Abstract
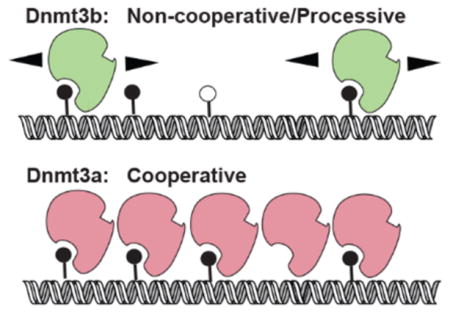
The catalytic domains of the de novo DNA methyltransferases Dnmt3a-C and Dnmt3b-C are highly homologous. However, their unique biochemical properties could potentially contribute to differences in the substrate preferences or biological functions of these enzymes. Dnmt3a-C forms tetramers through interactions at the dimer interface, which also promote multimerization on DNA and cooperativity. Similar to the case for processive enzymes, cooperativity allows Dnmt3a-C to methylate multiple sites on the same DNA molecule; however, it is unclear whether Dnmt3b-C methylates DNA by a cooperative or processive mechanism. The importance of the tetramer structure and cooperative mechanism is emphasized by the observation that the R882H mutation in the dimer interface of DNMT3A is highly prevalent in acute myeloid leukemia and leads to a substantial loss of its activity. Under conditions that distinguish between cooperativity and processivity, we show that in contrast to that of Dnmt3a-C, the activity of Dnmt3b-C is not cooperative and confirm the processivity of Dnmt3b-C and the full length Dnmt3b enzyme. Whereas the R878H mutation (mouse homologue of R882H) led to the loss of cooperativity of Dnmt3a-C, the activity and processivity of the analogous Dnmt3b-C R829H variant were comparable to those of the wild-type enzyme. Additionally, buffer acidification that attenuates the dimer interface interactions of Dnmt3a-C had no effect on Dnmt3b-C activity. Taken together, these results demonstrate an important mechanistic difference between Dnmt3b and Dnmt3a and suggest that interactions at the dimer interface may play a limited role in regulating Dnmt3b-C activity. These new insights have potential implications for the distinct biological roles of Dnmt3a and Dnmt3b.
Graphical Abstract
In mammals, DNA methylation at the C5 position of cytosine bases is catalyzed by three known DNA methyltransferases (Dnmts): Dnmt1, Dnmt3a, and Dnmt3b. Although mammalian Dnmts use a conserved catalytic mechanism that involves base flipping to methylate largely at CpG dinucleotides, these enzymes are structurally and functionally distinct.1–4 Dnmt1 is primarily responsible for the postreplication maintenance of DNA methylation by copying it from the parent to daughter strand.5 Dnmt3a and Dnmt3b, together with their catalytically inactive homologue, Dnmt3L, establish DNA methylation de novo.2 Dnmt3a and Dnmt3b have different functions, supported by the distinct phenotypes of their respective murine knockouts,6 their tissue specific expression patterns, and their unique roles in the development of cancer and other epigenetic disorders.7–14 However, the distinct biochemical properties of Dnmt3a and Dnmt3b that potentially contribute to their unique roles in vivo have not been fully examined.
A common feature of all mammalian Dnmts is the presence of a C-terminal catalytic domain that contains 10 motifs conserved among prokaryotic and eukaryotic C5 DNA methyltransferases. The C-terminal catalytic domains of Dnmt3a and Dnmt3b (Dnmt3a-C and Dnmt3b-C, respectively) share approximately 85% sequence similarity and are active methyltransferases in the absence of their respective N-terminal regulatory domains.15 Another member of the Dnmt3 family is Dnmt3L, in which the key catalytic residues required for binding of AdoMet and DNA are absent; Dnmt3L is therefore enzymatically inactive. Dnmt3L interacts with both Dnmt3a and Dnmt3b and allosterically stimulates their catalytic activity, therefore acting as a positive regulator of de novo DNA methylation.16,17 The crystal structure of the Dnmt3a catalytic domain (Dnmt3a-C) alone and complexed with Dnmt3L exhibits a heterotetrameric structure of Dnmt3a-C/3L or a homodimer of Dnmt3a-C that can self-tetramerize through two interaction surfaces, the R–D dimer interface and the F–F tetramer interface.18,19 The interactions at the dimer interface of Dnmt3a-C tetramers support its multimerization on DNA forming nucleoprotein filaments. This allows the enzyme to bind and methylate multiple CpG sites on DNA in a cooperative manner, thus increasing its activity.20–22 Although controversial, some evidence also supports a processive mechanism for Dnmt3a.15,23–25 Similar to the case for a processive enzyme, cooperative binding of multiple Dnmt3a molecules to DNA would allow it to methylate multiple sites on the same DNA molecule, complicating data analysis. However, the processivity of Dnmts is defined by their ability to diffuse along the DNA and methylate multiple sites before dissociation.26,27 Therefore, it is important to use methods that differentiate processivity and cooperative activity of Dnmts. Previous studies have shown that Dnmt3a-C methylates DNA in a distributive manner. Supporting its cooperative mechanism, its activity was shown to increase exponentially with a 2-fold increase in enzyme concentration, however only with long DNA substrates.24,28 The critical role of the tetramer formation and cooperative mechanism of Dnmt3a in its biological activity is apparent in acute myeloid leukemia (AML) cells that have a high frequency of mutation R882H present at the dimer interface of DNMT3A.27 In vitro studies show that the R882H mutant protein has only ~20% activity29,30 and is suggested to cause genomewide hypomethylation in AML cells.31,32
Multiple-sequence alignment reveals that most residues in the dimer interface are conserved between Dnmt3a and Dnmt3b. However, the role of the dimer interface and that of cooperativity of Dnmt3b-C has not been evaluated. An earlier investigation using methylation-dependent restriction protection analysis of a 430-mer DNA showed that Dnmt3b-C methylates its substrate in a processive manner.15 However, it is not clear from this study whether the activity of Dnmt3b-C was influenced by cooperativity in a manner similar to that reported for Dnmt3a-C. In the study presented here, we performed experiments that distinguish between cooperative and processive mechanisms. Our data show that in contrast to Dnmt3a-C, Dnmt3b-C methylates DNA in a noncooperative manner and by a processive mechanism on both unmethylated and hemimethylated substrates at physiological pH and ionic strength and confirms the processivity of the full length Dnmt3b enzyme. Experiments further characterizing the catalytic mechanism of Dnmt3b-C show that whereas preincubation of the enzyme with DNA reduces its activity, preincubation with AdoMet decreases its KM by 10-fold, indicating that the cofactor AdoMet-bound form has an increased specificity for target sites. To further confirm the noncooperative mechanism of Dnmt3b-C, we tested the role of the R829 residue of Dnmt3b-C that is analogous to dimer interface residue R882 of Dnmt3a-C. Our data here show that whereas the reduced activity of the Dnmt3a-C R882H variant could be partially attributed to the loss of its cooperativity, mutation of the conserved analogous arginine R829 had no effect on the activity or processivity of Dnmt3b-C, thus supporting its noncooperative mechanism. Similarly, disruption of interactions at the dimer interface of Dnm3a-C, with a decrease in the pH of buffer, causes a 2–3-fold loss of Dnmt3a-C activity;24,28 however, it has no effect on the DNA binding or catalytic activity of Dnmt3b-C. These data suggest that formation of tetramers may not be critical for the processive activity of Dnmt3b-C. Collectively, these data reveal important biochemical differences between Dnmt3a and Dnmt3b that can potentially impact their activity and function during development and in cancer cells.
EXPERIMENTAL PROCEDURES
Protein Purification
Mouse Dnmt3a-C, Dnmt3b-C, Dnmt3b-C E703A, Dnmt3a-C R878H, Dnmt3b-C R829H, and Dnmt3L cloned in pET28a with a six-His tag were expressed and purified using affinity chromatography as described previously.25 Briefly, transformed BL21-DE3 cells were induced with 1 mM IPTG and grown for 2 h at 32 °C. Harvested cells were washed with STE buffer [10 mM Tris-HCl (pH 8.0), 0.1 mM EDTA, and 0.1 M NaCl] and resuspended in buffer A [20 mM potassium phosphate (pH 7.5), 0.5 M NaCl, 10% (v/v) glycerol, 1 mM EDTA, 0.1 mM DTT, and 80 mM imidazole]. Cells were disrupted by sonication, followed by removal of cell debris by centrifugation. The clarified lysate was incubated with 0.75 mL of Ni-NTA agarose for 3 h at 4 °C. The protein-bound slurry was packed in a 2 mL Bio-Rad column and washed with 50 mL of buffer A. Protein (5–10 μM) was eluted using 200 mM imidazole in buffer A at pH 7.5 and then stored in 20 mM HEPES (pH 7.5), 40 mM KCl, 0.2 mM DTT, 1 mM EDTA, and 20% glycerol at –80 °C. The purity and integrity of recombinant proteins were checked by SDS–PAGE and Western blot analysis using a mouse monoclonal anti-His antibody (Invitrogen, MA1-21315). M.SssI methyltransferase was obtained from New England Biolabs.
