Abstract
Nicotine is a major addictive compound in tobacco and a component of smoking-related products, such as e-cigarettes. Once internalized, nicotine can perturb many cellular pathways and can induce alterations in proteins across different cell types, however the mechanisms thereof remain undetermined. We hypothesize that both tissue-specific and global protein abundance alterations result from nicotine exposure. As such, we present the first proteome analysis of multiple tissues from nicotine-exposed mice. We treat mice via oral administration of nicotine at 200μg/mL in drinking water for 21 days. We investigate 7 tissues (brain, heart, kidney, liver, lung, pancreas, and spleen) from treated (n=5) and untreated control (n=5) mice. For each tissue, a TMT10-plex experiment (5 versus 5) was assembled. We apply a minimalistic proteomics strategy was employed using TMT reagents efficiently and fractionating by centrifugation-based reversed-phase columns to streamline sample preparation. Combined, we quantified over 11,000 non-redundant proteins from over 138,000 different peptides in 7 TMT10-plex experiments. Between 7 and 126 proteins are significantly altered in tissues from nicotine-exposed mice. Among these proteins, only 11 are altered in two or more tissues, many classified as from extracellular exosomes or involved in lipid metabolism. Our data showcase the vast extent of nicotine exposure across murine tissue.
Keywords: SPS-MS3, Multi-Notch, TMT, Lumos
INTRODUCTION
Smoking affects the entire body, increasing the risk for life-threatening diseases, particularly cancer. Understanding protein function at the biomolecular level is essential to fully comprehend the intricacies of toxic stress on specific organs. Tobacco use is the leading cause of preventable illness and death in the United States. Tobacco products have been associated with multiple cancer types and chronic diseases affecting virtually all organ systems [1]. Cigarette smoking results in over 480,000 premature deaths in the United States each year and is linked to an additional 16 million people with a serious illness [2]. Preventing individuals from smoking would be ideal, however, such intervention is not feasible, and thus methods must be established to counteract the ensuing cellular damage.
We will focus on a single toxin found in tobacco products, nicotine. Aside from its presence in tobacco, nicotine is also a main component in smoking cessation products, notably the increasingly popular electronic cigarettes. E-cigarettes vaporize nicotine-containing solutions, thereby eliminating some byproducts resulting from burning tobacco. However, both the addictive and pathogenic effects of nicotine remain [3]. Growing evidence has linked nicotine to the disruption of cellular metabolic processes and its potential to be genotoxic and tumor-promoting [4]. Nicotine has been shown to induce cell proliferation and invasion in multiple lung and breast cancer lines [5], and alter the phosphorylation states of proteins in pancreatic stellate cells [6]. Previous studies link nicotine and diseases, but none have explored the proteome-wide effects on a mouse model system as examined here. In this analysis, we focus on seven tissues: brain, heart, kidney, liver, lung, pancreas, and spleen. Understanding the perturbations attributed to nicotine at the molecular, i.e. protein and phosphorylation, level before marked changes, is paramount to diagnosing, modifying, or retarding diseases before the development of end-stage complications.
Multiplexing strategies in mass spectrometry-based quantitative proteomics have significantly expanded the depth, efficiency, and throughput of comprehensive protein analyses. The use of tandem mass tags (TMT) is a well-established method for relative quantification of peptides and by inference, proteins [7]. Using this technique, the relative abundances of several thousand proteins can be measured at once in currently up to 11 different samples - equivalent to performing thousands of quantitative Western blotting analyses in a single experiment [8].
Here we employ a strategy using TMT10-plex reagents to enhance our ability to quantitatively analyze proteomic alterations resulting from nicotine treatment. Using a total of 10 mice, we treated five with nicotine in drinking water and the remaining five were controls. Following three weeks of nicotine treatment, seven tissues (brain, heart, kidney, liver, lung, pancreas, and spleen) were harvested from each mouse. For each tissue, a TMT10-plex experiment was performed comparing five untreated with five nicotine-treated mice. A minimalistic proteomics strategy was applied to streamline TMT sample preparation, whereby we use less TMT reagent and more time-efficient offline centrifugation-based fractionation. Our strategy enabled the quantification of >11,000 non-redundant proteins from which we classify and highlight those with significantly different abundance across tissues. The techniques used herein may be applied to investigate alterations resulting from other routes of nicotine administration, as well as cell type-specific alterations.
METHODS
Materials
Tandem mass tag (TMT) isobaric reagents were from Thermo Scientific (Rockford, IL). Nicotine was purchased from Sigma (St. Louis, MO). Water and organic solvents were from J.T. Baker (Center Valley, PA). Lys-C was from Wako (Richmond, VA). Unless otherwise noted, all other chemicals, including trypsin, were from ThermoFisher Scientific (Rockford, IL).
Mice
All animal experiments were performed according to procedures approved by Beth Israel Deaconess Medical Center’s Institutional Animal Care and Use Committee, animal protocol number: 104-2014. The mice (strain: B6.Cg-Lepob/J from Jackson Labs) were 8 weeks old when administered nicotine in drinking water (200μg/mL) for 21 days. Brain, heart, kidney, liver, lung, pancreas, and spleen from 10 mice (5 nicotine-treated and 5 controls) were dissected and flash frozen in liquid nitrogen.
