Abstract
Qualitative differences in the innate and adaptive responses elicited by different HIV vaccine candidates have not been thoroughly investigated. We tested the ability of the Aventis Pasteur live recombinant canarypox vector (ALVAC)–SIV, DNA–SIV and Ad26–SIV vaccine prime modalities together with two ALVAC–SIV + gp120 protein boosts to reduce the risk of SIVmac251 acquisition in rhesus macaques. We found that the DNA and ALVAC prime regimens were effective, but the Ad26 prime was not. The activation of hypoxia and the inflammasome in CD14+CD16− monocytes, gut-homing CCR5-negative CD4+ T helper 2 (TH2) cells and antibodies to variable region 2 correlated with a decreased risk of SIVmac251 acquisition. By contrast, signal transducer and activator of transcription 3 activation in CD16+ monocytes was associated with an increased risk of virus acquisition. The Ad26 prime regimen induced the accumulation of CX3CR1+CD163+ macrophages in lymph nodes and of long-lasting CD4+ TH17 cells in the gut and lungs. Our data indicate that the selective engagement of monocyte subsets following a vaccine prime influences long-term immunity, uncovering an unexpected association of CD14+ innate monocytes with a reduced risk of SIVmac251 acquisition.
Of the six independent HIV vaccine trials conducted so far1–4, only the RV144 vaccine trial demonstrated limited, but significant, efficacy (31.2%)5. In the RV144 trial, volunteers were vaccinated twice with the HIV recombinant Aventis Pasteur’s canarypox vector (ALVAC) and then two additional times with ALVAC–HIV in combination with two gp120 envelope (Env) proteins formulated in alum. The vaccine induced high-binding antibody titers to the HIV-1 Env proteins, Env-specific CD4+ T cells in nearly all vaccinees and negligible CD8+ T cell responses5. The titer of IgG antibodies to variable regions 1 and 2 (V1/V2) of the HIV Env protein was a primary correlate of risk, whereas CD4 polyfunctional cells and antibody-dependent cytotoxicity were secondary correlates of risk of HIV acquisition6–9. The decreased risk of HIV acquisition afforded by the ALVAC–SIV + gp120 alum regimen was limited and unsustained, requiring improvement5. Vaccination with a similar SIV-based vaccine regimen also significantly decreased the risk of SIVmac251 acquisition (44% efficacy) in macaques10, which was associated with the level of mucosal antibodies to V2, the frequency of mucosal natural cytoxicity receptor NKp44+ cells and RAS activation. Here, we used the identical SIVmac251 macaque model that recapitulated the results of the RV144 trial to assess whether priming modalities other than ALVAC–SIV could improve the efficacy of the ALVAC–SIV + gp120 alum vaccine regimen and possibly uncover additional correlates of risk of SIVmac251 acquisition.
We chose an intramuscular DNA–SIV prime with the intent of increasing CD4+ T cells, proven to strengthen anti-Env titers11, or an Ad26–SIV prime to increase mucosal Env antibody levels and CD8+ T cell responses12. The Ad26 prime regimen induced a higher innate proinflammatory profile and higher systemic and mucosal adaptive responses than the DNA prime regimen, but unexpectedly failed to prevent SIVmac251 acquisition. The DNA prime regimen induced an innate immunity signature, evidenced by the activation of the hypoxia13,14 and inflammasome15 pathways in CD14+CD16− monocytes, which correlated with a reduced risk of virus acquisition.
Results
Vaccine efficacy of the ALVAC–SIV, DNA–SIV or Ad26–SIV prime regimens
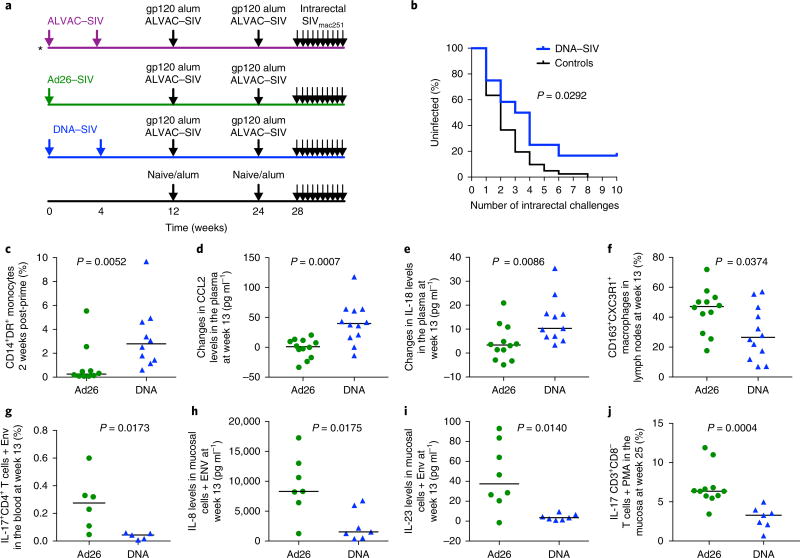
We report here a study that overlapped in execution with the ALVAC prime regimen10, in which we immunized two additional groups of 12 macaques each intramuscularly with either one dose of Ad26–SIV gag, pol and env (Ad26 prime; week 0) or with two doses of DNA–SIV gag, pol and env (DNA prime; weeks 0 and 4). At weeks 12 and 24, both groups were boosted intramuscularly with two doses of ALVAC–SIV combined with both SIVmac251 and SIVsmE660 gp120 proteins formulated in alum alhydrogel (Fig. 1a). The three groups of immunized macaques were exposed intrarectally to a weekly dose of the identical SIVmac251 stock, starting at 4 weeks after the last immunization. The rate of virus acquisition was compared between vaccinated and control animals (6 simultaneous controls and 35 historical controls)10,16,17. All 41 controls became infected after the ninth challenge (Fig. 1b), and no significant difference was observed between historical and concurrent controls (Supplementary Fig. 1a). The Ad26 prime did not significantly decrease the risk of SIVmac251 acquisition when compared to all controls (log-rank test: P = 0.47) or only concurrent controls (Supplementary Fig. 1b,c), whereas the DNA prime did (log-rank test: P = 0.0292; Fig. 1b). In the DNA prime, we only observed a trend towards decreased risk of acquisition when compared to the six concurrent controls, probably because of the small number of animals (see Methods and Supplementary Fig. 1d). The risk of acquisition did not differ between the two regimens as the study was not powered to compare vaccinated groups (Supplementary Fig. 1e). None of these regimens affected the number of transmitted variants, plasma virus levels or CD4+ T cell counts10 (Supplementary Fig. 1f–j).
Fig. 1. Study design and differences in monocytes in the DNA and Ad26 group.
a, A scematic of the study design (see. ref.10). Arrows represent time of vaccination (weeks 0–24) or challenges (week 28). b, Acquisition curve in the DNA-primed group (n = 12). The black line represents 53 controls (concurrent: n = 6 and historical n = 47), and the number of challenges before viral acquisition was assessed using the log-rank test of the discrete-time proportional hazards model (P = 0.029, vaccine efficacy = 52%). c, The frequency of total monocytes in Ad26-primed (n = 10) and DNA-primed (n = 10) animals measured in the blood 2 weeks after the prime with 1× Ad26–SIV or 2× DNA–SIV (weeks 2 and 6, respectively). d,e, Changes in CCL2 (pg ml−1) (d) and in IL-18 (e) plasma levels at week 13 (n = 12 animals each), after the first ALVAC–SIV + gp120 boost, compared to prevaccination levels. f, The percentage of CXC3R1-expressing macrophages (CD163+ cells) in peripheral lymph nodes at week 13 in the Ad26 group and the DNA group (n = 12 animals each). g, The percentage of IL-17+CD4+ T cells in the blood at week 13. h,i, The levels of IL-8 (h) and IL-23 (pg ml−1) (i) in rectal mucosa cell culture after stimulation with Env peptides (n = 7 each group). j, The percentage of CD8−CD3+ (CD4+) T cells in the mucosa at week 25 after phorbol 12-myristate 13-acetate (PMA) ionomycine stimulation. In panels c–j, the median (black horizontal line) is shown and the Mann–Whitney two-tailed test was used for statistical analysis.
The priming influences the inflammatory profile in tissues
The ALVAC, DNA and Ad26 prime vaccine regimens tested here differed only in their prime modalities, suggesting that innate immunity may have contributed to the different viral challenge outcome. Thus, we investigated the effect of the prime on innate myeloid cells by measuring the frequency of blood monocytes18. Two weeks after the prime, we observed differences in the frequency of total monocytes (Supplementary Fig. 2a–d) that resulted in a significant difference in CD14+DR+ monocytes in the blood between the DNA and Ad26 prime groups (P = 0.005, Mann–Whitney test; Fig. 1c). This difference was not maintained following the last ALVAC–SIV + gp120 boosts (Supplementary Fig. 2d). A higher count of the CD14+, but not the CD16+, monocyte subsets was observed in the blood in ALVAC-primed animals10 at 2 weeks following the prime than the DNA or Ad26 prime (Supplementary Fig. 2e,f). Following the ALVAC–SIV + gp120 boost (week 13), the DNA prime group had higher levels of plasma C-C motif chemokine ligand 2 (CCL2), a chemotactic chemokine for monocytes and memory T cells19 than the Ad26 group (P = 0.0007), with the absolute CCL2 levels being highest in the DNA prime (Fig. 1d and Supplementary Fig. 2g). Importantly, the plasma level of IL-18, a cytokine that is strictly regulated by the inflammasome, was higher in the DNA prime than in the Ad26 prime (P = 0.008; Fig. 1e). We reasoned that the lower frequency of blood CD14+CD16− monocytes in the Ad26 group (Fig. 1c) could either be due to cell death or egress from blood to tissues where, depending on the stimulus, they differentiate into macrophages or dendritic cells. Patrolling monocytes produce high levels of cytokines and chemokines, promote CD4+ T helper 17 (TH17) cell differentiation and express the CD16 marker and CX3 chemokine receptor 1 (CX3CR1)18. Indeed, the percentages of CX3CR1+CD163+ de novo–differentiated macrophages in the lymph nodes and Env-responsive TH17 cells in the blood were higher in the Ad26 group than in the DNA group following the first boost (P = 0.0374 and P = 0.0173, respectively; Fig. 1f,g). At the same timepoint (week 13), cells from the rectal mucosa that were stimulated or not stimulated with SIV Env peptides produced significantly higher levels of several cytokines and chemokines in the Ad26 group (Supplementary Table 1). Of note, the levels of IL-8 and IL-23, which are cytokines that recruit neutrophils and promote TH17 cell differentiation20, were higher in the Ad26 prime than in the DNA prime (P = 0.017 and P = 0.014; Fig. 1h,i). Strikingly, the difference in TH17 cell frequency between the DNA and the Ad26 prime groups persisted following the last immunization (week 25) in the rectal mucosa (P = 0.0004; Fig. 1j), suggesting that the Ad26 prime affected the long-term quality of the CD4+ T cell response. Thus, the Ad26 prime enhanced tissue migration and the differentiation of monocytes, increased the production of proinflammatory cytokines at mucosal sites and resulted in long-lasting, elevated counts of mucosal TH17 cells.