Enzyme Assays
Most methylation assays for determining kinetic parameters of recombinant enzymes were performed using 3H-labeled S-adenosylmethionine (AdoMet) as a cofactor and biotinylated oligonucleotides of varying sizes bound on avidin-coated high-binding Elisa plates (Corning) as described previously.33 The DNA methylation reaction was performed using either 250 nM 30-mer/32-mer DNA substrate or 150 nM 509-mer/719-mer DNA substrate in methylation buffer [20 mM HEPES (pH 7.5), 50 mM KCl, and 1 mM EDTA supplemented with 5 μg/mL BSA]. The methylation reaction included 0.76 μM [methyl-3H]AdoMet (PerkinElmer Life Sciences). Storage buffer was added to compensate for the different enzyme volumes in all reaction mixtures. The substrates consisted of biotinylated oligonucleotides, including (1) a 30 bp oligonucleotide containing one CpG site, (2) a 32 bp oligonucleotide containing two CpG sites, (3) a 509-mer with 58 CpGs (amplified from the human SUHW1 gene promoter), or (4) a 719-mer DNA fragment with 46 CpGs (amplified from the mouse Aprt gene promoter). Primers used to amplify these substrates are listed in Table 1.
Table 1.
DNA Oligomers Used for DNA Methylation Analysisa
| oligomer | sequence | no. of CpG sites |
|---|---|---|
| 30-mer F | /5BiosG/GAAGCTGGGACTTCCGGGAGGAGAGTGCAA | 1 |
| 30-mer R | TTGCACTCTCCTCCCGGAAGTCCCAGCTTC | 1 |
| 32-mer F | /5BiosG/TGGGACTTCCGGGAGCTTCCGGGAGGAGAGTG | 2 |
| 32-mer R | CACTCTCCTCCCGGAAGCTCCCGGAAGTCCCA | 2 |
| 509-mer F | /5BiosG/AGATTAGGGAAGGGGGTGTG | 58 |
| 509-mer R | AAGATCCTTTCAAGGCCTCAG | 58 |
| 719-mer F | /5BiosG/CCCATGCGCCTGCGCCGGGTGCC | 47 |
| 719-mer R | ATGCTCTAGACCTGCGATGTAGTTCGATC | 47 |
For the DNA binding experiments, the 30-mer oligomers used Cy5 5′ end labels in place of the biotin modification (/5BiosG/).
For steady state kinetic analysis, the larger amounts of the substrate could not be accommodated in the standard plate assay, which has a maximal binding capacity per well for DNA substrate of 2 μM. These methylation assays were, therefore, performed using a filter binding assay.10 Briefly, 10 μL of reaction mix was spotted on a 1.5 cm DE81 filter that was then washed three to five times in 0.2 M ammonium bicarbonate, followed by 100% ethanol, and air-dried. Incorporated radioactivity was quantified by scintillation counting.
Cooperativity Assay
To examine cooperativity, the following changes were made to the plate assay described above. Increasing concentrations of enzyme were preincubated with DNA substrate for 10 min at room temperature prior to the addition of AdoMet to start the reaction. AdoMet was a mixture of unlabeled and 0.76 μM 3H-labeled AdoMet, which yielded a final concentration of 2 μM. Methylation assays were performed using 30-mer, 509-mer, and 719-mer DNA substrates. On the basis of the structural studies of Dnmt3a-C, it was estimated that the 30-mer substrate is too short to allow potential multimerization of the enzyme18,21 and could therefore be used as a control. Additionally, cooperativity assays using 100 ng of the pUC19 plasmid as a substrate were performed using the filter binding assay described above. For these assays, unmethylated pUC19 was purified from the dam−/dcm− Escherichia coli strain (C2925I, NEB).
Stimulation of Dnmt3b-C by the Inactive Mutant E703A
For assays aiming to investigate the ability of an inactive mutant to stimulate wild-type (WT) Dnmt3b-C activity, the activity of a 1:1 μM mixture of Dnmt3b-C E703A mutant and WT Dnmt3b-C was compared to the activity of WT Dnmt3b-C (1 or 2 μM), using two different DNA substrates, 1 μM 30-mer substrate with 1 CpG or 150 nM 509-mer substrate with 58 CpGs. Reactions were initiated by addition of enzyme. The WT and/or E703A enzymes were mixed together and incubated for 5 min at room temperature before addition to the reaction mix. A mixture of unlabeled and 0.76 μM 3H-labeled AdoMet was used at a final concentration of 2 μM. Methylation rates were determined by using linear regression to analyze data. The fold change in methylation rate compared to that for 1 μM WT enzyme was plotted for each enzyme treatment.
Processivity Assay
Methylation kinetic analyses were performed using a range of enzyme concentrations and short oligonucleotide 30- and 32-mer substrates with one and two CpG sites, respectively. Low enzyme concentrations relative to DNA substrate concentrations were included to ensure that the reaction occurred under multiple-turnover conditions. Each DNA substrate was used at a concentration of 250 nM, and a 1:1 ratio of labeled and unlabeled AdoMet (final concentration of 1.5 μM) was used. To compare the identical CpG molarity, a parallel reaction using 500 nM 30-mer was also performed. Reactions were started by enzyme addition. M.SssI and Dnmt3a-C were used as positive and negative controls, respectively.
DNA Binding Assays
DNA binding of Dnmt3b-C was performed using Cy5-labeled 30-mer DNA containing one CpG site (Table 1) in nitrocellulose filter binding assays. Binding reactions were performed in 20 mM HEPES (pH 7.5), 100 mM KCl, 1 mM EDTA buffer, in the presence of 0.2 mM S-adenosylhomocysteine (AdoHcy) and 30 nM DNA, and increasing concentrations of Dnmt3b-C. Reaction mixtures were incubated at room temperature for 15 min prior to being spotted on the nitrocellulose membrane in a dot blot apparatus, followed by three washes with binding buffer. The enzyme-bound fraction of Cy5-DNA was quantified by fluorescence measurement (Typhoon).
Data Analysis
Data were analyzed using Prism software (GraphPad). For time-dependent kinetic measurements, values were fitted using linear regression of a nonlinear fit, which was weighted by 1/Y2. Each time point for methylation kinetics was an average and standard deviation of three to six experimental replicates. For secondary plots, a least-squares fitting method was used to plot the data, and the linear regression was not weighted. To determine the equilibrium binding constant, data were fitted to a one-site binding model with a Hill coefficient. Standard errors of the mean were calculated for three to six independent experiments, as described in the figure legends.
RESULTS
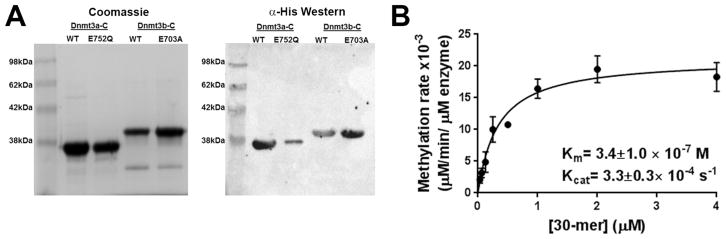
N-Terminally His-tagged Dnmt3b-C, Dnmt3b-C E703A, and Dnmt3L were overexpressed and purified on Ni-NTA agarose to 90–95% purity as estimated by the Coomassie-stained SDS gel (Figure 1A). The KM and the turnover rate constant (Kcat) value for purified Dnmt3b-C were determined to be (3.4 ± 1) × 10−7 M and (3.3 ± 0.3) × 10−4 s−1, respectively (Figure 1B), and catalytic efficiency Kcat/KM ~ 103 M−1 s−1. To confirm the catalytic robustness of the enzyme, we assessed the allosteric activation of Dnmt3b-C by Dnmt3L.16 Consistent with earlier observations, we observed ~6-fold activation of Dnmt3b activity in the presence of Dnmt3L16 (SI Figure 1A). This Dnmt3b-C protein was next used to determine the catalytic mechanism.
