Abstract
Non-syndromic cleft lip with or without cleft palate (NS-CL/P) is one of the most common human birth defects and is generally considered a complex trait. Despite numerous loci identified by genome-wide association studies, the effect sizes of common variants are relatively small, with much of the presumed genetic contribution remaining elusive. We report exome-sequencing results in 209 people from 72 multi-affected families with pedigree structures consistent with autosomal-dominant inheritance and variable penetrance. Herein, pathogenic variants are described in four genes encoding components of the p120-catenin complex (CTNND1, PLEKHA7, PLEKHA5) and an epithelial splicing regulator (ESRP2), in addition to the known CL/P-associated gene, CDH1, which encodes E-cadherin. The findings were also validated in a second cohort of 497 people with NS-CL/P, comprising small families and singletons with pathogenic variants in these genes identified in 14% of multi-affected families and 2% of the replication cohort of smaller families. Enriched expression of each gene/protein in human and mouse embryonic oro-palatal epithelia, demonstration of functional impact of CTNND1 and ESRP2 variants, and recapitulation of the CL/P spectrum in Ctnnd1 knockout mice support a causative role in CL/P pathogenesis. These data show that primary defects in regulators of epithelial cell adhesion are the most significant contributors to NS-CL/P identified to date and that inherited and de novo single gene variants explain a substantial proportion of NS-CL/P.
Keywords: cleft lip, cleft palate, cleft lip/palate, exome sequencing, adherens junction, epithelia, cell adhesion, cadherin, catenin, knockout
Introduction
Cleft lip with or without cleft palate (CL/P) is one of the most common human birth defects with an incidence of 1 in 700–1,000 people.1 Much of the insight into CL/P pathogenesis has come from the identification of genes harboring mutations that cause syndromic forms of CL/P, including van der Woude syndrome (IRF6 [MIM: 607199], GRHL3 [MIM: 608317]), Opitz GBBB syndrome (MID1 [MIM: 300552], SPECC1L [MIM: 614140]), ectrodactyly, ectodermal dysplasia, cleft lip/palate (EEC) syndrome (p63 [MIM: 603273]), branchio-oculo-facial syndrome (TFAP2A [MIM: 107580]), and familial gastric cancer with CL/P (CDH1 [MIM: 192090]).1 Additionally, variants in CTNND1 (MIM: 601045) and CDH1 were recently identified as the cause of blepharocheilodontic syndrome, of which CL/P is a variable feature of the syndrome.2 However, approximately 70% of case subjects are non-syndromic (NS-CL/P) in nature. Concordance rates in monozygotic twins of 25%–45%,3, 4 variable expressivity, and reduced penetrance in families has led NS-CL/P to be considered a complex trait due to genetic, environmental, and stochastic factors.5 GWAS and meta-GWA studies on NS-CL/P have implicated 40 genomic loci, each of relatively small effect size,5, 6, 7 and include only a few syndromic CL/P-associated genes, such as IRF6. However, other association studies have also found evidence of a contribution of single-nucleotide variants in CDH1 in NS-CL/P.8, 9, 10, 11 In addition, recent studies have identified pathogenic variants in ARHGAP29 (MIM: 610496) in familial cleft palate.12, 13, 14 While progress has been made in identifying genetic etiologies, a biological pathway for NS-CL/P has hitherto not been established.
Here, we report the results of a large whole-exome sequencing (WES) study in a NS-CL/P discovery cohort of 72 multigenerational families, the findings of which we have replicated in a second NS-CL/P cohort of 497 individuals from 444 families (both singleton and familial case subjects). Twenty pathogenic or likely pathogenic variants were identified in five functionally linked genes, encoding proteins regulating the assembly of the epithelial cadherin-catenin complex. Variants in four of these genes are now implicated as a cause of CL/P in humans. These studies reveal that dysregulation of the epithelial adhesion complex is a significant primary mechanism responsible for NS-CL/P.
Material and Methods
Recruitment and Sample Collection
Seventy-two multiplex families with non-syndromic cleft lip with or without cleft palate were ascertained by the J.C.M. and A.C.L. laboratories, as well as clinics in Sydney, Australia. Families were selected from a larger cohort by pedigree analysis for apparent segregation consistent with multigenerational autosomal-dominant inheritance, with or without reduced penetrance limited to one or two linking individuals. A smaller number of families with apparent autosomal-recessive or X-linked recessive pedigree structures were also enrolled. The WES cohort consisted of families from the Philippines (n = 41), Colombia (n = 15), European origins in the USA and Australia (n = 15), Hispanic (n = 1), and of unknown origins (n = 2). The replication cohort consisted of individuals from the Philippines (n = 356), USA (n = 135), Guatemala (n = 1), and of unknown origins (n = 5). Informed consent was obtained prior to sample testing and the procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national). DNA was extracted using standard protocols and sample quality-control steps were taken including co-efficient of inbreeding and XY genotyping.
Exome Sequencing and Genomic Data Generation
DNA samples from three affected individuals from each multi-generational family were selected for exome sequencing which was performed at the University of Washington, Center for Mendelian Genomics, University of Washington, Seattle. A Roche/Nimblegen SeqCap EZ v2.0kit was used for enrichment capture and samples sequenced using the Illumina HiSeq 2500. The raw sequences from 209 libraries, which met quality standards, were aligned to the human genome build hg19 using BWA software.15 Single-nucleotide variant (SNV) and insertion/deletion (indel) calls were performed using PICARD, SAMTOOLS, and GATK.16, 17, 18
Bioinformatics and Genomic Data Analysis
Files from each individual were joint-called using GATK to present all variant types within a single VCF file. SNVs and indels were analyzed in joint-called VCF files for each individual. As the GATK Unified Genotyper16 provides missing genotypes (i.e., “no calls”), homozygous reference sites were embedded in missing genotype sites by single-sample calling variants. This allowed variants at high frequency within the cohort to be de-prioritized as being unlikely to be the etiology in multiple families with Mendelian forms of CL/P. Variants were annotated with the Ensembl Variant Effect Predictor (VEP)19 and then filtered using GEMINI20 according to a standardized set of criteria to identify rare and pathogenic variants. The majority of families were assessed as having an autosomal-dominant (AD) form of CL/P and therefore a heterozygous model of inheritance was used for analysis. Variants in families where autosomal-recessive (AR) inheritance patterns were possible according to pedigree structure were also filtered for homozygous and compound heterozygous forms of inheritance. Candidate variants in each family were identified from a subset of variants derived using clinical and bioinformatics criteria, including filtering for population frequencies and in silico pathogenicity scores. As there is no database of genomic variants from Filipino control subjects, we used comparative data from closely related populations from East Asia, principally Han Chinese. After standard filters for variant quality, candidate variants were prioritized based on rarity in the general population, conservation of the affected sites, and predicted functional impact. In general, nonsense mutations, splice acceptor/donor site mutations, frameshift insertions, or deletions were considered to be loss-of-function variants. The likely pathogenicity of missense variants and their effects on protein function were predicted by using in silico tools such as PolyPhen-2, SIFT, and v.1.3 CADD, with a threshold score for the latter set at >15, which is the median value for all possible canonical splice site changes and non-synonymous variants. Gene tolerance to variation was calculated for all variants using the residual variation intolerance score (RVIS) and then combined with the PROVEAN scores for missense variants, which combines Grantham and conservation scores. Evolutionary conservation, effects on protein structure, very low frequency, absence in controls, the presence of pathogenic variants in the same gene in multiple families, and the nature of the mutation were taken into account to increase the priority of a candidate gene. External population control databases used included the 1000 Genomes database, the Seattle-based Exome Variant Server (EVS) database, and the BROAD-based Exome Aggregation Consortium (ExAC and gnomAD) (see Web Resources). Genes harboring candidate variants in more than one family were also identified and prompted reassessment of these genes for variants of interest in additional families where variants may not have met criteria for pathogenicity. The top candidate variants were then assessed for segregation within families using Sanger sequencing. All individuals for whom DNA was available were tested in the segregation analyses. Mutated protein models were constructed in HOPE and PyMOL.
Prioritization of Candidate Genes
Genes of interest were classified as likely or possible candidates through the following criteria: consistent zygosity for familial inheritance pattern, rarity, location in a functional protein domain, the availability of an animal model with a consistent phenotype, links to a known biological pathway for CL/P, and expression in embryonic orofacial tissues in mouse and humans. For analysis of mouse gene expression, the bioinformatics tool SysFACE (Systems tool for craniofacial expression-based gene discovery) was used to analyze microarray-based genome-level gene expression profiles from various dissected embryonic day 10.5 orofacial epithelia and neuroepithelia. The following Affymetrix Mouse Gene 1.0 ST array datasets, available in the FaceBase repository, were analyzed: medial neuroepithelium (FB00000106), central neural epithelium (FB00000349), flanking neural epithelium (FB00000350), lateral eminence neural epithelium (FB00000351), medial eminence neural epithelium (FB00000352), mandibular columnar epithelium (FB00000353), and maxillary columnar epithelium (FB00000354). Analysis was performed using an in-house script to obtain gene level normalized expression intensities.
