Abstract
Aminoacyl-tRNA synthetases (ARSs) are ubiquitously expressed enzymes that ligate amino acids onto tRNA molecules. Genes encoding ARSs have been implicated in phenotypically diverse dominant and recessive human diseases. The charging of tRNAPHE with phenylalanine is performed by a tetrameric enzyme that contains two alpha (FARSA) and two beta (FARSB) subunits. To date, mutations in the genes encoding these subunits (FARSA and FARSB) have not been implicated in any human disease. Here, we describe a patient with a severe, lethal, multisystem, developmental phenotype who was compound heterozygous for FARSB variants: p.Thr256Met and p.His496Lysfs*14. Expression studies using fibroblasts isolated from the proband revealed a severe depletion of both FARSB and FARSA protein levels. These data indicate that the FARSB variants destabilize total phenylalanyl-tRNA synthetase levels, thus causing a loss-of-function effect. Importantly, our patient shows strong phenotypic overlap with patients that have recessive diseases associated with other ARS loci; these observations strongly support the pathogenicity of the identified FARSB variants and are consistent with the essential function of phenylalanyl-tRNA synthetase in human cells. In sum, our clinical, genetic, and functional analyses revealed the first FARSB variants associated with a human disease phenotype and expand the locus heterogeneity of ARS-related human disease.
Keywords: Phenylalanyl-tRNA synthetase, FARSB, aminoacyl-tRNA synthetase, developmental syndrome, recessive disease, loss-of-function mutations
MAIN TEXT
Aminoacyl-tRNA synthetases (ARSs) are ubiquitously expressed enzymes that ligate tRNA molecules to cognate amino acids, which is the first step of protein translation (Antonellis and Green, 2008). Unsurprisingly, ablation of the function of any ARS enzyme in species ranging from mammals to bacteria is lethal, underscoring the essential role of these enzymes in cellular life. The human nuclear genome contains 37 ARS-encoding loci, which together encode 17 enzymes required for tRNA charging in the mitochondria, 17 enzymes required for tRNA charging in the cytoplasm, and three enzymes that charge tRNA in both cellular compartments (Antonellis and Green, 2008). ARS enzymes are named according to the single-letter code of the amino acid they recognize followed by ‘ARS’ and a variable ‘2’ for mitochondrial enzymes. For example, cytoplasmic tyrosyl-tRNA synthetase is referred to as ‘YARS’ while the corresponding mitochondrial enzyme is referred to as ‘YARS2’ (Antonellis and Green, 2008).
To date, 31 ARS loci have been implicated in dominant and recessive human disease phenotypes (Meyer-Schuman and Antonellis, 2017). First, each of the 17 ARS loci encoding a mitochondrial enzyme have been implicated in recessive diseases and the vast majority are consistent with stereotypical mitochondrial disease phenotypes. Second, 13 of the 20 ARS loci encoding a cytoplasmic or bifunctional enzyme have been implicated in severe, early onset, developmental phenotypes with mostly overlapping features (Meyer-Schuman and Antonellis, 2017). Finally, five ARS loci encoding either a cytoplasmic or bifunctional enzyme have been implicated in dominant, axonal Charcot-Marie-Tooth disease (Antonellis et al., 2003; Jordanova et al., 2006; Latour et al., 2010; Vester et al., 2012; Tsai et al., 2017). Importantly, for the recessive diseases associated with ARS mutations, the inheritance patterns, nature of the mutations, functional studies, and significant phenotypic overlap associated with each locus all point to a loss-of-function mechanism (Meyer-Schuman and Antonellis, 2017; Oprescu et al., 2017).
The charging of tRNA molecules with phenylalanine in the cytoplasm is accomplished via a tetrameric enzyme that contains two catalytic subunits encoded by phenylalanyl-tRNA synthetase alpha (FARSA; MIM# 602918) and two regulatory subunits encoded by phenylalanyl-tRNA synthetase beta (FARSB; MIM# 609690)(Rodova et al., 1999). To date, no variants in FARSA or FARSB have been implicated in human disease; however, loss-of-function mutations in the gene encoding mitochondrial phenylalanyl-tRNA synthetase (FARS2) have been implicated in recessive phenotypes including hereditary spastic paraplegia, Alpers syndrome, early onset epilepsy, global delay, dysarthria, and tremor (Elo et al., 2012; Shamseldin et al., 2012; Almalki et al., 2014; Vernon et al., 2015; Yang et al., 2015; Raviglione et al., 2016; Walker et al., 2016; Cho et al., 2017). Here, we describe a patient with a severe, lethal, multisystem, developmental phenotype associated with compound heterozygosity for loss-of-function FARSB variants. Consent was obtained for this research, which was approved by the Institutional Review Board of the University of Michigan.
