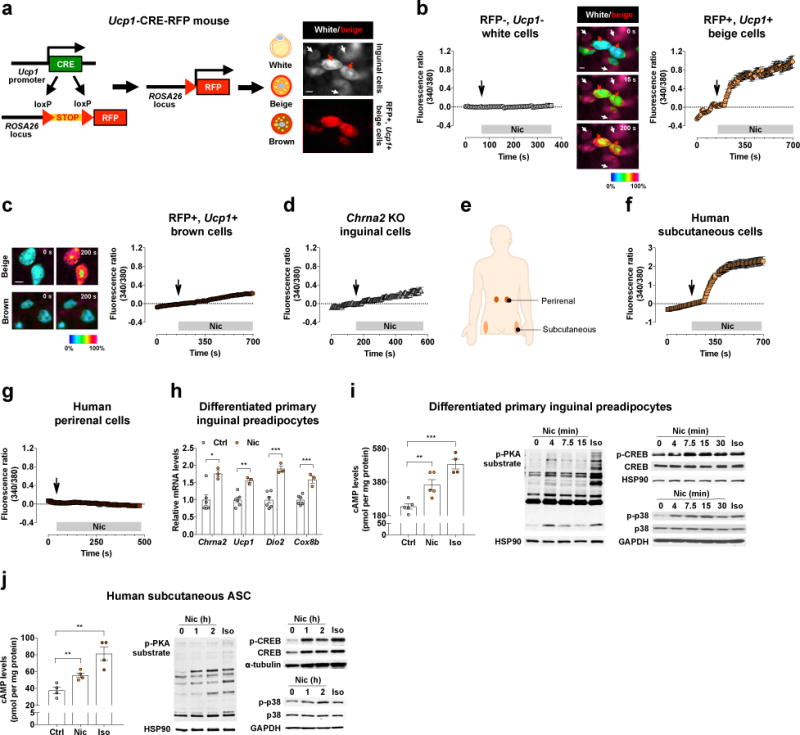
CHRNA2 signaling is beige adipocyte-selective. (a) Schematic of the generation of Ucp1-CRE-RFP mice. In differentiated inguinal preadipocytes, only beige adipocytes expressing RFP are visible at 550nm (red arrows), whereas white adipocytes are not (white arrows) (scale bar, 10 μm). (b) Intracellular Ca2+ rises in response to the CHRNA2 agonist nicotine (Nic, 500 μM) only in RFP+ beige adipocytes (red arrow) but not in white adipocytes (white arrow) within the subcutaneous fat cultures. (n = 6 for white, 7 for beige). Representative images are shown in a pseudo color scale at different time points (0, 15 and 200 s) (scale bar, 10 μm). (c) Brown adipocytes of Ucp1-CRE-RFP mice do not respond to nicotine stimulation (n = 13) (scale bar, 10 μm). (d) Calcium uptake in response to nicotine is absent in the differentiated inguinal preadipocytes from Chrna2 KO mice (n = 10). (e) Locations of the human subcutaneous and perirenal fat biopsy sites used in this study. (f, g) Differentiated human adipose stromal cells (ASC) isolated from the subcutaneous fat depot (n = 5) (f), but not those from the perirenal depot (n = 11) (g), mediate calcium uptake upon nicotine stimulation. (h) qPCR analyses of thermogenic markers in differentiated inguinal preadipocytes of wild type (WT) mice following treatment with vehicle control (Ctrl) or 2 mM nicotine (Nic) for 8 h (n = 6 for Ctrl, 3 for Nic). (i) cAMP levels of differentiated WT inguinal preadipocytes stimulated with 2 mM Nic or 10 μM isoproterenol (Iso) (left) (n = 5 for Ctrl and Nic, 4 for Iso). Immunoblot for phosphorylation of PKA substrate (middle), CREB, and p38 (right) in differentiated inguinal preadipocytes exposed to 2 mM nicotine for the indicated time or 10 μM Iso for 5 min as a positive control. (j) cAMP levels in differentiated human ASC from the subcutaneous depot treated with 2 mM nicotine or 10 μM Iso (n = 4 per group) (left). Immunoblots for phosphorylation of PKA substrate (middle), CREB and p38 (right) in differentiated human ASC treated with 2 mM nicotine for the indicated time or 10 μM Iso for 1 h as a positive control. Representative images or blots are shown. Data are presented as mean ± s.e.m. Data were analyzed by a two-tailed Student’s t-test (h) or a one-way ANOVA (i,j). *P<0.05, **P<0.01, ***P<0.001. (b–g) In some panels, error bars are almost invisible.
Figure 5.
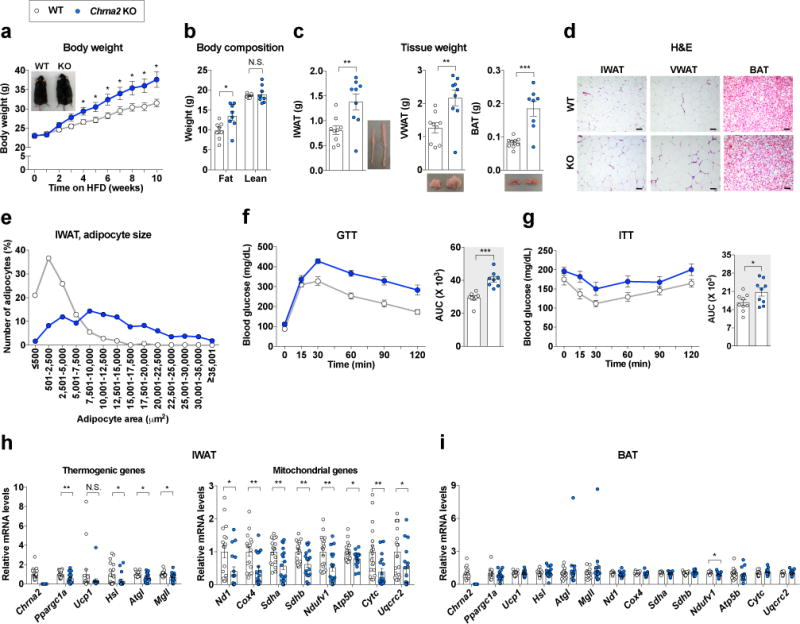
Loss of Chrna2 exacerbates diet-induced obesity. (a) Body weights of WT and Chrna2 KO mice during high-fat diet (HFD) feeding (n = 9 per group). (b) Body composition (n = 8 per group) and (c) fat mass (n = 9 per group) of WT and Chrna2 KO mice following 10 weeks on a HFD. (d, e) Representative images of H&E stained sections of IWAT, VWAT, and interscapular BAT (d, scale bar, 100 μm), and distribution of adipocyte size of IWAT (e) from WT and Chrna2 KO mice fed a HFD. (f, g) Intraperitoneal glucose tolerance (f, GTT with 1 g/kg glucose: n = 8 per group) and insulin tolerance (g, ITT with 0.5 U/kg insulin: n = 9 per group) test in WT and Chrna2 KO mice. AUC, area under the curve. (h, i) qPCR analyses of thermogenic gene expression in IWAT (n = 18 per group) (h) and BAT (n = 18 for WT, 19 for KO) (i) of WT and Chrna2 KO mice fed a HFD for 10 weeks. Data are presented as mean ± s.e.m. Data were analyzed by a two-tailed Student’s t-test (a–c,f-i) or Mann-Whitney U test (h,i). *P<0.05, **P<0.01, ***P<0.001. N.S., not significant.