Hypoxia and inflammasome activation in CD14+CD16− monocytes are correlates of decreased risk of SIVmac251 acquisition
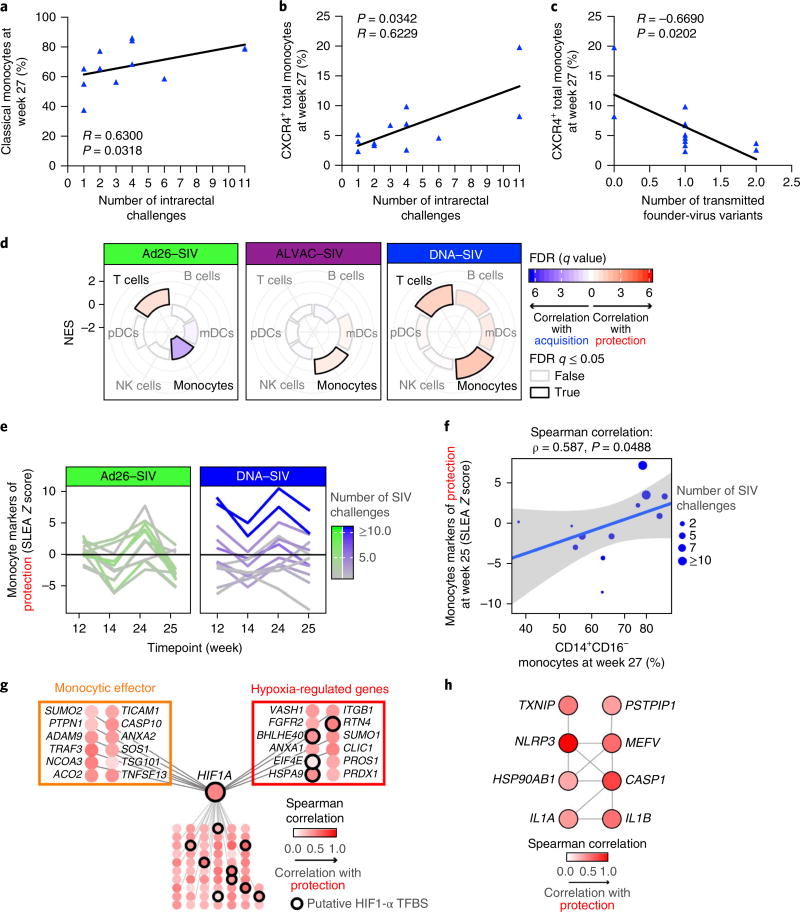
The temporospatial difference in the frequency of myeloid cells in the blood and tissues between the DNA and the Ad26 prime regimens prompted us to assess their role in affecting the risk of SIVmac251 acquisition. In the DNA prime, we found that the frequency of the CD14+CD16− monocytes in the blood correlated with a decreased risk of SIVmac251 acquisition (Spearman correlation R = 0.630, P = 0.031, adjusted P = 0.036; Fig. 2a and data not shown). CXCR4+ monocytes, which constitute transitional bone marrow premonocytes21, also correlated with a decreased risk of SIV acquisition and a decreased number of transmitted SIV variants (Fig. 2b,c). To further validate the role of monocyte-mediated innate immunity, we analyzed the total blood collected at 24 h and at 1 or 2 weeks after each ALVAC–SIV + gp120 boost (Fig. 1a) in the Ad26, DNA and ALVAC prime regimens using a Gene Set Enrichment Analysis (GSEA) tool to estimate and compare the contribution of immune cell subsets (including T cells, B cells, myeloid and plasmacytoid dendritic cells, monocytes and NK cells) to vaccine-conferred decreased risk of SIVmac251 acquisition10. This analysis revealed that markers of the monocyte subsets were over-represented among genes that correlated with protection in the ALVAC and DNA prime regimens (Fig. 2d and Supplementary Fig. 3a). In the DNA prime group, we observed a trend (week 25, P = 0.090; Fig. 2e) for higher expression of monocyte genes associated with a decreased risk of SIVmac251 acquisition than the Ad26 group. The expression of monocyte genes associated with decreased risk of SIVmac251 acquisition at week 25 correlated with a higher proportion of classical monocytes (CD14+CD16−) among the monocyte subsets at week 27 (Fig. 2f and Supplementary Fig. 3b). Multivariate analysis based on all immunological markers measured in this study (482 markers; Supplementary Table 3) was used to identify the optimal set of markers, allowing for an accurate prediction of the number of SIVmac251 challenges required to infect vaccinated macaques. The analysis revealed that the top multivariate model associated with the number of SIVmac251 challenges to infection in the DNA prime group included markers of both the monocyte subset and the frequency of TH2 cells measured at week 27 (R2 = 0.544; F test: P = 0.029; Supplementary Table 3). Thus, both phenotypical and gene expression analyses (Supplementary Table 2) validated the association of CD14+CD16− monocytes with a decreased risk of SIVmac251 acquisition and revealed that of TH2 cells.
Fig. 2. Differential contribution of monocytes to protection.
a,b, The frequency of classical (CD14+CD16−) (a) and CXCR4+ (b) monocytes measured in the blood (week 27) in the DNA group correlates with the risk of SIVmac251 acquisition (the number of challenges to infection; n = 12). c, The frequency of CXCR4+ monocytes in the DNA group correlates with the number of transmitted SIV variants in the animals that became infected. A Spearman correlation was used for statistical analysis in a–c. d, Radial plots showing the relative contribution of immune subsets in protection against SIV acquisition. The normalized enrichment score (NES) estimated by GSEA is plotted for each subset and separated by vaccine regimen (Ad26–SIV: n = 11 animals, ALVAC–SIV: n = 27 animals and DNA–SIV: n = 12 animals). An NES of >0 or NES of <0 corresponds to markers of immune subsets enriched among genes associated with protection or SIV acquisition, respectively. FDR, false discovery rate; mDC, myeloid dendritic cell; pDC, plasmacytoid dendritic cell. e, Line plot showing the normalized average expression of monocyte genes associated with protection in each macaque (y axis) as a function of the four timepoints after boosting with ALVAC–SIV + gp120 (x axis) for both the Ad26–SIV (n = 11 animals) and the DNA–SIV (n = 12 animals) vaccine regimens. SLEA, sample-level enrichment analysis. f, Scatter plot showing the normalized average expression of monocyte genes associated with protection in each macaque at week 25 (y axis) as a function of the fraction of classical monocytes (CD14+CD16−) among monocytes at week 27 (n = 12 animals). The linear regression fit (blue line) and the 95% confidence interval (gray region) are given on the plot. g,h, The GeneMANIA network of the monocyte genes associated with protection from SIV challenge (g) and the network of the monocyte genes associated with protection from SIV infection that are part of the inflammasome pathway of REACTOME (h). TFBS, transcription factor–binding site. Each node is colored by the Spearman correlation of the genes (24 h after the second boost) with the number of SIV challenges to infection of the DNA–SIV animals (n = 12 animals).
Monocyte markers associated with a decreased risk of SIVmac251 acquisition included genes implicated in IL-1β production and regulation (ANXA2, EIF4A and IL1RL2) and in the suppression of proinflammatory cytokines (TLR10 and TBK1)22,23. Gene network inference revealed the importance of hypoxia-inducible factor 1-α (HIF1-α) as a key transcriptional regulator of these protective genes13,14 (Fig. 2g). In addition, putative binding sites to HIF1-α were enriched in regulatory regions of the protective genes (HOMER: P = −0.0284). HIF1A gene (which encodes HIF1-α) expression correlated with a decreased risk of SIVmac251 acquisition at each time-point analyzed (Supplementary Table 2) and its mRNA expression was correlated with two transcriptional regulators of HIF1-α, EIF4E and BHLHE40 (ref. 24). Following the last immunization, a significant correlation was found between the risk of SIVmac251 acquisition and the genes encoding the NLRP3 component of the inflammasome, a multiprotein scaffold that promotes the cleavage of caspase 1 and processing of the IL-1β and IL-18 precursor proteins to initiate innate immune defense15 (Fig. 2h and Supplementary Table 4). The monocyte markers associated with inflammasome activation and vaccine efficacy are expressed at a higher level in sorted classical CD14+ monocytes than in non-classical CD16+ cells (Supplementary Fig. 3b). Quantitative reverse transcription PCR (RT–PCR) analysis validated the NLRP3 enrichment in sorted CD14+, but not CD14−, cells obtained from immunized or unimmunized macaques (Supplementary Fig. 3c).
The frequency of CD16+ monocytes (intermediate and non-classical) correlated with an increased risk of SIVmac251 acquisition (R = −0.617, P = 0.036, adjusted P = 0.036; Supplementary Fig. 4a), and analyses of genes associated with an increased risk of SIVmac251 acquisition in both groups (Supplementary Fig. 4b) revealed a correlation with CD16+ monocytes (Supplementary Fig. 4c), validating the phenotypic results in Supplementary Fig. 4a. Importantly, signal transducer and activator of transcription 3 (STAT3) was the key transcriptional regulator of this signature of acquisition (HOMER: P = 0.109; Supplementary Fig. 4d). The monocyte gene associated with acquisition included genes encoding STAT3 co-factors, such as CSF3R, EP38 and ILK, and canonical STAT3 targets, such as BCL3, DDAH2 and MED13 (ref. 25). Thus, an analysis of the vaccine-modulated expression of genes that correlated with a risk of SIVmac251 acquisition strongly supported the opposite roles of the different monocyte subsets, with the activation of hypoxia and inflammasome pathways in CD14+ monocytes associated with a decreased risk of SIVmac251 acquisition, and the activation STAT3 in CD16+ monocytes with an increased risk of acquisition. Monocyte subsets also display discordant roles in immunity against other infectious viruses, including Zika virus, Ebola virus, Dengue virus, Epstein–Barr virus and SIV/HIV26,27. We next investigated monocyte-mediated phagocytosis in the presence or absence of anti-SIV antibodies. We used sera collected at the end of immunization in either a THP-1 cell-based assay or an assay in which the phagocytic function of monocytes from macaques was measured with their matching sera. In the DNA and Ad26 vaccine regimens, we neither found a difference between the phagocytic activities of monocytes measured in the two assays (Supplementary Fig. 4e–g) nor a correlation with the risk of SIVmac251 acquisition in either regimen (data not shown).