Figure 1.
Expression of Dnmt3a-C, Dnmt3b-C, and their respective mutants. (A) Coomassie-stained SDS–PAGE gel and Western blot showing purified His-tagged WT Dnmt3a-C, inactive mutant Dnmt3a-C E752Q, WT Dnmt3b-C, and inactive mutant Dnmt3b-C E703A. The mouse monoclonal anti-His antibody was used to detect proteins on the Western blot. (B) Steady state kinetic analysis of Dnmt3b-C activity. Methylation reactions were performed for 10 min, and initial velocities were calculated as the number of methyl groups transferred per minute per micromolar enzyme for varying concentrations of 30-mer substrate ranging from 0.031 to 4 μM in the presence of 0.75 μM AdoMet. The reactions were started by addition of enzyme to the substrate cocktail. The data were fitted to the Michaelis–Menten equation to yield the kinetic parameters. The data are the average ± the standard error of the mean (n ≥ 3 independent experiments).
Investigating the Cooperativity of Dnmt3b-C
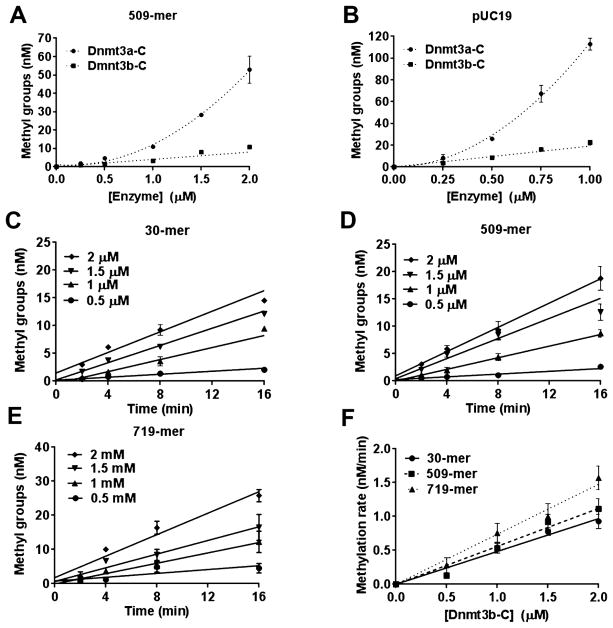
Dnmt3a-C monomers interact with each other through two interfaces, the Dnmt3a–Dnmt3L interface (F–F interface) and the Dnmt3a–Dnmt3a dimer interface (R–D interface), to form tetramers.18 Dnmt3a-C tetramers multimerize on DNA potentially through interactions along the dimer interface, which supports cooperativity of Dnmt3a.24 Many amino acids known to be critical for the dimer and tetramer interface-mediated interactions of Dnmt3a are conserved in Dnmt3b.18,19 However, the cooperativity of Dnmt3b has not been tested. The activity of a cooperative enzyme like Dnmt3a increases in a nonlinear manner at higher enzyme concentrations because of the allosteric effect of enzyme subunit interactions,24 in contrast to a noncooperative enzyme for which the activity is expected to increase in a linear manner as a function of enzyme concentration. Previous studies showing cooperativity of Dnmt3a-C used a 509 bp DNA substrate to allow the binding of multiple protein molecules at higher enzyme concentrations.24 To compare the cooperativity of Dnmt3a-C and Dnmt3b-C, DNA methylation analysis was performed using the same 509-mer DNA substrate. Increasing concentrations of the Dnmt3a-C and Dnmt3b-C enzymes ranging from 0 to 2 μM were preincubated with 509-mer DNA substrate for 10 min to allow the formation of nucleoprotein complexes. The methylation reaction was initiated by the addition of radio-labeled AdoMet and was monitored by incorporation of radioactivity into DNA for an additional 10 min. The methylation activity was plotted against Dnmt3a-C and Dnmt3b-C enzyme concentrations (Figure 2A). The data show that for Dnmt3a-C with an increase in concentration from 1 to 2 μM the activity increased ~5-fold, whereas for Dnmt3b-C, there was only ~2-fold increase in activity for every 2-fold increase in enzyme concentration. As shown previously,24 this exponential increase in methylation activity of Dnmt3a-C was specific for only the long DNA substrate (509-mer) and was absent for a short 30-mer DNA substrate (SI Figure 1B). On the basis of the structural studies of Dnmt3a-C, the 30-mer DNA substrate is not expected to accommodate more than one or two tetramers and thus cannot support multimerization and cooperativity.18,21,24 To test if Dnmt3b-C cooperativity could be supported on longer DNA substrates, we repeated the methylation assays using the pUC19 plasmid as a substrate. As shown in Figure 2B, whereas the activity of Dnmt3a-C was cooperative, the activity of Dnmt3b-C increased linearly with the increase in enzyme concentration similar to the data shown in Figure 2A. We also noticed that compared to the 509-mer, the plasmid substrate stimulated cooperativity of Dnmt3a-C at lower enzyme concentrations, suggesting cooperativity of Dnmt3a-C may be influenced by substrate length. To confirm the results for Dnmt3b-C, we performed a time course of DNA methylation to determine the initial rate of DNA methylation using three biotinylated substrates, a 30-mer with one CpG site at 250 nM, a 509 bp DNA substrate with 58 CpGs, and a 719 bp substrate with 46 CpG sites at concentrations of 150 nM each. Increasing concentrations of the Dnmt3b-C enzyme ranging from 0.5 to 2 μM were preincubated with the DNA, and the reaction was initiated via the addition of radiolabeled AdoMet and was monitored by the incorporation of radioactivity into the DNA (Figure 2C–E). Irrespective of the substrate length, the methylation rate increased linearly with the increase in Dnmt3b concentration (Figure 2F), with an ~2-fold increase in the rate of methylation for every 2-fold increase in enzyme concentration. We observed rates of methylation slightly lower than expected at the lowest enzyme concentration for all three substrates potentially because of the slow turnover under these conditions. At higher enzyme concentrations in which cooperative methylation was observed for Dnmt3a-C24 (Figure 2A), the absence of an exponential or nonlinear increase in the methylation rate for Dnmt3b-C suggests that Dnmt3b methylates DNA by a noncooperative mechanism.
Figure 2.
Catalytic mechanism of Dnmt3b-C. (A and B) Methylation activity of Dnmt3a-C and Dnmt3b-C enzymes measured for 10 min with the 509 bp and pUC19 substrates in the presence of 0.25–2 and 0.25–1 μM enzyme, respectively. The enzymes were preincubated with DNA for 10 min at room temperature, and the reaction was initiated by addition of AdoMet. Total methylation activity was plotted vs enzyme concentration using an average and standard deviation (n ≥ 3 independent experiments). (C–E) Time course of DNA methylation with 30-mer, 509-mer, and 719-mer DNA substrates in the presence of 0.5, 1, 1.5, or 2 μM Dnmt3b-C enzyme that was preincubated with DNA for 10 min at room temperature. The reaction was initiated by addition of AdoMet. The data were fitted by linear regression, weighted by 1/Y2. (F) Methylation rates for all three DNA substrates plotted vs enzyme concentrations using linear regression without weighting. Averages ± the standard error of the mean are shown (n ≥ 3 independent experiments).