Since the calcium imaging assay indicated that the CHRNA2-containing ion channel is functional in beige adipocytes, we next investigated how CHRNA2 signaling is mediated in differentiated primary inguinal preadipocytes. Thermogenic markers Ucp1, Dio2 and Cox8b were induced with nicotine treatment compared to control (Fig. 2h). It is worth noting that the Chrna2 receptor itself was also induced upon nicotine treatment. This is consistent with our previous data showing that Chrna2 was induced during the activation of beige fat cells (Fig. 1). We observed a clear elevation of cAMP, a second messenger molecule to thermogenic stimuli, and activation of protein kinase A (PKA) after nicotine stimulation of murine inguinal fat cells (Fig. 2i). We further showed that both CREB and p38 were activated in nicotine treated inguinal fat cells (Fig. 2i and Supplementary Fig. 6a). Similar activation of the cAMP and PKA pathway was observed in differentiated human subcutaneous adipose cultures stimulated with nicotine (Fig. 2j), and this activation could be blocked by the PKA inhibitor H-89 in both murine inguinal and human subcutaneous fat cultures (Supplementary Fig. 6b). This signal was dependent on CHRNA2 with no significant compensation from other nAChR family proteins since no activation of PKA, CREB or p38 was observed in nicotine-treated Chrna2 KO inguinal fat cells (Supplementary Fig. 6c, d). Similarly, the thermogenic marker Ucp1 was not induced by nicotine stimulation in the absence of Chrna2 (Supplementary Fig. 6e).
Immune cells in subcutaneous fat produce acetylcholine
To investigate whether the activation of CHRNA2 in subcutaneous fat is mediated by locally produced acetylcholine via paracrine/autocrine mechanisms, we analyzed GFP+ cells within the inguinal adipose tissue from ChATBAC-eGFP mice24, which express enhanced green fluorescent protein (eGFP) under the control of the transcriptional regulatory element for Choline acetyltransferase (ChAT), the rate limiting enzyme that mediates the biosynthesis of acetylcholine in vivo (Fig. 3a). qPCR and immunohistochemistry analysis revealed that GFP+ cells are indeed present in the inguinal adipose tissue, and consist of a subset of stromal vascular fraction (SVF) cells (Fig. 3b and Supplementary Fig. 7a). Enrichment of ChAT protein in inguinal SVF cells was further confirmed by flow cytometric analysis (Fig. 3c). The secretion of acetylcholine from the SVF was directly assayed and validated with mass spectrometry (Fig. 3d). We hypothesized that the local production of acetylcholine from ChAT expressing cells within the subcutaneous fat tissue may closely correlate with the induction of its receptor, namely CHRNA2, in activated beige adipocytes. Indeed both acetylcholine secretion and ChAT expression were higher in the SVF from cold exposed mice compared to those from mice housed at the ambient temperature (Fig. 3e). Conversely, ChAT expression in the SVF was lower in obese mice compared that of lean mice (Supplementary Fig. 7b). In order to directly assess the potential role of these acetylcholine-producing SVF cells in regulating the function of neighboring adipocytes, coculture experiments were carried out with a two-chamber transwell system (Fig. 3f). The expression of thermogenic markers was elevated in differentiated inguinal fat cultures that were exposed to freshly isolated SVF compared to control inguinal fat cultures (Fig. 3f) and an additional induction was detected when the coculture was performed in the presence of rivastigmine (inhibitor of acetylcholinesterase and butyrylcholinesterase), which prevents acetylcholine degradation (Fig. 3f), indicating that the effect was at least in part mediated through acetylcholine released from the SVF.
Figure 3.
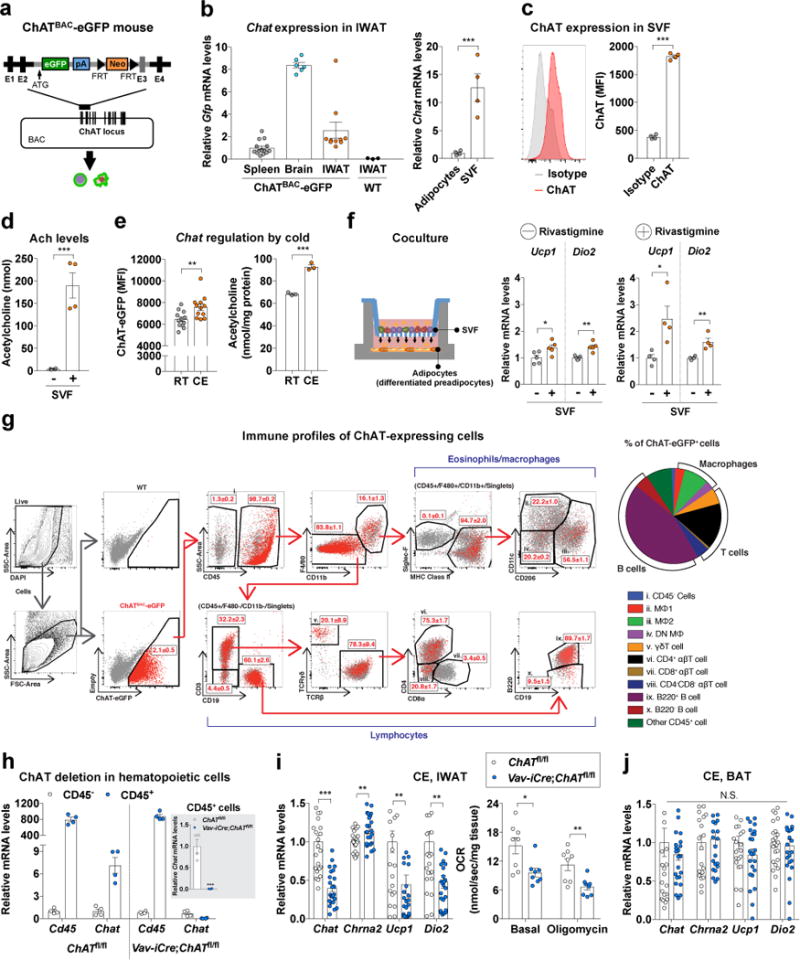
Acetylcholine-synthesizing cells of hematopoietic origin reside within inguinal subcutaneous fat tissue. (a) Schematic of the generation of ChATBAC-eGFP mice. (b) qPCR analyses of Gfp mRNA levels in the spleen (n = 14), the brain (n = 6) and IWAT (n = 10) of ChATBAC-eGFP mice and IWAT(n = 3) of C57BL/6J (WT) mice (negative control) (left). qPCR analyses of Chat mRNA levels in stromal vascular fraction (SVF) and mature adipocytes from IWAT of WT mice (n = 4 per group) (right). (c) Representative histograms and median fluorescence intensities (MFI) for ChAT protein expression in inguinal SVF of WT mice (n = 4 per group). (d) Quantification of acetylcholine levels secreted from the inguinal SVF of WT mice. PBS incubated with SVF and without SVF were labeled with “+” and “-”, respectively (n = 4 per group). (e) MFI of ChAT-eGFP in the GFP+ population from inguinal SVF of ChATBAC-eGFP mice housed at room temperature (RT) or 4°C for 4 h (CE). GFP+ cells and their MFI were monitored by flow cytometry (n = 12 per group) (left). Quantification of acetylcholine levels secreted from the inguinal SVF of WT mice housed at RT or CE (n = 3 per group) (right). (f) Adipocytes and SVF in a two-chamber coculture system using a bicompartmental Transwell plate (left). qPCR analyses of thermogenic markers in differentiated WT inguinal preadipocytes cocultured with or without freshly isolated inguinal SVF from WT mice for 4 h in the absence (n = 5 per group) or presence (n = 4 per group) of rivastigmine using a two-chamber coculture system (right). (g) Immune profiling of ChAT-expressing cells from inguinal SVF of ChATBAC-eGFP mice. Flow cytometric gating strategy for identifying immune cell subsets in ChAT-eGFP+ cells. All immune cell subsets were labeled with numerals i through x and reported as percentages of the parent population (n = 3 per group) (left). The relative frequencies of immune cell subsets (i-x) within ChAT-eGFP+ cell population are summarized and shown as a pie chart (right). (h) qPCR analyses of Chat mRNA levels in CD45− and CD45+ cell populations collected by FACS from inguinal SVF of ChATfl/fl and Vav-iCre;ChATfl/fl mice (n = 4 per group). (i) qPCR analyses of thermogenic markers in IWAT of ChATfl/fl and Vav-iCre;ChATfl/fl mice exposed at 4°C for 6 h (CE) (n = 24 per group) (left). Oxygen consumption rate (OCR) in freshly isolated inguinal fat tissue from cold exposed ChATfl/fl and Vav-iCre;ChATfl/fl mice in the absence (basal) or presence of oligomycin (n = 8 per group) (right). (j) qPCR analyses of thermogenic markers in the interscapular BAT from ChATfl/fl and Vav-iCre;ChATfl/fl mice following CE (n = 24 per group). Data are presented as mean ± s.e.m. Data were analyzed by a two-tailed Student’s t-test (b–f,h–j) or Mann-Whitney U test (i). *P<0.05, **P<0.01, ***P<0.001. N.S., not significant.