Classical monocytes directly correlate with the frequency of vaccine-induced CCR5-negative TH2 cells
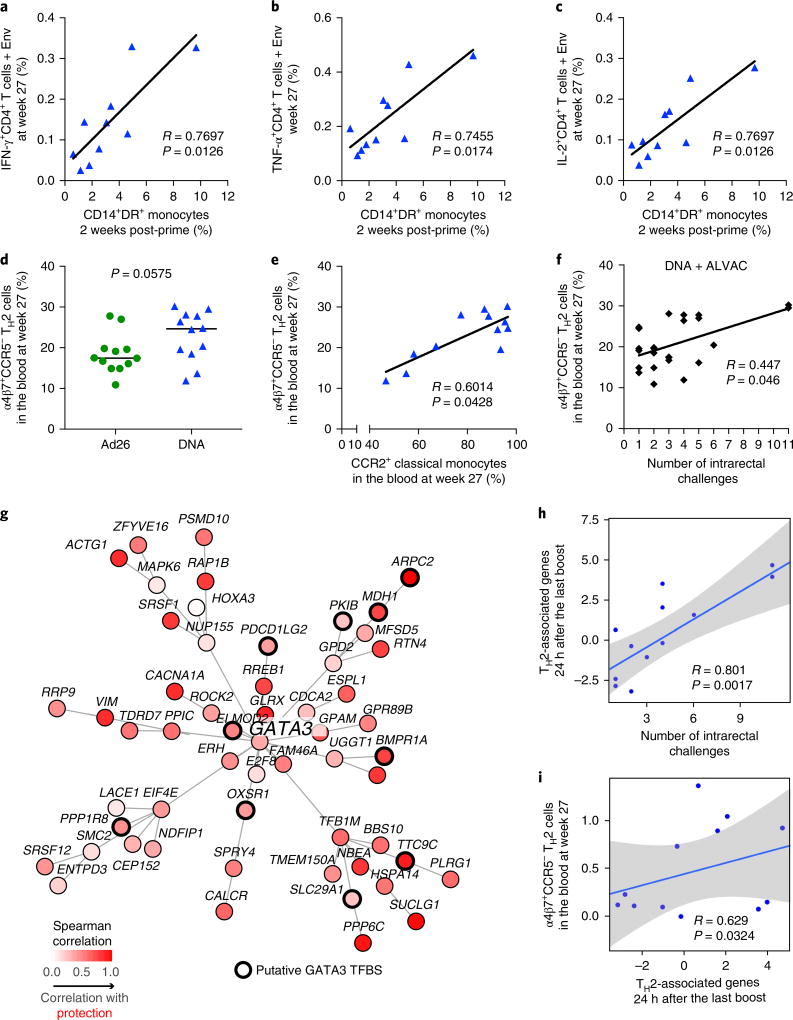
ALVAC preferentially infects CD14+ cells28 and causes dendritic cell maturation by inducing apoptosis and tumor necrosis factor-α (TNF-α) production29. Cytosolic DNA phagocytosed by monocytes activates cyclic GMP-AMP synthase (cGAS) and IF116 (refs30,31). Less is known about Ad26 target cells, but it is likely that Ad26 also infects CD16+ monocytes as the Ad26 receptor, CD46, is expressed at a high level in CD16+ cells32. Thus, we investigated whether the frequency of CD14+CD16− classical monocytes was associated with vaccine-induced T cell and NK cell responses. Samples were collected from the blood and mucosal compartments following the last immunization in the ALVAC prime regimen (week 25 or week 27). The blood counts of SIV-Gag-specific T cells producing IL-2, interferon-γ (IFN-γ) and TNF-α, and of IL-2- and IL-17-producing T cells in the bronchoalveolar lavage were higher in the Ad26 group than in the DNA group (Supplementary Figs. 5 and 6 and Supplementary Table 5). Importantly, the frequency of total monocytes (DR+CD14+) measured in the DNA prime regimen 2 weeks after priming correlated positively with that of SIV-Env-specific CD4+ T cells producing IL-2 (R = 0.76, P = 0.012), TNF-α (R = 0.74, P = 0.017) and IFN-γ (R = 0.76, P = 0.012) at the end of immunization (Fig. 3a–c). A similar association was not found in the Ad26 prime regimen. In the DNA prime, we found a significantly higher count of CD4+ T cells expressing GATA3 (P = 0.028), the master transcription factor for the differentiation of TH2 cells (week 10, 1 week prior to the ALVAC–SIV + gp120 boost), than in the Ad26 prime (Supplementary Fig. 7). Analysis of the Ki67+ vaccine-induced CD4+ T cells at the end of immunization (week 27) demonstrated a trend (P = 0.057) for higher counts of circulating CCR5-negative TH2 (CXCR3−CCR6−) cells expressing mucosal homing α4β7+ in the DNA prime group (Fig. 3d and Supplementary Fig. 8) and no difference in TH1 (CXCR3+CCR6−) or TH17 (CXCR3−CCR6+) cell numbers (data not shown)33. Importantly, however, the CCR5-negative α4β7+ TH2 cells were associated with the frequency of classical monocytes (R = 0.60, P = 0.042; Fig. 3e) and with a decreased risk of SIVmac251 acquisition (R = 0.44, P = 0.046; Fig. 3f). Low levels of CCR5 expression were also observed in vaccine-induced TH1 cells obtained from humans vaccinated with the ALVAC–HIV + gp120 vaccine platform in the RV144 trial34. We identified specific TH2 cell genes from existing data sets35 and observed a significant enrichment of TH2 transcriptomic markers among genes associated with a decreased risk of SIVmac251 acquisition in the DNA prime (24 h after the last immunization; GSEA: P < 0.033). Network inference based on the overlapping genes revealed GATA3 as the central transcription factor regulating TH2 genes associated with a reduced risk of virus acquisition (HOMER: P = 0.037; Fig. 3g). The TH2 transcriptomic markers that are controlled by GATA3 were significantly correlated with delayed SIVmac251 acquisition (Fig. 3h) and, importantly, with CCR5-negative α4β7+ TH2 cell counts (Fig. 3i). Thus, the transcriptomic analysis corroborates the flow cytometry data and points to the potential importance of the vaccine-induced CCR5-negative TH2 cell subset in decreasing the risk of SIVmac251 acquisition.
Fig. 3. Monocytes cross-talk with CD4+ T cells and NK cells.
a–c, A significant direct correlation between the frequency of total monocytes and the blood cells after the prime and env-specific T cell responses in the blood to Env peptides measured at week 27 in the DNA group (n = 11, Spearman correlation test, two-tailed, 95% confidence). d–f, The frequency of vaccine-induced TH2-type CD4+ T cell expressing α4β7, but not CCR5, was higher in the DNA group (n = 12 each, Mann–Whitney two-sided test, median represented by horizontal line) (d), and this cell subset directly correlated with CCR2+ classical monocytes in the blood at the same time (week 27) in the DNA group (e) and with the number of challenges to infection in both vaccines (f). The Jonckheere–Terpstra test was used in this analysis. g, The GeneMANIA network shows the TH2 genes associated with protection of the DNA–SIV animals (n = 12 animals) and GATA3, the main hub in the network. h,i, TH2 genes associated with protection 24 h after the second ALVAC boost. Scatters plots show the relative expression of the TH2 genes as a function of the number of SIV challenge to infection (h) and the relative expression of α4β7+CCR5− TH2 cells as a function of TH2 genes for the DNA-primed animals (n = 12 animals) (i). The linear regression fit (blue line) and the 95% confidence interval (gray region) are given on the plot. The Student’s t-test was used to assess the significance of the correlation.
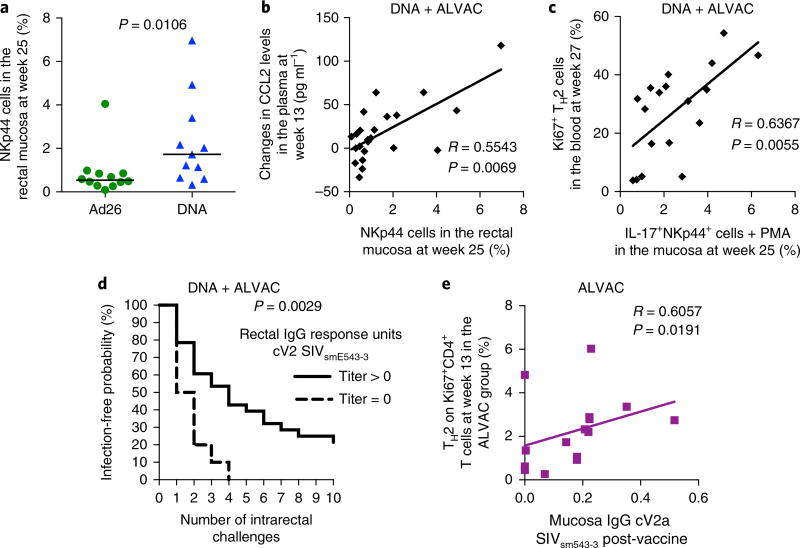
TH2 cells are associated with NKp44+ cells and antibody response to V2
We previously observed that mucosal NKp44+-producing IL-17 correlates with a decreased risk of SIVmac251 acquisition in the ALVAC prime regimen10. Hence, we measured the frequency of NKp44+ in the rectal mucosa at the end of the immunization regimens in the DNA and Ad26 prime regimens and found a significantly higher frequency of mucosal NKp44+ cells in the DNA prime than in the Ad26 prime (P = 0.010; Fig. 4a and Supplementary Fig. 8b,c). Interestingly, the plasma level of CCL2 correlated with the total number of mucosal NKp44+ cells (R = 0.55, P = 0.006; Fig. 4b), and vaccine-induced total TH2 cells correlated with the ability of NKp44+ cells to produce IL-17 (R = 0.63, P = 0.005; Fig. 4c). These data indicate a potential cross-talk among these immune cells that warrants further investigation. Analyses of anti-SIV Env humoral responses demonstrated that the Ad26 prime regimen elicited higher serum IgG and IgA titers to the gp120 and V1/V2 scaffolds of both viral strains (SIVmac251 and SIVsmE660) and higher serum titers of neutralizing antibodies to the tier 1 SIVmac251 than did the DNA prime (Supplementary Table 5 and Supplementary Fig. 9a–e). In rectal secretions, the levels of IgG to the SIVmac251 gp120 proteins and the corresponding V1/V2 scaffolds were higher in the Ad26 prime than in the DNA prime (Supplementary Fig. 9f–i), whereas the levels of mucosal IgG and IgA to the cyclic V2 did not differ significantly between the two groups (Supplementary Fig. 9j,k). Interestingly, however, the level of IgG to a conformational V2 peptide designated as cyclic V2 was associated with delayed SIVmac251 acquisition in the DNA and ALVAC prime regimens, but not in the Ad26 regimen (P = 0.002; Fig. 4d and data not shown). Generation of the mucosal antibody responses to cyclic V2 in the ALVAC prime correlated with the frequency of TH2 cells in the blood (R = 0.60, P = 0.019; Fig. 4e). Collectively, these data support that the DNA and ALVAC prime regimens elicited CD4+ TH2 cell responses that correlated with mucosal antibodies to V2 and the recruitment and ability of mucosal NKp44+ cells to produce IL-17. By contrast, the Ad26 prime was associated with the skewing of the CD4+ T cell response at mucosal sites towards a more inflammatory TH17 cell response, an event that was probably related to the increased frequency of CX3CR1+CD163+ macrophages and IL-23 in tissue, as observed in other inflammatory conditions36,37.