Tissue homogenization and cell lysis
Tissues were homogenized in lysis buffer 8 M urea, 200 mM EPPS pH 8.5, 1X Roche Protease Inhibitors, 1X Roche PhosphoStop phosphatase inhibitors) at a tissue concentration of approximately 10–15 mg/mL using a polytron tissue grinder. Next, the homogenized sample was passed 10 times through a 21-gauge (1.25 inches long) needle. The homogenate was sedimented by centrifugation at 21,000 × g for 5 min and the supernatant was transferred to a new tube. Protein concentrations were determined using the bicinchoninic acid (BCA) assay (ThermoFisher Scientific, Waltham, MA). 100 μg of protein was aliquoted from each fraction for subsequent reduction and alkylation. Proteins were subjected to disulfide bond reduction with 5 mM tris (2-carboxyethyl)phosphine (room temperature, 30 min) and alkylation with 10 mM iodoacetamide (room temperature, 30 min in the dark). Excess iodoacetamide was quenched with 10 mM dithiotreitol (room temperature, 15 min in the dark). Methanol-chloroform precipitation was performed prior to protease digestion. In brief, four parts of neat methanol were added to each sample and vortexed, one part chloroform was added to the sample and vortexed, and three parts water was added to the sample and vortexed. The sample was centrifuged at 14,000 rpm for 2 min at room temperature and subsequently washed twice with 100% methanol. Samples were resuspended in 200 mM EPPS, pH 8.5 and digested at room temperature for 13 h with Lys-C protease at a 100:1 protein-to-protease ratio. Trypsin was then added at a 100:1 ratio and the reaction was incubated 6 h at 37°C. We used the Pierce Quantitative Colorimetric Peptide Assay (cat. #. 23275) to quantify the digest and to accurately aliquot 20 μg of peptides per sample for labeling.
Tandem mass tag labeling
For labeling, a final acetonitrile concentration of ~30% (v/v) was added along with 2 μL of TMT reagent (20 ng/μL) to the 20 μg of peptides. Following incubation at room temperature for 1.5 h, the reaction was quenched with hydroxylamine to a final concentration of 0.3% (v/v) for 15 min. The TMT-labeled samples were pooled at a 1:1 ratio across all samples. The combined sample was vacuum centrifuged to near dryness and subjected to C18 solid-phase extraction (SPE) via Sep-Pak (Waters, Milford, MA).
Off-line basic pH reversed-phase (BPRP) fractionation
We fractionated the pooled TMT-labeled peptide sample using the Pierce High pH Reversed-Phase Peptide Fractionation Kit (cat. # 84868). Twelve fractions were collected using: 7.5%, 10%, 12.5%, 15%, 17.5%, 20%, 22.5%, 25%, 27.5%, 30%, 35%, and 60% acetonitrile. This method enabled the simultaneous processing of the 7 tissue samples. Samples were subsequently acidified with 1% formic acid and vacuum centrifuged to near dryness. Each fraction was desalted via StageTip, dried again via vacuum centrifugation, and reconstituted in 5% acetonitrile, 5% formic acid for LC-MS/MS processing.
Liquid chromatography and tandem mass spectrometry
Mass spectrometry data were collected using an Orbitrap Fusion Lumos mass spectrometer (Thermo-Fisher Scientific, San Jose, CA) coupled to a Proxeon EASY-nLC 1200 liquid chromatography (LC) pump (ThermoFisher Scientific, San Jose, CA). Peptides were separated on a 100 μm inner diameter microcapillary column packed with ~40 cm of Accucore150 resin (2.6 μm, 150 Å, ThermoFisher Scientific, San Jose, CA). For each analysis, we loaded ~2 μg onto the column and separation was achieved using a 2.5 h gradient of 7 to 27% acetonitrile in 0.125% formic acid at a flow rate of ~550 nL/min. Each analysis used an SPS-MS3-based TMT method [9, 10], which has been shown to reduce ion interference compared to MS2-based quantification [11]. The scan sequence began with an MS1 spectrum (Orbitrap; resolution 120,000; mass range 400–1400 m/z; automatic gain control (AGC) target 5 × 105; maximum injection time 100 ms). Precursors for MS2/MS3 analysis were selected using a Top10 method. MS2 analysis consisted of collision-induced dissociation (quadrupole ion trap; AGC 2 × 104; normalized collision energy (NCE) 35; maximum injection time 150 ms). Following acquisition of each MS2 spectrum, we collected an MS3 spectrum using our recently described method in which multiple MS2 fragment ions were captured in the MS3 precursor population using isolation waveforms with multiple frequency notches [10]. MS3 precursors were fragmented by high energy collision-induced dissociation (HCD) and analyzed using the Orbitrap (NCE 65; AGC 1 × 105; maximum injection time 150 ms, resolution was 50,000 at 200 Th).
Data analysis
Mass spectra were processed using a Sequest-based pipeline [12]. Spectra were converted to mzXML using a modified version of ReAdW.exe. Database searching included all entries from the mouse UniProt database (April 20, 2016). This database was concatenated with one composed of all protein sequences in the reversed order. Searches were performed using a 50 ppm precursor ion tolerance for total protein level profiling. Pancreas searches were performed with semi-tryptic specificity, while proteins from the remaining tissues required fully-tryptic specificity. The product ion tolerance was set to 0.9 Da. These wide mass tolerance windows were chosen to maximize sensitivity in conjunction with Sequest searches and linear discriminant analysis [12, 13]. TMT tags on lysine residues and peptide N termini (+229.163 Da) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) was set as a variable modification. Peptide-spectrum matches (PSMs) were adjusted to a 1% false discovery rate (FDR) [14]. PSM filtering was performed using a linear discriminant analysis, as described previously [12], while considering the following parameters: XCorr, ΔCn, missed cleavages, peptide length, charge state, and precursor mass accuracy. For TMT-based reporter ion quantitation, we extracted the summed signal-to-noise (S/N) ratio for each TMT channel and found the closest matching centroid to the expected mass of the TMT reporter ion. PSMs were identified, quantified, and collapsed to a 1% peptide false discovery rate (FDR) and then collapsed further to a final protein-level FDR of 1%. Moreover, protein assembly was guided by principles of parsimony to produce the smallest set of proteins necessary to account for all observed peptides.