Flow cytometric analysis revealed that about 2.1% of the inguinal SVF of the ChATBAC-eGFP mice were GFP positive, showing significantly higher GFP median fluorescence intensity (MFI) and enriched expression of Chat compared to GFP− cells (Fig. 3g and Supplementary Fig. 7c). Immune profiling by flow cytometric analysis (Supplementary Table 1) revealed that the majority of ChAT-eGFP+ cells were of the CD45+ hematopoietic lineage (98.7±0.2%). Among hematopoietic cells, B220+ B cells made up the largest proportion of immune cell subsets with 44.5±2.5%, followed by CD4+ αβT cells (15.6±1.1%) and M2 macrophages (8.5±0.4%). Of note, there were almost no ChAT-eGFP+ cells that expressed the eosinophil marker Siglec-F (Fig. 3g and Supplementary Table 1). It has previously been reported that certain types of B cells or T cells can produce acetylcholine25,26. It is of note that ~ 9% of the total ChAT-eGFP+ cells were characterized as alternatively activated M2 macrophages, which have been implicated to produce catecholamines that activate thermogenic fat cells27. The upregulation of thermogenic gene expression by coculture with SVF was detected even in the absence of β-adrenergic signaling28, indicating that non-catecholamine stimulant(s) exerts this activating effect (Supplementary Fig. 7d). This is consistent with recent reports that have challenged the secretion of catecholamine from M2 macrophages29,30.
To investigate the physiological impact of acetylcholine-producing immune cells during beiging, we generated mice in which the Chat gene was deleted in CD45+ hematopoietic cells (Vav-iCre;ChATfl/fl; Fig. 3h and Supplementary Fig. 8a). A significant lower Chat expression was observed in inguinal white adipose tissue (IWAT) of Vav-iCre;ChATfl/fl mice compared to that of control mice, but not in the interscapular brown adipose tissue (BAT), which is consistent with the notion that the ChAT-CHRNA2 axis is beige-selective (Supplementary Fig. 8b). The phenotype of Vav-iCre;ChATfl/fl mice was comparable to littermate ChATfl/fl controls when housed at room temperature (Supplementary Fig. 8c–g). However, hematopoietic ablation of Chat caused thermogenic defects in IWAT upon acute cold exposure with a significant lower induction in thermogenic marker expression and oxygen consumption rate compared to those of the control mice (Fig. 3i), whereas not in BAT (Fig. 3j). We performed coculture experiments and observed that the induction of thermogenic markers was trending lower (albeit not statistical significant) in the inguinal fat cells cocultured with the SVF from Vav-iCre;ChATfl/fl mice compared to that of littermate ChATfl/fl controls (Supplementary Fig. 8h). It is worth noting that thermogenic gene expression in IWAT was comparable upon chronic cold exposure between Vav-iCre;ChATfl/fl mice and littermate controls (Supplementary Fig. 8i–o) suggesting that systemic acetylcholine may compensate for the loss of local acetylcholine production when cold stimulus persists.
Loss of CHRNA2 affects whole-body metabolism
Our results so far demonstrate that an acetylcholine-CHRNA2 signaling axis exists within the subcutaneous adipose depot, and immune cells closely regulate beige fat activation, likely through a paracrine mechanism (Supplementary Fig. 8p and 9). Low environmental temperature greatly activates beige fat recruitment within the subcutaneous white adipose depot so that organisms have additional thermogenic capacity to maintain core body temperature9. Cold exposure induced Chrna2 expression in inguinal fat (Fig. 1d), suggesting that adaptation to cold challenges may be mediated through CHRNA2. Chrna2 KO mice showed no gross abnormality when housed at ambient temperature31 (Supplementary Fig. 10a–c). After chronic cold exposure, adipose tissues of WT mice underwent drastic tissue remodeling including reduced tissue mass, which led to the reduction of overall body weight (Fig. 4a). In comparison, Chrna2 KO mice showed significantly less body weight loss after being housed in the cold chamber (Fig. 4a). Adipose tissue mass of the visceral white fat depot (VWAT, perigonadal depot) was significantly bigger in Chrna2 KO mice compared to WT following cold exposure. A similar but not statistically significant trend was observed in the inguinal depot, whereas no mass difference in the interscapular BAT was found between the two genotypes (Fig. 4b). Histology and immunohistochemistry analyses revealed that after cold exposure, fewer UCP1+ multilocular adipocytes were present in inguinal fat tissues of Chrna2 KO mice compared to WT mice, yet no differences in brown fat were observed (Supplementary Fig. 10d,e).
Figure 4.
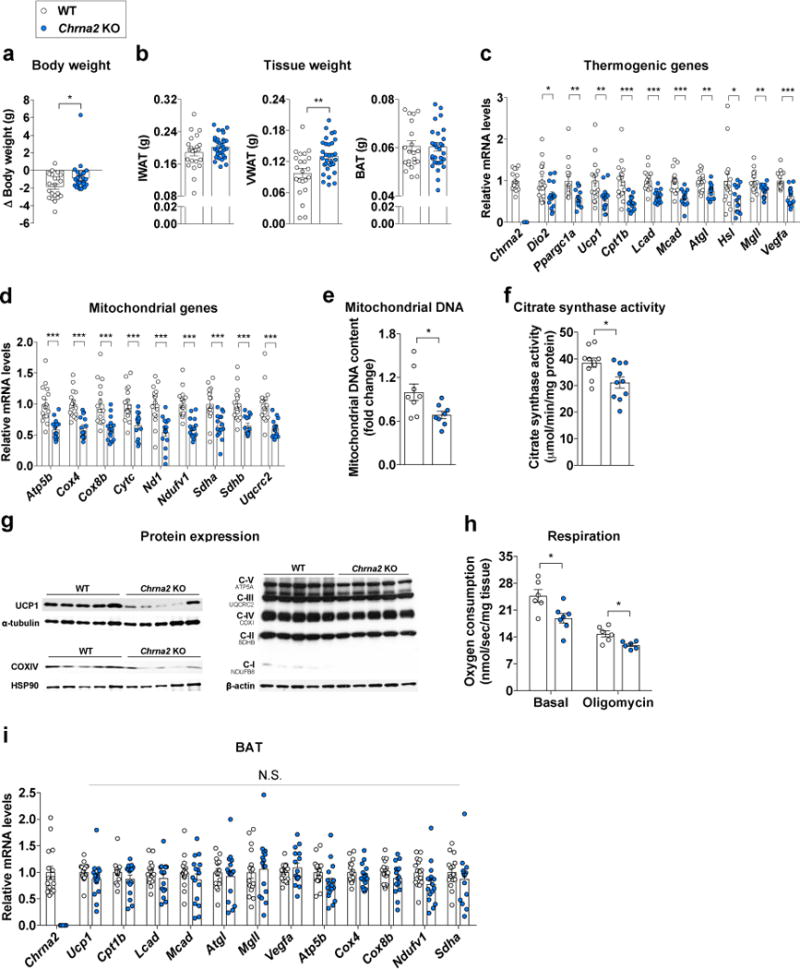
Loss of Chrna2 reduces the adaptive thermogenic capacity of inguinal adipose tissue. (a) Changes in body weight of wild type (WT) and Chrna2 KO mice following 2 weeks cold exposure (CE) at 10°C (n = 22 for WT, 25 for KO). (b) Inguinal (IWAT), visceral (VWAT), and interscapular brown (BAT) fat mass in WT and Chrna2 KO mice after CE (n = 22 for WT, 31 for KO). (c, d) qPCR analyses of thermogenic genes (c) and mitochondrial genes (d) in IWAT from WT and Chrna2 KO mice after CE (n = 16 for WT, 15 for KO). (e) mitochondrial DNA content in IWAT of WT and Chrna2 KO mice following CE (n = 8 per group). (f) Citrate synthase activity in IWAT of cold exposed WT and Chrna2 KO mice (n = 10 per group). (g) Immunoblot analyses of UCP1, the mitochondrial marker COXIV and OxPhos components in IWAT of WT and Chrna2 KO mice after CE. Representative images are shown. (h) Oxygen consumption rate (OCR) in freshly isolated inguinal fat tissue from cold exposed WT and Chrna2 KO mice in the absence (basal) or presence of oligomycin (n = 6 for WT, 7 for KO). (i) qPCR analyses of thermogenic markers in the interscapular BAT from WT and Chrna2 KO mice following 2 weeks CE (n = 16 per group). Data are presented as mean ± s.e.m. Data were analyzed by a two-tailed Student’s t-test (a–f,h,i). *P<0.05, **P<0.01, ***P<0.001. N.S., not significant.