Fig. 4. TH2 cells are associated with NKp44+ cells and antibody response to V2.
a, The DNA vaccine induced mucosal recruitment of NKp44+ cells at significantly higher counts than the Ad26 group (week 25; n = 12 and n = 11, respectively, Mann–Whitney two-sided test). The median (black horizontal line) is shown. b,c, Correlation between the numbers of NKp44+ cells measured in the mucosa in both vaccines and the levels of CCL2 in the plasma (week 13) (b) or with the percentage of vaccine-induced (Ki67+) TH2 (CXCR3−CCR6−) cells in the blood at week 27 (c). d, The positivity for mucosal antibody to the cyclic V2 (cV2) to E660 in the ALVAC + gp120 alum and the DNA-primed ALVAC + gp120 alum strategy showed an association with protection (log-rank test). e, Association between the frequency of TH2 cell responses induced by the ALVAC + gp120 alum strategy (ALVAC primed) and protective mucosal responses to the cyclic V2 SIVsmE600 (n = 16). A Spearman correlation test (two-tailed, 95% confidence) was used for statistical analysis in b–e.
Recurrent gene signatures in HIV vaccine candidates associated with a decreased risk of SIVmac251 acquisition
We next evaluated the expression of monocytes and T cell genes in two independent data sets in which animals were vaccinated with an ALVAC prime regimen or gp96-SIV + gp120 alum and exposed to the same SIVmac251 challenge stock10,16.
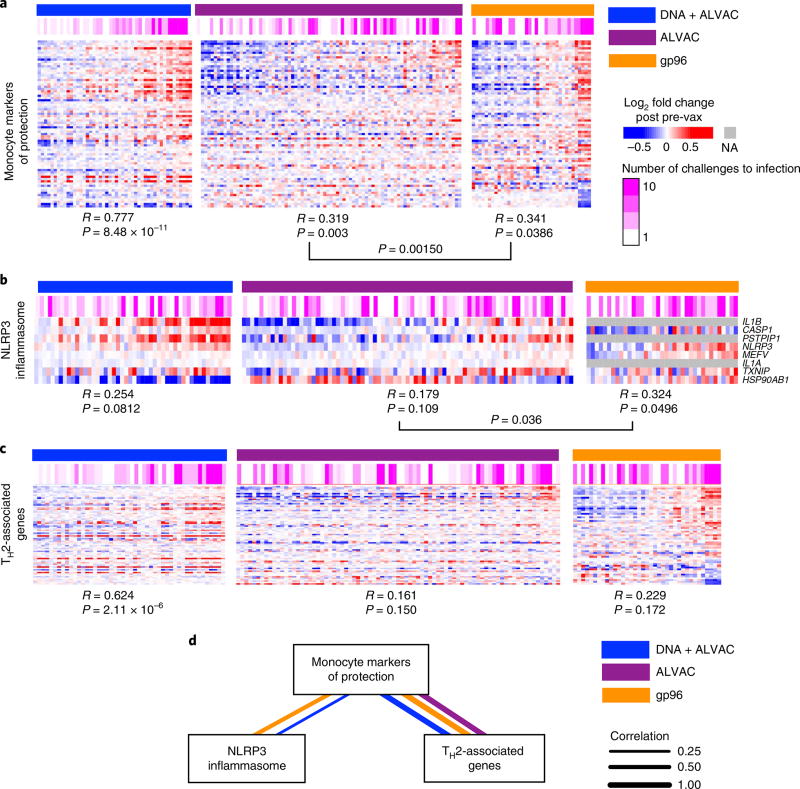
Independent analyses of the ALVAC–SIV and gp96-SIV prime groups revealed 88 monocyte genes associated with a decreased risk of SIVmac251 in all three vaccine regimens (DNA–SIV, ALVAC–SIV and gp96-SIV; Fisher combined test: P = 0.001; Fig. 5a). Similarly, the inflammasome gene signature correlated with a decreased risk of SIVmac251 acquisition in both the ALVAC and gp96 prime regimens (Fisher combined test: P = 0.033; Fig. 5b), whereas the TH2-associated genes did not (P = 0.120). Although we could not find a direct association between TH2 and a decreased risk of SIVmac251 acquisition in the two independent data sets, we observed a strong positive correlation of the 88 monocyte markers and TH2-associated genes (P < 1.23 × 10−8, R > 9.673; Fig. 5c,d), suggesting that TH2 may still be relevant in the ALVAC–SIV and gp96-SIV primes, as it was in the DNA–SIV prime regimen.
Fig. 5. Monocyte markers of protection identified in the present study are associated with the number of SIV challenges to infection in previous studies with ALVAC–SIV or gp96 SIV prime.
a, Heatmaps showing the expression of the 88 monocytes markers of protection after DNA (n = 12 animals), ALVAC (n = 24 animals) or gp96 prime inoculation (n = 12 animals). All transcriptomic data collected after immunization were used to perform this analysis (DNA: 24 h and 1 or 2 weeks after immunization; ALVAC: 24 h and 1 week after immunization; gp96: 1, 7 and 17 weeks after immunization). Samples were ordered from left to right by increasing mean expression of the 88 monocytes markers. A Pearson correlation and t-test were performed to statistically evaluate the association between the markers of protection and the number of SIV challenges to infection. NA, value not available. b,c, A similar analysis (as in a) for inflammasome (b) and TH2-associated (c) genes. d, The expression of the 88 monocytes markers of protection, the NLRP3 inflammasome and TH2-associated genes are correlated with each other. A Student’s t-test was used to test the significance of the correlation. Only correlations associated with P < 0.05 are presented in the network.
Discussion
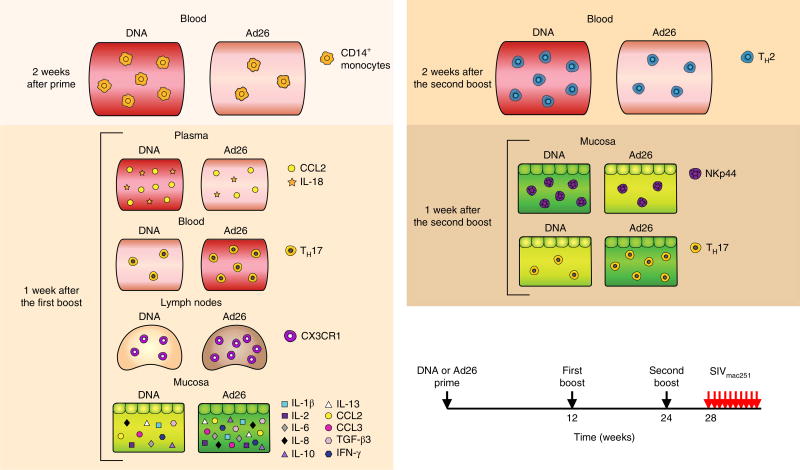
We report here an integrated study of immune parameters, validated by changes in gene signatures within myeloid and T cell subsets, associated with the risk of mucosal acquisition of the highly pathogenic SIVmac251. By using three different priming methods with SIV recombinant DNA, ALVAC or Ad26, followed by two identical boosts with ALVAC–SIV + gp120 alum, we identified temporospatial differences (Fig. 6) in the frequency of myeloid subsets in the blood and tissues that associated with qualitative differences in innate and adaptive responses. The DNA, ALVAC and alum components of the DNA prime vaccine regimen all activate the inflammasome through the engagement of NLRP3, cGAS, IF116 and AIM2 (refs30,31,38–40). ALVAC preferentially infects CD14+ cells28 and induces 50-fold higher levels of IL-1β within 24 h of immunization than does Ad26 (refs41,42) (Supplementary Table 6). This early burst of IL-1β, a cytokine that is tightly regulated by the inflammasome, may provide the ‘emergency’ signal43 that is necessary for engagement of the myeloid compartment and the generation of a memory innate response following ALVAC immunization. This is a well-characterized phenomenon in which hypoxia and inflammasome activation in monocytes causes a metabolic switch from oxidative phosphorylation to glycolysis, epigenetic reprogramming that results either in increased (training) or decreased (tolerance) cytokine and chemokine responsiveness upon restimulation43–46 and involves several cellular pathways, including RAS. Future studies on the CD14+ monocytes epigenome will be needed to clarify the nature (training or tolerance) and duration of the innate immunity elicited by the DNA–ALVAC-gp120 vaccine platform. In a previous study10, we found that the RAS pathway also correlated with a decreased risk of SIVmac251 acquisition and, interestingly, the mitogen-activated protein kinase (MAPK)–extracellular signal-regulated kinase (ERK) pathway also contributes to inflammasome function47. Furthermore, hypoxia activates the inflammasome by increasing the levels of NLRP3, AIM2 and pro-ILβ in monocytes48. In the DNA prime regimen, adaptive responses such as the CCR5-negative TH2 subset of gut-homing CD4+ TH cells correlated directly with CD14+ monocytes (Fig. 3d), consistent with the finding that the level of CCL2, a chemokine that regulates TH2 polarization, was higher in the DNA prime than in the Ad26 prime49. Importantly, TH2 cells negative for the expression of CCR5, the SIV/HIV co-receptor, correlated with a decreased risk of SIVmac251 acquisition (Fig. 3f), suggesting that the elicitation of CD4+ T cells that are resistant to virus infection may be important, as previously suggested by others50–52. The antibodies to cyclic V2 correlated with a decreased risk of SIVmac251 acquisition in the ALVAC- and DNA-primed animals, but not in the Ad26 prime regimen despite there being no significant difference in their magnitude. This finding suggests that mucosal antibodies to V2 may not be sufficient to decrease the risk of virus acquisition alone but may tip the balance towards reduced acquisition when paired with the innate immunity induced by DNA or ALVAC priming. Alternate possibilities are that antibodies to V2 do not impact the risk of virus acquisition, are a surrogate of another host response or differ functionally in distinct vaccine settings. In the Ad26 prime, the accumulation of CD16+ monocyte-derived de novo-differentiated CD163+CX3CR1+ macrophages20 in the lymph nodes and the high counts of TH17 cells in the mucosa may have created an inflammatory innate response that negated the protective effect of the mucosal antibodies to V2. The association of CD16+ monocytes and increased TH17 cell differentiation has also been observed in other inflammatory conditions36,37,53. Ad26 uses CD46 as a receptor whose engagement induces autophagy54, a strategy used by adenoviruses to increase their replication55,56. However, autophagy inhibits inflammasome activation and limits IL-1β production32,56,57. In contrast to our findings, an Ad26–SIV prime followed by a single SIV recombinant poxvirus modified vaccinia virus Ankara (MVA) boost protected macaques from SIVmac251 challenge in a 2012 study by Barouch et al.58. Substantial differences between these studies may account for the discordant results. First, ALVAC and MVA poxviruses differ in their inflammatory profiles: the MVA prime does not induce the early burst in plasma IL-1β levels as observed with the ALVAC prime42 (summarized in Supplementary Table 6). Second, we exposed macaques to SIVmac251 at 1 month following the last immunization (versus 6 months in the Barouch study58) to explore a possible compound effect of increased target cells and protective responses. Third, the genetic complexity of the viral challenge used in our study is considerably higher59 than that used by Barouch et al.58. Finally, our study used a SIVmac251 challenge stock propagated in macaque cells and vaccines grown in chicken cells (ALVAC), hamster cells (gp120) and bacteria (DNA). By contrast, both the SIVmac251 challenge stock and the Ad26-based vaccines used in Barouch et al.58 were propagated in human cells, raising the possibility that a potent xenoresponse may have contributed to protection in that specific macaque model, as observed in other studies60. Based on our data presented here, we hypothesize that monocyte innate immunity contributes to the vaccine-associated decreased risk of SIVmac251 acquisition either directly by causing inflammation or indirectly by influencing cellular (TH1, TH2 and NKp44+ cells) and humoral (antibody to V2) adaptive responses. The true effector mechanism (or mechanisms) that reduces virus acquisition remains to be elucidated.