Proteins were quantified by summing reporter ion counts across all matching PSMs, as described previously [12]. PSMs with poor quality, MS3 spectra with more than eight TMT reporter ion channels missing, MS3 spectra with TMT reporter summed signal-to-noise ratio less than 100, or no MS3 spectra were excluded from quantification [8]. Protein quantitation values were exported for further analysis in Microsoft Excel or SAS JMP. Each reporter ion channel was summed across all quantified proteins and normalized assuming equal protein loading of all 10 samples. We defined statistically significant proteins as those having a p-value <0.05 as determined using a student’s two-sided t-test. A second threshold based on a log2 fold change of greater than 1.5-fold or less than −1.5 fold was chosen so as to focus the data analysis on a small set of proteins with the largest alterations in abundance [15].
Data access
RAW files will be made available upon request. Supplemental Table 1 lists proteins quantified in this dataset, while Supplemental Table 2 provides the peptides quantified in this dataset.
RESULTS and DISCUSSION
Sample processing was efficient and suitable for high-throughput analyses
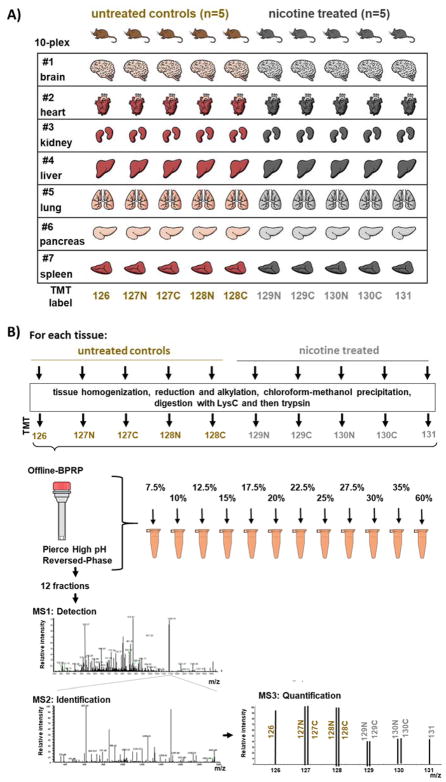
The samples used in this experiment are shown in Figure 1A and the workflow is illustrated in Figure 1B. Although we are not limited by protein amount, as milligrams can be extracted from these tissues, conservative sample processing can better utilize reagents and time. We aimed to perform a large-scale analysis of mouse tissue while efficiently processing samples. Previously, protein level TMT-based mass spectrometry protocols [6, 16, 17] recommended 50–100 μg of peptide per condition, however, only one-fifth to one-tenth of the sample is typically analyzed by the mass spectrometry, the remaining is either discarded or used for re-analysis in the event that errors occurred during the initial analysis. Here, we labeled only one-fifth of the typical starting material (20 μg), and thus use the proportional amount of the labeling reagent. Notably, aside from instrument time, the labeling reagent is typically the costliest component of a TMT experiment. Moreover, we fractionated via a step gradient over spin columns, allowing multiplexed processing of pooled samples and thereby permitting us to simultaneously fractionate our seven separate TMT10-plex experiments (70 total samples) in under 1 hr. The techniques used herein can be applied in experiments where protein amount is limited, thereby circumventing the requirement of 50–100 μg of protein per channel for deep proteome analysis.
Figure 1. Samples analyzed and experimental TMT10-plex design.
A) Seven tissues were selected from five controls and five nicotine-treated mice. One TMT10-plex experiment was performed per tissue. B) Samples were processed following a modified TMT-based protocol. Tissues were homogenized and precipitated proteins were digested with LysC and trypsin prior to TMT labeling. Pooled peptides were fractionated via offline, spin column-based basic pH reversed-phase (BPRP) liquid chromatography into 12 fractions (sequential elution using 7.5%, 10%, 12.5%, 15%, 17.5%, 20%, 22.5%, 25%, 27.5%, 30%, 35%, and 60% acetonitrile) prior to SPS-MS3 analysis.
In total, over 11,516 non-redundant proteins were quantified in the seven TMT10-plex experiments
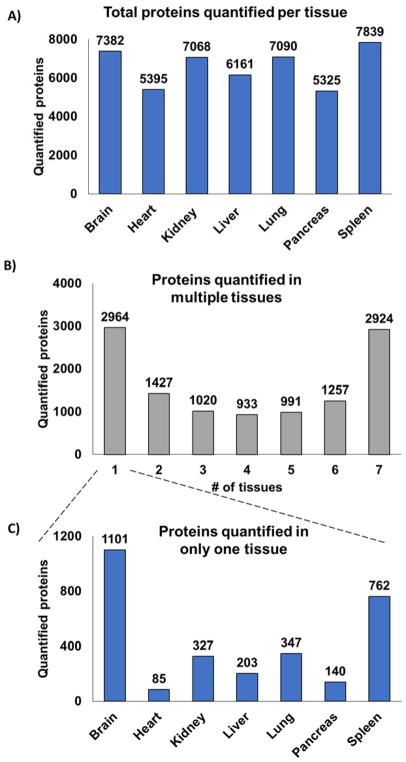
The number of proteins quantified in each tissue ranged from 5,325 to 7,839 at a 1% false discovery rate (FDR) for each tissue: pancreas (5,325 proteins), heart (5,395 proteins), liver (6,161 proteins), kidney (7,068 proteins), lung (7,090 proteins), brain (7,382 proteins), and spleen (7,839 proteins), as illustrated in Figure 2A. These numbers approached those from previous studies [17], which used substantially more starting material. As expected, not all proteins were quantified in all tissues. In fact, a bimodal distribution is observed, as almost equally as many proteins were identified in one tissue as were so in all seven TMT10-plexes (Figure 2B). A total of 2,964 proteins were quantified in a single tissue, while 2,900 were so in all tissues. Of the 2,964 total proteins quantified in a single tissue, brain had the most unique proteins with 1,101, and heart with the least (only 85 proteins) (Figure 2C).
Figure 2. Proteins quantified for all seven tissues.
A) Thousands of non-redundant proteins were quantified per tissue. B) A bar chart illustrating the number of proteins identified in multiple organs showed that most were quantified in either only one tissue or were common across all seven tissues. C) We separated further the proteins identified in only one tissue.