Dnmt3b-C Activity Is Not Stimulated by the Catalytically Inactive Mutant
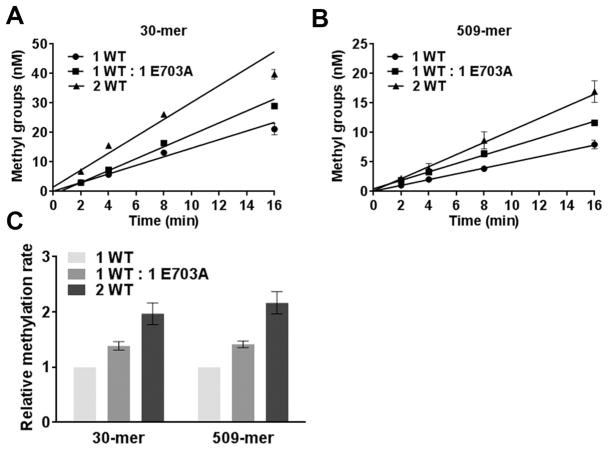
Addition of a catalytically inactive mutant to WT Dnmt3a-C was shown to stimulate its catalytic activity because it contributes to the cooperativity of Dnmt3a on long DNA substrates, resulting in an allosteric effect.24 This variant carries a mutation in conserved motif IV (ENV) that is required for catalysis, but this mutation does not affect the DNA binding activity of the methyltransferase enzymes.24 The corresponding mutant Dnmt3b-C E703A has very low residual activity (SI Figure 2). As an additional test of cooperative stimulation, we tested the influence of the Dnmt3b-C E703A mutant on the activity of WT Dnmt3b-C. Methylation kinetic reactions were performed with two biotinylated substrates at near saturating concentrations: a 30 bp substrate with one CpG site at 1 μM and a 509 bp substrate at 150 nM using either 1 μM WT enzyme or 1 μM WT mixed with 1 μM inactive mutant (Figure 3A,B). The fold change in the rates of DNA methylation compared to that of 1 μM WT enzyme alone was then plotted (Figure 3C). A 2-fold increase in activity was observed when the enzyme concentration was increased from 1 to 2 μM for both substrates. The 1:1 WT/inactive mutant mixture yielded a small increase in activity, however, with both short (30-mer) and long (509-mer) DNA substrates. Because the short DNA substrate cannot bind multiple enzyme units,18,21,24 this increase in activity is not likely due to cooperativity but may be due to interactions at the tetramer interface (F–F interface) as shown previously for Dnmt3a-C.16,24 These data provide further support for the non-cooperative mechanism of Dnmt3b activity.
Figure 3.
Dnmt3b-C is not cooperatively stimulated by addition of inactive Dnmt3b-C E703A. (A and B) Methylation kinetics of 30-mer and 509-mer substrates, respectively, by WT (Dnmt3b-C) and a mutant (catalytically inactive Dnmt3b-C E703A). Methylation was conducted using 1 μM WT, 1 μM WT and 1 μM E703A, and 2 μM WT with either 1 μM 30-mer or 150 nM 509-mer substrate. The reaction was initiated by addition of enzyme to the substrate mix, and data were fitted by linear regression, weighted by 1/Y2. (C) Methylation rates were measured from the slopes in panels A and B. The rate of methylation for each substrate was normalized to the rate for 1 μM enzyme and plotted in the bar graph with the normalized error. The average ± the standard error of the mean is shown (n ≥ 4 independent experiments).
Dnmt3b-C and Full Length Dnmt3b Methylate DNA in a Processive Manner
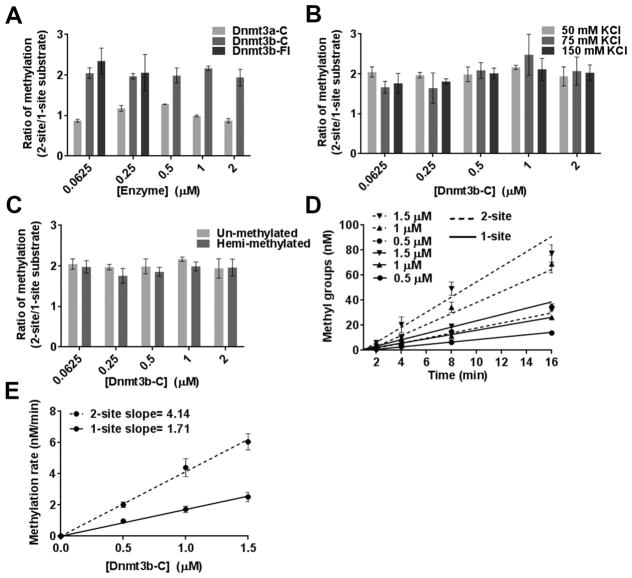
Unlike a previous method that used methylation-dependent restriction protection to test the processivity of Dnmt3b,15 here we assayed the processivity using two DNA substrates of the same sequence, 30 and 32 bp in length, except that one substrate contained one CpG and the second contained two CpGs.25 In a processive reaction, the second site on the two-site DNA substrate is expected to be methylated faster, so the methylation rate for the two-site substrate would be expected to be ~2-fold higher than that for the one-site substrate. This effect should be prominent at a low enzyme:DNA ratio, in which each DNA molecule is occupied by only one enzyme molecule, in other words, under multiple-turnover conditions. Therefore, methylation reactions were performed using a range of Dnmt3b-C enzyme concentrations at near saturating substrate (DNA and AdoMet) concentrations. Unlike the cooperativity assays, the enzyme was not preincubated with DNA, and the methylation reactions were initiated by addition of enzyme to a buffer/substrate cocktail. This allowed the enzyme to bind to its preferred substrate first and catalysis to proceed at maximal efficiency. Short oligonucleotides were used as substrates to eliminate any potential cooperativity from occurring, because they cannot bind more than a few molecules of the enzyme. Assays of Dnmt3a-C and Dnmt3b-C (0.0625–2 μM) were performed with 0.25 μM one-site and two-site substrates (0.5 and 1 μM CpG sites). As previously reported, Dnmt3a-C methylated one-and two-site substrates to the same degree, indicating a distributive mechanism of DNA methylation.15,24 In contrast, Dnmt3b-C methylated the two-site substrate with a 2-fold higher efficiency (Figure 4A). Because this effect was evident at the lowest concentration of enzyme assayed, the data indicate that the Dnmt3b enzyme methylates DNA in a processive fashion. We also demonstrated the processive activity of Dnmt3b by using the full length Dnmt3b enzyme (Dnmt3b-Fl) in the assays described above (Figure 4A and SI Figure 3A). Our data show that similar to the truncated Dnmt3b-C, at lower enzyme concentrations Dnmt3b-Fl methylates DNA in a processive manner. Because of its large size and tendency to precipitate, Dnmt3b-Fl could be purified at a maximal concentration of 0.5–1 μM; consequently, the assays at higher concentrations could not be performed. A similar effect was observed for the known processive methyltransferase M.SssI (SI Figure 3B). To determine if these results were influenced by the presence of a 2-fold higher CpG molarity of the two-site substrate, we performed a parallel reaction at an equal CpG molarity by comparing methylation of 0.5 μM one-site substrate and 0.25 μM two-site substrate. The ratio of DNA methylation at either 0.25 or 0.5 μM Dnmt3b-C remained at 2, confirming its processive mechanism of DNA methylation (SI Figure 3C). We next tested the effect of ionic strength on the processivity of the Dnmt3b-C enzyme. Figure 4B shows that increasing the ionic strength from 50 to 150 mM KCl had no effect on the degree of methylation of one- or two-site substrate, indicating that Dnmt3b can methylate DNA processively at a physiological ionic strength ranging from 100 to 150 mM. Salt concentrations of >250 mM disrupted enzyme activity. Dnmt3b has been shown to collaborate with Dnmt1 for the maintenance of DNA methylation.34 We therefore tested the processivity of Dnmt3b-C on one- or two-site hemimethylated substrates. Similar to the unmethylated substrate, Dnmt3b methylated the two-site hemimethylated substrate with an efficiency 2-fold higher than that of the one-site substrate (Figure 4C).
Figure 4.
Dnmt3b-C exhibits processivity on a DNA substrate with two CpG sites. (A) Steady state DNA methylation levels were measured for 1 h at various concentrations of Dnmt3b-C, Dnmt3b-Fl, or Dnmt3a-C enzymes using 1.5 μM 3H-labeled AdoMet (1:1 mixture of labeled and unlabeled) and 250 nM one-site or two-site substrate. For each enzyme concentration, the methylation of the two-site substrate was normalized to that of the one-site substrate to show the relative change in methylation level. (B and C) Relative DNA methylation as described in panel A at physiologically relevant salt concentrations with 250 nM hemimethylated one-site and two-site substrates. (D) Initial velocity of methylation measured at early time points (2, 4, 8, and 16 min) for one-site (—) and two-site (---) substrates. The concentration of Dnmt3b-C ranged from 0.5 to 1.5 μM, and the data were fitted to linear regression, weighted by 1/Y2. (E) DNA methylation rates measured in panel D were plotted vs enzyme concentration, and the slopes were determined from the linear regression without weighting. Data are the average ± the standard error of the mean (n ≥ 3 independent experiments).