Sanger Sequencing
Sanger sequencing was used to validate identified variants in affected and unaffected relatives. The accession numbers for the genes described in this study were GenBank: NM_001085460.1 (CTNND1), NM_175058.4 (PLEKHA7 [MIM: 612686]), NM_019012.5 (PLEKHA5 [MIM: 607770]), NM_024939.2 (ESRP2 [MIM: 612960]), and NM_004360.3 (CDH1). Primers around the genomic region of the variants of interest were designed using Primer 3 with primer sequences and conditions are available upon request. PCR products were sent for sequencing using an ABI 3730XL (Functional Biosciences). Chromatograms were transferred to a UNIX workstation, base-called with PHRED (v. 0.961028), assembled with PHRAP (v. 0960731), scanned by POLYPHRED (v. 0.970312), and viewed with CONSED program (v. 4.0).
Targeted, Biotinylated Probe Hybridization-Based Capture
The five genes described in this study were targeted for sequencing in the validation cohort of 497 people. This included 296 singletons with CL/P and no family history as well as a further 201 people, one each from a multi-affected family. Sample libraries for genes of interest were constructed using the KAPA HyperPlus kit (KAPA Biosystems). Reactions were assembled using a SciClone G3 (Perkin-Elmer) liquid handling robot with 500 ng of input DNA for each sample using a laboratory-adapted version of the manufacturer’s manual protocol. All samples were dual-indexed with standard Illumina-compatible adapters. Libraries were pooled (up to 24 per pool) prior to target enrichment. Custom xGen (IDT) probes were designed and purchased to target the exons and putative cis-regulatory regions. Following enrichment, pools were combined to produce a sequencing template containing up to 192 libraries. Paired-end sequencing (2 × 100 bp) of the templates was performed using Rapid Run v2.0 (Illumina) chemistry on a HiSeq 2500 (Illumina) sequencer according to the manufacturer’s recommended protocol. Resulting sequences were aligned to the human genome reference (hg19) using the Burrows-Wheeler Aligner (BWA) and variants identified with the Genome Analysis Tool Kit (GATK). A joint VCF was generated for subsequent annotation using the same bioinformatic platform as described for WES analysis in this study.
Tissue Collection and Immunohistochemistry
Human fetal tissues were recovered, with the age of specimens estimated by gestational ultrasound and/or fetal foot measurements.21 Specimens ranging in age from 57 to 70 days after conception were used as these represent the time period during which the secondary palate fuses. Younger specimens spanning the time of primary palatal fusion were not available. Embryonic tissues were embedded in paraffin and 4-micron sections cut in the coronal plane. Sections were then stained with various primary antibodies—CDH1/E-cadherin (BD Biosciences #610181), CTNND1/p120Ctn (isoform 1: SantaCruz #sc23873 [6H11]; pan-isoform: SantaCruz #sc23872 [15D2]), PLEKHA5 (SantaCruz #sc390311), PLEKHA7 (OneWorldLabs #bs13730R), and ESRP2 (Novus Biologicals #NBP2-13972)—followed by an HRP-conjugated secondary antibody of the appropriate species as previously described.22
Generation of Expression Constructs
Full-length mouse Ctnnd1 (p120Ctn) cDNA (isoform 1A) was a kind gift of Dr. Konstantin Birukov (University of Chicago) with permission from Dr. Al Reynolds (Vanderbilt University). The complete Ctnnd1-1A open reading frame was amplified and cloned into pCR8-TOPO (Invitrogen). Each CTNND1 mutation identified in the initial cohort of families was found to affect residues conserved between humans and mice, and so each of these variants was independently introduced into the full-length mCtnnd1-pCR8-TOPO clone using either overlap PCR or by swapping out a restriction fragment from a synthesized fragment containing the specific mutation of interest (Genscript). Primer sequences used for all amplifications are available upon request. Each pCR8-TOPO clone was then transferred into the following Gateway vectors: pcDNA3.1 (no tag), pDEST53 (N-terminal GFP tag), and pDEST-myc (N-terminal myc tag). All clones were validated by sequencing.
Cell Culture and Transfection
Human SW-48 epithelial cells was obtained from ATCC (CCL-231) and maintained in L-15 Medium (30-2008) containing 10% FCS without carbon dioxide. Lipofectamine 3000 (Invitrogen) was used to transfect SW-48 cells with p120Ctn wild-type and mutant expression constructs, according to manufacturer instructions.
Cells were washed with cold PBS twice and harvested in 1% digitonin lysis buffer (50 mM Tris [pH 7.4], 100 mM NaCl, 1 mM EDTA [pH 8.0]) with protease/phosphatase inhibitors. The lysates were centrifuged at 14,000 rpm for 10 min to remove insoluble cellular debris. Protein was quantified by BCA Protein Assay (Pierce) and normalized. 500 μg of total protein was pre-cleared with Protein A/G Magnetic Beads (Pierce) at 4°C for 30 min to remove nonspecific binding proteins for each transfectant and respective controls. Co-immunoprecipitation was performed using Anti-c-Myc Magnetic Beads (Pierce) for 4 hr at 4°C. Magnetic beads were washed twice with lysis buffer without digitonin, eluted with 100 mM glycine (pH 2.0), and neutralized with 1 M Tris (pH 8.5). Immuno analysis of whole-cell lysate and co-immunoprecipitated eluate was performed and quantified using capillary electrophoresis on a WesSimple Western (ProteinSimple) automated platform using the following antibodies; C-Myc [9E10] (1:100 Abcam Ab32), E-cadherin (1:500 BD Biosciences #610181), p120Ctn (isoform 1:1:500 Abcam Ab11508 [6H11]; pan-isoform: 1:250 SantaCruz #sc23872 [15D2]). E-cadherin followed by an HRP-conjugated secondary antibody of the appropriate species (ProteinSimple). Experiments were repeated in full three times.
Statistical Analyses
Data are reported as mean E-cadherin:c-Myc tagged p120Ctn ratios +SD. Statistical significance was determined by two-tailed unpaired t test with equal variance. Statistical significance was set at p < 0.05.
Mouse Conditional Knockout Generation
Ctnnd1 floxed mice (Ctnnd1fl/fl) were a gift from Dr. Al Reynolds (Vanderbilt University) via Dr. Jennifer Bailey (University of Texas Houston).23 Epithelial-specific Cre driver mice (Crect), in which the Cre recombinase is driven by a TFAP2A enhancer, were a gift from Dr. Trevor Williams (University of Colorado).24 Female Ctnnd1fl/fl mice were initially bred with male Crect mice to generate heterozygous conditional null mice (Ctnnd1cKO/+; Crect). Male offspring were then backcrossed to female Ctnnd1fl/fl mice and embryos harvested as described below. A few pregnant dams were allowed to litter down to examine phenotypes at birth. All mice were genotyped using the relevant primers sets; embryonic and neonatal sex was determined by genotyping for Sry.
Embryo Collection and Tomographic Imaging
Pregnant mice were euthanized at between 10 and 18 days after detection of a vaginal plug—defined as 0.5 days post-coitum (dpc) on the morning the plug was detected. Embryos were dissected from the uterus into ice-cold phosphate-buffered saline (PBS), then fixed in 4% paraformaldehyde overnight at 4°C. Embryos older than 14.5 dpc were decapitated immediately following dissection. For the pregnant dams that gave birth, pups that were found dead at birth were promptly removed from the cages and the mandibles removed for palatal inspection. The heads were subsequently placed in 4% paraformaldehyde. Embryos (or embryonic heads) were then embedded in low-melting-point agarose and cleared for optical projection tomography (OPT). The dissected neonatal heads were imaged using micro-computed tomography (microCT) using standard protocols. OPT and microCT imaging was conducted in the Small Animal Tomographic Analysis (SANTA) Facility at the Seattle Children’s Research Institute using a Bioptonics 3001M and Skyscan 1076 scanners, respectively. Raw imaging data from both modalities were reconstructed using NRecon v1.6.9.4 software (Skyscan, Belgium), then imported into Drishti v2.6.1 for 3D rendering and image capture.
ESRP2 Functional Studies
Expression vectors for the wild-type and the p.Arg520∗ truncation mutant of ESRP2 were generated by PCR amplification of a cDNA corresponding to MGC clone BC030146. The wild-type clone was amplified with primers For_XhoI_hESRP2 and Rev_BamHI-hESRP2 and the truncation mutant with primers For_XhoI_hESRP2 and Rev_BamHI-R520Ter. The PCR products were cloned into the BamH1 and Xho1 sites of the previously described pIBX-C-FF-B vector for expression as C-terminal FLAG tagged proteins. Plasmids were transfected in 293T cells using Mirus 293T according to the manufacturer’s protocol and RNA and protein were harvested 48 hr later using Trizol reagent (Thermo) and RIPA lysis buffer (Santa Cruz Biotechnology). Reverse-transcription PCR was performed to analyze splicing patterns of ESRP regulated exons as previously described.25
Results
Whole-Exome Sequencing and Targeted, Biotinylated Probe Hybridization-Based Capture Reveal a Role of the Epithelial Adhesion Complex in NS-CL/P
Ten pathogenic or likely pathogenic alleles were identified in five genes by WES in the Mendelian discovery cohort (14% of families; Table 1). Variant pathogenicity was determined following the ACMG/Sherloc framework26 (see Material and Methods). These included CTNND1 (5 families, 6.9%), PLEKHA7 (1 family, 1.4%), PLEKHA5 (1 family, 1.4%), and ESRP2 (1 family, 1.4%), as well as CDH1 (2 families, 2.8%). Notably, three genes (CTNND1, PLEKHA5, PLEKHA7) encode core components of the p120-catenin (p120Ctn) complex that bind to two other known CL/P proteins, E-cadherin and Nectin1.27, 28 ESRP2 encodes an epithelial-specific mRNA splicing factor, which regulates epithelial adhesion through target genes including CTNND1.29, 30 Re-sequencing of these five genes in a second multinational NS-CL/P cohort of 497 individuals from 444 families (296 singletons and 201 familial) validated these findings by identifying a further 10 independent pathogenic or likely pathogenic alleles (CTNND1, 4 alleles; PLEKHA7, 2 alleles; ESPR2 1 allele; and CDH1, 3 alleles) (2.25%) with each variant present in only one family (Table 1). An additional 9 class 3C and 2 class 3B VOUS were also identified in the validation cohort (CTNND1, 2 alleles; PLEKHA5, 5 alleles; PLEKHA7, 1 allele; and ESPR2, 3 alleles).