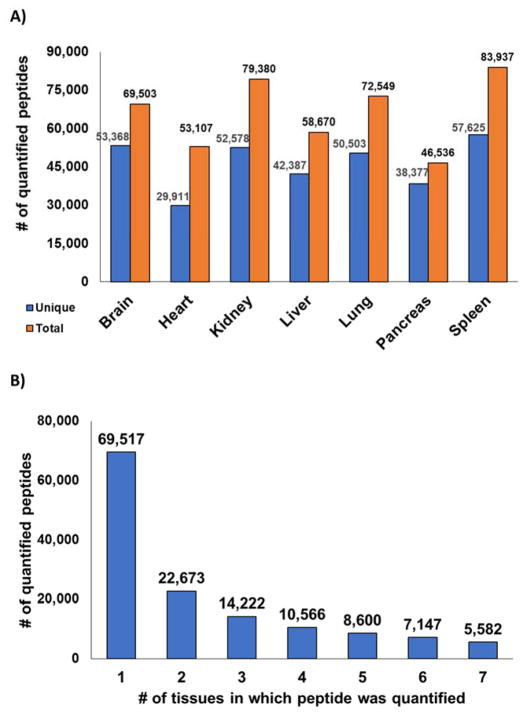
Tens of thousands of peptides were identified in the seven TMT experiments. The number of total peptides identified in each experiment ranged from 46,536 in pancreas to 83,937 in spleen at a 1% false discovery rate. Considering only unique peptides per tissue resulted in a range from 29,911 peptides in heart to 57,625 in spleen (Figure 3A). In the complete dataset of the seven tissues, a total of 138,307 non-redundant peptides were quantified. As performed for the protein level analysis, we tallied the number of tissues in which a peptide was quantified (Figure 3B). This analysis revealed that the majority of peptides (69,517) were identified in only 1 tissue. These data were not entirely surprising due to incomplete and stochastic sampling of the ionized peptide population. Also, cells from specific tissues may process proteins differently regarding splice variants and/or post-translational modifications (PTMs). In addition to variation in protein abundances among tissues, differential PTMs and splice variants will contribute to a diverse peptide population, which is not considered in typical database searching.
Figure 3. Peptides quantified for all seven tissues.
A) Thousands of non-redundant peptides were quantified per tissue. B) A bar chart illustrates the number of peptides identified in multiple organs.
Several tissue-specific proteins demonstrated significant abundance alterations in nicotine-exposed mice
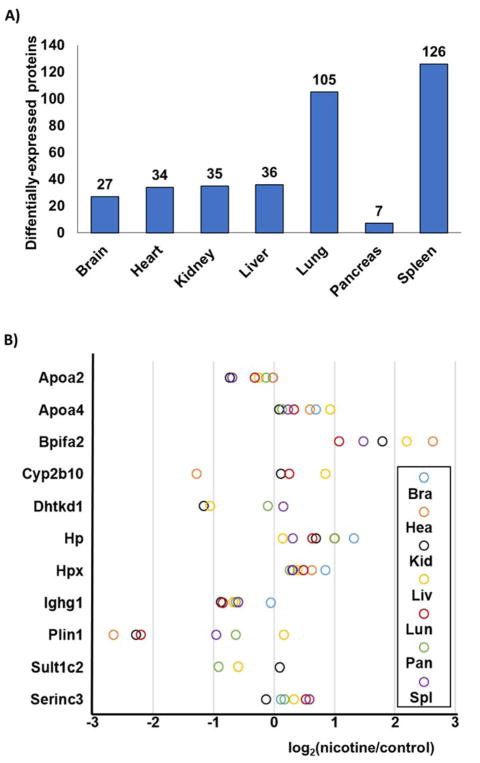
We defined statistical significance as an absolute fold-change in abundance exceeding 1.5 having a p-value <0.05. In total, 340 non-redundant proteins were altered by nicotine treatment across the 7 murine tissues (Supplemental Table 3). Of these, between 7 and 126 proteins were significantly altered in tissues originating from nicotine exposed mice (Figure 4A). The tissues ranked from the least to the highest number of altered proteins were as follows: pancreas (7 proteins), brain (27 proteins), heart (34 proteins), kidney (35 proteins), liver (36 proteins), lung (105 proteins) and spleen (126 proteins). These numbers did not directly correlate with the total proteins quantified for each tissue. For example, brain tissue demonstrated the second fewest number of differentially regulated proteins, but represented the tissue with the second most proteins quantified. Lists of these significantly altered proteins for each tissue were analyzed via DAVID to explore potential enrichment in gene ontology and UniProt classification terms. Table 1 summarizes the top categories in which proteins quantified in only one tissue were classified. A high percentage of proteins classified as “secreted” were identified in several tissues, specifically brain, heart, kidney, and liver. In contrast, lung and spleen showed enrichment in acetylated and nucleotide-binding proteins. Overall, no significant global effect of nicotine on protein abundance in a specific gene ontology category was observed.
Figure 4. Proteins significantly altered following nicotine treatment.
A) The number of statistically significant proteins (defined as displaying at least a 1.5-fold change and as having a p-value <0.05) were tallied for each tissue. B) Proteins demonstrating statistical significance in two or more tissues were illustrated in a modified dot plot.
Table 1.