The male proband was born to a 31-year-old G2P0 mother at 35 and 4/7 weeks gestation and had a history of intrauterine growth restriction and oligohydramnios; his parents are non-consanguineous. The family history was significant for a previous pregnancy loss at 37 weeks secondary to placental abruption. The proband was delivered by urgent cesarean section due to recurrent fetal heart rate with variable (and a few prolonged) decelerations. Apgar scores were 8 and 9 at 1 and 5 minutes, respectively. Birth weight was 1.927 kg (2%); length was 44 cm (8%); and head circumference was 32 cm (31%). Grunting was noted ten minutes after birth, but with normal oximetry, and then recurred once at 5 hours after birth and resolved. The proband passed his newborn hearing and metabolic screens and pre- and post-ductal pulse oximetry. Physical examinations, other than being small for gestation age, were normal. He developed mild indirect hyperbilirubinemia on day of life 2 that responded to phototherapy appropriately. He was discharged home and was initially bottle fed to gain weight, but was transitioned to breast feeding. He had frequent emesis greater than 20 minutes after feeds.
At 5 months (4 months corrected) his pediatrician referred him back to the University of Michigan for evaluation of failure to thrive, developmental delay, left inguinal hernia, tachypnea and a chronic cough since birth occurring throughout the day and night. He was hospitalized for 2 months. Growth parameters had moved below the 3rd centile for weight and length and to 5th-10th centile for head circumference. Resting heart rate was in the 130s and he had baseline tachypnea (respiratory rate in the 70s). Physical examination was significant for sunken eyes, prominent cheeks, high palate, increased range of motion of joints, truncal hypotonia with marked head lag on pull to sit, and a small scrotum and phallus. Because of respiratory distress, he was transferred to the pediatric intensive care unit, where he required intubation for 4 days. Chest radiograph showed diffuse patchy airspace opacities with peribronchial cuffing. Multiple medical problems were identified including gastroesophageal reflux disease, profound vitamin D and A deficiency, severe hypocalcemia (requiring, at times, continuous calcium infusion up to 25 mg/kg/hour to maintain ionized calcium ≥1.2), supplementation with ergocalciferol or cholecalciferol, and hypoalbuminemia (albumin 2.4, protein 3.9) with anasarca and ascites, hungry bone syndrome with osteopenia and rickets, elevated liver transaminases (AST:100–300; ALT:80–150) and cholestasis (total bilirubin 3–4; direct bilirubin 2–3), yet normal INR. He had a non-anion gap metabolic acidosis, which resolved. CPK was 46, iron 39, ferritin 23.3, alpha-1-antitrypsin 173, and ceruloplasmin and cystic fibrosis (CF) sweat testing were normal. Numerous problems ensued during this hospitalization. He developed urinary tract infections with Enterobacter cloacae and Enterococcus feacalis, and had a normal VCUG. He also developed Candida parapsilosis bacteremia from his PICC line. He subsequently developed a DVT in the axillary vein above the PICC line. He had a seizure presumed secondary to hypoglycemia and hypocalcemia. Echocardiograms were performed on multiple occasions and revealed normal cardiac anatomy and good systolic function. Abdominal ultrasound revealed ascites without portal hypertension, as well as diffuse and increased parenchymal echogenicity suggestive of hepatocellular disease, and diffusely increased echogenicity of the kidneys, suggestive of renal disease. His abdominal distension improved after albumin infusion and he required diuretics for fluid overload. He did not have diarrhea and he had normal stool alpha-1-antitrypsin and stool elastase. He required post-pyloric tube feeds. Due to anemia and pancytopenia, a bone marrow biopsy was performed and flow cytometry was normal. He had a normal swallow study. MRI of the abdomen revealed hepatic steatosis but no increased liver iron. Hepatitis viral serologies were negative. Head ultrasound identified possible abnormal periventricular white matter and bilateral basal ganglia echogenicity, and follow-up brain MRI at 5 months corrected age, showed cerebral volume loss and incomplete closure of the Sylvian fissures (Fig. 1A, B); MR spectroscopy showed no lactate peak. Myelination was appropriate for age given his prematurity. He had an episode of dysconjugate gaze and abnormal tracking eye movements and was found to have hypoglycemia to 37 mg/dL after being fasted for 2.5 hours in preparation for a fasting blood specimen; at that time head CT and MRI were nonspecific. One week later, he had a seizure-like event with left upward eye gaze and symmetric jerking of all extremities associated with ionized calcium of 0.88 mmol/L, which responded to supplementation. He developed worsening tachypnea with increasing ascites, usually associated with hypoalbuminemia (a chronic problem during and after this hospitalization). After extubation, he was placed on CPAP and then weaned to high flow nasal cannula and then room air over the course of a week. He remained intermittently tachypneic. He was discharged after two months (corrected age 6 months) and had a heart rate of 150–160 per minute, respiratory rate of ~40 per minute, discharge weight 4.86 kg (≪3%; 50% for a 1.5 month old), length 60 cm (<3%; 50% for a 2.75 month old), and head circumference 42 cm (10%).