Table 1.
Variant and Clinical Summary
| Gene | Variant | Domain | Family | Origin | Inheritance | Cohort | gnomAD | Interpretation |
|---|---|---|---|---|---|---|---|---|
| CTNND1 | chr11:g.57559005C>G; p.Gln19Glu | CC domain | 1133 | USA | multi-affected AD | WES | 9 NFE | likely pathogenic; FGF8 p.Asp120Asn likely pathogenic |
| chr11:g.57564445_57564446del; p.Asp313Profs∗9 | before ARM1 | 20001528 | Philippines | singleton | replication | – | pathogenic, de novoa | |
| chr11:g.57569255G>A; p.Trp336∗ | before ARM1 | 114 | Colombia | multi-affected AD | WES | – | pathogenic, non-penetrant | |
| chr11:g.57571168A>G; p.Asp499Gly | ARM4 | 20000128 | Philippines | multi-affected AD | WES | – | likely pathogenic, non-penetrant, reduced E-cadherin binding. New donor site | |
| chr11:g.57572202C>T; p.Leu558Phe | ARM5 | 528 | USA | multi-affected AD | WES | 16 NFE, 2 L, 1 O | likely pathogenic, reduced E-cadherin binding | |
| chr11:g.57573381C>T; p.Arg584Trp | ARM6 | 368 | Philippines | multi-affected AD | WES | – | likely pathogenic, abolishes binding to E-cadherin, | |
| chr11:g.57575761G>T; p.Trp690Cys | ARM7 | 20021556 | USA | singleton | replication | – | likely pathogenic, de novoa | |
| chr11:g.57576939G>T; c.2417+1G>T | splice – after ARM10 | 20031592 | USA | singleton | replication | – | pathogenic - segregation not possible; canonical splice | |
| chr11:g.57578892C>T; p.Arg852∗ | after ARM10 | 4450 | Philippines | multi-affected AD | replication | – | likely pathogenic | |
| PLEKHA5 | chr12:g.19440414A>G; p.Tyr590Cys | p120Ctn binding domain | 20010429 | Philippines | multi-affected AD, de novo | WES | 1 L | pathogenic, de novoa |
| PLEKHA7 | chr11:g.16838582C>T; p.Gly544Asp | p120Ctn binding domain | 20040409 | Philippines | multi-affected AD | WES | 1 NFE | likely pathogenic |
| chr11:g.16838676G>A; p.Arg513Trp | – | 6160 | USA | singleton | replication | 3 NFE, 1 SA | class 3C VOUS, or likely pathogenic, bi-parental inheritance | |
| chr11:g.16834682T>C; p.Asp662Gly | p120Ctn binding domain | 6160 | USA | singleton | replication | – | likely pathogenic, bi-parental inheritance | |
| ESRP2 | chr16:g.68266284C>T; p.Arg315His | RRM1 | 4798 | Philippines | multi-affected AD | WES | 3 L | likely pathogenic |
| chr16:g.68265234G>A; p.Arg520∗ | RRM3 | 4763 | Philippines | multi-affected AD | replication | 1 FE, 3 NFE | likely pathogenic | |
| CDH1 | chr16:g.68844164C>T; p.Thr251Met | EC1-2 linker | 20013522 | USA | singleton | replication | 1 EA | likely pathogenic |
| chr16:g.68844172G>A; p.Asp254Asn | EC1-2 linker | 6128 | USA | singleton | replication | – | likely pathogenic, de novoa | |
| chr16:g.68844180T>A; p.Asn256Lys | EC1-2 linker | SA | Australia | multi-affected AD | WES | – | likely pathogenic | |
| chr16:g.68849586G>A; p.Glu497Lys | EC4-5 linker | 20021591 | Philippines | singleton | replication | – | likely pathogenic | |
| chr16:g.68855958A>T; p.Asn589Ile | EC4-5 linker | 4991 | Philippines | multi-affected AD | replication | – | likely pathogenic, de novoa | |
| chr16:g.68863687_68863688del; p.Asn809Ilefs∗3 | beta-catenin binding domain | 1001 | USA | Multi-affected AD | WES | – | likely pathogenic |
Genomic positions are from GRCh37. Abbreviations: NFE, Non-Finnish European; L, Latino; O, other; SA, South Asian; FE, Finnish European; AD, autosomal-dominant.
GWA/TDT and allele studies consistent with family structure
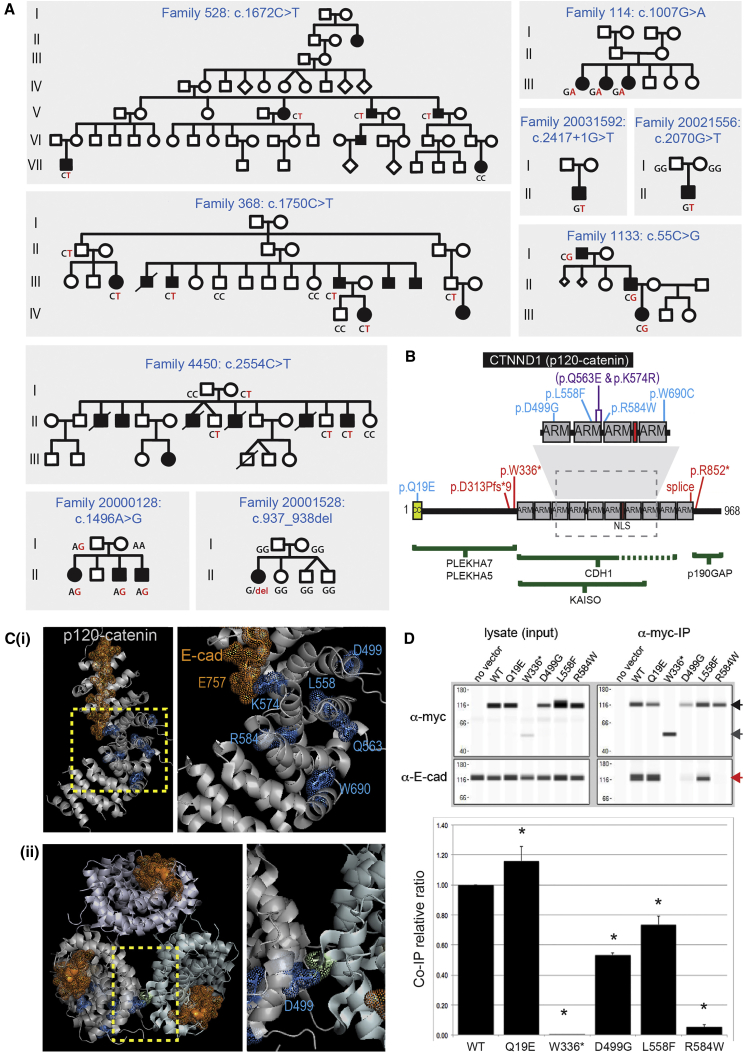
In total, nine likely pathogenic variants and two class 3C were identified in CTNND1, which encodes p120Ctn (Figure 1A), a critical E-cadherin binding partner that stabilizes adherens junction strength.31 Review of the clinical presentations of these individuals confirmed a diagnosis of NS-CL/P. Of the pathogenic variants, two nonsense (c.1007G>A [p.Trp336∗] and c.2554C>T [p.Arg852∗]), a frameshift (c.937_938del [p.Asp313Profs∗9]), and a splice site mutation (c.2417+1G>T) (Figure 1B) are predicted to result in nonsense-mediated mRNA decay. Four missense variants (c.1496A>G [p.Asp499Gly]; c.1672C>T [p.Leu558Phe]; c.1750C>T [p.Arg584Trp]; and c.2070G>T [p.Trp690Cys]) cluster in the ARM domains, responsible for binding the E-cadherin tail (Figures 1B and 1C). The p.Gln19Glu (c.55C>G), identified in a three-generation NS-CL/P-affected family (1133) also exhibiting hearing loss, hypodontia, and enamel hypoplasia, is located near the N terminus of the long isoform of p120Ctn (Figure 1B). A second variant that also co-segregated in this family and met the criteria for likely pathogenicity was identified in FGF8 (MIM: 600483; GenBank: NM_033163.3; c.359A>G [p.Asp120Gly]). FGF8 variants have previously been implicated in CL/P pathogenesis as well as tooth agenesis.32, 33, 34 Therefore, further studies may be warranted to determine which rare variant—FGF8 p.Asp120Gly or CTNND1 p.Gln19Glu—is causal or whether both contribute to the clinical presentation in this family. Of note, the two class 3C variants, c.1721A>G (p.Lys574Arg) and c.1687C>G (p.Gln563Glu), were identified in a singleton family (2328) and were located in cis (Table S1). The p.Lys574Arg variant impacts an amino acid that interacts directly with the E-cadherin juxtamembrane core region via glutamate 757, which is conserved in all mammalian cadherins that bind p120Ctn. This variant and p.Gln563Glu both reside in same ARM domain. We surmise that one or both variants may be causative and could act as a complex allele with potential additive effects.