Gene ontology category enrichment of proteins quantified in only one tissue.
| Tissue | Category | No. proteins | % totala |
|---|---|---|---|
| Brain | |||
| Secreted | 16 | 55.2% | |
| Glycoprotein | 15 | 51.7% | |
| Heart | |||
| Signal | 15 | 42.9% | |
| Secreted | 14 | 40.0% | |
| N-linked glycosylation | 13 | 37.1% | |
| Kidney | |||
| Secreted | 13 | 34.2% | |
| Lipid metabolism | 5 | 13.2% | |
| Liver | |||
| Alternative splicing | 12 | 35.3% | |
| Secreted | 7 | 20.6% | |
| Oxidation-reduction | 6 | 17.6% | |
| Lung | |||
| Cytoplasm | 40 | 38.1% | |
| Acetylation | 24 | 22.9% | |
| Methylation | 18 | 17.1% | |
| Nucleotide-binding | 16 | 15.2% | |
| Calcium ion binding | 12 | 11.4% | |
| Pancreas | |||
| None | N/A | N/A | |
| Spleen | |||
| Acetylation | 54 | 41.5% | |
| Cytoplasm | 53 | 40.8% | |
| Nucleus | 52 | 40.0% | |
| Nucleotide-binding | 20 | 15.4% | |
| Mitotic nuclear division | 19 | 14.6% | |
% total relates to the percentage of proteins that are classified under a given category. As more than one category can be assigned to a protein, the sum of the % total for each organ can exceed 100%.
Eleven proteins were significantly altered in two or more tissues, many of which were identified in other nicotine-related studies
In addition to highlighting proteins that are altered in specific tissues, we also explored the more global effect of nicotine by investigating the proteins that change significantly in abundance in several tissues. We examined the expression profiles of proteins with statistically significant (p-value <0.05; fold change>|1.5|) differences in abundance in two or more tissues due to nicotine treatment. Of the 340 non-redundant proteins that were dysregulated in one of the seven tissues, eleven were significantly altered in two or more tissues. The expression profiles of these representative proteins (Apoa2, Apoa4, Bpifa2, Cyp2b10, Dhtkd1, Hp, Hpx, Ighg1, Plin1, Sult1c2, Serinc3) were illustrated in Figure 4B and described further below.
Six of these differentially-regulated proteins are involved in lipid metabolism: Apoa2, Apoa4, Bpifa2, Plin1, Serinc3, and Cyp2b10. Apoa2 (apolipoprotein A-II) has a role in high-density lipoprotein (HDL) metabolism [18]. In contrast, another apolipoprotein, Apoa4 (apolipoprotein A-IV) has a role in lipid-containing chylomicrons and very-low-density lipoprotein (VLDL) secretion and catabolism [19]. In addition, Plin1 (perilipin 1) is a known modulator of adipocyte lipid metabolism [20]. Likewise, Serinc3 (serine incorporator 3) is involved in sphingolipid metabolic process [21]. Serinc3 was also up-regulated in nicotine-treated cells in various cell lines including, pancreatic stellate cells [6], HEK293 [22], HeLa [22], SH-SY5Y [22], HPNE [23] and Panc1 [23] cells. Bpifa2 (BPI fold A2) lipopolysaccharide binding protein participates in lipid transport [24], and shows a significant increase in abundance in five tissues: lung, spleen, kidney, liver, and heart. Cyp2b10 (cytochrome P450 2B10) is a liver enzyme that oxidases a variety of compounds including steroids and fatty acids [25]. Cyp2b10 also has xenobiotic activity and is a member of a family of proteins known to breakdown nicotine and its derivatives. This protein was significantly up-regulated in liver tissue, as expected. Another xenobiotic enzyme, Sult1c2 (sulfotransferase 1C2), catalyzes the sulfate conjugation of drugs, hormones, and neurotransmitters [26] and was significantly down-regulated in liver and pancreas tissue.
In addition to proteins with roles in lipid metabolism, two proteins involved in iron metabolism (Hp and Hpx) were up-regulated in several tissues. Hp (haptoglobin) is an antioxidant that captures hemoglobin for hepatic recycling of iron to prevent kidney damage [27]. We determined Hp to be up-regulated in all organs, and particularly so in brain and pancreas. Likewise, Hpx (hemopexin) is a secreted protein that transports heme to the liver for iron recovery [28]. Like Hp, Hpx was significantly up-regulated in brain tissue. Other proteins significantly altered in abundance had unrelated functions. Dhtkd1 (dehydrogenase E1 and transketolase domain containing 1) is a mitochondrial protein that is a member of a larger complex involved in the degradation pathways of several amino acids [29]. One particularly interesting protein is Ighg1 (immunoglobulin mu binding protein 2), which is down-regulated in all tissues. Ighg1is a transcriptional regulator (helicase) that unwinds RNA and DNA and has been implicated in pancreatic carcinomas [30]. These proteins may serve as a basis to dissect further the role of nicotine in pathway modulation and to target specific mechanisms that lead to or are consequences of the observed protein alterations. Further assays using the TMT-based strategy outlined herein testing for the effects of nicotine metabolites or breakdown products may be used to clarify if the effect is indeed nicotine-specific.
Tissue-specific and pathway-based nicotine-induced protein changes have implications in future translational investigations
We used DAVID functional analysis to characterize further those eleven proteins that were significantly altered in abundance in more than one tissue when mice were exposed to nicotine. The majority of those proteins were classified as secreted or as being components of extracellular exosomes [31]. Six proteins (Apoa2, Apoa4, Bpifa2, Hp, Hpx, and Sult1c2) were associated with extracellular exosomes. Exosomes are of growing interest in clinical applications particularly for prognosis, for therapy, and as biomarkers [32]. Exosomes – which may contain RNA, proteins, lipids and metabolites - can transfer molecules from one cell to another via membrane vesicle trafficking. Cell culture-based assays support the role of exosomes in signaling pathways, such as Akt, ERK, and STAT3 [33]. Similarly, the six proteins classified as secreted were: Apoa2, Apoa4, Bpifa2, Hp, Hpx, and Ighg1. These proteins are typically present in body fluids and their levels are often indicative of disease [34]. As such, the dysregulation of these proteins may be a consequence of a response, not specific to nicotine, but rather a general response to an external stimulus or drug.