Gene expression analyses revealed that cold-induced activation of the thermogenic program was defective in Chrna2 KO IWAT, including thermogenic markers (Dio2, Ppargc1a and Ucp1), fatty acid oxidation genes (Cpt1b, Lcad and Mcad), lipolytic genes (Atg, Hsl and Mgll), the angiogenic gene Vegfa and mitochondrial genes (Atp5b, Cox4, Cox8b, Cytc, Nd1, Ndufv1, Sdha, Sdhb and Uqcrc2) (Fig. 4c,d). Citrate synthase is the initial enzyme of the TCA cycle and its activity often serves as a sensitive marker for mitochondrial function. Mitochondrial DNA content and citrate synthase activity were lower in the inguinal fat tissue of the cold challenged Chrna2 KO mice compared to WT mice (Fig. 4e,f). In line with these observations, less induction of protein levels of UCP1, the mitochondrial marker COXIV and representative subunits of the respiratory complexes I-V were detected in IWAT of Chrna2 KO mice after cold exposure compared to WT mice (Fig. 4g). These led to a significant lower level of both basal and uncoupled respiration in IWAT of Chrna2 KO mice compared with that of WT mice (Fig. 4h). Notably, the impaired adaptive thermogenic capacity caused by Chrna2 ablation was limited to IWAT. The induction of thermogenic gene expression was not defective in the interscapular BAT from Chrna2 KO mice after cold exposure compared to WT mice (Fig. 4i). However, the defective inguinal thermogenic capacity did not cause hypothermia under the cold challenge, presumably due to a compensatory induction of skeletal muscle shivering10,32,33 in Chrna2 KO mice (Supplementary Fig. 10f–j).
Since impaired energy expenditure in IWAT is closely associated with the development of obesity34, we examined the physiological acclimation of Chrna2 KO mice to nutrient overload. Chrna2 KO mice did not show differences from WT mice during chow diet feeding (Supplementary Fig. 11a–g). The mutant mice displayed greater weight gain on a high-fat diet (HFD) with similar food intake (Fig. 5a and Supplementary Fig. 11h). This was accompanied by higher fat content as determined by NMR (Fig. 5b), and larger tissue mass in all three major fat depots, IWAT, VWAT, and BAT (Fig. 5c). Analysis of adipocyte size distribution showed that Chrna2 KO mice have a lower proportion of small adipocytes but a greater proportion of large adipocytes in both IWAT and VWAT compared to WT mice after HFD challenge (Fig. 5d,e and Supplementary Fig. 11i). Along with exacerbated weight gain, the fasting blood glucose and serum insulin levels were significantly higher in Chrna2 KO mice than in controls, indicating that insulin resistance was developed in the absence of Chrna2 signaling (Supplementary Fig. 11j,k). Consistently, the mutant mice had worsened glucose tolerance and insulin sensitivity (Fig. 5f,g). HFD feeding resulted in a lower thermogenic gene expression in the inguinal fat of Chrna2 KO mice compared to the controls, with a minimal effect in the BAT (Fig. 5h,i), and a lower whole-body energy expenditure in Chrna2 KO mice (Supplementary Fig. 11l). Visceral fat of Chrna2 KO mice fed on HFD demonstrated significantly higher mRNA levels of macrophage-related inflammatory genes and was worse in oxidative stress through NADPH oxidase activation, compared to those of control mice (Supplementary Fig. 11i). However, Chrna2 ablation did not have a significant impact on the skeletal muscle following HFD feeding. There were no differences in locomotor activities between the two genotypes and the effects of Chrna2 ablation were minimal in mRNA expression of mitochondrial oxidative phosphorylation-related genes (Ppargc1a, Atp5b, Nd1, Sdha, Sdhb and Ndufv1), known to be affected by HFD challenge in the skeletal muscle35 (Supplementary Fig. 11m,n). Collectively, these results suggest that CHRNA2 signaling in beige adipocytes may provide essential protection against HFD-induced obesity and associated metabolic disorders, and that even partial loss of beige fat regulation may lead to serious detrimental consequences in systemic metabolism.
DISCUSSION
Understanding the molecular mechanisms and signaling networks that regulate beige fat activation is of fundamental importance towards the goal of inducing it to protect against obesity in humans. We have identified a unique signaling pathway within activated beige adipocytes, which is conserved between mice and humans. The beige-selective pathway is mediated through a novel immune-adipose communication, and this previously unrecognized acetylcholine-dependent CHRNA2 signaling coordinates the integrated thermogenic response in activated beige fat. Lack of CHRNA2 signaling leads to a compromised adaptation to chronic cold challenge, dysregulates whole body metabolism and exacerbates diet-induced obesity. It is well documented that upon the onset of cold challenge, skeletal muscle and classical brown fat play an essential role to counteract hypothermia. Indeed, snap-shot evaluation of whole-body energy expenditure under cold did not reveal statistically significant differences between Chrna2 KO mice and the controls. It is also worth pointing out that the canonical β-adrenergic regulation of beige fat remains intact within these Chrna2 mutant mice. It has been reported that mice with deletion of beige adipocyte function are vulnerable to metabolic challenges34. The thermogenic defects after chronic cold exposure and metabolic dysfunction upon HFD observed in Chrna2 KO mice, a model with only one branch of beige fat regulation missing, not only further confirms the physiological significance of beige adipocytes as an important metabolic regulator but also highlights the essential role of catecholamine-independent, acetylcholine-CHRNA2 signaling in energy metabolism.
It has been well reported that immune cells residing within the adipose tissue niche closely interact with fat cells to mediate local and systemic metabolic homeostasis36,37. The novel signaling described here demonstrated a dynamic interaction between acetylcholine secreted from immune cells and beige fat-specific CHRNA2, and this signaling directly influences organismal fitness upon environmental challenges, such as cold and obesity. Future investigations will reveal if one subpopulation of the ChAT-expressing immune cells within the subcutaneous fat tissue plays a dominant role regulating CHRNA2-conducted beige fat function. The identification of such a subset, (if it exists), will help to incorporate the acetylcholine-beige fat signaling to the previously known signaling network between immune cells and adipocytes and place this newly defined pathway within the interconnected metabolic matrix.
Nicotine, the main component of tobacco, has been implicated in the effects of smoking on body weight, presumably through central regulation on appetite38. It has also been observed that daily injections of nicotine for 6 months led to decreased food intake, weight loss, and induction of UCP1 in white adipose tissue in mice39. The activation of the thermogenic program in WAT could be, at least partially, mediated through cell-autonomous activation of CHRNA2-signaling in beige adipocytes. Meanwhile, it has been reported that changes in nerve activity influence systemic energy balance and promote WAT browning40,41. Therefore, it is also conceivable that chronic exposure of nicotine activates certain types of cholinergic neurons in the central and/or peripheral nervous systems, elevating energy expenditure and increasing UCP1 expression. No statistically significant differences were observed in food intake between Chrna2 KO and control mice in our studies. Future investigations should reveal whether CHRNA2-mediated central regulation may also affect systemic metabolism.