Figure 1.
CTNND1 Variants
(A) Pedigrees showing segregation of CTNND1 variants. Segregation of indicated variants was performed by Sanger sequencing in all individuals for whom DNA was available.
(B) CTNND1 (p120Ctn) protein domain structure showing variants (blue, pathogenic missense variants; purple, missense variants of unknown significance; red, nonsense/frameshift variants). Regions required for binding partner interaction are shown below as green bars.
(C) X-ray structure of the p120Ctn ARM domains (silver) complexed with the E-cadherin tail juxtamembrane core (gold dots and sticks) (PDB: 3l6y). The dashed box in (B) and the left image in (i) highlight the enlarged image to the right in (i). Mutated residues are identified in blue (dots/sticks). The E-cadherin E757 juxtamembrane core residue, which interacts with p120Ctn Lys574, is indicated in gold. (ii) Top-down view of the packing of the p120Ctn monomer structure. Asp499 is positioned at the monomer interaction surface. The unlabeled residue (chartreuse dots and sticks) on the p120Ctn monomer contacts Asp499.
(D) Co-immunoprecipitation of endogenous E-cadherin with myc-tagged ectopically expressed wild-type and mutant p120Ctn proteins (top). Western blots showing each ectopically expressed p120Ctn protein (detected using an anti-myc antibody) and endogenous E-cadherin in each lysate and following p120Ctn immunoprecipitation. Full-length p120Ctn (black arrow), p.Trp336∗ protein (dark gray arrow), and E-cadherin (red arrow, lower panel). The relative amounts of E-cadherin pulled down are shown relative to the amount of the respective immunoprecipitated p120Ctn proteins, following normalization to the wild-type p120Ctn:E-cadherin ratio. Data are reported as mean E-cadherin:c-Myc tagged p120Ctn ratios + SD; ∗p < 0.05 (n = 3).
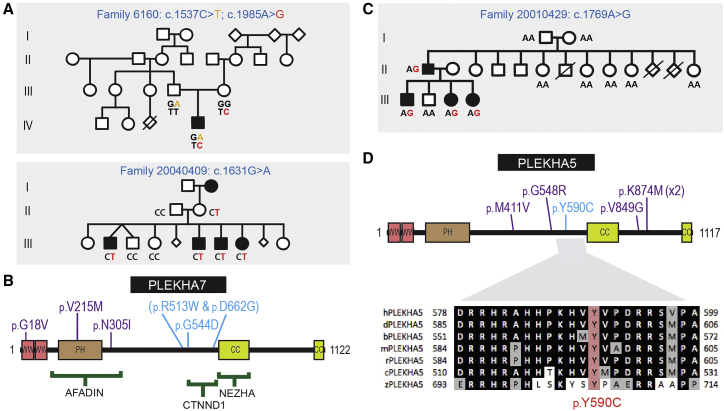
A likely pathogenic missense variant in PLEKHA7 (c.1631G>A [p.Gly544Asp]) was identified in a family with four affected siblings (20040409). Two additional rare missense variants in PLEKHA7 (c.1537C>T [p.Arg513Trp]; c.1985A>G [p.Asp662Gly]) were identified in a singleton (6160) in the validation cohort. These variants were inherited from separate parents (see Figure 2A) and therefore could represent an additive or autosomal-recessive disease mechanism. The p.Gly544Asp and p.Asp662Gly variants reside in the p120Ctn binding region of PLEKHA7, with the p.Arg513Trp variant residing just outside the minimal interaction region.35 Three additional class 3C variants, which localized to N-terminal PLEKHA7 domains, were identified in other families (Figure 2B, Table S1). A rare variant in PLEKHA5 (a close paralog of PLEKHA7), c.1769A>G (p.Tyr590Cys), arose de novo in an affected male (family 20010429) and then segregated with the phenotype, providing compelling support for a role of PLEKHA5 in NS-CL/P (Figure 2C). This variant resides in the region homologous to the PLEKHA7 p120Ctn interaction domain in which the p.Gly544Asp and p.Asp662Gly variants are located. Five rare PLEKHA5 class 3C variants were identified in six additional families (Figure 2D, Table S1). While there is support for some PLEKHA5 and PLEKHA7 variants being pathogenic, others may represent alleles that contribute to susceptibility in a polygenic model for cleft lip and palate. PLEKHA5 and to a lesser degree PLEKHA7 should therefore be considered as strong candidate genes for NS-CL/P.
Figure 2.
Rare Variants Identified in PLEKHA7 and PLEKHA5
(A) Pedigrees showing segregation of PLEKHA7 variants. Segregation of indicated variants was performed by Sanger sequencing in all individuals for whom DNA was available.
(B) PLEKHA7 protein structure showing variants (blue, pathogenic missense variants; purple, missense variants of unknown significance).
(C) Family 20010429 showing de novo PLEKHA5 c.1769A>G variant (p.Tyr590Cys) in the second generation and subsequent segregation (PDB: 3q2v).
(D) PLEKHA5 protein structure showing variants (blue, pathogenic missense variants; purple, missense variants of unknown significance). Lower panel: Evolutionary conservation of Tyr590, a phosphorylation site. Sequences shown from human (h), dog (d), cow (b), mouse (m), rat (r), chick (c), and zebrafish (z).
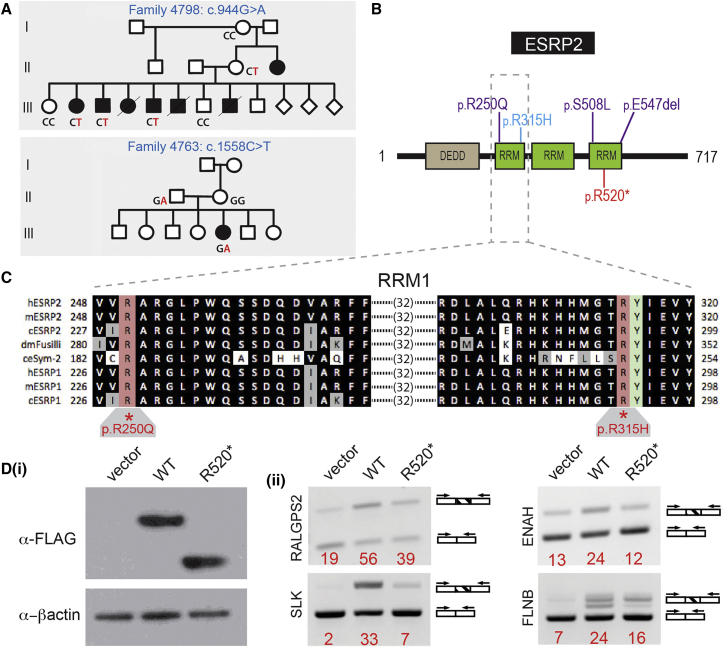
Two variants were identified in a fourth gene, ESRP2, which encodes a regulator of epithelial mRNA splicing (Figure 3). A missense variant, c.944G>A (p.Arg315His) (family 4798), localizes to the first of three RNA recognition motifs (RRMs). One additional allele, a stop-gain variant (c.1558C>T [p.Arg520∗]) located in the third RRM (Figure 3B), was identified in the validation cohort. Three class 3C variants were also identified in additional families, including one in the first RRM and two in the third RRM (Figure 3B, Table S1). The variant of unknown significance in the first RRM, c.749G>A (p.Arg250Gln), involves a residue that is invariant in all ESRP2 and ESRP1 proteins down to C. elegans (Figure 3C). As with PLEKHA5 and PLEKHA7, some variants in ESRP2 may represent alleles that contribute to susceptibility in the respective individuals.
Figure 3.
ESRP2 Variants
(A) Pedigrees showing segregation of ESRP2 variants. Segregation of indicated variants was performed by Sanger sequencing in all individuals for whom DNA was available.
(B) ESRP2 protein structure variants (blue, pathogenic missense variants; purple, missense variants of unknown significance; red, nonsense variant).
(C) Evolutionary conservation of the first RNA recognition motif (RRM1) in ESRP1 and ESRP2 with the orthologs of ESRP2 in D. melanogaster and C. elegans. The locations and conservation of two residues mutated in NS-CL/P, Arg250 and Arg315, are shown. Arg315 is adjacent the invariant Tyr316 that is critical for RNA recognition.
(D) Expression of the ESRP2 nonsense mutant, p.Arg520∗, in HEK293T cells alters the amount of splice isoforms of ESRP2 target genes (ii) compared to cells expressing comparable ectopic wild-type ESRP2 protein (i).