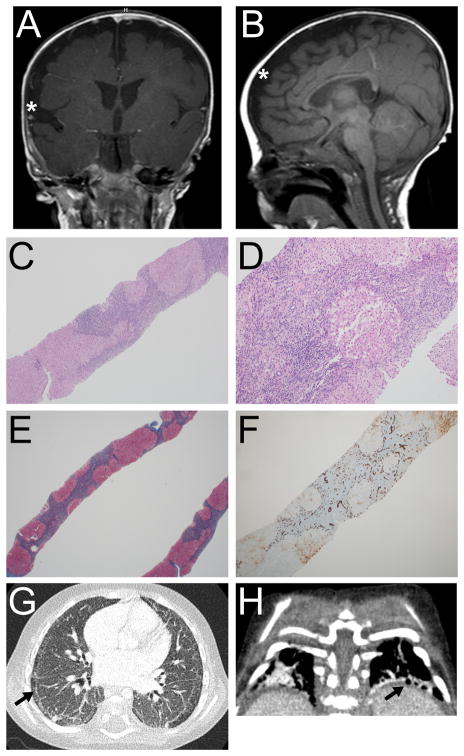
Figure 1.
Head MRI, liver pathology, and chest CT of the proband. A and B: Coronal and sagittal T1-weighted head MRI images obtained at 5 months corrected age showing incomplete closure of the Sylvian fissures (asterisk in A), and bilateral frontoparietal cerebral volume loss (asterisk in B). C–F: Liver pathology. Architectural distortion (4X) with nodular formation (C); portal chronic inflammation with focal steatosis (10X) (D); Masson trichrome stain (2X) with well-developed cirrhosis and bridging fibrosis (E); and CK7 immunostain (4X) showing extensive bile ductular reaction (F). G and H: Chest CT (18 months) showing subpleural cystic changes (arrows) and interstitial disease.
Interstitial pulmonary disease, liver cirrhosis (hypoalbuminemia) and portal hypertension with ascites, esophageal varices with GI bleeding, developmental delay, and failure to thrive continued to be problems. G-tube placement and percutaneous left lobe liver biopsy were performed at 14 months. The liver parenchyma showed architectural distortion with nodular formation and bridging fibrosis by trichrome (Fig. 1C, E). Focal mild steatosis was observed (Fig. 1D). CK7 immuno-staining revealed bile ductular proliferation (Fig. 1F). Moderate chronic portal inflammatory infiltrates were noted with neutrophils associated with the ductular reaction. There is was patchy intra-hepatocyte cholestasis, without significant intracanalicular cholestasis or bile plugging. No clear lobular hepatitic pattern or interface activity (such as plasma cells at the limiting plate) were appreciated. Iron stain was negative. There were no viral cytopathic effects identified. The overall findings were consistent with well-established cirrhosis of unknown etiology. Electron microscopy (Supp. Figure S1) was performed and hepatocytes showed reactive expansion of smooth ER, focal cystic dilation of rough ER containing sparse delicate material of no specific appearance, focal accumulation of glycogen granules, common autophagic bodies, bulky accumulations of non-membrane bound material consistent with bile residue in hepatocyte cytoplasm, and universally normal appearing mitochondria. Many hepatocytes contained abundant microvesicular lipid that appeared to be non-membrane bound and thus free in the cytoplasm. No abnormal storage of material was identified within the lysosomes or cytoplasm.
Chest CT at 1.5 years and chest radiographs at 22 months, showed subpleural cystic changes in the bilateral lungs consistent with interstitial lung disease (Fig 1G, H). Bronchoscopy showed only moderate dynamic collapse in the left main stem and lingual regions; milder collapse in the LLL segments. Bronchoalveolar lavage was negative for alveolar proteinosis and malignancy. Because of the pulmonary findings, a left nasal brushing biopsy was performed and showed motile cilia, and electron microscopy revealed normal respiratory ciliary ultrastructure. He was found to have pulmonary hypertension at 22 months at the time of an acute respiratory exacerbation requiring high frequency oscillatory ventilation, milrinone, and inhaled nitrous oxide. At that time, he had a normal bubble study negative for hepatopulmonary syndrome and normal biventricular function. He developed a subacute DVT in the distal external iliac and common femoral veins and was anticoagulated. He developed a right-sided intraparenchymal brain hemorrhage, presumed secondary to lovenox therapy. He received a ventriculostomy at 23 months; continued bleeding was noticed in the right parietal region. He developed hydrocephalus, which worsened with intraparenchymal hemorrhage and ultimately received a VP shunt at 24 months.