Our discovery that nicotine activates the thermogenic program in beige adipocytes through CHRNA2 may suggest novel targets through peripheral pathways to circumvent weight gain associated with smoking cessation. Collectively, our findings suggest that the Chnra2 pathway could present new targets for beige fat activation to curb obesity and its related metabolic diseases in humans.
ONLINE METHODS
Reagents
Dexamethasone (D4902), insulin (I5500), 3-IsoButyl-1-MethylXanthine (IBMX, I7018), biotin (B4639), D-pantothenic acid hemicalcium salt (P5155), isoproterenol (I6504), CL-316,243 (C5976), norepinephrine (A7257), dibutyryl-cAMP (D0260), triiodothyronine (T5516), acetylcholine chloride (A2661), nicotine (N3876), (1R,2S,5R)-(−)-menthol (M2780-25G-A) and Fura 2-AM (F0888) were acquired from Sigma Aldrich. Rosiglitazone (71740), GW9662 (70785), pioglitazone (71745), rivastigmine (129101-54-8), and H-89 (10010556) were obtained from Cayman chemical. Collagenase D (11088882001), collagenase B (11088831001), and dispase II (04942078001) were purchased from Roche. Recombinant human BMP4 (314-BP-010) was acquired from R&D Systems. All the cell culture media, including DMEM/F12 GlutaMAX (10565-042), DMEM (11995-073) and MesenPRO RS™ Medium (12746-012), and BODIPY 493/503 (4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene; D3922) were purchased from Life Technologies.
Animals
All animal studies were conducted according to the protocol reviewed and approved by the University Committee on Use and Care of Animals at the University of Michigan. All mice were housed under 12-h light/12-h dark cycle (6 a.m.–6 p.m.) with a standard rodent chow diet unless otherwise specified. The 129SVE and Balb/c mice were purchased from Taconic Farms Inc. The Ucp1-CRE transgenic mice, the Ai14 reporter mice, C57BL/6J, ob/ob, ChATBAC-eGFP, ChATfl/fl, Vav-iCre, B6.SJL and Chrna2 knockout mice were obtained from the Jackson Laboratory (Stocks #024670, #007914, #000664, #000632, #007902, #016920, #008610, #002014 and #005797 respectively). β-less mice were provided by Brad Lowell (Beth Israel Deaconess Medical Center, Boston). WT mice were treated with either vehicle (0.25% methylcellulose) or 20 mg/kg/d rosiglitazone via oral gavage for 2 weeks. Hematopoietic cell-specific ChAT-deleted mice (Vav-iCre;ChATfl/fl) were generated by crossing ChATfl/fl and Vav-iCre mice. Ten to twelve week old Chrna2 KO mice and either wild type C57BL/6J or littermate WT controls were used in cold exposure and high-fat diet (HFD) studies. Similar results were obtained with either control group. For cold exposure studies, mice were single housed in prechilled cages in a 10 °C environmental chamber for 2 weeks unless otherwise specified. Age-matched WT and Chrna2 KO mice were single-housed and maintained on either a chow diet or a HFD consisting of 45% of calories from fat (D12451, Research Diets) for 10 weeks to perform the HFD-induced obesity study. Body weight and food intake were measured weekly for 10 weeks. The food intake was determined by subtracting the weight of remaining food from that of provided food for singly housed mice.
Primary cell culture
The isolation, culture and differentiation of primary SVF from adipose depots were performed as described previously1. Briefly, fat tissues were dissected, minced and digested in collagenase (1.5 U/ml) (collagenase D for inguinal fat, and collagenase B for interscapular brown fat) and dispase II (2.4 U/ml) solution supplemented with 10 mM CaCl2 for 15 to 20 min in a 37 °C water bath with agitation. Digested tissues were filtered through a 100 μm cell strainer and centrifuged at 300–500 × g for 5 min to pellet cells. The floating mature adipocytes were collected when they were needed for gene expression analyses. The cell pellet was resuspended and passed through a 40 μm cell strainer and centrifuged as above. The cell pellet was resuspended with culture medium (DMEM/F12 GlutaMAX containing 15 % FBS and penicillin/streptomycin) and plated onto a collagen-coated 10 cm cell culture dish. For adipocyte differentiation, confluent cultures of cells were exposed to induction medium (DMEM/F-12 GlutaMAX supplemented with 10% FBS, penicillin/streptomycin, 0.5 μg/ml insulin, 5 μM dexamethasone, 1 μM rosiglitazone and 0.5 mM IBMX). Two days after induction, cells were cultured in maintenance medium containing 10% FBS, penicillin-streptomycin and 0.5 μg/ml insulin until they were ready for analyses.
Human adipose precursor cells from the subcutaneous adipose tissue were acquired from liposuction performed on healthy adults (a gift from Dr. Jeffrey M. Gimble at Tulane University, New Orleans, Louisiana, USA) and human perirenal adipose precursor cells were isolated from donors postmortem (a gift from Dr. William Rainey at the University of Michigan, Ann Arbor, Michigan, USA). All specimens were collected under the protocols reviewed and approved by the Western Institutional Review Board (Puyallup, WA) or the University of Michigan Medical School Institutional Review Board (IRBMED). Cells were cultured in MesenPRO RS™ medium supplemented with penicillin/streptomycin and stimulated to differentiate in DMEM/F12 GlutaMAX supplemented with 10% FBS, dexamethasone (5 μM), insulin (0.5 μg/ml), IBMX (0.5 mM), rosiglitazone (5 μM), biotin (33 μM), pantothenic acid (17 μM), and BMP4 (20 ng/ml) for 3 d. On the fourth day, cells were cultured in maintenance medium (DMEM/F12 GlutaMAX containing 10% FBS, 0.5 μg/ml insulin, 5 μM dexamethasone, 33 μM biotin, and 17 μM pantothenic acid) until they were fully differentiated.
Transcriptional profiling
Total RNA was isolated from differentiated inguinal SVF treated with rosiglitazone (1 μM for 4 d) or a vehicle control and used for microarray analysis. Array hybridization and scanning were performed by the Dana-Farber Cancer Institute Core Facility using affymetrix Gene Chip Mouse Genome 430 2.0 arrays according to established methods.
Gene expression analysis
Total RNA from adipose tissue and cultured cells were extracted using the TRIzol method. For quantitative real-time PCR (qPCR) analysis, an equal amount of RNA was used to synthesize cDNA according to the instruction for M-MLV Reverse Transcriptase (Life Technologies). qPCR reactions were performed in a 384-well format using SYBR Green (Thermo Fisher Scientific). Relative mRNA levels were calculated using the ΔΔCT method and normalized to Tbp. The primer sequence information is shown in Supplementary Table 2.
Luciferase reporter assay
To construct the luciferase reporter with the promoter of the murine Chrna2 gene, the −1053 to +31 region of mouse Chrna2 gene was amplified by PCR using mouse genomic DNA. The amplified fragment was digested with MluI and XhoI and was cloned into the pGL3 basic vector (Promega) upstream of the firefly luciferase-encoding region. HEK293 cells (ATCC, CRL-1573) were plated onto a 12-well plate and transiently transfected using the polyethylenimine (Thermo Fisher Scientific) method with 909 ng of PPARγ, 90.9 ng of the reporter and 10 ng of renilla (internal control) plasmids. Cells were cultured for 48 h before harvest. Luciferase activity was measured by the Dual-Luciferase Assay kit (Promega) according to the manufacturer’s recommendations with a SpectraMax L (Molecular Devices) plate reader. Firefly luciferase activity was normalized to renilla luciferase activity.
Chromatin immunoprecipitation (ChIP) assay
Differentiated primary inguinal preadipocytes were cross-linked with 1% formaldehyde at room temperature for 7 min and lysed with ChIP cell lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, protease inhibitor cocktail) and ChIP nuclear lysis buffer (50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 1% SDS, protease inhibitor cocktail). The cell lysates were sonicated to shear the chromatin, and immunoprecipitated with antibodies for PPARγ (#sc-7196, Santa Cruz) or IgG (#2729, Cell Signaling). The immunoprecipitants were isolated using protein A agarose beads (Invitrogen), and washed and eluted with 1% SDS in 0.1 M NaHCO3. After reverse cross-linking at 65 °C overnight and proteinase K digestion, immunoprecipitated DNA fragments and input DNA were recovered by a PCR purification kit (Qiagen). The purified DNA was used to amplify the PPARγ regulatory element on the mouse Chrna2 promoter by a PCR reaction. The PCR products were visualized by electrophoresis on an agarose gel containing ethidium bromide under UV light and quantified using ImageJ software. Primers directed at 7 kb upstream of the binding site were used as a negative control.