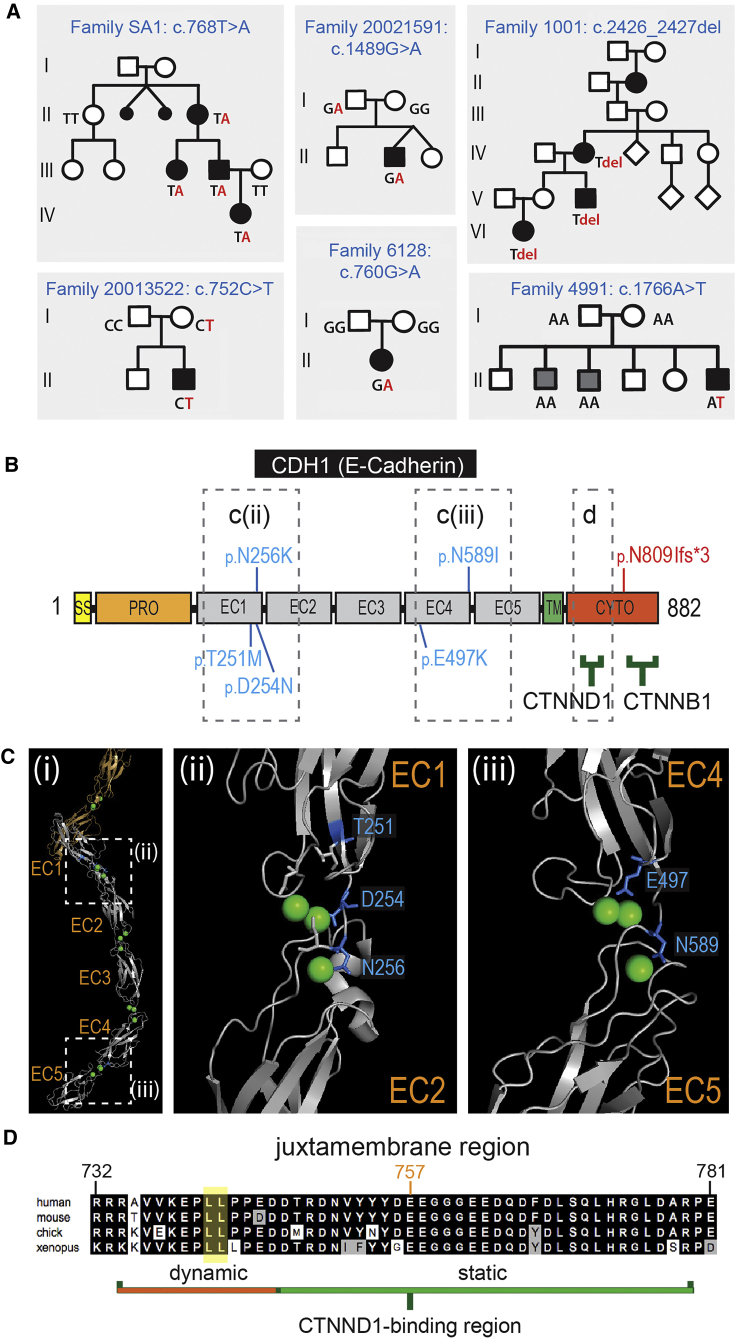
Six likely pathogenic variants were identified in CDH1 (Figure 4). Five missense variants (c.752C>T [p.Thr251Met]; c.760G>A [p.Asp254Asn]; c.768T>A [p.Asn256Lys]; c.1489G>A [p.Glu497Lys]; c.1766A>T [p.Asn589Ile]) cluster in the calcium-binding hinge regions between the extracellular cadherin (EC) domains: three to the EC1-EC2 hinge region and two to the EC4-EC5 hinge region (Figures 4B and 4C). The sixth variant, classified as pathogenic, is a frameshift mutation (c.2426_2427del [p.Asn809Ilefs∗3]) that is located immediately after the C-terminal region, with the mRNA expected to undergo nonsense-mediated decay.
Figure 4.
CDH1 Variants
(A) Pedigrees showing segregation of CDH1 variants. Segregation of indicated variants was performed by Sanger sequencing in all individuals for whom DNA was available. Family 4991 shows a de novo pathogenic variant in the individual presenting with CL/P. Two siblings (gray squares) have secondary palate clefting and do not carry the CDH1 variant; sibling phenotype due to a second unidentified cause.
(B) CDH1 (E-cadherin) protein structure showing variants (blue, missense variants; red, frameshift variant). Binding partner protein regions shown as green bars. The dashed boxes in (B) and (Ci) are represented in the X-ray structure shown in (Cii) and (Ciii), and the alignment in (D).
(C) (i) The X-ray structure of the extracellular region of E-cadherin. Dashed boxes mark the regions enlarged to the right: (ii) the Ca2+-binding EC1-EC2 hinge region, and (iii) the Ca2+-binding EC4-EC5 hinge region. In all cases the CL/P missense mutations involve residues (blue) surrounding the Ca2+ ions (green spheres) with some residues participating in chelation.
(D) Sequence conservation of the E-cadherin juxtamembrane core region essential for binding to CTNND1/p120Ctn. The dileucine endocytic motif (yellow) and the conserved Glu757, which interacts with p120Ctn Lys574 in the X-ray structure (Figure 1), are indicated.
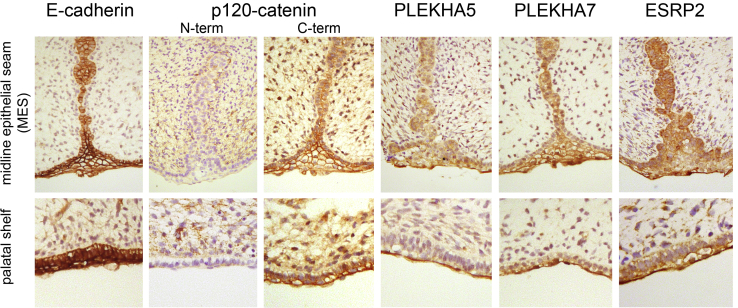
p120Ctn, PLEKHA5, PLEKHA7, ESRP2, and E-Cadherin Are Co-localized in Developing Human and Mouse Palatal Epithelia
A prediction from these findings is that the genes reported here would be expressed in palatal tissues at the time of fusion. Immunohistochemistry was performed to detect the protein products for each of these candidates on sectioned human fetal palatal tissue from the time of secondary palate fusion (67–72 days gestation). Two antibodies were used to distinguish between p120Ctn isoforms, which have distinct roles in epithelia and mesenchyme. Staining using an N-terminally directed isoform 1-specific p120Ctn antibody showed weak staining in the palatal shelf epithelia and the medial epithelial seam (MES) but prominent levels in the facial mesenchyme. In contrast, a C-terminally directed pan-isoform p120Ctn antibody showed strong staining in all oral and nasal epithelia (basal and peridermal layers), including the MES in fusing palatal shelves and less prominent staining in the facial mesenchyme (Figure 5). These data are consistent with the main short isoform (isoform 3) having a prominent role in epithelia and the longer isoform (isoform 1) in non-adherent cell types. The staining for both PLEKHA5 and PLEKHA7 indicated higher levels in the MES and periderm compared to basal epithelia. ESRP2 also stained all epithelia, but slightly stronger staining was apparent in the peridermal layer (Figure 5). Each of the five genes described here were also analyzed using SysFACE and shown to be prominently expressed in E10.5 maxillary epithelia in mice, similar to other known CL/P-associated genes (Table S2).
Figure 5.
Immunohistochemical Detection of CL/P Candidate Proteins in 67–72 day Human Embryonic Oral and Secondary Palatal Epithelia
Strong E-cadherin staining seen in all oral epithelia (basal and periderm layers). p120Ctn (pan-isoform antibody; C-term), PLEKHA5, PLEKHA7, and ESRP2 are also seen in all oral epithelia but show stronger staining in the peridermal layer of the palatal shelves as well as throughout the midline epithelial seam (MES). Staining of p120Ctn isoform 1 (N-term) is seen throughout the oral mesenchyme and weakly in the basal epithelial layer but is absent from the peridermal layer of the palate and MES.
NS-CL/P-Associated Mutations Disrupt the E-Cadherin-p120Ctn-PLEKHA Complex
Previous functional studies of E-cadherin hinge region variants are consistent with these mutations reducing E-cadherin adhesiveness36 and so functional assessments focused on pathogenic variants identified in CTNND1. Myc-tagged versions of each of the mutant p120Ctn proteins and the wild-type protein were ectopically expressed in SW-48 cells, which lack endogenous p120Ctn.37 p.Trp336∗ was included as a negative control since it lacks the domains required for E-cadherin binding. Co-immunoprecipitation assays showed that both the p.Trp336∗ and p.Arg584Trp variants were unable to bind endogenous E-cadherin (Figure 1D). The p.Asp499Gly and p.Leu558Phe variants retained some ability to bind E-cadherin, although this was significantly reduced compared to wild-type (Figure 1D). The p.Gln19Glu variant, which is encoded only by the p120Ctn long isoform that promotes a mesenchymal-like cellular phenotype, showed a statistically significant increase in co-precipitated E-cadherin.