Fig. 6.
Schematic of correlates of protection and the risk of acquisitions measured after inoculation of prime or each of the boosts in different compartments. TGF-β3, transforming growth factor-β3. Arrows represent time of vaccination (weeks 0–24) or challenges (week 28).
Methods
Animals, vaccines and SIVmac251 challenge
All animals used in this study were colony-bred rhesus macaques (Macaca mulatta) obtained from Covance Research Products. The animals were housed and handled in accordance with the standards of the Association for the Assessment and Accreditation of Laboratory Animal Care International. Both the care and the use of the animals were in compliance with all relevant institutional (US National Institutes of Health (NIH)) guidelines. The protocol (AUP 491) was approved by the Advanced BioScience Laboratories’ Institutional Animal Care and Use Committee.
In the present study, 24 juvenile macaques were randomized into two groups based on their major histocompatibility, status and sex. Two MamuA01+ animals were included in each of the vaccinated groups. Although the investigators were unblinded, the animal handlers were blinded to the vaccine groups. With the assumption of an infection rate in control macaques of 0.40 based on historical data, the study design with 12 macaques and 50% efficacy in each vaccinated group provided 84% power in tests of the infection rate of each group against combined concurrent and historical control animals. Twelve macaques, 6 female and 6 male, were immunized with 3 × 1010 viral particles (vp) of Ad26-expressing SIVsmE543 Gag, Pol and Env (1 × 1010 vp for each construct), formulated in PBS and sucrose (15 ml) and administered intramuscularly in 0.75-ml doses in the quadriceps61 (week 0; Fig. 1). Twelve macaques, 6 female and 6 male, were immunized twice with DNA–SIV at weeks 0 and 4, intramuscularly, as previously described62. Each vaccination contained a total of 6 mg DNA in 1.5 ml PBS. The animals were given the following DNA constructs: 206S SIV p57gagmac239 (1 mg), 209S MCP3-p39gagmac239 (1 mg), 221S SIVmacM766 gp160 (2 mg) and 103S LAMP-Polmac239 (2 mg). At weeks 12 and 24, all 24 macaques were boosted with intramuscular inoculations of 108 p.f.u. of recombinants ALVAC (vCP2432), expressing SIVmac251 gag-pro and gp120TM (Sanofi Pasteur), and with 200 µg SIVmac251-M766 and SIVsmE660-CG7V gp120-gD proteins formulated in alum alhydrogel (InvivoGen), as previously described63. The two proteins were administered intramuscularly in the opposite thigh. The 24 immunized macaques and 6 control macaques were challenged intrarectally with 10 low, repeated doses of pathogenic SIVmac251 (120 TCID50 (50% tissue culture infective dose)) once a week. The stock of SIVmac251 was propagated in macaque cells. Challenge was initiated at 4 weeks following the last immunization (week 28). The concurrent group included 6 male macaques, of which 4 were MamuA01+ animals. At the beginning of the study, it was decided that 35 non-contemporaneous controls, challenged in the same facility by the same operators with the same virus stock, would be added to the 6 concurrent controls. Of the 35 historical controls, 6 were naive, 17 received two inoculations with alum alhydrogel and 12 received gp96-Ig16,17. Of these controls, 12 were challenged within 1 month from the time of challenge of the six concurrent controls. Observed hazard ratios in the proportional hazards model from 0.36 to 4.4 would have been acceptable for the equivalence of the concurrent control group with the historical controls, using a log-rank test at the P = 0.05 level.
No significant differences were observed in the acquisition rates of adjuvanted versus non-treated controls or historical versus concurrent controls (log-rank test; Supplementary Fig. 1), supporting the rationale for combining them.
Single-genome amplification
Transmitted founder viruses and their progeny were identified by single-genome amplification of SIV RNA from plasma at 2–4 weeks from the infection. SIV RNA was extracted, and limiting-dilution PCR of newly synthesized cDNA was performed63,64. Transmitted founder-virus lineages were identified by low-diversity sequence and by single sequences with unique mutations. Phylogenetic trees were generated using ClustalW.
Viral RNA and DNA and CD4+ T cell count
SIVmac251 in plasma was quantified by nucleic acid sequence-based amplification as previously described65. Transmitted founder variants were measured in the plasma at 2 weeks post-infection as previously described64. CD4+ T cell counts were periodically determined from whole blood by flow cytometry as previously described66.
FACS analysis
NK cell counts were measured in the mucosa in 11 macaques from the DNA–SIV group and 13 Ad-ALVAC animals at 1 week following the last immunization (week 25). Mononuclear cells were isolated from rectal biopsies and 3 × 106 cells were used for phenotypic characterization. Flow cytometry staining was carried out for cell surface and intracellular molecules using anti-human fluorochrome-conjugated monoclonal antibodies that are known to cross-react with rhesus macaques: Alexa-Fluor 700 anti-CD3 (SP34-2; 557917, 0.2 mg ml−1); PE-C594 anti-IFN-γ (B27; 562392, 5 µl); PerCPCy5.5 anti-CD4 (L200; 552838, 5 µl) from BD Biosciences; V450 anti-TNF-α (Mab11; 502920, 5 µl); NKp44 (P44-8; 325110, 5 µl); PE-Cy7 anti-IL-2 (17H12; 500325, 50 µg ml−1); 605NC anti-CD20 (2H7; 302334, 50 µg ml−1) from BioLegend; PE-APC eFlour 780 anti-IL-17 (eBio64DEC17; 47–7179, 0.125 mg); FITC anti-CD107a (eBioH4A3; 11-1079-42, 5 µl, 0.5 µg); 605NC anti-CD8 (RPA-T8; 93–0088, 5 µl) from eBioscience; and PE anti-NKG2A (Z199; PN A60797, 5 µl) from Beckman Coulter. The yellow and aqua LIVE/DEAD viability dyes (Invitrogen; L34959, 1 µl) were used to exclude dead cells. Mucosal NK cells were gated on live lymphocytes negative for CD20 and CD3 and further classified based on the expression of NKG2A and NKp44 surface markers.
To identify monocytic myeloid cells, peripheral blood mononuclear cells (PBMCs; 5–10 × 106 cells) were stained with PE-Cy7 anti-CD3 (SP34-2; 563916, 2.0 µl, BD Biosciences), PE-Cy7 anti-CD20 (2H7; 560735, 1.0 µl, BD Biosciences), BV786 anti-NHP-CD45 (D058-1283; 563861, 3.0 µl, BD Biosciences), APC anti-CD14 (M5E2; 561390, 7.5 µl, BD Biosciences), HLA-DR-APC-Cy7 (L243; 307618, 5.0 µl, BioLegend), FITC anti-CD16 (3G8; 555406, 5.0 µl, BD Biosciences), BV421 anti-CD192 (CCR2) (48607; 564067, 3.0 µl, BD Biosciences) and PE-CF594 anti-CD184 (CXCR4) (12G5; 562389, 5.0 µl, BD Biosciences), and the Aqua LIVE/DEAD kit (L34966, 3.0 µl, Invitrogen) was used to exclude dead cells. All myeloid cell populations were gated as CD45+Lin− (CD3 and CD20). Monocyte populations were identified and differentiated by the expression of CD14 and CD16. Classical monocytes were identified as Lin−CD45+CD14+CD16−HLA-DR+, intermediate as Lin−CD45+CD14+CD16+HLA-DR+ and non-classical as Lin−CD45+CD14−CD16+HLA-DR+. Acquisition was performed on an LSRII (BD Biosciences) with a minimum of 500,000 events collected, and marker expression was examined using the FACSDiva software (BD Biosciences). Data were further analyzed using FlowJo version 10.1 (Treestar, Inc.). For all samples, data were acquired on an LSRII flow cytometer using the FACSDiva 8.0 software (BD Biosciences). FACS analysis was also performed using the FlowJo software.
The counts of subsets of CD4+ T cells were measured in the blood at week 27. PBMCs were stained with PerCPCy5.5 anti-CD4 (L200; 552838, 5 µl), Alexa-Fluor 700 anti-CD3 (SP34-2, 557917, 0.2 mg ml−1), BV650 anti-CCR5 (3A9, 5 µl) and PE-Cy5 anti-CD95 (DX2, 559773, 5 µl) from BD Biosciences; PE-eFluor 610 anti-CD185 (CXCR5) (MU5UBEE, 61-9185-42, 5 µl, eBioscience), APC Cy7 anti-CXCR3 (G025H7, 353721, 5 µl, BioLegend), BV605 anti-CCR6 (G034E3, 353419, 5 µl, BioLegend), FITC anti-Ki67 (556026, 10 µl) and APC anti-α4β7 provided by the NIH Nonhuman Primate Reagent Resource (R24 OD010976 and National Institute of Allergy and Infectious Diseases (NIAID) contract HHSN 272201300031C). Gating was done on live CD3+CD4+ cells and on vaccine-induced Ki67+ cells. CXCR3 and CCR6 expression was used to identify TH1 or TH2 populations, as depicted in Supplementary Fig. 6. For circulating follicular helper T cells, CXCR5 was used and subsets were identified in the same manner.
Intracellular cytokine and transcription factor staining assay
PBMCs (1–3 × 106 cells) were stimulated with 2 µg ml−1 of the cognate peptide pools for 6 h in RPMI containing 10% human serum in the presence of 5 µg ml−1 of GolgiPlug (10 µg ml−1, BD Biosciences). Negative controls received an equal concentration of DMSO. Cells were stained with the following surface-marker-specific antibodies for 30 min at 4 °C: APC Cy7 anti-CD3 (SP34.2; BD Biosciences), BV421 anti-CD4 (OKT4; BioLegend) and CD8-BV570 (clone RPA-T8; BioLegend). Following fixation and permeabilization, cells were stained with ECD anti-CD69 (clone TP1.55.3; Beckman Coulter), PE anti-IL-2 (MQ1-17H12; BD Biosciences), IFNγ-APC (B27; BD Biosciences) and FITC anti-TNF-α (Mab11; BD Biosciences) or GATA3, RORgT and Blimp1. The Aqua LIVE/DEAD kit (Invitrogen) was used to exclude dead cells. Samples were acquired on an LSRII flow cytometer and analyzed using FlowJo version 9.6.3 (Treestar, Inc.). Polyfunctional responses were analyzed using SPICE software v4.2.3 (provided by M. Roederer).