BODIPY (493/503) staining
To stain lipid droplets, differentiated inguinal preadipocytes of Ucp1-CRE-RFP mice were fixed in 10% formalin, washed with PBS, and incubated in PBS supplemented with 0.01 mg/mL fluorescent neutral lipid dye, BODIPY 493/503, at 4°C for 30 min in the dark. The cells were washed with PBS and imaged with a fluorescent inverted Leica DMIRB microscope.
Calcium imaging
SVF isolated from mouse inguinal, mouse interscapular brown, human subcutaneous and human perirenal depots were seeded on collagen-coated glass bottom culture dishes (MatTek Corporation) and induced to differentiate. Fully differentiated adipocytes were loaded with 10 μM of Fura 2-AM for 30 min at 37 °C. After 30 min they were washed twice with standard tyrode’s solution (135 mM NaCl, 4 mM KCl, 10 mM glucose, 10 mM HEPES, 2 mM CaCl2, and 1 mM MgCl2, pH 7.4) at room temperature. Calcium imaging was performed on an Olympus BX51WI Axiovert microscope under a 40× objective. Fluorescence images were documented upon sequential excitation with 340 nm followed by 380 nm with a Roper CoolSnap CCD camera. After establishing a baseline 340/380 nm ratio, the CHRNA2 agonist (500 μM nicotine or 100 μM acetylcholine) was perfused onto the cells. Images were processed with MetaFlour software (Olympus). At 0 s, the basal pseudo color of beige adipocytes and white adipocytes are different, (blue versus pink), reflecting that the color in beige fat cells is the combination of calcium sensing fluorescent dye Fura 2-AM and RFP, whereas white fat cells only have the Fura 2-AM.
ShRNA-mediated gene knockdown
Adenovirus-delivered shRNA targeting human CHRNA2 was generated by cloning designed shRNA into pAdTrack-H1 vector and constructing adenovirus using AdEasy system as described previously42. To make the adenoviral construct encoding CHRNA2 shRNA, the primers used were as follows: forward, 5′-GATCCCCCGTTCCTAGCTGGAATGATCTTTCAAGAGAAGATCATTCCAGCTAGGAACGTTTTTA-3′; reverse, 5′-AGCTTAAAAACGTTCCTAGCTGGAATGATCT TCTCTTGAAAGATCATTCCAGCTAGGAACGGGG-3′. The generated adenovirus expressing shCHRNA2 also expressed GFP, which allowed us to verify and visually monitor the infection. To knockdown CHRNA2 in human subcutaneous adipocytes, differentiated human adipose precursor cells were transduced with adenovirus expressing shRNA against CHRNA2 and further cultured for 3 d. Adenovirus encoding GFP was used as a control.
Cellular cAMP levels
cAMP was extracted with 0.1 M HCl from differentiated primary preadipocytes and then quantified using a competitive enzyme immunoassay kit according to manufacturer’s instructions (Cayman chemical).
Immunoblotting
Total protein from fat tissues and differentiated primary preadipocytes was prepared in ice-cold lysis buffer (50 mM Tris-HCl, pH 7.5, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride) supplemented with a protease inhibitor cocktail (Roche) and phosphatase inhibitors (10 mM NaF, 60 mM β-glycerolphosphate, pH 7.5, 2 mM sodium orthovanadate, 10 mM sodium pyrophosphate). Proteins were subjected to SDS-PAGE, and were then transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were probed with following antibodies: from Abcam: UCP1 (ab10983), MitoProfile total OXPHOS (ab110413); from Cell Signaling: COXIV (#4850), phospho-PKA substrateS/T (#9621), phospho-CREBS133 (#9198), CREB (#9212), phospho-p38T180/Y182 (#9215), p38 (#9197) α-tubulin (#2144), β-actin (#8457), GAPDH (#5174), and HSP90 (#4874). Phosphorylated CREB and p38 normalized to total CREB and p38, respectively, were quantified using ImageJ software.
Immunofluorescence staining
Inguinal fat and brain tissues were fixed in formalin (10%) after dissection. After washing with PBS, tissues were dehydrated in 15% sucrose for 2 d and in 30% sucrose for another 3 d. Following embedding in OCT (Sakura), tissues were sectioned at 16–20 μm. The sections were blocked in 2% normal donkey serum, 3% BSA, and 0.2% Triton X-100 in PBS for 1 h and incubated with a rabbit polyclonal GFP antibody (A-6455, Life Technologies) at a 1:400 dilution overnight. Following incubation with an Alexa Fluor 488-conjugated secondary antibody (ab150073, Abcam) for 1 h at room temperature, slides were mounted with ProLong Gold antifade reagent with DAPI (P36935, Thermo Fisher Scientific) and imaged with a LEICA DM2000.
Acetylcholine quantification
Inguinal SVF was isolated from C57BL/6J mice as described above and washed with PBS. The SVF was incubated in PBS supplemented with 150 μM rivastigmine for 40 min at room temperature and then centrifuged to collect supernatant containing acetylcholine (Ach) produced from the SVF. Ach was measured using a previously described LC-MS/MS assay for neurotransmitters43. Briefly, d4Ach (C/D/N isotopes, Pointe-Claire, Canada) was added to samples as an internal standard and resulting samples separated by LC using the Thermo Scientific TSQ Quantum LCMS system. Mobile phase A was 10 mM ammonium formate and 0.15% (v/v) formic acid in water. Mobile phase B was acetonitrile. The gradient was: initial, 5% B; 0.1 min, 19% B; 1 min, 26% B; 1.5 min, 75% B; 2.50 min, 100% B; 3.0 min, 100% B; 3.1 min, 5% B; 3.5 min, 5% B; 4.0 min, 0% B. Ach and d4-Ach were measured by MS/MS using the following transitions (m/z) Ach (product: 146, precursor: 87) and d4-Ach (product: 150, precursor: 91). Quantification was made by using ratio of the peak areas of Ach that were divided by the area of the internal standard for Ach (d4Ach) in comparison to calibration curves.
Adipocyte and SVF coculture assay
Adipocytes and SVF were cocultured in a bicompartmental system using Corning Transwell plates (Corning). Isolated inguinal SVF was seeded and differentiated in the lower compartment of Transwell plates as described above. When adipocytes in the lower compartment were fully differentiated, freshly harvested inguinal SVF was introduced in the upper compartment. The cells were cocultured in DMEM/F12 containing 10% FBS, 1% penicillin/streptomycin and 0.5 μg/ml insulin, with a polycarbonate membrane (0.4 μm pores) in between, for 4 h at 37 °C in 5% CO2. The adipocytes were then collected from the lower compartment to analyze gene expression by qPCR.