Conditional Ablation of Ctnnd1 in Developing Oral Epithelia Results in Clefting
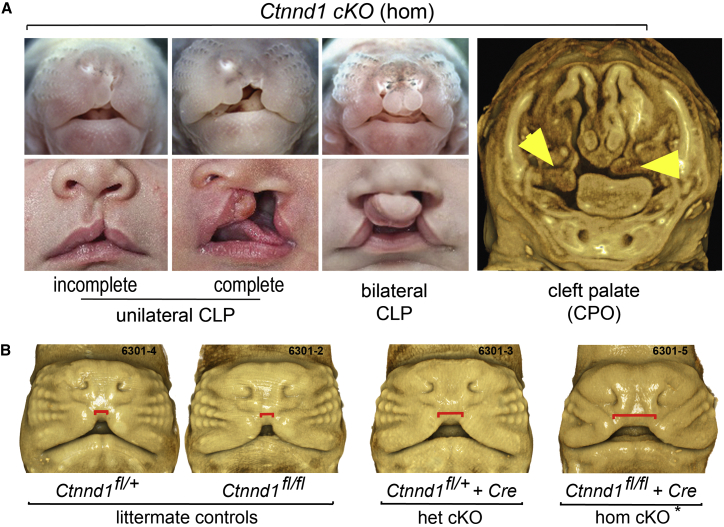
The role of Ctnnd1 in palate development was also assessed in vivo using a conditional gene knockout approach in mice. Mice carrying floxed alleles of mCtnnd1 were bred with a Cre driver line in which recombinase expression is driven by an oral epithelial-specific enhancer of TFAP2.24 Analysis of embryos at gestational ages 10.5–18.5 dpc showed the full spectrum of CL/P presentations in homozygotes (Figure 6). Overt clefts were observed in ∼47% (7/15) of homozygous embryos. 3D imaging of the remaining embryos using optical projection tomography detected obvious nasal airway asymmetry in approximately half the animals studied and delayed maxillary prominence growth in others (Figure 6B). Although no heterozygotes with an overt cleft were identified (n = 25), 3D imaging of embryos at 11.5 dpc was also able to identify heterozygotes by delayed medial growth of the maxillary labial processes (Figure 6B), consistent with dosage-dependent sensitivity to p120Ctn in the facial epithelia.
Figure 6.
Conditional Ablation of Ctnnd1 in the Embryonic Mouse Oral Epithelium Results in CL/P
(A) Mouse embryos homozygous (hom) for a Ctnnd1 oral epithelial-specific deletion displayed variable clefts—unilateral CL only (top left), unilateral CL/P (top center), bilateral CL/P (top right), or cleft secondary palate only (far right)—similar to the human spectrum (lower panel).
(B) 3D OPT-imaged littermates of different genotypes. Conditional heterozygotes and non-cleft presenting conditional homozygotes (∗) are distinguished by a delay in medial growth of the maxillary prominences shown by the increased gap marked by the red line. Non-cleft presenting homozygotes showed dysmorphic nasal tips and nares.
ESRP2 Disruption Leads to Aberrant Splicing of Epithelial Isoforms of Multiple Target Genes
Selected variants in ESRP2 were investigated for disruption of epithelial RNA splicing. cDNAs for the wild-type and two mutant ESRP2 proteins (p.Arg520∗ and p.Arg315His) were transfected into 293T cells and RNA was analyzed for splicing effects as previously described. 293T cells do not express endogenous ESRP1 (MIM: 612959) or ESRP2 and thus transfections of these cells with cDNA for either splicing factor is sufficient to result in a switch from mesenchymal toward epithelial splicing patterns.30 As shown in Figure 3D, transfection of wild-type ESRP2 produced the expected splicing switches in RALGPS2 (MIM: 617819), SLK (MIM: 616563), FLNB (MIM: 603381), and ENAH (MIM: 609061) transcripts compared to the control. Transfection of either mutant cDNA induced variable amounts of splicing switch toward the epithelial pattern that was less robust than that of the wild-type construct (Figure 3Dii for p.Arg520∗).
Discussion
The genes underlying the known syndromic forms of CL/P encode proteins involved in diverse cellular processes.1, 38 GWA studies have implicated 40 chromosomal loci, each of small effect size, with common variants in only a few of the syndromic CL/P genes contributing to NS-CL/P.5, 6, 7 The hypothesis for this study was that Mendelian gene and de novo etiologies for CL/P, which are not testable through GWA studies, would be identified by WES in multi-affected families. Consistent with this hypothesis, we report the identification of pathogenic or likely pathogenic variants in five genes in 14% of the multigenerational families in which NS-CL/P segregated in an apparent autosomal-dominant fashion with moderate to high penetrance. These data were validated by the finding of pathogenic or likely pathogenic variants in the same five genes in 2.25% of a replication cohort. This replication cohort consisted of representative affected individuals from 201 smaller families (3 with pathogenic or likely pathogenic variants, 1.5%) and 296 singletons (8 with pathogenic or likely pathogenic variants, 2.7%). The difference in variant frequencies between singletons and smaller affected families is not significant (Fisher’s exact test one tailed p = 0.283). Notably, additional rare variants of unknown significance in these genes were found in a further 3% of the replication cohort. Some of these could be pathogenic alleles, but it is also possible that the additional variants of unknown significance could contribute to NS-CL/P under a polygenic susceptibility model. Three of these genes (CTNND1, PLEKHA5, and ESRP2) represent previously unreported human NS-CL/P-associated genes, PLEKHA7 represents a strong candidate gene, and newly identified variants in CDH1 significantly increase the number reported to be associated with NS-CL/P and provide a clear mechanism of action. In support of pathogenicity for variants in these genes, we demonstrate that (1) each protein is localized almost exclusively in the oral and palatal epithelia at fusion, (2) investigated p120Ctn variants disrupt E-cadherin interaction, and (3) conditional Ctnnd1 ablation in mouse oral epithelia results in a spectrum of CL/P phenotypes that mimics that seen in individuals with CL/P. This establishes dysfunction in the regulators of epithelial adhesion dynamics as a significant cause of NS-CL/P, with major implications for molecular diagnostics, recurrence risk, and reproductive counseling for individuals with CL/P.
Recently, variants in CTNND1 and CDH1 have been identified as the cause of blepharocheilodontic syndrome, a rare syndrome in which CL/P is a variable feature.2 In this study we have demonstrated the role of variants in CTNND1, CDH1, and their interacting partner proteins in NS-CL/P. The involvement of CTNND1 is supported by the diversity in variant type (nonsense, frameshift, splicing, and missense) as well as the clustering of missense variants within the protein. Three protein-truncating variants and a variant affecting splicing (c.2417+1G>T) are highly likely to result in mRNA nonsense-mediated decay and haploinsufficiency. It is also striking that the majority (four of the five) of the pathogenic missense mutations and both variants of unknown significance are located within ARM repeats 3 through 6, which provide the residues that contact the critical juxtamembrane core of the E-cadherin tail31, 39, 40 (colored gold in Figures 1Ci and 1D). Substitution at arginine 584 alters the positively charged inner surface of the ARM domains that interface with the negatively charged E-cadherin juxtamembrane region. In support of pathogenicity, we show that p.Arg584Trp disrupts the ability of p120Ctn to bind E-cadherin (Figure 1D). Of note are two rare variants of unknown significance (p.Gln563Glu and p.Lys574Arg) identified in one affected individual, with the latter directly interacting with the negatively charged E-cadherin tail via the highly conserved Glu757 (Figures 1Ci and 4D). In contrast, variants not situated on the interaction interface (p.Asp499Gly and p.Leu558Phe) impact E-cadherin binding to a lesser but still significant degree. Each missense variant is therefore expected to be a loss-of-function or partial loss-of-function allele. Previous studies have shown that either reduction in p120Ctn levels or uncoupling E-cadherin interaction result in increased cadherin endocytosis, reduced cell-cell adhesion, and changes in epithelial morphology and behavior.40, 41, 42 p120Ctn is proposed to multimerize to facilitate E-cadherin clustering and stabilization. Although it is unknown whether the p120Ctn X-ray structure stacking mirrors the expected multimerization in vivo, it is noteworthy that aspartate 499 is the contacting residue between the monomers (Figure 1Cii). We speculate that p.Asp499Gly could also impact E-cadherin clustering independent of its effect on the interaction with the E-cadherin tail.
The essential role for the p120Ctn complex in fusion of the lip and palate in vivo was confirmed by conditional ablation of Ctnnd1 in oral epithelia in mice. Ctnnd1 ablation caused a spectrum of facial clefting phenotypes and various nasal airway asymmetries. In contrast to human phenotypes, conditional loss of one Ctnnd1 allele in mice delayed maxillary growth but did not cause overt clefting. This difference in sensitivity may be due to protective effects of specific inbred mouse models, that mutations in p120Ctn in humans also impact its mesenchymal function, thus compounding the effect on oral epithelium, or other as yet unidentified non-genetic influences as is widely accepted as contributing to NS-CL/P. Nevertheless, the variety and frequency of mutations, together with functional and animal studies, support loss of function of p120Ctn as the most significant cause of NS-CL/P reported to date.
Additional evidence for the role of the p120Ctn-complex in CL/P is provided by co-segregating variants in PLEKHA7 and its close paralog, PLEKHA5. PLEKHA7 was first identified as a microtubule-linked p120Ctn interactor critical for the regulation of surface expression of E-cadherin.35 PLEKHA7 has also been shown to bind Afadin, which in turn binds the C terminus of Nectin1 (encoded by another CL/P-associated gene, PVRL1), to initiate epithelial adherens junction formation.43 The finding of multiple variants in the paralogous PLEKHA5 in individuals with CL/P, including one (c.1769A>G [p.Tyr590Cys]) that arose de novo in one affected individual and then co-segregated with CL/P, provides compelling evidence of pathogenicity for this second member of the PLEKHA family. Co-expression of both PLEKHA proteins with E-cadherin and p120Ctn in the oral periderm and the regulation of endocytosis by PLEKHA544 are also consistent with this role. Each of these PLEKHA5/7 variants is predicted to lead to reduced E-cadherin surface levels or stability similar to that seen with loss of p120Ctn and knockdown of PLEKHA7.35, 45, 46 However, further functional studies and modeling in mice is required to determine whether all variants in these two genes are causal or whether they contribute to susceptibility in a polygenic model for CL/P.