For the bronchoalveolar lavage and the rectal mucosa, cells were separated by Ficoll and enzymatic digestion, respectively, and stained with Alexa-Fluor 700 anti-CD3 (SP34-2; 557917, 0.2 mg ml−1) and PE-C594 anti-IFN-γ (B27; 562392, 5 µl) from BD Biosciences; V450 anti-TNF-α (Mab11; 502920, 5 µl), NKp44 (P44-8; 325110, 5 µl), PE-Cy7 anti-IL-2 (17H12; 500325, 50 µg ml−1) and 605NC anti-CD20 (2H7; 302334, 50 µg ml−1) from BioLegend; PE-APC eFlour 780 anti-IL-17 (eBio64DEC17; 47-7179, 0.125 mg), FITC anti-CD107a (eBioH4A3; 11-1079-42, 5 µl, 0.5 µg) and 605NC anti-CD8 (RPA-T8; 93-0088, 5 µl) from eBioscience; and PE anti-NKG2A (Z199; PN A60797, 5 µl) from Beckman Coulter.
Cellular RNA extraction and RT–PCR
Total RNA was extracted from sorted CD14+ and CD14− live cells from the blood of three naive non-vaccinated and three vaccinated animals in the DNA group after the first boost with ALVAC in combination with the gp120 alum. In samples with > 105 cells, the RNeasy Plus Mini Kit (Qiagen) and the QuantiTect Reverse Transcription Kit (Qiagen) were used per manufacturer’s instructions. For samples with < 105 cells, TRIzol reagent (Thermo Fisher Scientific) was used according to the manufacturer’s instructions. The concentration of the RNA was assessed using a Nanodrop Spectrophotometer. For quantitative PCR, primers specific for NLRP3 (5′ -AGCTGCCTCCTGCAGAACCT-3′ and 5′ -GGTCAGCTCAGGCTTTTCTTCT-3′), ARG1 (5′ -ATTGAGAAAGGCTGGTCTGC-3′ and 5′ -CATTAGGGATGTCAGCAAAGG-3′), CXCR4 (5′ -GCATCTGGAGAACCAGCG-3′ and 5′ -GAAACAGGGTTCCTTCATGG-3′) and TSG101 (5′ -CTTGAGAAGGGGCGTGATAGAC-3′ and 5′ - GCCCTCAGCTGGAACTGTTTAC-3′) were used. The relative expression of target genes was calculated using delta delta threshold cycles (ΔΔTC) as ratios between the target gene and the GAPDH housekeeping gene.
CCL2 and IL-18 detection in plasma
CCL2 plasma levels were measured using a Rhesus Macaque MCP-1 ELISA commercial kit (Sigma-Aldrich) per the manufacturer’s instructions. Briefly, 100 µl plasma were added in singlet to the coated plate and incubated for 2.5 h. The detection antibody was incubated for 1 h and the streptavidin solution for 45 min at room temperature. After each incubation period, the wells were washed four times with wash buffer. The tetramethyl benzidine solution was then added and incubated for 30 min. The reaction was stopped by adding the stop solution. The plate was immediately read in a photometric plate reader at 450 nm. Plasma IL-18 quantification was performed with the Human IL-18 ELISA Kit (Code 7620, Lot. 096FA, Medical & Biological Laboratories), following the manufacturer’s instructions. Plasma was diluted to a ratio of 1:2.
Luminex
Cells were transferred, washed and resuspended in R10 media at 2 × 106 cells per ml in a 96-well U-bottom plate. Cells were unstimulated or stimulated with Env peptide pool (final concentration of 2 µg ml−1) in the presence of purified mouse anti-human CD28 antibody, at a final concentration of 0.5 µg ml−1 (556620, BD Biosciences) for 48 h in R10 media (RPMI 1640, 10% FBS and 1× Anti-Anti). At the end of the incubation, the cells were pelleted by centrifuging at 1,000g at 4 °C, and supernatants were collected and stored at −80 °C.
Cryopreserved supernatants were analyzed using three MILLIPLEX Non-Human Primate Multiplex assays (EMD Millipore). The following targets were assayed following the manufacturer’s instructions: IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-13, IL-17, IFN-γ, MCP-1, MIP-1a (PRCYTOMAG-40K-11), IL-21, IL-22, IL-23, RANTES (PRCYT2MAG-40K-04), and TGF-β, TGF-β2 and TGF-β3 (TGFBMAG-64K). Briefly, after thawing the samples on ice, 25 µl of each supernatant was loaded into the well and mixed with 25 µl assay buffer and 25 µl magnetic beads. The plates were incubated under agitation at 4 °C for 18 h. After washing, 25 µl detection antibody was added to each well and incubated for 1 h at room temperature. Next, 25 µl streptavidin-PE were added to each well and incubated for 30 min at room temperature. Finally, wells were washed and 150 µl sheath fluid was added. Samples were acquired on a Bio-Plex 200 System (Bio-Rad).
SIV Env-specific IgG- and IgA-binding antibody assay
The SIV Env-specific IgG and IgA antibodies in the serum and the rectal mucosa were determined by custom SIV bAb multiplex assays (SIV-BAMA) as previously described9. Positive control for each antigen was tracked via Levey–Jennings charts. Antibodies against V1/V2 epitopes were quantified by binding assays against scaffolded SIV V1/V2 antigens expressed as gp70 fusion proteins produced in the same way as the CaseA2 V1/V2 antigen used in the RV144 correlate study (provided by A. Pinter, State University of New Jersey, USA)1. These proteins contained the glycosylated, disulfide-bonded V1/V2 regions of SIVmac239, SIVmac251 and SIVsmE660 (amino acids 120–204 of HXB2 Env), fused to residue 263 of the SU (gp70) protein of Fr-MuLV. Rectal swab samples were eluted, filtered and concentrated to ~600 µl each. All samples were inspected to be free of notable blood contamination. Total IgG and IgA concentrations were measured by a custom total macaque total IgG or IgA ELISA, with purified IgG from a SIV-infected macaque as the standard to calculate SIV antibody concentration. Specific activity for mucosal samples was calculated by the median fluorescence intensity (MFI; the linear range of the standard curve) × dilution per µg per ml total IgG.
Cyclic V2 IgG1-binding antibodies in the serum
To detect binding to cyclic V2, ELISA plates were coated overnight with 100 ng per well of streptavidin (Sigma) at 4 °C. Contents were dumped and 2.8 ng cyclic V2 in bicarbonate buffer were added to each well. This quantity was calculated to obtain the same number of epitopes per well as if each well contained 50 ng gp120, accounting for the molecular weight of the molecules (gp120 is 18-times heavier than cyclic V2). Cyclic V2 was incubated for binding to streptavidin for 1 h at 37 °C. The contents were again dumped and 300 µl of 1% PBS–BSA were added to each well. To detect binding to gp120, ELISA plates were coated overnight with 50 ng per well of monomeric SIVmac251-gp120 and blocked overnight at 4 °C. Following this, 40 µl of serial 1:2 dilutions of monkey plasma starting from 1:100 were dispensed in each well and incubated for 1 h at 37 °C. Each plate also included a 1:5 serial dilution of the monoclonal antibodies NCI05 and NCI09, starting with 20 µl ml−1 in the first two columns of each plate. Plates were washed using a plate washer and 40 µl of 1:5,000-diluted anti-monkey IgG1 + horseradish peroxidase (clone 7H11) were dispensed in each well and incubated for 1 h at 37 °C. After washing, 40 µl ELISA Ultra tetramethyl benzidine substrate (Thermo Fisher) were added to each well and incubated at room temperature for 10 min. The color development was stopped with the addition of 1 M sulfuric acid and read with a VICTOR-3 at 450 nm (Perkin Elmer). Plasma dilutions in the linear range of the curve were compared to the binding curve of a known anti-gp120 IgG1 standard to interpolate IgG1 plasma concentration values.
Reagents and surface plasmon resonance
Biotinylated cyclic SIV E543 and biotinylated cyclic Mac 251 V2 peptides were synthesized by JPT Peptide Technologies as previously described10. CM5 chips and the Biacore amine coupling kit were purchased from GE Healthcare. Surface plasmon resonance measurements were conducted with the Biacore 4000 (GE Healthcare) using the CM5 chip, as described previously17. Streptavidin (Invitrogen) was immobilized onto the chip (1 µM in 10 mM sodium acetate, pH 4.5; with 10 min contact time, 10 µl min−1 flow rate) using the amine coupling kit as directed by the immobilization wizard packaged within the Biacore 4000 control software to obtain a total of 4,000–4,700 reported response units (RU). The biotinylated peptide was allowed to flow (at 10 µl min−1) over the streptavidin-coated surface until 1,000–1,200 RU of the respective SIV cyclic V2 peptides were captured. The prepared mucosal samples (prepared by Duke University and sent to us) were diluted 10-fold in 10 mM HEPES, 300 mM NaCl, 3 mM EDTA and 0.005% Tween-20, and then analyzed on the Biacore 4000. The diluted mucosal samples (4 replicates for each sample) were passed over the chip surface at a flow rate of 10 µl min−1 for 3 min followed by a 45-s dissociation period. Affinity-purified γ-chain- or α-chain-specific goat anti-monkey IgG or IgA antibody (20 µg ml−1; Rockland Immunochemicals) was then passed over the peptide-coated Ig-bound surface for 150 s at a flow rate of 10 µl min−1. After a 70-s dissociation period, the chip surface was regenerated with 125 mM HCl followed by a 70-s injection of buffer. The RU for the V2 IgG- or IgA-specific values are the difference between the baseline after the first injection (diluted rectal samples) and the end of second injection (goat anti-monkey IgG or IgA). The data were double subtracted (reference spot and multi-buffer/negative-control injections). The data are presented as dot plots for individual mucosal samples and were analyzed using the BIA evaluation 4.1 software.
Total IgG or IgA antibodies in the rectal samples were measured using goat anti-monkey IgG or goat anti-monkey IgA immobilized (4,000–4,470 RU) on a CM5 sensor chip. Purified monkey IgG or IgA (0–32 µg ml−1 of each) or rectal samples (diluted 1:10; 8 replicates for each sample) were passed over the chip surface at a flow rate of 10 µl min−1 for 3 min, followed by a 45-s dissociation period. The chip was regenerated as described above. The relative amount of monkey IgG or IgA in the rectal sample was determined based on a standard curve that was generated using purified monkey IgG or IgA and four parameter fitting equations. Data analysis was performed as described above. Chemstrips were used to determine the blood contamination in mucosal samples. Mucosal supernatant samples (10 µl) were spotted onto a Chemstrip 5 OB Urine Test Strip (11893467-160, Roche) in accordance with the manufacturer’s color chart.