Medical Genetics consultation revealed normal results for N-glycan and O-glycan profiles, CPK, lysosomal enzymes including acid maltase, quantitative plasma and CSF amino acids, very long chain fatty acids, acylcarnitine profiles, SNP-based chromosomal microarray, methylation PCR for Prader-Willi/Angelman syndromes, 7-dehydrocholesterol, and ATP7A (MIM# 300011) sequencing. Urine organic acid analysis revealed 4-hydroxyphenylacetic, 4-hydroxyphenylactic, and 4-hydroxyphenylpyruvate, consistent with liver dysfunction. Right thigh skeletal muscle biopsy by standard histological analysis showed non-specific findings with slightly increased lipid droplets within fibers. Transmission electron microscopy showed non-specific, mild mitochondrial and myofibrillar irregularities. Muscle mitochondrial electron transport chain (ETC) enzyme analysis demonstrated increased citrate synthase (CS) control enzyme activity, with deficiency (15% of control after normalization for CS activity) of rotenone sensitive complex I + III activity, fulfilling one major modified Walker criterion for the diagnosis of a respiratory chain disorder. Several other ETC activities were reduced with relative preservation of complex II. Venous and arterial lactate was measured multiple times and was not consistently elevated. Because of the enzyme deficiencies noted, a 164 nuclear gene and mitochondrial next-generation sequencing panel was obtained at 24 months. This analysis revealed heterozygosity for a maternally inherited NDUFS2 (MIM# 602985) variant of uncertain significance (VUS), p.H380D (a variant with a frequency of 0.15% in the NHLBI ESP and 0.06% in ExAC, with no homozygotes), and hemizygosity for a maternally inherited PHKA2 (MIM# 300798) VUS, p.L413S (not seen in ESP; 0.008% in ExAC with 5 hemizygotes)(Lek et al., 2016). No second variant was identified in NDUFS2, which would be required to consider this recessive disorder (Loeffen et al., 2001). PHKA2 deficiency is unlikely as there was no liver glycogenosis observed on biopsy and the other clinical features of the proband are much more severe than typical patients with this disorder (Hendrickx et al., 1995). Thus, neither the NDUFS2 nor PHKA2 variants accounted for the severe clinical manifestations of the proband, who was readmitted multiple times for respiratory failure and hypoalbuminemia requiring intensive care, and passed away at 32 months. No autopsy was obtained.
To further explore the genetic underpinnings of the phenotype identified in our proband, we performed whole-exome sequencing (WES) analysis on DNA isolated from blood from the proband and his unaffected parents. Trio WES studies performed at GeneDx (Gaithersburg, MD) revealed the aforementioned maternally inherited variants of unknown significance in NDUFS2 and PHKA2. The only other candidate pathogenic variants identified in these studies that met the inheritance pattern and allele frequency criteria were two variants of unknown significance in phenylalanyl-tRNA synthetase beta (FARSB): c.767C>T, which predicts a p.Thr256Met missense variant; and c.1486delCinsAA, which predicts a p.His496Lysfs*14 protein-truncating or null allele. The unaffected father is heterozygous for p.Thr256Met, the unaffected mother is heterozygous for p.His496Lysfs*14, and the proband was compound heterozygous for both FARSB variants (Fig. 2A). Please note that these variants are annotated on NM_005687.4 and each was validated using Mutalyzer (https://mutalyzer.nl). The variants and associated phenotype have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/): p.Thr256Met accession no. SCV000676382; p.His496Lysfs*14 accession no. SCV000676381.
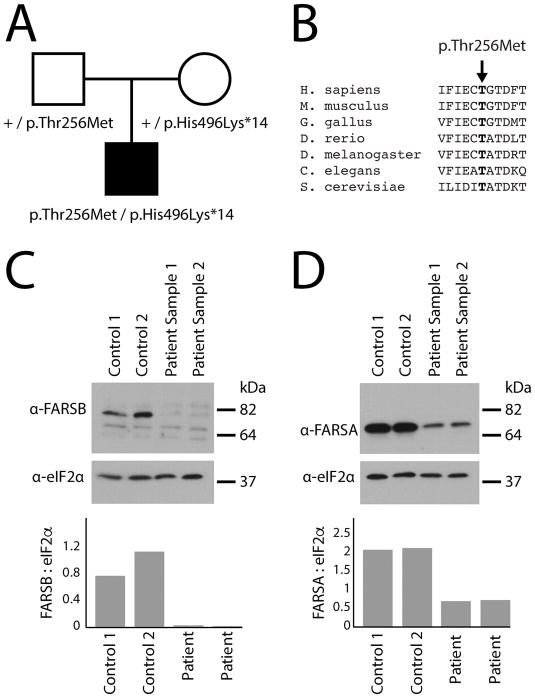
Figure 2.
Characterization of the FARSB variants identified in the proband. A: A simplex pedigree is shown with squares representing males and circles representing females. Genotypes are indicated under each symbol for the father, mother, and male proband (filled square). The proband is the only affected individual in the pedigree. B: The position of the p.Thr256Met variant is shown along with surrounding amino acid sequences for multiple, evolutionarily diverse species. Species are indicated along the left and the position of the affected residue is indicated by an arrow and bold text. C: Western blot analyses were performed using total protein lysates from fibroblasts isolated from two control individuals and from the patient described in this study, the latter performed in duplicate (Sample 1 and Sample 2). An anti-FARSB or anti-eIF2α antibody was employed to test for the effect of the FARSB variants on protein levels and to control for protein loading, respectively. Sample names are provided across the top of the panel and sizes in kDa are indicated on the right. D: Similar western blot analyses as described in panel C using an anti-FARSA antibody. In panels C and D, the ratio of FARSB or FARSA to eIF2α was determined by dividing the optical density of each FARS-specific band by that of the corresponding eIF2α-specific band.