The epithelial mRNA splicing regulators ESRP1 and ESRP2 have been implicated in CL/P pathogenesis through mouse studies and are well-characterized regulators of epithelial adhesion.29, 47 Here, the findings in humans of pathogenic variants in ESRP2 confirm the critical role of this splice regulator in the oral epithelium and suggest that ESRP2 may play the more critical role in humans. Two ESRP2 variants meet formal criteria of pathogenicity, with p.Arg315His localized in the RRM1 RNA recognition motif adjacent to the key tyrosine residue (Tyr316) required for RNA binding and the p.Arg520∗ variant predicted to undergo mRNA nonsense-mediated decay. Additional RNA recognition motif variants were conservatively classed as variants of uncertain significance (VUS). In particular, p.Arg250Gln lies within the first RNA recognition motif that is conserved in all ESRP2 and ESRP1 proteins as well as other closely related RNA splicing factors (Figure 3C).
Truncating and missense mutations in CDH1 have been described in families with hereditary gastric and breast cancer, with 5% of those families also having at least one affected family member with CL/P. The E-cadherin variants reported here were present in individuals with isolated CL/P but with neither gastric nor breast cancer, other than in one family (SA1) where one person was recently diagnosed with gastric cancer. Many of the hereditary diffuse gastric cancer-linked E-cadherin missense mutations are located in the extracellular cadherin (EC) domain hinge regions and are associated with reduced epithelial cell-cell adhesion.48 Each of the CL/P-associated missense variants reported here also localize to hinge regions: the EC1-EC2 hinge (p.Thr251Met, p.Asp254Asn, and p.Asn256Lys) or EC4-EC5 hinge (p.Glu497Lys and p.Asn589Ile), with some involved directly in calcium chelation. A missense mutation of Asn256 has been reported in an individual with gastric cancer without CL/P,36 while p.Asp254Asn is one of the few CL/P-associated E-cadherin variants previously described.8 Functional and structural studies in E-cadherin have confirmed that calcium binding to this hinge region stabilizes the extracellular cadherin loop structure to facilitate high-affinity transdimerization and strong epithelial adhesion.49, 50, 51 It remains to be elucidated why some CDH1 variants cause NS-CL/P while others give rise to gastric cancer or broader developmental phenotypes.
The identification of these genes highlights the utility of a Mendelian approach to gene identification in multi-affected families for phenotypes conventionally regarded as sporadic or multifactorial. It is noteworthy that numerous additional variants in these five genes were observed in other NS-CL/P-affected families in this study, representing a further 3% of the cohort (Table S1). Although not able to be formally categorized as pathogenic using the ACMG/Sherloc framework,26 many of these variants could still be disease causing or at least contributors to susceptibility. These include CTNND1 p.Lys574Arg (impacts direct contact with E-cadherin), PLEKHA5 p.Gly548Arg (located within the p120Ctn interaction domain and also predicted to impact RNA splicing), and ESRP2 p.Ser508Leu (hydrogen bond disruption).
The functional links between the five genes identified in this study and their high mutant frequency in two NS-CL/P cohorts are striking, consistent with the epithelial cadherin-catenin complex as an important biological pathway for CL/P in humans. NS-CL/P was previously considered to be a complex trait in most affected individuals but our findings provide compelling evidence for genes of major Mendelian effect in NS-CL/P. Pathogenic variants were not detected in any of the previously identified genes associated with Mendelian CL/P including ARHGAP29, IRF6, MSX1, MAFB, SATB2, and TP63 emphasizing the importance of the p120Ctn-E-cadherin complex. A regulator of the epithelial adhesive state (ESRP2) provides additional evidence that deregulation of epithelial adhesion is also a primary mechanism in Mendelian forms of human NS-CL/P. The majority of families had a single pathogenic allele in the genes described in this study, consistent with a Mendelian etiology with incomplete penetrance in a subset. A second potentially pathogenic allele was identified in four families, raising the possibility of an oligogenic model, including family 4798 (FGF8 p.Asp120Gly, likely pathogenic), family 20021591 (ESRP2 p.Glu547del class 3C VUS), and family 2328 where two missense variants (p.Lys574Arg and p.Gln563Glu [class 3C VUS]) in the ARM domains of p120Ctn were identified. This study therefore opens up new avenues to investigate gene interactions in determining CL/P penetrance and indicates that a subset of NS-CL/P-affected case subjects may be principally monogenic in origin, implying that a proportion of isolated cases are the result of de novo mutations. We recommend that detailed clinical phenotyping should be combined with consideration of diagnostic testing for the p120-Ecadherin gene complex and previously identified CL/P-associated genes such as IRF6, TP63, SATB2, and MSX1 in multiplex families presenting with NS-CL/P as a first line assessment. Molecular testing through a qualified medical genetic service could also be considered as a component of precision medicine in singletons with NS-CL/P to provide a personalized rather than a population-derived empiric recurrence risk.
Acknowledgments
We thank all families that put their faith in our research program for without them, none of this would be possible. We are grateful for the technical assistance of Bruce Bedell (University of Iowa), Ryan Anderson (Seattle Children’s Research Institute), and Theresa Naluai-Cecchini (Birth Defects Research Laboratory, University of Washington). Many nurses, physicians, community health care workers, and laboratory staff who both provide care to the families and facilitated sample collection were essential to this study including Sandy Daack-Hirsch and Operation Smile. This work was supported by a project grant from the Australian National Health & Medical Research Council (AU/1/BA51117) to T.R., M.F.B., H.Z., H.v.B., T.C.C., J.C.M., D.A.N., E.K., and A.C.L., the Laurel Foundation for Pediatric Craniofacial Research (T.C.C.), NIH R01-DE014667 (A.C.L.), NIH R37-DE008559 (J.C.M.), the March of Dimes Basil O’Connor award #FY 98-0718 and research grant #6-FY01-616 (A.C.L.), NIH R24-HD000836 (I.A.G., T.C.C., M.D.), NIH R03-DE024776 (S.A.L.), and NIH UM1-HG006493 (D.A.N., M.J.B.). We would like to thank Aaron Quinlan for devising and assisting with the bioinformatics analysis platform, GEMINI. Mice were housed at the Seattle Children’s Research Institute and all work approved by the Animal Care and Use Committee (#14614). Human fetal tissues were recovered under an approved protocol through the University of Washington (IRB# STUDY00000380). Families were enrolled into the study under the respective institutional review board protocols (AU/1/BA51117), Genetics of CL/P: a multicenter international consortium (PIROSTUDY10777), and the University of Iowa IRB (200109094).
Published: May 24, 2018
Footnotes
Supplemental Data include three tables (which contain respectively additional class 3 variants which were not assessed to be definitively pathogenic, CL/P gene expression in mouse epithelial tissues, and detailed in silico assessment of variants in Table 1) and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.04.009.
Contributor Information
Timothy C. Cox, Email: tccox@uw.edu.