Processivity of the Initial Phase of the Reaction
To confirm our conclusion from the steady state experiments described above, we next examined the processivity of Dnmt3b-C during the initial phase of the reaction. The initial rate of catalysis of a processive enzyme is also expected to be 2-fold higher with a two-site substrate than with a one-site substrate. DNA methylation using different enzyme concentrations was measured as a function of time, and initial rates were calculated from the slope after fitting the data by linear regression (Figure 4D). Methylation rates increased linearly with enzyme concentration, indicting multiple-turnover conditions. As shown in the secondary plot, DNA methylation rates for the two-site substrate were 2-fold higher than those for the one-site substrate, strongly suggesting that Dnmt3b-C operates in a processive fashion by methylating multiple CpGs on the same molecule of DNA before dissociation (Figure 4E).
Taken together, our data here confirm the processive mechanism of Dnmt3b and validate an important difference between the catalytic mechanism of Dnmt3a and Dnmt3b.
Effect of Preincubation with DNA on Catalytic Activity
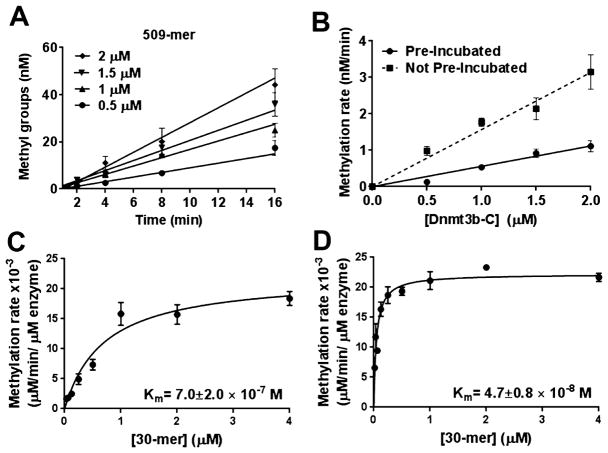
Processivity is expected to increase the catalytic activity of a methyltransferase toward long DNA substrates with multiple target sites. However, for experiments in which Dnmt3b-C was preincubated with DNA, we did not observe a substantial increase in the methylation rate for long DNA substrates compared to that with the 30-mer (Figure 2E). Although preincubation with DNA may simply lead to a decrease in the active fraction of the enzyme, it is also possible that in the absence of AdoMet, nonspecific DNA binding by Dnmt3b-C keeps it in a nonproductive complex. This can potentially slow the methylation reaction and influence the processivity of the enzyme. To investigate the effect of preincubation with DNA on the processivity of Dnmt3b, the rate of DNA methylation for the 509 bp DNA substrate was compared under two conditions. The first condition, in which various concentrations of Dnmt3b-C were preincubated with DNA substrate and the methylation reaction was initiated with AdoMet, is represented in Figure 2D. For the second condition, no preincubation was performed and methylation was initiated by addition of enzyme to the substrate cocktail. Kinetics of DNA methylation at each enzyme concentration was performed, and the initial methylation rates were determined by linear regression (Figure 5A). At all the tested enzyme concentrations, methylation rates were significantly slower when DNA was preincubated with enzyme than when it was not (Figure 5B), confirming a negative effect of preincubation with DNA on the catalytic activity of Dnmt3b-C. This is not intuitive for a processive enzyme, because during multiple reactions the exchange of AdoHcy with AdoMet should not require the enzyme to be dislodged from the DNA. We speculate that in the absence of a cofactor (AdoMet or AdoHcy), the enzyme preincubated with DNA binds strongly to nonspecific sites, which slows catalysis suggesting that the enzyme prefers to bind AdoMet for the first turnover.
Figure 5.
Preincubation with DNA decreases the methylation activity of Dnmt3b-C. (A) Methylation activity of Dnmt3b-C for the 509-mer was measured without preincubation with DNA for a range of enzyme concentrations. The reaction was initiated by addition of enzyme to the substrate cocktail. The data, which are the average ± the standard error of the mean (n ≥ 3 independent experiments), were fitted to linear regression, weighted by 1/Y2. (B) Rates of DNA methylation calculated from panel A plotted vs enzyme concentration and compared to the rates obtained from assays with the preincubated enzyme from Figure 1D. (C and D) Steady state kinetic analysis of methylation by Dnmt3b-C. Methylation reactions were performed for 10 min, and initial velocities were calculated as the number of methyl groups transferred per minute per micromolar enzyme for varying concentrations of the 30-mer substrate ranging from 0.031 to 4 μM and 0.75 μM AdoMet. (C) The enzyme was preincubated with DNA for 15 min, and the reaction was started with AdoMet. (D) The enzyme was preincubated with Adomet for 15 min, and the reaction was initiated with DNA. The data, which are the average ± the standard error of the mean (n ≥ 3 independent experiments), were fitted to the Michaelis–Menten equation, and the significance was estimated using a Student’s paired t test yielding a p value of 0.014.
Order of Addition for Steady State Kinetic Analysis
To determine the effect of preincubation with the substrate and cofactor, we analyzed the steady state methylation activity of Dnmt3b-C. The avidin plate assay used for methylation assays has a maximal DNA binding capacity of 2 μM;33 therefore, we used a filter binding assay for this experiment.35 Using a 30-mer substrate with one CpG site and 0.5 μM enzyme, steady state kinetic experiments were performed, and data were fitted to the Michaelis–Menten equation. Three different experimental conditions were used, one in which the enzyme was preincubated with DNA or AdoMet for 15 min before the reaction was started by addition of AdoMet or DNA, respectively, or the reaction was initiated by adding enzyme to a substrate/cofactor mix. All data sets fit well to the hyperbolic curve (Figures 1B and 5C,D). However, the data set for preincubation with DNA (Figure 5D) also fitted to a sigmoidal curve, indicating a slower reaction at lower substrate concentrations that recovers at higher enzyme concentrations. The comparison using Akaike’s Information Criteria (AICc) or the extra sum of squares F-test suggests that the hyperbolic Michaelis–Menten model is a better fit with a 71.5% higher probability of being correct. The estimated Vmax under all the conditions showed no significant change. Under these three conditions, the KM value was estimated to be (3.4 ± 1) × 10−7 M for the reaction with non-preincubated enzyme (Figure 1B) and (7.0 ± 2) × 10−7 and (4.7 ± 0.8) × 10−8 M when the enzyme was preincubated with DNA and AdoMet, respectively (Figure 5C,D). A 10-fold decrease in the KM of the enzyme upon preincubation with AdoMet demonstrates that the cofactor-bound Dnmt3b-C enzyme exists in a conformation that favors catalysis, potentially by enhancing the specific interaction of Dnmt3b-C with DNA. On the basis of these results, we propose that the binding of Dnmt3b-C to DNA in the absence of AdoMet may lead to the formation of a nonspecific DNA–enzyme binary complex in a conformation that limits or restricts binding of the cofactor AdoMet, thus slowing the reaction. This may have a stronger influence on initial or pre-steady state rates, thus influencing the processivity of the enzyme at earlier time points as seen in Figure 5B. We propose that in vivo most enzyme exists in the AdoMet-bound state and that during processive DNA methylation, cofactor exchange takes place without dissociation of the enzyme from the DNA.
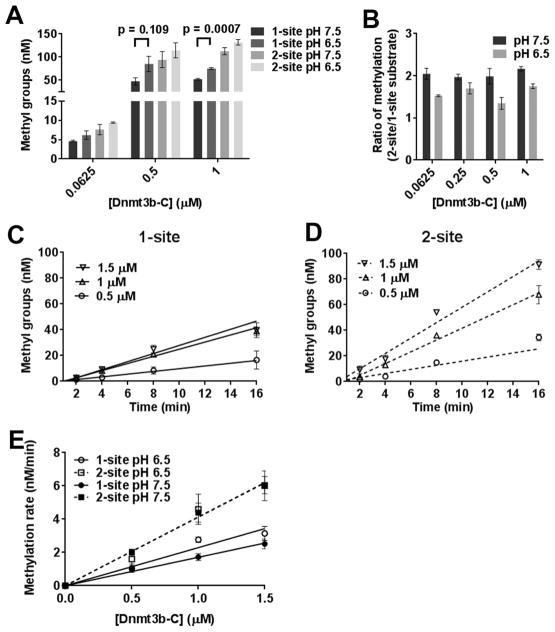
Activity and Processivity of Dnmt3b-C Are Not Affected by pH
Under various physiological conditions, changes in the intracellular pH can influence protein–protein and protein–DNA interactions.36 Previous studies have shown that the interaction of Dnmt3a through its dimer interface can be disrupted by a change in pH from 7.5 to 6.5, which results in a decrease in its activity and cooperativity.28 Our data showing that Dnmt3b methylates DNA in a noncooperative manner suggest that the activity of Dnmt3b may not be strongly dependent on interactions involving the dimer interface. We tested this by assessing the effect of pH on the processivity of Dnmt3b-C. Methylation assays were performed at pH 7.5 and 6.5 for one- and two-CpG site substrates for 60 min with varying concentrations of enzyme. As shown in Figure 6A, the steady state level of DNA methylation using a one-site substrate was slightly higher at pH 6.5 than at pH 7.5. As a consequence, the ratios of methylation between one- and two-site substrates were reduced to <2 (Figure 6B). We checked whether this difference in methylation activity was due to the impact of pH on the initial rate of DNA methylation. Methylation kinetics at various enzyme concentrations was performed with one- and two-site substrates at pH 6.5 and 7.5, and initial rates were measured (Figure 6C,D). The rates of DNA methylation for the one-site substrate at various enzyme concentrations were slightly higher at pH 6.5 compared to those at pH 7.5 (Figure 6E). We speculate that this higher methylation rate under pre-steady state conditions could be responsible for the difference observed at the steady state levels. However, the rates of methylation of the two-site substrate remained ~2-fold higher than those for the one-site substrate at all enzyme concentrations tested at pH 7.5 and 6.5, indicating that the activity and processivity of Dnmt3b-C are not affected by lower pH. This behavior is in contrast to that of Dnmt3a-C, for which lowering the pH to 6.5 decreased its methylation activity by disruption of the dimer interface.28
Figure 6.