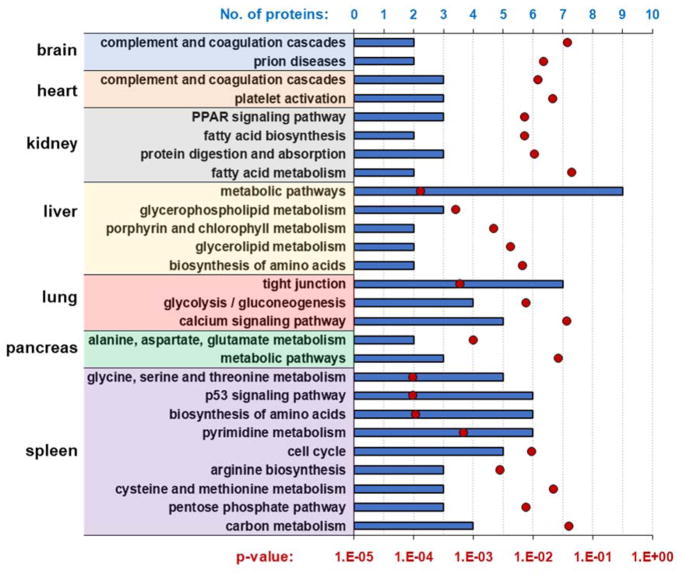
In addition to these more globally altered proteins, we also examined the subset of proteins showing significant changes in abundance for each individual tissue (Figure 2C). We subjected these lists of proteins to a gene set enrichment analysis web server, Enrichr [35], to investigate KEGG pathways that were enriched by nicotine (Figure 5). As may be expected, several of the categories extracted were relevant to nicotinic acetylcholine receptor (nAChR) function or have been associated previously with nicotinic receptors. For example, prion disease-related proteins were identified in brain tissues and nAChR have been associated with APP [36]. The nAChR is a well-studied calcium channel and our data show that calcium signaling pathways were modulated by nicotine in murine lung tissue. Other studies have shown that nicotine can activate cellular pathways in lung cells, although the precise mechanisms, including the role of calcium, have not been defined [37]. In the kidney, proteins related to fatty acid metabolism and PPAR gamma pathways were modulated by nicotine. This effect of nicotine on PPAR gamma had been established previously in smoking studies, and may be instrumental in suppressing the release of pro-inflammatory cytokines [38]. Of the tissues studied, the spleen had the most nicotine-altered proteins, and coincidently the widest variety of altered KEGG pathways. Several proteins affected by nicotine were cell cycle-related. Although nicotine has been associated with cell cycle regulation [39], it has yet to be investigated in the background of the spleen. Also, in brain and heart, the abundance of proteins with roles in complement and coagulation pathways were altered. Nicotine treatments have been shown previously to decrease blood coagulation in animal models and human [40]. Similarity in the heart, proteins involved in platelet activation were altered. Recent studies have demonstrated that nicotine alters platelet activation when using either traditional or electronic cigarettes [41]. Overall, our data highlight the vast extent of nicotine exposure across murine tissues, the biological implications of which may lead to novel clinical applications.
Figure 5. KEGG pathways that were enriched by nicotine.
We subjected these lists of proteins to a gene set enrichment analysis web server, Enrichr [35], to investigate the KEGG pathways that were enriched in each tissue resulting from nicotine treatment. Blue bars represent the number of proteins in that category, while red circle represent p-value of the Fisher’s exact test to test for significance of the enrichment.
As the proteomes of different tissues vary substantially, we expect that tissue-specific protein changes will be observed in response to nicotine treatment, as we discussed above. We note that α7 nAChR activation by nicotine increases calcium influx into cells. In our dataset, we observe that several of altered proteins, and associated pathways, were associated with functions that may be modulated by calcium. For example, platelet activation is strongly dependent on an increase of intracellular Ca2+ concentration [42]. Similarly, in K562 cells, exosome release is regulated by a calcium-dependent mechanism [43]. In addition, calcium is also related to prion protein processing, for example, calcium binding promotes prion protein fragment 90-231 conformational change [44]. Moreover, α7 nAChR-mediated calcium influx may have a role in calcium signaling induced by prion protein interaction with stress-inducible protein 1 [45]. Our data suggest that the biological consequences of observed protein alterations may be a result of changes in cellular calcium concentrations with a distinct effect on different tissues.
Conclusion
We presented the first proteome analysis of multiple tissues from nicotine-treated mice and corresponding controls. The quantification of over 11,000 non-redundant proteins was inferred from over 138,000 different peptides in seven TMT10-plex experiments. Surprisingly, no proteins or classes were identified as significantly altered in all organs, indicating the predominantly local, rather than global, effect of nicotine. Our data revealed 11 proteins to be differentially expressed upon exposure to nicotine in at least two tissues, and dozens more that were tissue-specific that can be used as a starting point for more in-depth studies of the associated pathway and mechanisms. These proteins may be key candidates for further targeted experiments and validation with orthogonal techniques, such as western blotting, immunofluorescence or mass spectrometry-based targeted MS2, specifically parallel reaction monitoring (PRM). The experimental design outlined herein can be modified to expand further the scope of this investigation.
Here we studied the effect of orally-dosed nicotine, however, the route of nicotine administration may be altered to represent a more pharmacologically-relevant delivery of nicotine. In lieu of oral administration, nicotine may be subcutaneously administered via osmatic minipumps [46], which mimics nicotine patch smoking secession treatment, or via inhalation chambers. Also, the procedures outlined herein can be extended to investigations of other drugs. Here, nicotine is the insult, however, analogous experiments using ethanol, nicotinic metabolites, and other cigarette toxins may be performed using similar techniques. The design of the TMT experiment can be also modified to allow for comparisons across tissue types. We chose a five versus five TMT10-plex to enable better statistical analysis. However, recently, an eleventh channel has been added to the TMT10-plex reagent set. The “bridge channel” concept was used previously when comparing triplicates of three conditions with the tenth channel comprised of an equal mix of all other channels [16, 17, 47]. The TMT11 reagent allows for comparison across TMT10-plexes, while retaining the statistical advantages of a five versus five experimental design.