Examination of each FARSB variant on the genome aggregate database browser (gnomAD; http://gnomad.broadinstitute.org)(Lek et al., 2016) revealed the presence of p.Thr256Met FARSB at a very low frequency (12 out of 276,122 alleles with no homozygous individuals) and the absence of p.His496Lysfs*14 FARSB. While p.His496Lysfs*14 FARSB is very likely to result in a null allele or a non-functional protein, the importance of p.Thr256 in enzyme function is less clear. Multiple-species conservation analysis revealed that p.Thr256 is conserved among diverse species ranging from human to yeast (Fig. 2B). Furthermore, resolving the structure of human cytoplasmic phenylalanyl-tRNA synthetase revealed that threonine 256 resides in the editing domain and mutating this residue to alanine reduces the ability to distinguish between tyrosine and phenylalanine, resulting in enzyme mischarging (Sasaki et al., 2006; Finarov et al., 2010).
Since p.His496Lysfs*14 FARSB causes a frameshift mutation that leads to a premature stop codon, the variant is predicted to ablate the expression of full-length FARSB, which would predict a ~50% decrease in full-length protein levels in patient cells. To address this, we performed western blot analysis on proteins isolated from cultured patient fibroblasts and two control fibroblast lines (i.e., from individuals without FARSB variants). Briefly, protein samples (20 micrograms per sample) were subjected to electrophoresis, transferred onto a PVDF membrane, and incubated with an anti-FARSB antibody (Sigma-Aldrich; HPA061398) or an anti-eIF2α antibody (Cell Signaling Technology; 9722) to control for protein loading. Interestingly, our results revealed a dramatic reduction of full-length FARSB protein levels (~97% reduction) in patient fibroblast cells compared to control cell lines (Fig. 2C). Since FARSB forms an α2β2 hetero-tetramer with FARSA (Rodova et al., 1999), we hypothesized that severely reduced FARSB levels may destabilize and reduce FARSA levels. To address this, we performed the same western blot analyses described above using an antibody designed against FARSA (Sigma-Aldrich; HPA001911). These analyses revealed a significant decrease in FARSA levels (~66% reduction) in patient cells compared to the control cell lines (Fig. 2D).
The observation of dramatically reduced FARSB protein levels in patient cells requires further consideration of the p.Thr256Met FARSB missense variant. We computationally predicted the pathogenicity of this variant using CADD (Kircher et al., 2014), PolyPhen2 (Adzhubei et al., 2010), and MUPro (Cheng et al., 2006), and all three algorithms provided a prediction of “pathogenic”: CADD score = 33; PolyPhen2 score = 1; and MUPro score = −0.4315118. Importantly, MUPro scores that are less than 0 predict a decrease in protein stability that is consistent with our western blot analyses. Thus, even though p.Thr256Met FARSB resides in the editing domain (see above), this variant likely impacts protein stability that would result in reduced tRNA charging. Indeed, the lack of a phenotype in the father that carries p.Thr256Met FARSB and previous reports showing that Chinese hamster ovary cells are tolerant of misincorporations of phenylalanine for tyrosine (Raina et al., 2014) indicate that altered editing is not likely to be responsible for the phenotype observed in the proband. We therefore conclude that compound heterozygosity for p.Thr256Met and p.His496Lysfs*14 FARSB results in a severe reduction of FARSB protein levels that causes a downstream reduction in FARSA levels. These reduced protein levels likely result in a severe reduction in phenylalanyl-tRNA synthetase activity that is responsible for the observed recessive disease phenotype in our patient. In support of this notion, we are aware of another family with two affected siblings, both with novel, bi-allelic FARSB variants including a predicted loss-of-function variant and a predicted deleterious missense variant, and very similar clinical features (Wendy Chung, personal communication).
A detailed comparison of the phenotypes of our proband (described above) and the phenotypes of previously reported patients with ARS-associated recessive disease (Meyer-Schuman and Antonellis, 2017) revealed striking overlap (Supp. Table S1). For example, the following phenotypes that we observed in our patient have been reported in patients with other ARS-associated recessive syndromes: (1) intrauterine growth restriction in patients with AARS (MIM# 601065), GARS (MIM# 600287), IARS (MIM# 600709), MARS (MIM# 156560), MARS2 (MIM# 609728), QARS (MIM# 603727), VARS (MIM# 192150), and YARS2 (MIM# 610957) variants; (2) failure to thrive in patients with AARS, AARS2 (MIM# 612035), CARS2 (MIM# 612800), EARS2 (MIM# 612799), IARS, LARS (MIM# 151350), MARS, MARS2, SARS2 (MIM# 612804), YARS (MIM# 603623), and YARS2 variants; (3) developmental delay in patients with AARS, FARS2 (MIM# 611592), GARS, IARS, KARS (MIM# 601421), LARS, MARS, MARS2, NARS2 (MIM# 612803), PARS2 (MIM# 612036), QARS, RARS2 (MIM# 611524), SARS2, VARS, and YARS variants; (4) hypotonia in patients with CARS2, EARS2, HARS2 (MIM# 600783), IARS, KARS, LARS, MARS, MARS2, QARS, RARS (MIM# 107820), RARS2, SARS2, TARS2 (MIM# 612805), and YARS variants; (5) abnormal or decreased white matter in patients with AARS, CARS2, DARS (MIM# 603084), DARS2 (MIM# 610956), EARS2, GARS, IARS, KARS, MARS2, NARS2, QARS, RARS, RARS2, and YARS variants; and (6) liver dysfunction in patients with EARS2, FARS2, IARS, LARS, LARS2 (MIM# 604544), MARS, and YARS variants.