Neutralizing antibodies
Neutralization was measured as a reduction in luciferase reporter gene expression after a single round of infection in TZM-b1 cells as described previously67. TZM-b1 cells were obtained from the NIH AIDS Research and Reference Reagent Program as contributed by J. Kappes and X. Wu. Two hundred TCID50 of virus were incubated with serial 3-fold dilutions of test sample in duplicate, in a total volume of 150 µl for 1 out of 18 h at 37 °C, as instructed, in 96-well flat-bottom culture plates. Freshly trypsinized cells (10,000 cells in 100 µl of growth medium containing 75 µg ml−1 DEAE-dextran) were added to each well. One set of control wells received cells and virus (virus control) and another set received cells only (background control). Cells were transferred to 96-well black solid plates (Costar) for measurements of luminescence using the Britelite luminescence reporter gene assay system (PerkinElmer Life Sciences). Neutralization titers are defined as the dilution at which relative luminescence units were reduced by 50% compared to that in virus control wells after subtraction of background relative luminescence units. Assay stocks of molecularly cloned Env-pseudo-typed viruses (SIVmac251 CS.41, SIVmac239 0.23, SIVmac251 and WY:30) were prepared by transfection in 293T cells and were titrated in TZM-bl cells, as previously described68. Assay stocks of uncloned TCLA-SIVmac251 and SIVmac251 CS/2002 letvin were produced in H9 and human PBMCs, respectively, and were titrated in TZM-bl cells.
Bead phagocytosis
For all phagocytosis assays, 10 µm BB fluorescent polystyrene beads (Polysciences) were coated with SIV-gp120 (Advanced BioScience Laboratories) according to the manufacturer’s protocol. SIV-gp120-coated beads were then opsonized with experimental or control sera diluted 1:100 in serum-free RPMI for 1 h at 37 °C on a shaker. To measure autologous phagocytic activity, PBMCs from the same macaques whose sera were used to opsonize the beads were added at a ratio of 16 cells/1 bead in serum-free RPMI and incubated for 30 min at 37 °C on a shaker. Ice-cold PBS was added to stop the phagocytic reaction and cells were incubated for 15 min in Accutase (Stemcell) at 37 °C to remove surface-bound beads. Cells were then stained for monocyte-specific surface markers using the protocol outlined in Autissier et al.69 with modifications. Briefly, the cells were stained with Aqua or Yellow LIVE/DEAD Kit (Invitrogen) and then with the following antibody panel: PE-CD3 (SK7) and PE-CD20 (2H7) from BioLegend, and APC-CD14 (TÜK4), FITC-HLA-DR, -DP and -DQ (REA332) from Miltenyi Biotec in FACS buffer (10% FBS in PBS). Cells were fixed with 4% paraformaldehyde for 30 min at 4 °C and analyzed on the Acea Novocyte 3000 (Acea Biosciences). Monocytes were identified as CD14+HLA-DR+CD3−CD20−. A phagocytic score was calculated as follows: % bead positive monocytes × MFI bead positive monocytes. Each animal’s PBMCs were assayed twice in duplicate. To measure the serum phagocytic activity with THP-1 cells, the above protocol was replicated except that cells were incubated at a ratio of 2 THP-1 cells/1 bead, and cells were fixed after the Accutase wash without surface staining. THP-1 cells were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID and were routinely tested for mycoplasma contamination with a Sigma Mycoplasm PCR Kit. Because sera were assayed in multiple runs, results were made comparable by adjusting for the assay-to-assay variability using a positivecontrol serum. Each serum was assayed twice in triplicate.
Gene expression arrays analysis
Twelve macaques from the DNA prime arm and 11 (out of 12) macaques from the Ad26 prime arm were included in a gene expression profiling study. One macaque (R270) was not included for lack of samples for transcriptomic analysis. Microarray analysis was conducted using biotinylated cRNA hybridized to Human HT-12 version 4 BeadChips (Illumina). The arrays were scanned using iSCAN (Illumina) and quantified using Genome Studio (Illumina). Analysis of the Genome Studio output data was conducted using R/Bioconductor software packages. Bead arrays were read and missing values (> 0.01%) were imputed using the nearest-neighbor method as implemented in the R package impute. Quantile normalization and log2 transformation for variance stabilization were then applied to raw intensities.
The number of SIV challenges to infection of macaques that were not infected after 10 SIV challenges was recoded as 11. For each gene, a linear regression model with the number of SIV challenges to infection as an independent variable and gene expression as a dependent variable was fit using the R package LIMMA70, and a moderated t-test was used to test that the coefficient of regression was statistically different from 0. The Benjamini–Hochberg method was used to correct the P values for multiple testing (adjusted P values). Genes with an adjusted P value below 5% were defined as differentially expressed genes. GSEA was used to evaluate the gene sets (that is, pathways) associated with the number of SIV challenges to infection71.
In GSEA, the most variable probes across samples were used to remove redundant probes annotated to the same gene. Genes were preranked by LIMMA t statistic, and GSEA was used to assess the enrichment of gene sets from the Molecular Signatures Database gene sets (version 5.1) and blood cell markers72. The GSEA Java desktop program was downloaded from the Broad Institute (http://www.broadinstitute.org/gsea/index.jsp) and used with GSEA Pre-Ranked module parameters (number of permutations: 1,000; enrichment statistic: weighted; seed for permutation: 111; 15 ≤ gene set size ≤ 2,000). Sample-level enrichment analysis was used to investigate the enrichment of pathways in the different samples73. Briefly, the expression of all the genes in a specific pathway were averaged across samples and compared to the average expression of 1,000 randomly generated gene sets of the same size. The resulting Z score was then used to reflect the overall perturbation of a pathway in a sample.
GeneMANIA version 3.3.1 was used to infer gene networks and to identify relations (co-localization, co-expression and pathways) between genes of interest74. To that end, the human orthologs and homologs of the macaques’ genes included in the classifier were obtained from the NCBI Gene and homoloGene portal. The human homologs were then imported into GeneMANIA, and a network was generated with equal weight of the network and no added nodes to consolidate the network as parameters. Motif discovery in a 2-kb window around the transcription start site of genes of interest was done using the application HOMER version 4.9 using default parameters75.
Statistical analysis
The number of challenges before viral acquisition was assessed using the log-rank (Mantel–Cox) test of the discrete-time proportional hazards model. The Kruskal–Wallis test (non-parametric ANOVA) was used to determine differences between three independent samples of equal or different sample sizes and was corrected for multiple comparisons. The non-parametric Dunn’s multiple comparison test was applied to determine which specific means were significant from the others.
The Mann–Whitney–Wilcoxon non-parametric, two-tailed test was used to compare continuous factors between two matched samples. Correlation analysis was performed using the non-parametric Spearman rank correlation method (two-tailed, 95% confidence) using exact permutation P values. The Fisher combined test was performed to assess the significance of the correlation across microarray data sets.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Code availability
All the source codes used are available at https://bitbucket.org/fouslim/v43dna.
Data availability
The microarray data have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GEO: GSE108011.
Supplementary Material
Acknowledgments
We thank D. Ahern for editorial and graphical support and all of the staff at Advanced BioScience Laboratories for helping with the execution of the animal study. We thank J. Lucas, J. Peel and Y. Lin for specific binding and total antibody assays and G. Overman and N. Yates for assay and technical assistance. We thank D. Barouch (Harvard Medical School) for providing the Ad26–SIV recombinant vaccine. This work was mostly supported with federal funds from the intramural program of the National Cancer Institute, NIH, including contract no. HHSN261200800001E (G.F.). Contributions were made by the extramural NIAID program (HHSN27201100016C; D.M.), the Henry M. Jackson Foundation, the US Department of Defense and the Collaboration for Aids Vaccine Discovery (CAVD) grants OPP1032325 (R.A.K.) and OPP1147555 (R.A.K.) from the Bill and Melinda Gates Foundation. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Footnotes
Author contributions
G.F. designed and coordinated the study with M.V. and S.N.G., interpreted the data and wrote the manuscript. S.F. and R.P.S. analyzed the gene expression data, performed the correlates of risk analyses, prepared the figures and helped to write the manuscript. D.R.B. performed the flow cytometry for monocytes in the blood. M.V. performed the studies on TH cell types and analyzed the antibody data and prepared some of the figures together with S.N.G. M.B., L.S., I.S.d.C. M.N.D. performed the analysis of cytokines in the serum. V.G., M.O. and D.F. performed the PCR analysis and RNA extraction. G.G. and N.P.M.L. measured the serum IgG titers and NK cells in the mucosa. H.V.T. and M. Rao measured and analyzed responses to cyclic V2. K.M.M. helped with sorting of monocytes. K.E.F., M. Roederer and R.A.K. performed the intracellular cytokine analysis. B.F.K. measured the number of transmitted variants. X.S., G.D.T., M.P.W., K.J.M., J.S.G., D.N.F., D.C.M. and M. Rosati studied humoral responses in the serum. D.J.V. assisted with statistical analyses. B.K.F., M. Rosati and G.N.P. provided the SIV DNAs.
Competing interests
The US Government in conjunction with Sanofi Pasteur holds Patent 5766598: A Recombinant Attenuated ALVAC Canarypox virus Expression Vectors Containing Heterologous DNA Segments Encoding Lentiviral Gene, inventors E. Paoletti, J. Tartaglia and W. I. Cox, issued 16 June 1998, for the ALVAC vaccine. The US Government also holds Patent 7094408: Improved Immunogenicity Using a Combination of DNA and Vaccinia Virus Vector Vaccines, inventors G. Franchini, Z. Hel and G. Pavlakis, issued 22 August 2006. This patent is for the combination DNA and ALVAC poxvirus vaccines.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41591-018-0025-7.