pH sensitivity of Dnmt3b-C processive catalysis. (A) Methylation at one- and two-site substrates by Dnmt3b-C was measured for 1 h at pH 6.5 or 7.5. (B) Ratio of the methylation levels calculated as the fold change at two-site vs one-site substrate. (C and D) Time course of DNA methylation by Dnmt3b-C at pH 6.5 for one- and two-site substrates. For each substrate, the time course of DNA methylation reaction was determined in the presence of 0.5–2 μM Dnmt3b-C. The reaction was started by the addition of enzyme, and the data were fitted by linear regression, which was weighted by 1/Y2. (E) The calculated methylation rates at pH 6.5 were plotted vs enzyme concentration and compared to those at pH 7.5. Data are the average ± the standard error of the mean (n ≥ 4 independent experiments).
Influence of pH on the Dissociation Constant
To test the effect of lower pH on DNA binding by Dnmt3b-C, we performed DNA binding assays using Cy5-labeled 30-mer DNA with one CpG site. An increasing amount of Dnmt3b-C ranging from 0.5 to 2 μM was incubated with 30 nM DNA substrate, and the binding assay was performed using a dot blot assay. The binding was measured by trapping the protein–DNA complexes on a nitrocellulose membrane. The intensity of the spots was measured, and data were fitted to a one-site specific binding saturation curve. Binding curves were hyperbolic, and the data could also be fitted well with the Hill coefficient constrained to 1 (Figure 7A,B). The dissociation constants at pH 7.5 and 6.5 showed no significant difference, indicating that the binding of Dnmt3b-C to DNA is not sensitive to lower pH and there is potentially a limited contribution of the dimer interface to the activity of Dnmt3b-C.
Figure 7.
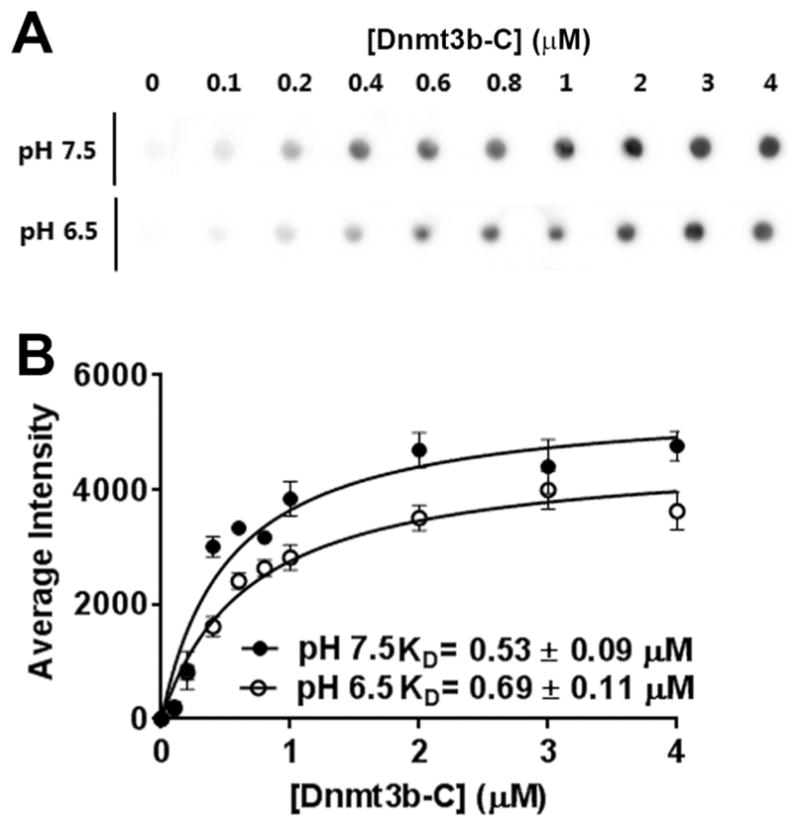
Effect of pH on DNA binding by Dnmt3b-C. For DNA binding analysis, the Dnmt3b-C enzyme was incubated with 30 nM Cy5-labeled 30-mer DNA in the presence of 0.2 mM AdoHcy (S-adenosylhomocysteine) in binding buffer at pH 6.5 or 7.5. (A) Representative blot for DNA binding. (B) The signal intensity on the blot was quantified using ImageQuant software, and data (average ± the standard error of the mean, n ≥ 3 independent experiments) were fitted to a one-site binding saturation model and binding constants determined. The binding constants are within an error of <20%.
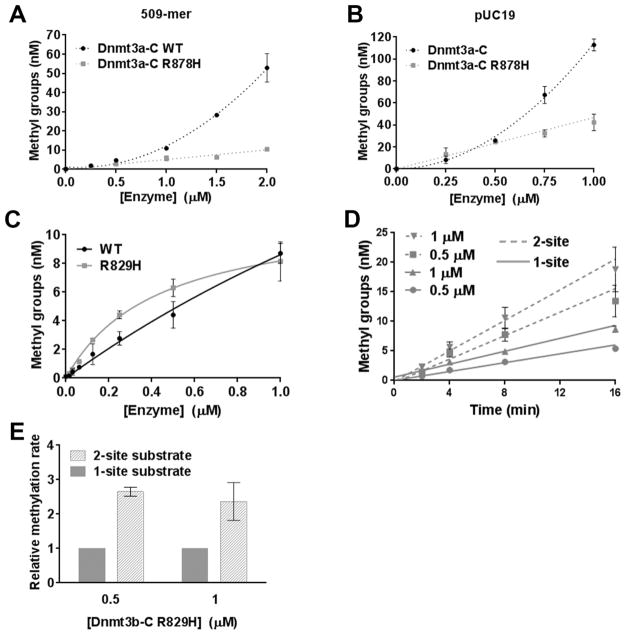
The Processivity of Dnmt3b-C Is Not Affected by Mutation of the R829 Residue
The R882H mutation, which is close to the dimer interface of Dnmt3a, is highly prevalent in human AML cells. This mutation, as well as the equivalent R878H mutation in mouse, disrupts tetramer formation and leads to a significant decrease in enzymatic activity.29,31,32 Although this residue is implicated in tetramer formation, its effect on the cooperativity of Dnmt3a has not been tested. We performed the cooperativity assays as described above using various concentrations of WT Dnmt3a-C and Dnmt3a-C R878H enzyme (SI Figure 4A). Methylation assays were performed using 32-mer, 509-mer, and pUC19 DNA as substrates of various lengths. Loss of 64% activity for the 32-mer substrate at two tested enzyme concentrations (0.5 and 1 μM) confirms the previous observation that the mutation affects tetramer formation and that the tetrameric form is the most catalytically active form of Dnmt3a-C30,37(SI Figure 4B,C). Interestingly, on longer DNA substrates, although the activity was rescued at a lower enzyme concentration (0.5 μM) (SI Figure 4D), at a higher enzyme concentration, the enzymatic activity did not demonstrate an exponential increase that is expected because of cooperativity (Figure 8A,B). These data support our observation for the WT enzyme that the length of DNA positively affects the cooperative behavior of Dnmt3a. Destabilization of the tetramer structure due to the absence of R878 can be partially rescued by long DNA substrates, however not to the extent that it can restore cooperativity. Loss of cooperativity resulted in 78 and 63% losses of activity for 509-mer and pUC19, respectively, at higher enzyme concentrations. These data suggest that the human R882H mutation of Dnmt3a affects its cooperative mechanism at multiple CpG sites, which would have a substantial effect on its activity in vivo. We next tested the effect of the analogous mutation of the conserved R829 in Dnmt3b-C. On the basis of our data showing that the noncooperative and processive mechanism of Dnmt3b-C is not affected by acidification, we predicted that the mutation of R829 will not affect the processivity of Dnmt3b-C. Our data in Figure 8C–E demonstrate that the variant enzyme R829H has activity and processivity comparable to those of the WT enzyme.