The minimalistic proteomics strategy applied herein efficiently used TMT reagents and offline centrifugation-based fractionation to streamline sample preparation, thereby requiring less starting protein. As such, different cell types may be isolated from tissues, which increases analytical depth and allows for comparisons between tissues and within tissue-derived cell types. Consequently, post-translational modifications play a very important role in signal transduction pathways and may have cell type-specific roles. Moreover, future analyses can interrogate the nicotine-induced phosphoproteomic alterations via TMT10-plex methodologies, as described previously [48]. Comparing across tissues or cell types may also be of interest to some researchers. In summary, we have highlighted the effects of nicotine exposure on the proteome of several mouse tissues in efforts to understand better the tissue-specific effects of the toxin. Moreover, the techniques used herein can be applied to investigate drug-specific protein alterations resulting from other routes of nicotine administration, as well as tissue-based and cell type-specific applications.
Supplementary Material
Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), protein description/name (Description), number of peptides quantified per protein (peptides), and the normalized summed signal-to-noise for each of the 10 channels (126 to 131). Each tab is labeled with the corresponding organ.
Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), description/name (Description), redundancy, peptide sequence (peptide sequence), number of quantified peptides (num_quant), and the summed signal to noise for each of the 10 channels (126 to 131). Each tab is labeled with the corresponding organ.
Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), description/name (Description), redundancy, peptide sequence (peptide sequence), number of quantified peptides (num_quant), and the summed signal to noise for each of the 10 channels (126 to 131).
Acknowledgments
We would like to thank the members of the Gygi Lab at Harvard Medical School. This work was funded in part by an NIH/NIDDK grant K01 DK098285 (J.A.P.) and GM97645 (S.P.G.).
References
- 1.U. D. o. H. a. H. Services, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health. 2014 [Google Scholar]
- 2.C. f. D. C. a. Prevention. 2014 [Google Scholar]
- 3.McKee M, Capewell S. Lancet. 2015;386:1239. doi: 10.1016/S0140-6736(15)00146-4. [DOI] [PubMed] [Google Scholar]; McKee M, Chapman S, Daube M, Glantz S. Lancet. 2014;384:2107. doi: 10.1016/S0140-6736(14)62366-7. [DOI] [PubMed] [Google Scholar]
- 4.Grando SA. Nat Rev Cancer. 2014;14:419. doi: 10.1038/nrc3725. [DOI] [PubMed] [Google Scholar]
- 5.Dasgupta P, Rastogi S, Pillai S, Ordonez-Ercan D, Morris M, Haura E, Chellappan S. J Clin Invest. 2006;116:2208. doi: 10.1172/JCI28164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paulo JA, Gaun A, Gygi SP. Journal of proteome research. 2015;14:4246. doi: 10.1021/acs.jproteome.5b00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Analytical chemistry. 2008;80:2921. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]; Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Analytical chemistry. 2003;75:1895. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]; Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Mol Cell Proteomics. 2004;3:1154. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD, Gygi SP. Analytical chemistry. 2012;84:7469. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ting L, Rad R, Gygi SP, Haas W. Nat Methods. 2011;8:937. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP. Analytical chemistry. 2014;86:7150. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulo JA, O’Connell JD, Gygi SP. Journal of the American Society for Mass Spectrometry. 2016;27:1620. doi: 10.1007/s13361-016-1434-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. Cell. 2010;143:1174. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. Nature biotechnology. 2006;24:1285. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 14.Elias JE, Gygi SP. Methods Mol Biol. 2010;604:55. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Elias JE, Gygi SP. Nat Methods. 2007;4:207. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 15.Podwojski K, Stephan C, Eisenacher M. Methods in molecular biology (Clifton, NJ) 2012;893:3. doi: 10.1007/978-1-61779-885-6_1. [DOI] [PubMed] [Google Scholar]
- 16.Paulo JA, Gygi SP. Proteomics. 2015;15:474. doi: 10.1002/pmic.201400155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paulo JA, McAllister FE, Everley RA, Beausoleil SA, Banks AS, Gygi SP. Proteomics. 2015;15:462. doi: 10.1002/pmic.201400154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsao YK, Wei CF, Robberson DL, Gotto AM, Jr, Chan L. J Biol Chem. 1985;260:15222. [PubMed] [Google Scholar]
- 19.Karathanasis SK, Oettgen P, Haddad IA, Antonarakis SE. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:8457. doi: 10.1073/pnas.83.22.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchette-Mackie EJ, Londos C. J Biol Chem. 1991;266:11341. [PubMed] [Google Scholar]
- 21.Bossolasco M, Lebel M, Lemieux N, Mes-Masson AM. Mol Carcinog. 1999;26:189. [PubMed] [Google Scholar]
- 22.Paulo JA, Gygi SP. Proteomics. 2017:17. doi: 10.1002/pmic.201600319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulo JA. JOP. 2014;15:465. doi: 10.6092/1590-8577/2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prokopovic V, Popovic M, Andjelkovic U, Marsavelski A, Raskovic B, Gavrovic-Jankulovic M, Polovic N. Arch Oral Biol. 2014;59:302. doi: 10.1016/j.archoralbio.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Honkakoski P, Moore R, Gynther J, Negishi M. J Biol Chem. 1996;271:9746. doi: 10.1074/jbc.271.16.9746. [DOI] [PubMed] [Google Scholar]