In addition to the overlap with individual features of other ARS-related recessive syndromes, the phenotype of our patient displayed remarkable overlap with features previously associated with homozygosity or compound heterozygosity for loss-of-function methionyl-tRNA synthetase (MARS) variants including: intrauterine growth restriction; failure to thrive; developmental delay; inguinal hernia; hypotonia; recurrent infections; interstitial lung disease; and liver dysfunction (van Meel et al., 2013; Hadchouel et al., 2015; Sun et al., 2017). Similarly, our patient shared many clinical features previously associated with homozygosity or compound heterozygosity for loss-of-function tyrosyl-tRNA synthetase (YARS) variants including: intrauterine growth restriction; failure to thrive; developmental delay; emesis; deep-set eyes; prominent cheeks; high palate; joint hypermobility; hypotonia; abnormal white matter; and liver dysfunction (Nowaczyk et al., 2017; Tracewska-Siemiątkowska et al., 2017). Importantly, the phenotypic overlap between our patient and other patients with recessive ARS-related diseases strongly supports the pathogenicity of the FARSB variants reported here. Furthermore, this phenotypic overlap also indicates that the mutations have a direct effect on the primary function of the enzyme: charging tRNA with phenylalanine for protein translation. In support of this latter notion, many of the clinical manifestations reported here have been associated with mutations that disrupt the cytoplasmic or mitochondrial protein translational machinery (Scheper et al., 2007), including liver dysfunction—arguably the most severe phenotype observed in our patient—associated with EIF2AK3 (MIM# 604032), GFM1 (MIM# 606639), TUFM (MIM# 602389), and GCN2 (MIM# 609280) perturbations (Delépine et al., 2000; Coenen et al., 2004; Guo and Cavener, 2007; Valente et al., 2007).
In summary, we describe clinical, genetic, and functional data that implicate FARSB mutations in a severe, lethal, multisystem, recessive, developmental phenotype. These data warrant careful assessments for loss-of-function FARSB–or FARSA–variants in patients with a similar clinical syndrome, especially when liver dysfunction is described. Our study expands the locus heterogeneity of diseases associated with aminoacyl-tRNA synthetases (Meyer-Schuman and Antonellis, 2017) and emphasizes the importance of detailed clinical comparisons and relevant functional studies when implicating ARS variants in disease pathogenesis (Oprescu et al., 2017).
Supplementary Material
Acknowledgments
CONTRACT GRANT SPONSORS: National Institute of General Medical Sciences (GM118647, GM007863, and GM007315) and National Institute of Neurological Disease and Stroke (NS092238).
The authors thank the family for their patience and involvement in the study, and their interest and dedication to this research. We also wish to acknowledge the efforts of a large army of physicians, nurses, and other caretakers in the life of this patient. Finally, we thank Dr. Doug Quint for help with the head MRI interpretation and Dr. Wendy Chung for sharing knowledge of a second family with FARSB variants. JWI would like to acknowledge support from the Morton S. and Henrietta K. Sellner Professorship in Human Genetics. A. Amalfitano acknowledges the Osteopathic Heritage Foundation and the Michigan State University Foundation for support.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have no conflicts to declare.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almalki A, Alston CL, Parker A, Simonic I, Mehta SG, He L, Reza M, Oliveira JMA, Lightowlers RN, McFarland R, Taylor RW, Chrzanowska-Lightowlers ZMA. Mutation of the human mitochondrial phenylalanine-tRNA synthetase causes infantile-onset epilepsy and cytochrome c oxidase deficiency. Biochim Biophys Acta. 2014;1842:56–64. doi: 10.1016/j.bbadis.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin S-Q, Jordanova A, Kremensky I, Christodoulou K, Middleton LT, Sivakumar K, Ionasescu V, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Green ED. The role of aminoacyl-tRNA synthetases in genetic diseases. Annual review of genomics and human genetics. 2008;9:87–107. doi: 10.1146/annurev.genom.9.081307.164204. [DOI] [PubMed] [Google Scholar]