Figure 8.
R829H mutation of Dnmt3b-C that does not reduce its activity and processivity. (A and B) Methylation activity of the WT and R878H mutant Dnmt3a-C enzymes was measured for 10 min with the 509 bp or pUC19 substrate in the presence of 0.25–2 and 0.25–1 μM enzyme, respectively. Enzymes were preincubated with DNA for 10 min at room temperature, and the reaction was initiated by addition of AdoMet. The total methylation activity was plotted vs enzyme concentration using an average and standard deviation (n ≥ 3 independent experiments). (C) Methylation activity was measured for 10 min with the 30-mer substrate at enzyme concentrations ranging from 0.015 to 1 μM for the WT and mutant R829H Dnmt3b-C enzymes. (D) The initial velocity of methylation was measured for one-site (—) and two-site (---) substrates, in the presence of 0.5 and 1 μM Dnmt3b-C R829H mutant enzyme. The data were fitted to linear regression, weighted by 1/Y2. (E) Methylation rates for the one-site substrate were normalized to 1, and the relative change in the rate of methylation for the two-site substrate at different enzyme concentrations was plotted with a normalized error. For all the experiments, the average ± the standard error of the mean was derived (n ≥ 4 independent experiments).
Taken together, these data show that Dnmt3b-C methylates DNA in a noncooperative and processive manner and suggest that Dnmt3b-C may not require the interaction of Dnmt3b monomers or dimers along the dimer interface for optimal activity.
DISCUSSION
DNA methylation together with specific histone modifications plays an important role in regulating chromatin structure and thereby controlling gene expression. DNA methylation has a major role in regulation of epigenetic processes, including genomic imprinting, X-chromosome inactivation, nuclear reprogramming, and carcinogenesis.7,38 Recent studies of the functions of the mammalian de novo methyltransferases Dnmt3a and Dmnt3b reveal both redundant and specific functions, highlighting the importance of differentiating their individual contributions to normal and diseased epigenomes.39 Whereas several biochemical studies have elucidated the structure–function relationship of Dnmt3a, less is known about the biochemical properties of Dnmt3b. Studies characterizing the enzymatic properties of Dnmt3a-C have provided critical information that facilitates our understanding of the biochemical basis of Dnmt3a function in vivo. This is underscored by the recent finding that ~20% of acute myeloid leukemia (AML) patients have mutations in Dnmt3a of which many are present in the dimer or tetramer interface of the enzyme.30,31,37,40,41 These mutations were shown to disrupt oligomerization of Dnmt3a and alter its catalytic properties in vitro and in vivo. Likewise, a high frequency of Dnmt3b mutations is found in ICF syndrome, which is a rare genetic disease causing immunodeficiency, centromeric instabilities, and facial abnormalities. Many of these mutations are present in the Dnmt3b catalytic domain and result in reduced catalytic activity in vitro, potentially linking the mutations to hypomethylation of DNA in the diseased state.6,15,42–45 Some of these mutations are present in the potential DNA binding region of Dnmt3b, and these residues are conserved in the dimer interface of Dnmt3a. In the absence of a Dnmt3b-C crystal structure, the detailed characterization of the catalytic mechanism of Dnmt3b-C here provides important insights into the impact of these mutations on the catalytic mechanism of Dnmt3b compared to that of Dnmt3a and highlights its properties that are distinct from those of Dnmt3a. Our findings suggest that the reduced activity of Dnmt3b mutants in ICF syndrome may not be because of the dimer-interface disruption.
Processivity of Dnmt3b-C has been reported previously; however, it was not clear whether the results reffected cooperative methylation of multiple sites by a multimerized enzyme or one enzyme unit processively methylating multiple sites.15 By using DNA substrates of various lengths, our assays in this study distinguish processivity from cooperativity and show that Dnmt3b-C methylates DNA by a processive mechanism in a noncooperative manner. We also show that Dnmt3b can methylate both unmethylated and hemimethylated sites processively, consistent with its collaborative function with Dnmt1 during the maintenance of DNA methylation. The noncooperative mechanism is further supported by our data showing that on the longer DNA substrates, the catalytic activity of Dnmt3b is not enhanced by the addition of its inactive mutant. Surprisingly, in our assays using DNA substrates of various lengths to examine the cooperativity of Dnmt3b-C, the methylation rate was not higher for the long DNA, which would be expected from a processive enzyme. Because these experiments were performed by preincubating the enzyme with DNA, this could be explained if the enzyme prefers binding AdoMet before binding to DNA. Our data demonstrating a 10-fold decrease in the KM of Dnmt3b-C when it is preincubated with AdoMet support this conclusion. Our data are consistent with the model in which the binding of the enzyme to specific sites on DNA is favored by AdoMet binding, and during the processive catalysis, Dnmt3b is capable of cofactor exchange without dissociating from DNA.
Perturbations in the intracellular microenvironment including pH has a regulatory effect in several DNA binding proteins and enzymes. These regulatory mechanisms are expected to be prominent during tumor progression and also during development. Acidification has been shown to disrupt cooperativity and reduce the catalytic activity of Dnmt3a. Interestingly, again in contrast to that of Dnmt3a, the catalytic activity of Dnmt3b was not significantly affected at pH 6.5 and instead showed a slight increase in activity for a one-site substrate. We speculate that this may be because Dnmt3b-C has a slightly higher KD value at pH 6.5. This difference between Dnmt3a and Dnmt3b mechanisms could be explained by the differences in their amino acid sequences, especially in the DNA binding region and the dimer interface. The interaction of several DNA binding proteins involves either water-mediated or direct H-bond interactions between the Arg and His residues and the DNA backbone. The protonation of His residues is pH-sensitive, which can alter its interaction with DNA and affect the activity of the protein. The activity of Dnmt3a decreases at lower pH because of the disruption of its dimer interface involving H821 and/or H873.28 However, in Dnmt3b, the residues at the equivalent positions are replaced by Y821 and L847, respectively, both of which are also a part of DNA binding loop L218,19 (SI Figure 5). Compared to Dnmt3a, this makes the enzymatic activity of Dnmt3b potentially less dependent on protein–protein interaction at the dimer interface and less sensitive to changes in pH. A high level of expression of Dnmt3b has been implicated in the initiation and progression of several cancers and in some cases contributes to CpG island promoter methylation.11 Its ability to function under low-pH conditions can be exploited by cancer cells in which the intracellular environment is typically more acidic than that of normal cells.
Further supporting a noncooperative mechanism, our data here show for the first time that the Dnmt3a R878H mutation disrupts the cooperativity of the enzyme whereas the analogous R829H mutation in Dnmt3b-C has no effect on its processivity. Although the SNP causing the R to H mutation in human DNMT3A was shown to be present at a very high frequency in AML cells,46 the analogous mutation is not reported for DNMT3B in any epigenetic diseases. Therefore, we speculate that a smaller contribution of the dimer interface would promote the processivity of Dnmt3b, which may work efficiently for enzymes that do not form oligomers on DNA. These observations also emphasize that the minor difference in the amino acid sequence of Dnmt3a and Dnmt3b catalytic domains dictates a processive versus a cooperative mechanism.
Supplementary Material
Acknowledgments
Funding
This work was supported by the Institutional Start up Fund of the Department of Biochemistry and Small Grants Program from the Purdue University Center for Cancer Research (P30 CA023168) and Purdue Showalter Trust Grant. A graduate research fellowship for L.A. is provided by the Saudi Arabian Cultural Mission.
We thank Dr. Albert Jeltsch for providing Dnmt3a and Dnmt3b variants and Gowher lab members for their contributions via discussion.