- 26.Her C, Kaur GP, Athwal RS, Weinshilboum RM. Genomics. 1997;41:467. doi: 10.1006/geno.1997.4683. [DOI] [PubMed] [Google Scholar]
- 27.Wassell J. Clin Lab. 2000;46:547. [PubMed] [Google Scholar]
- 28.Tolosano E, Altruda F. DNA Cell Biol. 2002;21:297. doi: 10.1089/104454902753759717. [DOI] [PubMed] [Google Scholar]
- 29.Xu W, Zhu H, Gu M, Luo Q, Ding J, Yao Y, Chen F, Wang Z. FEBS Lett. 2013;587:3587. doi: 10.1016/j.febslet.2013.08.047. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Ni R, Chen J, Liu Z, Xiao M, Jiang F, Lu C. Pancreas. 2011;40:753. doi: 10.1097/MPA.0b013e318213d51b. [DOI] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Nat Protoc. 2009;4:44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]; Huang da W, Sherman BT, Lempicki RA. Nucleic acids research. 2009;37:1. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keller S, Sanderson MP, Stoeck A, Altevogt P. Immunol Lett. 2006;107:102. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]; van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R. Pharmacological reviews. 2012;64:676. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 33.Silva AM, Teixeira JH, Almeida MI, Goncalves RM, Barbosa MA, Santos SG. Eur J Pharm Sci. 2017;98:86. doi: 10.1016/j.ejps.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 34.Csosz E, Kallo G, Markus B, Deak E, Csutak A, Tozser J. J Proteomics. 2017;153:30. doi: 10.1016/j.jprot.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 35.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A. Nucleic acids research. 2016;44:W90. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabbagh MN, Walker DG, Reid RT, Stadnick T, Anand K, Lue LF. Neurosci Lett. 2008;448:217. doi: 10.1016/j.neulet.2008.10.004. [DOI] [PubMed] [Google Scholar]; Seo J, Kim S, Kim H, Park CH, Jeong S, Lee J, Choi SH, Chang K, Rah J, Koo J, Kim E, Suh Y. Biol Psychiatry. 2001;49:240. doi: 10.1016/s0006-3223(00)01124-0. [DOI] [PubMed] [Google Scholar]
- 37.Zanetti F, Giacomello M, Donati Y, Carnesecchi S, Frieden M, Barazzone-Argiroffo C. Free radical biology & medicine. 2014;76:173. doi: 10.1016/j.freeradbiomed.2014.08.002. [DOI] [PubMed] [Google Scholar]; Carlisle DL, Liu X, Hopkins TM, Swick MC, Dhir R, Siegfried JM. Pulm Pharmacol Ther. 2007;20:629. doi: 10.1016/j.pupt.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Amoruso A, Bardelli C, Gunella G, Fresu LG, Ferrero V, Brunelleschi S. Life Sci. 2007;81:906. doi: 10.1016/j.lfs.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 39.Lee HJ, Guo HY, Lee SK, Jeon BH, Jun CD, Lee SK, Park MH, Kim EC. Journal of oral pathology & medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2005;34:436. doi: 10.1111/j.1600-0714.2005.00342.x. [DOI] [PubMed] [Google Scholar]; Chu M, Guo J, Chen CY. J Biol Chem. 2005;280:6369. doi: 10.1074/jbc.M408947200. [DOI] [PubMed] [Google Scholar]
- 40.Blann AD, Steele C, McCollum CN. Thromb Haemost. 1997;78:1093. [PubMed] [Google Scholar]; Wenzel DG, Singh J. J Pharm Sci. 1962;51:875. doi: 10.1002/jps.2600510913. [DOI] [PubMed] [Google Scholar]
- 41.Hom S, Chen L, Wang T, Ghebrehiwet B, Yin W, Rubenstein DA. Platelets. 2016;27:694. doi: 10.3109/09537104.2016.1158403. [DOI] [PubMed] [Google Scholar]; Girdhar G, Xu S, Bluestein D, Jesty J. Nicotine & tobacco research: official journal of the Society for Research on Nicotine and Tobacco. 2008;10:1737. doi: 10.1080/14622200802443528. [DOI] [PubMed] [Google Scholar]
- 42.Davlouros P, Xanthopoulou I, Mparampoutis N, Giannopoulos G, Deftereos S, Alexopoulos D. Med Chem. 2016;12:131. doi: 10.2174/157340641202160208195923. [DOI] [PubMed] [Google Scholar]
- 43.Savina A, Furlan M, Vidal M, Colombo MI. J Biol Chem. 2003;278:20083. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 44.Sorrentino S, Bucciarelli T, Corsaro A, Tosatto A, Thellung S, Villa V, Schinina ME, Maras B, Galeno R, Scotti L, Creati F, Marrone A, Re N, Aceto A, Florio T, Mazzanti M. PLoS One. 2012;7:e38314. doi: 10.1371/journal.pone.0038314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beraldo FH, Arantes CP, Santos TG, Queiroz NG, Young K, Rylett RJ, Markus RP, Prado MA, Martins VR. J Biol Chem. 2010;285:36542. doi: 10.1074/jbc.M110.157263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dickson PE, Rogers TD, Lester DB, Miller MM, Matta SG, Chesler EJ, Goldowitz D, Blaha CD, Mittleman G. Psychopharmacology (Berl) 2011;215:631. doi: 10.1007/s00213-010-2159-2. [DOI] [PubMed] [Google Scholar]
- 47.Paulo JA, O’Connell JD, Gaun A, Gygi SP. Molecular biology of the cell. 2015;26:4063. doi: 10.1091/mbc.E15-07-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Possemato AP, Paulo JA, Mulhern D, Guo A, Gygi SP, Beausoleil SA. Journal of proteome research. 2017;16:1506. doi: 10.1021/acs.jproteome.6b00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), protein description/name (Description), number of peptides quantified per protein (peptides), and the normalized summed signal-to-noise for each of the 10 channels (126 to 131). Each tab is labeled with the corresponding organ.
Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), description/name (Description), redundancy, peptide sequence (peptide sequence), number of quantified peptides (num_quant), and the summed signal to noise for each of the 10 channels (126 to 131). Each tab is labeled with the corresponding organ.
Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), description/name (Description), redundancy, peptide sequence (peptide sequence), number of quantified peptides (num_quant), and the summed signal to noise for each of the 10 channels (126 to 131).