Flow cytometric analysis
Isolated SVF from inguinal tissues was blocked in a mixture of normal rat and mouse serum on ice for 15 min and incubated with the following antibodies for immune profiling: CD45 (30-F11, BioLegend), F4/80 (BM8, BioLegend), CD11b (M1/70, Biolegend), MHC Class II (M5/114.15.2, BioLegend), Siglec-F (E50-2440, BD Biosciences), CD11c (N418, BioLegend), CD206 (MR5D3, BioLegend), CD3 (17A2, BioLegend; 17A2, BioLegend), TCRγδ (UC7-13D5, BioLegend), TCRβ (H57-597, BioLegend), CD4 (GK1.5, BioLegend), CD8 (53-6.7, BioLegend), CD19 (1D3/CD19, BioLegend) and B220 (Ra3-6B2, BioLegend). Dead cells were initially excluded based on DAPI (4,6-diamidino-2-phenylindole) uptake and then debris were eliminated using side scatter (SSC)-area (A) vs. forward scatter (FSC)-A. Inguinal SVF of WT mice served as a negative control to identify the ChAT-eGFP+ cell population. The ChAT-eGFP+ cells were gated on hematopoietic lineage (CD45) and subsequently myeloid cells (F4/80 and CD11b). Singlets were separately selected from ChAT-eGFP+ myeloid cells (CD45+, F4/80+, CD11b+) and ChAT-eGFP+ non-myeloid cells (CD45+, F4/80-, CD11b-) using SSC-A vs. SSC-width (not shown) to identify eosinophil/macrophage subsets (Siglec-F, eosinophil; MHC Class II, pan-macrophages; CD11c, M1; CD206, M2) and lymphocyte subsets (CD3, TCRγδ, TCRβ, CD4 and CD8α, T cells; CD19 and B220, B cells), respectively. For intracellular analysis of ChAT expression, the SVF was fixed and permeabilized using BD Cytofix/Cytoperm kit (BD Biosciences) and then incubated with a rabbit monoclonal ChAT antibody (ab181023, Abcam) or isotype IgG control followed by BV421-conjugated anti-rabbit secondary antibody (BioLegend). Dead cells were excluded using Zombie Aqua Fixable Viability kit (BioLegend). Flow cytometry acquisition was performed on a 3-laser Fortessa flow cytometer (Becton Dickinson) and data were analyzed with FlowJo software (TreeStar).
Bone marrow transplantation
Bone marrow cells (1×106) from adult Chrna2 KO and littermate WT control mice (CD45.2) were transplanted into lethally irradiated (2 × 550 cGy, 4 hours apart) recipient B6.SJL mice (CD45.1). After 6 weeks to allow for full reconstitution by the transplanted bone marrow, we confirmed that myeloid and B cells showed a complete turnover in both WT and Chrna2 KO bone marrow transplant (BMT) recipients mice44.
Histology
Adipose tissues were dissected, and fixed in 10% formalin overnight at 4 °C. Paraffin embedding and Hematoxylin & Eosin staining were performed by the University of Michigan Comprehensive Cancer Center Research Histology & Immunoperoxidase Laboratory. Adipocyte size was analyzed with images of H&E sections of WT and Chrna2 KO IWAT and VWAT (3 mice per genotype), using ImageJ software (Adiposoft45).
Respiration
Freshly isolated inguinal tissues were weighed and minced in respiration buffer (2.5 mM glucose, 50 μM palmitoryl-L-carnitine hydrochloride, 2.5 mM malate, 120 mM NaCl, 4.5 mM KCl, 0.7 mM Na2HPO4, 1.5 mM NaH2PO4, 0.5 mM MgCl2, pH 7.4). Tissue oxygen consumption in the respiration buffer was recorded for 1–2 min at each stage (basal or uncoupled) using a Clark electrode (Strathkelvin Instruments). 4 mg/ml oligomycin (Sigma) was acutely added to the respiration chamber to measure the uncoupled respiration.
Mitochondrial DNA copy number analysis
Total DNA was isolated from mouse inguinal fat tissue using Tri-reagent (Sigma) according to the manufacturer’s instructions. To estimate mitochondrial DNA copy number, the relative amounts of nuclear and mitochondrial DNA were determined by qPCR with primers specific to the CoxI region of the mitochondrial genome and Rip140 as a nuclear marker gene.
Citrate synthase activity
Inguinal tissues were homogenized in extraction buffer (50 mM Tris-HCl, 1 mM MgCl2, 100 mM KCl, 250 mM sucrose and 30 mM 2-mercaptoethanol) and centrifuged at 1,000 × g for 15 min at 4 °C. Citrate synthase activity in the supernatant was measured with a citrate synthase activity assay kit according to manufacturer’s instructions (Cayman chemical).
Immunohistochemistry
Excised inguinal and brown adipose tissues were fixed in 10% formalin for 24 h, and then were embedded in paraffin and sectioned at 5 μm by the University of Michigan Comprehensive Cancer Center Research Histology & Immunoperoxidase Laboratory. Sections were deparaffinized in xylene, rehydrated in ethanol and distilled water, and then microwaved in 10 mM sodium citrate buffer (pH 6.0) for antigen retrieval. Endogenous peroxidase was quenched with hydrogen peroxide in methanol for 30 min. Sections were blocked with 3% BSA for 1 h and incubated with rabbit polyclonal anti-UCP1 antibody (1:200; ab10983, Abcam) in 3% BSA overnight at 4°C. PBS-washed sections were incubated with biotinylated anti-rabbit antibody for 1 h and exposed to 3,3′-diaminobenzidine (DAB, Vector Laboratories) for 2 min to develop the signal. Hematoxylin was used to counterstain sections. The images were visualized and captured randomly from the stained sections using a LEICA DM2000 microscope.
Metabolic phenotyping
Blood glucose and serum insulin levels of 4 h-fasted mice were measured using OneTouch Ultra Glucometer (Lifescan) and an insulin ELISA kit (Crystal Chem Inc), respectively. For the glucose tolerance test (GTT), overnight-fasted mice were injected intraperitoneally with 1 g/kg glucose. For the insulin tolerance test (ITT), mice were fasted for 4 h and then received an intraperitoneal injection of 0.5 U/kg insulin (Humulin R, Lilly). GTT and ITT were done 9 and 8 weeks after HFD feeding, respectively. Glucose levels were measured in tail blood prior to and at the indicated times following the injection. The area under the curve (AUC) was calculated for the GTT and ITT with Prism 6 (GraphPad Software, Inc.). University of Michigan Animal Phenotyping Core determined the body composition (quantification of fat and lean mass) using nuclear magnetic resonance analysis. Oxygen consumption (VO2), carbon dioxide production (VCO2) and spontaneous locomotor activity (at x-axis and y-axis) were monitored for 48 h using a Comprehensive Laboratory Monitoring System (Columbus Instruments) equipped with photo beam sensors in the University of Michigan Animal Phenotyping Core. Energy expenditure was estimated with VO2 and VCO246. Body temperature was monitored using a RET-3 mouse rectal probe (World Precision Instruments).
Statistical analyses
All results are presented as mean ± s.e.m. and graphed using Prism 6. Data are representative of 2–4 independent experiments. Sample sizes are biological replicates and were chosen based on preliminary data or previously published reports. We first performed normality tests using the Shapiro-Wilk test (3 ≤ n ≤ 7) or the D’Agostino–Pearson omnibus test (n ≥ 8). Normally distributed data were further analyzed by parametric tests including a two-tailed Student’s t-test for two-group comparisons or a one-way analysis of variance (ANOVA) for multiple comparisons involving one independent variable. When groups followed a non-normal distribution, Mann–Whitney U test was used. Data for whole-body energy expenditure in HFD study were analyzed using analysis of covariance (ANCOVA) with body weight as a covariate since body weight was significantly different between WT and Chrna2 KO mice fed a HFD. p<0.05 was considered to be statistically significant and was noted as *(p<0.05), **(p<0.01), ***(p<0.001).
Data availability
Accession code for the deposited microarray is GSE79031. The data set generated in the current study is available from the corresponding author on reasonable request. Uncropped immunoblot images available in Supplementary Fig. 12. A Life Sciences Reporting Summary is available.
Supplementary Material
Acknowledgments
We thank Drs. B.M. Spiegelman and J.D. Lin for their advice and discussion throughout this work. Supported by the Human Frontier Science Program (RGY0082/14), the Edward Mallinckrodt Jr. Foundation, the American Diabetes Association (1-18-IBS-281) and the National Institutes of Health (R01DK107583, P30-DK020572, P30-DK089503) to J.W., (F31DK112625) to M.P.E., a postdoctoral fellowship from the American Heart Association to D.K. (17POST33060001) and grants from the NIH (R37EB003320, DK046960) to R.T.K., (R01-AI091627) to I.M. and (T32DA007268) to A.G.Z.