ABBREVIATIONS
- AdoMet
S-adenosylmethionine, also known as SAM
- AdoHcy
S-adenosylhomocysteine, also known as SAH
- Aprt
adenine phosphoribosyltransferase
- CpG
dinucleotide of cytosine followed by guanine
- Dnmt
DNA methyltransferase
- DDT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- ENV
three amino acids in conserved motif IV, involved in AdoMet binding
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- MT
mutant enzyme
- Ni-NTA
nickel-charged affinity resin
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SUHW1
zinc finger protein 280A
- WT
wild type
Footnotes
Author Contributions
A.B.N. and C.J.P. contributed equally to this work. A.B.N., L.W., C.J.P., and L.A. performed the experiments. A.N., C.P., S.R., and H.G. performed data analysis and wrote the manuscript.
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.6b00964.
Five figures showing data that further supports the mechanism proposed herein (PDF)
References
- 1.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 2.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. ChemBioChem. 2011;12:206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 3.Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- 4.Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol Life Sci. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishiyama A, Yamaguchi L, Nakanishi M. Regulation of maintenance DNA methylation via histone ubiquitylation. J Biochem. 2016;159:9–15. doi: 10.1093/jb/mvv113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 7.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 8.Chen BF, Chan WY. The de novo DNA methyltransferase DNMT3A in development and cancer. Epigenetics. 2014;9:669–677. doi: 10.4161/epi.28324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat Rev Cancer. 2015;15:152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong M, Goodell MA. New answers to old questions from genome-wide maps of DNA methylation in hematopoietic cells. Exp Hematol. 2014;42:609–617. doi: 10.1016/j.exphem.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walton EL, Francastel C, Velasco G. Dnmt3b Prefers Germ Line Genes and Centromeric Regions: Lessons from the ICF Syndrome and Cancer and Implications for Diseases. Biology. 2014;3:578–605. doi: 10.3390/biology3030578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan F, Cui S, Song C, Dai L, Zhao X, Zhang X. Systematic evaluation of cancer risk associated with DNMT3B polymorphisms. J Cancer Res Clin Oncol. 2015;141:1205–1220. doi: 10.1007/s00432-014-1894-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 14.Challen GA, Sun D, Mayle A, Jeong M, Luo M, Rodriguez B, Mallaney C, Celik H, Yang L, Xia Z, Cullen S, Berg J, Zheng Y, Darlington GJ, Li W, Goodell MA. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell stem cell. 2014;15:350–364. doi: 10.1016/j.stem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gowher H, Jeltsch A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J Biol Chem. 2002;277:20409–20414. doi: 10.1074/jbc.M202148200. [DOI] [PubMed] [Google Scholar]
- 16.Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- 17.Kareta MS, Botello ZM, Ennis JJ, Chou C, Chedin F. Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J Biol Chem. 2006;281:25893–25902. doi: 10.1074/jbc.M603140200. [DOI] [PubMed] [Google Scholar]
- 18.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo X, Wang L, Li J, Ding Z, Xiao J, Yin X, He S, Shi P, Dong L, Li G, Tian C, Wang J, Cong Y, Xu Y. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2014;517:640–644. doi: 10.1038/nature13899. [DOI] [PubMed] [Google Scholar]
- 20.Jurkowska RZ, Rajavelu A, Anspach N, Urbanke C, Jankevicius G, Ragozin S, Nellen W, Jeltsch A. Oligomerization and binding of the Dnmt3a DNA methyltransferase to parallel DNA molecules: heterochromatic localization and role of Dnmt3L. J Biol Chem. 2011;286:24200–24207. doi: 10.1074/jbc.M111.254987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurkowska RZ, Anspach N, Urbanke C, Jia D, Reinhardt R, Nellen W, Cheng X, Jeltsch A. Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res. 2008;36:6656–6663. doi: 10.1093/nar/gkn747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holz-Schietinger C, Matje DM, Harrison MF, Reich NO. Oligomerization of DNMT3A controls the mechanism of de novo DNA methylation. J Biol Chem. 2011;286:41479–41488. doi: 10.1074/jbc.M111.284687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holz-Schietinger C, Reich NO. The inherent processivity of the human de novo methyltransferase 3A (DNMT3A) is enhanced by DNMT3L. J Biol Chem. 2010;285:29091–29100. doi: 10.1074/jbc.M110.142513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emperle M, Rajavelu A, Reinhardt R, Jurkowska RZ, Jeltsch A. Cooperative DNA binding and protein/DNA fiber formation increases the activity of the Dnmt3a DNA methyltransferase. J Biol Chem. 2014;289:29602–29613. doi: 10.1074/jbc.M114.572032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gowher H, Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J Mol Biol. 2001;309:1201–1208. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- 26.Renbaum P, Razin A. Mode of action of the Spiroplasma CpG methylase M.SssI. FEBS Lett. 1992;313:243–247. doi: 10.1016/0014-5793(92)81201-v. [DOI] [PubMed] [Google Scholar]
- 27.Vilkaitis G, Suetake I, Klimasauskas S, Tajima S. Processive methylation of hemimethylated CpG sites by mouse Dnmt1 DNA methyltransferase. J Biol Chem. 2005;280:64–72. doi: 10.1074/jbc.M411126200. [DOI] [PubMed] [Google Scholar]
- 28.Holz-Schietinger C, Reich NO. De novo DNA methyltransferase DNMT3A: Regulation of oligomeric state and mechanism of action in response to pH changes. Biochim Biophys Acta, Gen Subj. 2015;1850:1131–1139. doi: 10.1016/j.bbagen.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Gowher H, Loutchanwoot P, Vorobjeva O, Handa V, Jurkowska RZ, Jurkowski TP, Jeltsch A. Mutational analysis of the catalytic domain of the murine Dnmt3a DNA-(cytosine C5)-methyltransferase. J Mol Biol. 2006;357:928–941. doi: 10.1016/j.jmb.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 30.Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, Meyer MR, Erdmann-Gilmore P, Townsend RR, Wilson RK, Ley TJ. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25:442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O’Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim SJ, Zhao H, Hardikar S, Singh AK, Goodell MA, Chen T. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood. 2013;122:4086–4089. doi: 10.1182/blood-2013-02-483487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth M, Jeltsch A. Biotin-avidin microplate assay for the quantitative analysis of enzymatic methylation of DNA by DNA methyltransferases. Biol Chem. 2000;381:269–272. doi: 10.1515/BC.2000.035. [DOI] [PubMed] [Google Scholar]
- 34.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, Baylin SB, Vogelstein B. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 35.Rubin RA, Modrich P. EcoRI methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1977;252:7265–7272. [PubMed] [Google Scholar]
- 36.Smallbone K, Gavaghan DJ, Gatenby RA, Maini PK. The role of acidity in solid tumour growth and invasion. J Theor Biol. 2005;235:476–484. doi: 10.1016/j.jtbi.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Holz-Schietinger C, Matje DM, Reich NO. Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation. J Biol Chem. 2012;287:30941–30951. doi: 10.1074/jbc.M112.366625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiedemann RL, Putiri EL, Lee JH, Hlady RA, Kashiwagi K, Ordog T, Zhang Z, Liu C, Choi JH, Robertson KD. Acute depletion redefines the division of labor among DNA methyltransferases in methylating the human genome. Cell Rep. 2014;9:1554–1566. doi: 10.1016/j.celrep.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 41.Walter MJ, Ding L, Shen D, Shao J, Grillot M, McLellan M, Fulton R, Schmidt H, Kalicki-Veizer J, O’Laughlin M, Kandoth C, Baty J, Westervelt P, DiPersio JF, Mardis ER, Wilson RK, Ley TJ, Graubert TA. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 43.Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehrlich M, Buchanan KL, Tsien F, Jiang G, Sun B, Uicker W, Weemaes CM, Smeets D, Sperling K, Belohradsky BH, Tommerup N, Misek DE, Rouillard JM, Kuick R, Hanash SM. DNA methyltransferase 3B mutations linked to the ICF syndrome cause dysregulation of lymphogenesis genes. Hum Mol Genet. 2001;10:2917–2931. doi: 10.1093/hmg/10.25.2917. [DOI] [PubMed] [Google Scholar]
- 45.Hamidi T, Singh AK, Chen T. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics. 2015;7:247–265. doi: 10.2217/epi.14.80. [DOI] [PubMed] [Google Scholar]
- 46.Miller CA, Wilson RK, Ley TJ. Genomic landscapes and clonality of de novo AML. N Engl J Med. 2013;369:1473. doi: 10.1056/NEJMc1308782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.