- Cheng J, Randall A, Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62:1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- Cho JS, Kim SH, Kim HY, Chung T, Kim D, Jang S, Lee SB, Yoo SK, Shin J, Kim J-I, Kim H, Hwang H, et al. FARS2 mutation and epilepsy: Possible link with early-onset epileptic encephalopathy. Epilepsy Res. 2017;129:118–124. doi: 10.1016/j.eplepsyres.2016.11.022. [DOI] [PubMed] [Google Scholar]
- Coenen MJH, Antonicka H, Ugalde C, Sasarman F, Rossi R, Heister JGAMA, Newbold RF, Trijbels FJMF, van den Heuvel LP, Shoubridge EA, Smeitink JAM. Mutant mitochondrial elongation factor G1 and combined oxidative phosphorylation deficiency. N Engl J Med. 2004;351:2080–2086. doi: 10.1056/NEJMoa041878. [DOI] [PubMed] [Google Scholar]
- Delépine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat Genet. 2000;25:406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- Elo JM, Yadavalli SS, Euro L, Isohanni P, Götz A, Carroll CJ, Valanne L, Alkuraya FS, Uusimaa J, Paetau A, Caruso EM, Pihko H, et al. Mitochondrial phenylalanyl-tRNA synthetase mutations underlie fatal infantile Alpers encephalopathy. Hum Mol Genet. 2012;21:4521–4529. doi: 10.1093/hmg/dds294. [DOI] [PubMed] [Google Scholar]
- Finarov I, Moor N, Kessler N, Klipcan L, Safro MG. Structure of human cytosolic phenylalanyl-tRNA synthetase: evidence for kingdom-specific design of the active sites and tRNA binding patterns. Structure. 2010;18:343–353. doi: 10.1016/j.str.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Guo F, Cavener DR. The GCN2 eIF2alpha kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab. 2007;5:103–114. doi: 10.1016/j.cmet.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Hadchouel A, Wieland T, Griese M, Baruffini E, Lorenz-Depiereux B, Enaud L, Graf E, Dubus JC, Halioui-Louhaichi S, Coulomb A, Delacourt C, Eckstein G, et al. Biallelic Mutations of Methionyl-tRNA Synthetase Cause a Specific Type of Pulmonary Alveolar Proteinosis Prevalent on Réunion Island. 2015;96:826–831. doi: 10.1016/j.ajhg.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx J, Coucke P, Dams E, Lee P, Odièvre M, Corbeel L, Fernandes JF, Willems PJ. Mutations in the phosphorylase kinase gene PHKA2 are responsible for X-linked liver glycogen storage disease. Hum Mol Genet. 1995;4:77–83. doi: 10.1093/hmg/4.1.77. [DOI] [PubMed] [Google Scholar]
- Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, Dierick I, Jacobs A, De Vriendt E, Guergueltcheva V, Rao CV, Tournev I, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet. 2006;38:197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latour P, Thauvin-Robinet C, Baudelet-Méry C, Soichot P, Cusin V, Faivre L, Locatelli M-C, Mayençon M, Sarcey A, Broussolle E, Camu W, David A, et al. A major determinant for binding and aminoacylation of tRNA(Ala) in cytoplasmic Alanyl-tRNA synthetase is mutated in dominant axonal Charcot-Marie-Tooth disease. 2010;86:77–82. doi: 10.1016/j.ajhg.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffen J, Elpeleg O, Smeitink J, Smeets R, Stöckler-Ipsiroglu S, Mandel H, Sengers R, Trijbels F, van den Heuvel L. Mutations in the complex I NDUFS2 gene of patients with cardiomyopathy and encephalomyopathy. Annals of neurology. 2001;49:195–201. doi: 10.1002/1531-8249(20010201)49:2<195::aid-ana39>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Meyer-Schuman R, Antonellis A. Emerging mechanisms of aminoacyl-tRNA synthetase mutations in recessive and dominant human disease. Hum Mol Genet. 2017;26:R114–R127. doi: 10.1093/hmg/ddx231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowaczyk MJM, Huang L, Tarnopolsky M, Schwartzentruber J, Majewski J, Bulman DE, Hartley T, Boycott KM FORGE Canada Consortium, Care4Rare Canada Consortium. A novel multisystem disease associated with recessive mutations in the tyrosyl-tRNA synthetase (YARS) gene. Am J Med Genet A. 2017;173:126–134. doi: 10.1002/ajmg.a.37973. [DOI] [PubMed] [Google Scholar]
- Oprescu SN, Griffin LB, Beg AA, Antonellis A. Predicting the pathogenicity of aminoacyl-tRNA synthetase mutations. Methods. 2017;113:139–151. doi: 10.1016/j.ymeth.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina M, Moghal A, Kano A, Jerums M, Schnier PD, Luo S, Deshpande R, Bondarenko PV, Lin H, Ibba M. Reduced amino acid specificity of mammalian tyrosyl-tRNA synthetase is associated with elevated mistranslation of Tyr codons. J Biol Chem. 2014;289:17780–17790. doi: 10.1074/jbc.M114.564609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviglione F, Conte G, Ghezzi D, Parazzini C, Righini A, Vergaro R, Legati A, Spaccini L, Gasperini S, Garavaglia B, Mastrangelo M. Clinical findings in a patient with FARS2 mutations and early-infantile-encephalopathy with epilepsy. Am J Med Genet A. 2016;170:3004–3007. doi: 10.1002/ajmg.a.37836. [DOI] [PubMed] [Google Scholar]