References
- 1.Pitisuttithum P, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J. Infect. Dis. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 2.Gray GE, et al. Safety and efficacy of the HVTN 503/Phambili study of a clade-B-based HIV-1 vaccine in South Africa: a double-blind, randomised, placebo-controlled test-of-concept phase 2b study. Lancet Infect. Dis. 2011;11:507–515. doi: 10.1016/S1473-3099(11)70098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchbinder SP, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammer SM, et al. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N. Engl. J. Med. 2013;369:2083–2092. doi: 10.1056/NEJMoa1310566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 6.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zolla-Pazner S, et al. Vaccine-induced IgG antibodies to V1V2 regions of multiple HIV-1 subtypes correlate with decreased risk of HIV-1 infection. PLoS ONE. 2014;9:e87572. doi: 10.1371/journal.pone.0087572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin L, et al. COMPASS identifies T-cell subsets correlated with clinical outcomes. Nat. Biotechnol. 2015;33:610–616. doi: 10.1038/nbt.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomaras GD, et al. Vaccine-induced plasma IgA specific for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG. Proc. Natl Acad. Sci. USA. 2013;110:9019–9024. doi: 10.1073/pnas.1301456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaccari M, et al. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat. Med. 2016;22:762–770. doi: 10.1038/nm.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaccari M, et al. Reduced protection from simian immunodeficiency virus SIVmac251 infection afforded by memory CD8+ T cells induced by vaccination during CD4+ T-cell deficiency. J. Virol. 2008;82:9629–9638. doi: 10.1128/JVI.00893-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baden LR, et al. Induction of HIV-1-specific mucosal immune responses following intramuscular recombinant adenovirus serotype 26 HIV-1 vaccination of humans. J. Infect. Dis. 2015;211:518–528. doi: 10.1093/infdis/jiu485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi LZ, et al. HIF1α-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winning S, Fandrey J. Dendritic cells under hypoxia: how oxygen shortage affects the linkage between innate and adaptive immunity. J. Immunol. Res. 2016;2016:5134329. doi: 10.1155/2016/5134329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 16.Strbo N, et al. Cutting edge: novel vaccination modality provides significant protection against mucosal infection by highly pathogenic simian immunodeficiency virus. J. Immunol. 2013;190:2495–2499. doi: 10.4049/jimmunol.1202655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pegu P, et al. Antibodies with high avidity to the gp120 envelope protein in protection from simian immunodeficiency virus SIVmac251 acquisition in an immunization regimen that mimics the RV-144 Thai trial. J. Virol. 2013;87:1708–1719. doi: 10.1128/JVI.02544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carr MW, et al. Monocyte chemoattractant protein 1 acts as a T-lymphocyte chemoattractant. Proc. Natl Acad. Sci. USA. 1994;91:3652–3656. doi: 10.1073/pnas.91.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yago T, et al. IL-23 and Th17 disease in inflammatory arthritis. J. Clin. Med. 2017;6:81. doi: 10.3390/jcm6090081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chong SZ, et al. CXCR4 identifies transitional bone marrow premonocytes that replenish the mature monocyte pool for peripheral responses. J. Exp. Med. 2016;213:2293–2314. doi: 10.1084/jem.20160800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao W, et al. Sodium salicylate modulates inflammatory responses through AMP-activated protein kinase activation in LPS-stimulated THP-1 cells. J. Cell. Biochem. 2018;119:850–860. doi: 10.1002/jcb.26249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung YH, Kim DH, Lee WW. Monosodium urate crystal-induced pro-interleukin-1β production is post-transcriptionally regulated via the p38 signaling pathway in human monocytes. Sci. Rep. 2016;6:34533. doi: 10.1038/srep34533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan S, et al. The eIF4E/eIF4G interaction inhibitor 4EGI-1 augments TRAIL-mediated apoptosis through c-FLIP down-regulation and DR5 induction independent of inhibition of cap-dependent protein translation. Neoplasia. 2010;12:346–356. doi: 10.1593/neo.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kung CP, Raab-Traub N. Epstein–Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor through effects on Bcl-3 and STAT3. J. Virol. 2008;82:5486–5493. doi: 10.1128/JVI.00125-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwissa M, et al. Dengue virus infection induces expansion of a CD14+CD16+ monocyte population that stimulates plasmablast differentiation. Cell Host Microbe. 2014;16:115–127. doi: 10.1016/j.chom.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludtke A, et al. Ebola virus disease is characterized by poor activation and reduced levels of circulating CD16+ monocytes. J. Infect. Dis. 2016;214:S275–S280. doi: 10.1093/infdis/jiw260. [DOI] [PubMed] [Google Scholar]
- 28.Yu Q, et al. Comparative analysis of tropism between canarypox (ALVAC) and vaccinia viruses reveals a more restricted and preferential tropism of ALVAC for human cells of the monocytic lineage. Vaccine. 2006;24:6376–6391. doi: 10.1016/j.vaccine.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Ignatius R, et al. Canarypox virus–induced maturation of dendritic cells is mediated by apoptotic cell death and tumor necrosis factor alpha secretion. J. Virol. 2000;74:11329–11338. doi: 10.1128/jvi.74.23.11329-11338.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandes-Alnemri T, et al. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 32.Rao SP, et al. Human peripheral blood mononuclear cells exhibit heterogeneous CD52 expression levels and show differential sensitivity to alemtuzumab mediated cytolysis. PLoS ONE. 2012;7:e39416. doi: 10.1371/journal.pone.0039416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivino L, et al. Chemokine receptor expression identifies pre-T helper (Th)1, pre-Th2, and nonpolarized cells among human CD4+ central memory T cells. J. Exp. Med. 2004;200:725–735. doi: 10.1084/jem.20040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auclair SL, et al. Distinct susceptibility of HIV vaccine vector-induced CD4 T cells to HIV infection. PLoS Pathog. 2018;14:e1006888. doi: 10.1371/journal.ppat.1006888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossol M, et al. The CD14brightCD16+ monocyte subset is expanded in rheumatoid arthritis and promotes expansion of the Th17 cell population. Arthritis Rheum. 2012;64:671–677. doi: 10.1002/art.33418. [DOI] [PubMed] [Google Scholar]
- 37.Zhu H, et al. CD16+ monocyte subset was enriched and functionally exacerbated in driving T-cell activation and B-cell response in systemic lupus erythematosus. Front. Immunol. 2016;7:512. doi: 10.3389/fimmu.2016.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eisenbarth SC, et al. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu F, et al. Priming and activation of inflammasome by canarypox virus vector ALVAC via the cGAS/IFI16–STING–type I IFN pathway and AIM2 sensor. J. Immunol. 2017;199:3293–3305. doi: 10.4049/jimmunol.1700698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suschak JJ, et al. Identification of Aim2 as a sensor for DNA vaccines. J. Immunol. 2015;194:630–636. doi: 10.4049/jimmunol.1402530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teigler JE, Iampietro MJ, Barouch DH. Vaccination with adenovirus serotypes 35, 26, and 48 elicits higher levels of innate cytokine responses than adenovirus serotype 5 in rhesus monkeys. J. Virol. 2012;86:9590–9598. doi: 10.1128/JVI.00740-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teigler JE, et al. The canarypox virus vector ALVAC induces distinct cytokine responses compared to the vaccinia virus-based vectors MVA and NYVAC in rhesus monkeys. J. Virol. 2014;88:1809–1814. doi: 10.1128/JVI.02386-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitroulis I, et al. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell. 2018;172:147–161.e12. doi: 10.1016/j.cell.2017.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng SC, et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Netea MG, et al. Trained immunity: a program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saeed S, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghonime MG, et al. Inflammasome priming by lipopolysaccharide is dependent upon ERK signaling and proteasome function. J. Immunol. 2014;192:3881–3888. doi: 10.4049/jimmunol.1301974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panchanathan R, Liu H, Choubey D. Hypoxia primes human normal prostate epithelial cells and cancer cell lines for the NLRP3 and AIM2 inflammasome activation. Oncotarget. 2016;7:28183–28194. doi: 10.18632/oncotarget.8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu L, et al. Control of TH2 polarization by the chemokine monocyte chemoattractant protein-1. Nature. 2000;404:407–411. doi: 10.1038/35006097. [DOI] [PubMed] [Google Scholar]
- 50.Cecchinato V, et al. Altered balance between Th17 and Th1 cells at mucosal sites predicts AIDS progression in simian immunodeficiency virus-infected macaques. Mucosal Immunol. 2008;1:279–288. doi: 10.1038/mi.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fouts TR, et al. Balance of cellular and humoral immunity determines the level of protection by HIV vaccines in rhesus macaque models of HIV infection. Proc. Natl Acad. Sci. USA. 2015;112:E992–E999. doi: 10.1073/pnas.1423669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qureshi H, et al. Low-dose penile SIVmac251 exposure of rhesus macaques infected with adenovirus type 5 (Ad5) and then immunized with a replication-defective Ad5-based SIV gag/pol/nef vaccine recapitulates the results of the phase IIb step trial of a similar HIV-1 vaccine. J. Virol. 2012;86:2239–2250. doi: 10.1128/JVI.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reinhardt-Heller K, et al. Increase of intermediate monocytes in graft-versus-host disease: correlation with MDR1+ Th17.1 levels and the effect of prednisolone and 1α,25-dihydroxyvitamin D3. Biol. Blood Marrow Transplant. 2017;23:2057–2064. doi: 10.1016/j.bbmt.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 54.Joubert PE, et al. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009;6:354–366. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez-Rocha H, et al. Adenoviruses induce autophagy to promote virus replication and oncolysis. Virology. 2011;416:9–15. doi: 10.1016/j.virol.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi CS, et al. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat. Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Byrne BG, et al. Inflammasome components coordinate autophagy and pyroptosis as macrophage responses to infection. mBio. 2013;4:e00620–12. doi: 10.1128/mBio.00620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barouch DH, et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012;482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strickland SL, et al. Significant genetic heterogeneity of the SIVmac251 viral swarm derived from different sources. AIDS Res. Hum. Retroviruses. 2011;27:1327–1332. doi: 10.1089/aid.2011.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stott EJ. Anti-cell antibody in macaques. Nature. 1991;353:393. doi: 10.1038/353393a0. [DOI] [PubMed] [Google Scholar]
- 61.Barouch DH, et al. Characterization of humoral and cellular immune responses elicited by a recombinant adenovirus serotype 26 HIV-1 Env vaccine in healthy adults (IPCAVD 001) J. Infect. Dis. 2013;207:248–256. doi: 10.1093/infdis/jis671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosati M, et al. Increased immune responses in rhesus macaques by DNA vaccination combined with electroporation. Vaccine. 2008;26:5223–5229. doi: 10.1016/j.vaccine.2008.03.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keele BF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl Acad. Sci. USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keele BF, et al. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 2009;206:1117–1134. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Romano JW, et al. NASBA technology: isothermal RNA amplification in qualitative and quantitative diagnostics. Immunol. Invest. 1997;26:15–28. doi: 10.3109/08820139709048912. [DOI] [PubMed] [Google Scholar]
- 66.Vaccari M, et al. Vaccine-induced CD8+ central memory T cells in protection from simian AIDS. J. Immunol. 2005;175:3502–3507. doi: 10.4049/jimmunol.175.6.3502. [DOI] [PubMed] [Google Scholar]
- 67.Schiffner T, et al. Immune focusing and enhanced neutralization induced by HIV-1 gp140 chemical cross-linking. J. Virol. 2013;87:10163–10172. doi: 10.1128/JVI.01161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Autissier P, Soulas C, Burdo TH, Williams KC. Immunophenotyping of lymphocyte, monocyte and dendritic cell subsets in normal rhesus macaques by 12-color flow cytometry: clarification on DC heterogeneity. J. Immunol. Methods. 2010;360:119–128. doi: 10.1016/j.jim.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gundem G, Lopez-Bigas N. Sample-level enrichment analysis unravels shared stress phenotypes among multiple cancer types. Genome Med. 2012;4:28. doi: 10.1186/gm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Montojo J, et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GEO: GSE108011.