Footnotes
AUTHOR CONTRIBUTIONS
H.J., H.Y., and J.W. designed the experiments and wrote the manuscript. H.J., H.Y., J.G., J.J., X.Q, E.P., D.K., M.P.E., A.G.Z., J.C. and J.W. performed the experiments. H.J., H.Y., J.G., E.P., A.G.Z, J.L., R.T.K, I.M, S.X. and J.W. analyzed the data.
COMPLETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharp LZ, et al. Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS One. 2012;7:e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lidell ME, et al. Evidence for two types of brown adipose tissue in humans. Nat Med. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- 4.Shinoda K, et al. Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat Med. 2015;21:389–394. doi: 10.1038/nm.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee P, et al. Temperature-acclimated brown adipose tissue modulates insulin sensitivity in humans. Diabetes. 2014;63:3686–3698. doi: 10.2337/db14-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanssen MJ, et al. Short-term cold acclimation improves insulin sensitivity in patients with type 2 diabetes mellitus. Nat Med. 2015;21:863–865. doi: 10.1038/nm.3891. [DOI] [PubMed] [Google Scholar]
- 7.Yoneshiro T, et al. Recruited brown adipose tissue as an antiobesity agent in humans. J Clin Invest. 2013;123:3404–3408. doi: 10.1172/JCI67803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chondronikola M, et al. Brown adipose tissue improves whole-body glucose homeostasis and insulin sensitivity in humans. Diabetes. 2014;63:4089–4099. doi: 10.2337/db14-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kajimura S, Spiegelman BM, Seale P. Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell Metab. 2015;22:546–559. doi: 10.1016/j.cmet.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 11.Le Novere N, Corringer PJ, Changeux JP. The diversity of subunit composition in nAChRs: evolutionary origins, physiologic and pharmacologic consequences. J Neurobiol. 2002;53:447–456. doi: 10.1002/neu.10153. [DOI] [PubMed] [Google Scholar]
- 12.Changeux JP. Nicotine addiction and nicotinic receptors: lessons from genetically modified mice. Nat Rev Neurosci. 2010;11:389–401. doi: 10.1038/nrn2849. [DOI] [PubMed] [Google Scholar]
- 13.Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 2012;15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiang L, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Ppargamma. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson-Fritch L, et al. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest. 2004;114:1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartesaghi S, et al. Thermogenic activity of UCP1 in human white fat-derived beige adipocytes. Mol Endocrinol. 2015;29:130–139. doi: 10.1210/me.2014-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elsen M, et al. BMP4 and BMP7 induce the white-to-brown transition of primary human adipose stem cells. Am J Physiol Cell Physiol. 2014;306:C431–440. doi: 10.1152/ajpcell.00290.2013. [DOI] [PubMed] [Google Scholar]
- 18.Kong X, et al. IRF4 is a key thermogenic transcriptional partner of PGC-1alpha. Cell. 2014;158:69–83. doi: 10.1016/j.cell.2014.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svensson PA, et al. Characterization of brown adipose tissue in the human perirenal depot. Obesity (Silver Spring) 2014;22:1830–1837. doi: 10.1002/oby.20765. [DOI] [PubMed] [Google Scholar]
- 22.Nagano G, et al. Activation of classical brown adipocytes in the adult human perirenal depot is highly correlated with PRDM16-EHMT1 complex expression. PLoS One. 2015;10:e0122584. doi: 10.1371/journal.pone.0122584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma S, et al. Activation of the cold-sensing TRPM8 channel triggers UCP1-dependent thermogenesis and prevents obesity. J Mol Cell Biol. 2012;4:88–96. doi: 10.1093/jmcb/mjs001. [DOI] [PubMed] [Google Scholar]
- 24.Tallini YN, et al. BAC transgenic mice express enhanced green fluorescent protein in central and peripheral cholinergic neurons. Physiol Genomics. 2006;27:391–397. doi: 10.1152/physiolgenomics.00092.2006. [DOI] [PubMed] [Google Scholar]
- 25.Rosas-Ballina M, et al. Acetylcholine-synthesizing T cells relay neural signals in a vagus nerve circuit. Science. 2011;334:98–101. doi: 10.1126/science.1209985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reardon C, et al. Lymphocyte-derived ACh regulates local innate but not adaptive immunity. Proc Natl Acad Sci U S A. 2013;110:1410–1415. doi: 10.1073/pnas.1221655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen KD, et al. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature. 2011;480:104–108. doi: 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachman ES, et al. betaAR signaling required for diet-induced thermogenesis and obesity resistance. Science. 2002;297:843–845. doi: 10.1126/science.1073160. [DOI] [PubMed] [Google Scholar]
- 29.Fischer K, et al. Alternatively activated macrophages do not synthesize catecholamines or contribute to adipose tissue adaptive thermogenesis. Nat Med. 2017;23:623–630. doi: 10.1038/nm.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang H, Ding X, Cao Y, Wang H, Zeng W. Dense Intra-adipose Sympathetic Arborizations Are Essential for Cold-Induced Beiging of Mouse White Adipose Tissue. Cell Metab. 2017;26:686–692e683. doi: 10.1016/j.cmet.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Phenotypic analysis of CHRNA2 KO mice. Deltagen Knockout Mice Phenotypic Data Summary, Mouse Genome Informatics, Jax Mice; http://www.informatics.jax.org/external/ko/deltagen/1658.html. [Google Scholar]
- 32.Rowland LA, Bal NC, Kozak LP, Periasamy M. Uncoupling protein 1 and sarcolipin are required to maintain optimal thermogenesis, and loss of both systems compromises survival of mice under cold stress. J Biol Chem. 2015;290:12282–12289. doi: 10.1074/jbc.M115.637603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruton JD, et al. Increased fatigue resistance linked to Ca2+-stimulated mitochondrial biogenesis in muscle fibres of cold-acclimated mice. J Physiol. 2010;588:4275–4288. doi: 10.1113/jphysiol.2010.198598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen P, et al. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. 2014;156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sparks LM, et al. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes. 2005;54:1926–1933. doi: 10.2337/diabetes.54.7.1926. [DOI] [PubMed] [Google Scholar]
- 36.Brestoff JR, Artis D. Immune regulation of metabolic homeostasis in health and disease. Cell. 2015;161:146–160. doi: 10.1016/j.cell.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J Clin Invest. 2011;121:2111–2117. doi: 10.1172/JCI57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williamson DF, et al. Smoking cessation and severity of weight gain in a national cohort. N Engl J Med. 1991;324:739–745. doi: 10.1056/NEJM199103143241106. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida T, et al. Nicotine induces uncoupling protein 1 in white adipose tissue of obese mice. Int J Obes Relat Metab Disord. 1999;23:570–575. doi: 10.1038/sj.ijo.0800870. [DOI] [PubMed] [Google Scholar]
- 40.Dodd GT, et al. Leptin and insulin act on POMC neurons to promote the browning of white fat. Cell. 2015;160:88–104. doi: 10.1016/j.cell.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Owen BM, et al. FGF21 acts centrally to induce sympathetic nerve activity, energy expenditure, and weight loss. Cell Metab. 2014;20:670–677. doi: 10.1016/j.cmet.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo J, et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 43.Song P, Mabrouk OS, Hershey ND, Kennedy RT. In vivo neurochemical monitoring using benzoyl chloride derivatization and liquid chromatography-mass spectrometry. Anal Chem. 2012;84:412–419. doi: 10.1021/ac202794q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldschneider I, Komschlies KL, Greiner DL. Studies of thymocytopoiesis in rats and mice. I. Kinetics of appearance of thymocytes using a direct intrathymic adoptive transfer assay for thymocyte precursors. J Exp Med. 1986;163:1–17. doi: 10.1084/jem.163.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galarraga M, et al. Adiposoft: automated software for the analysis of white adipose tissue cellularity in histological sections. J Lipid Res. 2012;53:2791–2796. doi: 10.1194/jlr.D023788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weir JB. New methods for calculating metabolic rate with special reference to protein metabolism. 1949. Nutrition. 1990;6:213–221. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession code for the deposited microarray is GSE79031. The data set generated in the current study is available from the corresponding author on reasonable request. Uncropped immunoblot images available in Supplementary Fig. 12. A Life Sciences Reporting Summary is available.