- Rodova M, Ankilova V, Safro MG. Human phenylalanyl-tRNA synthetase: cloning, characterization of the deduced amino acid sequences in terms of the structural domains and coordinately regulated expression of the alpha and beta subunits in chronic myeloid leukemia cells. Biochem Biophys Res Commun. 1999;255:765–773. doi: 10.1006/bbrc.1999.0141. [DOI] [PubMed] [Google Scholar]
- Sasaki HM, Sekine S-I, Sengoku T, Fukunaga R, Hattori M, Utsunomiya Y, Kuroishi C, Kuramitsu S, Shirouzu M, Yokoyama S. Structural and mutational studies of the amino acid-editing domain from archaeal/eukaryal phenylalanyl-tRNA synthetase. Proc Natl Acad Sci USA. 2006;103:14744–14749. doi: 10.1073/pnas.0603182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper GC, van der Knaap MS, Proud CG. Translation matters: protein synthesis defects in inherited disease. Nat Rev Genet. 2007;8:711–723. doi: 10.1038/nrg2142. [DOI] [PubMed] [Google Scholar]
- Shamseldin HE, Alshammari M, Al-Sheddi T, Salih MA, Alkhalidi H, Kentab A, Repetto GM, Hashem M, Alkuraya FS. Genomic analysis of mitochondrial diseases in a consanguineous population reveals novel candidate disease genes. J Med Genet. 2012;49:234–241. doi: 10.1136/jmedgenet-2012-100836. [DOI] [PubMed] [Google Scholar]
- Sun Y, Hu G, Luo J, Fang D, Yu Y, Wang X, Chen J, Qiu W. Mutations in methionyl-tRNA synthetase gene in a Chinese family with interstitial lung and liver disease, postnatal growth failure and anemia. J Hum Genet. 2017;48:337. doi: 10.1038/jhg.2017.10. [DOI] [PubMed] [Google Scholar]
- Tracewska-Siemiątkowska A, Haer-Wigman L, Bosch DGM, Nickerson D, Bamshad MJ, van de Vorst M, Rendtorff ND, Möller C, Kjellström U, Andréasson S, Cremers FPM, et al. University of Washington Center for Mendelian Genomics. An Expanded Multi-Organ Disease Phenotype Associated with Mutations in YARS. Genes (Basel) 2017;8:381. doi: 10.3390/genes8120381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P-C, Soong B-W, Mademan I, Huang Y-H, Liu C-R, Hsiao C-T, Wu H-T, Liu T-T, Liu Y-T, Tseng Y-T, Lin K-P, Yang U-C, et al. A recurrent WARS mutation is a novel cause of autosomal dominant distal hereditary motor neuropathy. Brain. 2017;140:1252–1266. doi: 10.1093/brain/awx058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L, Tiranti V, Marsano RM, Malfatti E, Fernandez-Vizarra E, Donnini C, Mereghetti P, De Gioia L, Burlina A, Castellan C, Comi GP, Savasta S, et al. Infantile encephalopathy and defective mitochondrial DNA translation in patients with mutations of mitochondrial elongation factors EFG1 and EFTu. Am J Hum Genet. 2007;80:44–58. doi: 10.1086/510559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meel E, Wegner DJ, Cliften P, Willing MC, White FV, Kornfeld S, Cole FS. Rare recessive loss-of-function methionyl-tRNA synthetase mutations presenting as a multi-organ phenotype. BMC Med Genet. 2013;14:106. doi: 10.1186/1471-2350-14-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon HJ, McClellan R, Batista DAS, Naidu S. Mutations in FARS2 and non-fatal mitochondrial dysfunction in two siblings. Am J Med Genet A. 2015;167A:1147–1151. doi: 10.1002/ajmg.a.36993. [DOI] [PubMed] [Google Scholar]
- Vester A, Velez-Ruiz G, McLaughlin HM, Lupski JR, Talbot K, Vance JM, Züchner S, Roda RH, Fischbeck KH, Biesecker LG, Nicholson G, et al. NISC Comparative Sequencing Program. A Loss-of-Function Variant in the Human Histidyl-tRNA Synthetase ( HARS) Gene is Neurotoxic In Vivo. Hum Mutat. 2012;34:191–199. doi: 10.1002/humu.22210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MA, Mohler KP, Hopkins KW, Oakley DH, Sweetser DA, Ibba M, Frosch MP, Thibert RL. Novel Compound Heterozygous Mutations Expand the Recognized Phenotypes of FARS2-Linked Disease. Journal of Child Neurology. 2016;31:1127–1137. doi: 10.1177/0883073816643402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu W, Fang Z, Shi J, Che F, He C, Yao L, Wang E, Wu Y. A Newly Identified Missense Mutation in FARS2Causes Autosomal-Recessive Spastic Paraplegia. Hum Mutat. 2015;37:165–169. doi: 10.1002/humu.22930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.