Tony Roscioli, Email: tony.roscioli@health.nsw.gov.au.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
CADD v.1.3, http://cadd.gs.washington.edu/
ExAC Browser, http://exac.broadinstitute.org/
FaceBase, https://www.facebase.org
Genic Intolerance, http://genic-intolerance.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Primer3, http://bioinfo.ut.ee/primer3-0.4.0/
PyMOL, https://pymol.org/2
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Supplemental Data
References
- 1.Dixon M.J., Marazita M.L., Beaty T.H., Murray J.C. Cleft lip and palate: understanding genetic and environmental influences. Nat. Rev. Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghoumid J., Stichelbout M., Jourdain A.S., Frenois F., Lejeune-Dumoulin S., Alex-Cordier M.P., Lebrun M., Guerreschi P., Duquennoy-Martinot V., Vinchon M. Blepharocheilodontic syndrome is a CDH1 pathway-related disorder due to mutations in CDH1 and CTNND1. Genet. Med. 2017;19:1013–1021. doi: 10.1038/gim.2017.11. [DOI] [PubMed] [Google Scholar]
- 3.Rahimov F., Jugessur A., Murray J.C. Genetics of nonsyndromic orofacial clefts. Cleft Palate Craniofac. J. 2012;49:73–91. doi: 10.1597/10-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell L.E., Risch N. Mode of inheritance of nonsyndromic cleft lip with or without cleft palate: a reanalysis. Am. J. Hum. Genet. 1992;51:323–332. [PMC free article] [PubMed] [Google Scholar]
- 5.Beaty T.H., Marazita M.L., Leslie E.J. Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow’s opportunities. F1000Res. 2016;5:2800. doi: 10.12688/f1000research.9503.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mostowska A., Gaczkowska A., Zukowski K., Ludwig K.U., Hozyasz K.K., Wojcicki P., Mangold E., Bohmer A.C., Heilmann-Heimbach S., Knapp M. Common variants in DLG1 locus are associated with non-syndromic cleft lip with or without cleft palate. Clin. Genet. 2017;93:784–793. doi: 10.1111/cge.13141. [DOI] [PubMed] [Google Scholar]
- 7.Yu Y., Zuo X., He M., Gao J., Fu Y., Qin C., Meng L., Wang W., Song Y., Cheng Y. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat. Commun. 2017;8:14364. doi: 10.1038/ncomms14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brito L.A., Yamamoto G.L., Melo S., Malcher C., Ferreira S.G., Figueiredo J., Alvizi L., Kobayashi G.S., Naslavsky M.S., Alonso N. Rare variants in the epithelial cadherin gene underlying the genetic etiology of nonsyndromic cleft lip with or without cleft palate. Hum. Mutat. 2015;36:1029–1033. doi: 10.1002/humu.22827. [DOI] [PubMed] [Google Scholar]
- 9.Hozyasz K.K., Mostowska A., Wójcicki P., Lasota A., Offert B., Balcerek A., Dunin-Wilczyńska I., Jagodziński P.P. Nucleotide variants of the cancer predisposing gene CDH1 and the risk of non-syndromic cleft lip with or without cleft palate. Fam. Cancer. 2014;13:415–421. doi: 10.1007/s10689-014-9727-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ittiwut R., Ittiwut C., Siriwan P., Chichareon V., Suphapeetiporn K., Shotelersuk V. Variants of the CDH1 (E-Cadherin) gene associated with oral clefts in the Thai population. Genet. Test. Mol. Biomarkers. 2016;20:406–409. doi: 10.1089/gtmb.2015.0325. [DOI] [PubMed] [Google Scholar]
- 11.Rafighdoost H., Hashemi M., Narouei A., Eskanadri-Nasab E., Dashti-Khadivaki G., Taheri M. Association between CDH1 and MSX1 gene polymorphisms and the risk of nonsyndromic cleft lip and/or cleft palate in a southeast Iranian population. Cleft Palate Craniofac. J. 2013;50:e98–e104. doi: 10.1597/12-144. [DOI] [PubMed] [Google Scholar]
- 12.Liu H., Busch T., Eliason S., Anand D., Bullard S., Gowans L.J.J., Nidey N., Petrin A., Augustine-Akpan E.A., Saadi I. Exome sequencing provides additional evidence for the involvement of ARHGAP29 in Mendelian orofacial clefting and extends the phenotypic spectrum to isolated cleft palate. Birth Defects Res. 2017;109:27–37. doi: 10.1002/bdra.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leslie E.J., Mansilla M.A., Biggs L.C., Schuette K., Bullard S., Cooper M., Dunnwald M., Lidral A.C., Marazita M.L., Beaty T.H., Murray J.C. Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Res. A Clin. Mol. Teratol. 2012;94:934–942. doi: 10.1002/bdra.23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savastano C.P., Brito L.A., Faria A.C., Setó-Salvia N., Peskett E., Musso C.M., Alvizi L., Ezquina S.A., James C., GOSgene Impact of rare variants in ARHGAP29 to the etiology of oral clefts: role of loss-of-function vs missense variants. Clin. Genet. 2017;91:683–689. doi: 10.1111/cge.12823. [DOI] [PubMed] [Google Scholar]
- 15.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paila U., Chapman B.A., Kirchner R., Quinlan A.R. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput. Biol. 2013;9:e1003153. doi: 10.1371/journal.pcbi.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shepard T.H. Normal and abnormal growth patterns. In: Gardner L.I., editor. Endocrine and Genetic Diseases of Childhood. W.B. Saunders; Philadelphia: 1975. pp. 1–6. [Google Scholar]
- 22.Tamasas B., Cox T.C. Massively increased caries susceptibility in an Irf6 cleft lip/palate model. J. Dent. Res. 2017;96:315–322. doi: 10.1177/0022034516679376. [DOI] [PubMed] [Google Scholar]
- 23.Davis M.A., Reynolds A.B. Blocked acinar development, E-cadherin reduction, and intraepithelial neoplasia upon ablation of p120-catenin in the mouse salivary gland. Dev. Cell. 2006;10:21–31. doi: 10.1016/j.devcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Schock E.N., Struve J.N., Chang C.F., Williams T.J., Snedeker J., Attia A.C., Stottmann R.W., Brugmann S.A. A tissue-specific role for intraflagellar transport genes during craniofacial development. PLoS ONE. 2017;12:e0174206. doi: 10.1371/journal.pone.0174206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y., Park J.W., Bebee T.W., Warzecha C.C., Guo Y., Shang X., Xing Y., Carstens R.P. Determination of a comprehensive alternative splicing regulatory network and combinatorial regulation by key factors during the epithelial-to-mesenchymal transition. Mol. Cell. Biol. 2016;36:1704–1719. doi: 10.1128/MCB.00019-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nykamp K., Anderson M., Powers M., Garcia J., Herrera B., Ho Y.Y., Kobayashi Y., Patil N., Thusberg J., Westbrook M., Topper S., Invitae Clinical Genomics Group Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet. Med. 2017;19:1105–1117. doi: 10.1038/gim.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anastasiadis P.Z., Reynolds A.B. The p120 catenin family: complex roles in adhesion, signaling and cancer. J. Cell Sci. 2000;113:1319–1334. doi: 10.1242/jcs.113.8.1319. [DOI] [PubMed] [Google Scholar]
- 28.Pulimeno P., Bauer C., Stutz J., Citi S. PLEKHA7 is an adherens junction protein with a tissue distribution and subcellular localization distinct from ZO-1 and E-cadherin. PLoS ONE. 2010;5:e12207. doi: 10.1371/journal.pone.0012207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bebee T.W., Park J.W., Sheridan K.I., Warzecha C.C., Cieply B.W., Rohacek A.M., Xing Y., Carstens R.P. The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. eLife. 2015;4:4. doi: 10.7554/eLife.08954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warzecha C.C., Jiang P., Amirikian K., Dittmar K.A., Lu H., Shen S., Guo W., Xing Y., Carstens R.P. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010;29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yap A.S., Niessen C.M., Gumbiner B.M. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J. Cell Biol. 1998;141:779–789. doi: 10.1083/jcb.141.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phan M., Conte F., Khandelwal K.D., Ockeloen C.W., Bartzela T., Kleefstra T., van Bokhoven H., Rubini M., Zhou H., Carels C.E. Tooth agenesis and orofacial clefting: genetic brothers in arms? Hum. Genet. 2016;135:1299–1327. doi: 10.1007/s00439-016-1733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riley B.M., Mansilla M.A., Ma J., Daack-Hirsch S., Maher B.S., Raffensperger L.M., Russo E.T., Vieira A.R., Dodé C., Mohammadi M. Impaired FGF signaling contributes to cleft lip and palate. Proc. Natl. Acad. Sci. USA. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riley B.M., Murray J.C. Sequence evaluation of FGF and FGFR gene conserved non-coding elements in non-syndromic cleft lip and palate cases. Am. J. Med. Genet. A. 2007;143A:3228–3234. doi: 10.1002/ajmg.a.31965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng W., Mushika Y., Ichii T., Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 2008;135:948–959. doi: 10.1016/j.cell.2008.09.040. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y.S., Cho Y.S., Lee G.K., Lee S., Kim Y.W., Jho S., Kim H.M., Hong S.H., Hwang J.A., Kim S.Y. Genomic profile analysis of diffuse-type gastric cancers. Genome Biol. 2014;15:R55. doi: 10.1186/gb-2014-15-4-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ireton R.C., Davis M.A., van Hengel J., Mariner D.J., Barnes K., Thoreson M.A., Anastasiadis P.Z., Matrisian L., Bundy L.M., Sealy L. A novel role for p120 catenin in E-cadherin function. J. Cell Biol. 2002;159:465–476. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leslie E.J., Marazita M.L. Genetics of cleft lip and cleft palate. Am. J. Med. Genet. C. Semin. Med. Genet. 2013;163C:246–258. doi: 10.1002/ajmg.c.31381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daniel J.M., Reynolds A.B. The tyrosine kinase substrate p120cas binds directly to E-cadherin but not to the adenomatous polyposis coli protein or alpha-catenin. Mol. Cell. Biol. 1995;15:4819–4824. doi: 10.1128/mcb.15.9.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishiyama N., Lee S.H., Liu S., Li G.Y., Smith M.J., Reichardt L.F., Ikura M. Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell. 2010;141:117–128. doi: 10.1016/j.cell.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 41.Peifer M., Yap A.S. Traffic control: p120-catenin acts as a gatekeeper to control the fate of classical cadherins in mammalian cells. J. Cell Biol. 2003;163:437–440. doi: 10.1083/jcb.200310090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thoreson M.A., Anastasiadis P.Z., Daniel J.M., Ireton R.C., Wheelock M.J., Johnson K.R., Hummingbird D.K., Reynolds A.B. Selective uncoupling of p120(ctn) from E-cadherin disrupts strong adhesion. J. Cell Biol. 2000;148:189–202. doi: 10.1083/jcb.148.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurita S., Yamada T., Rikitsu E., Ikeda W., Takai Y. Binding between the junctional proteins afadin and PLEKHA7 and implication in the formation of adherens junction in epithelial cells. J. Biol. Chem. 2013;288:29356–29368. doi: 10.1074/jbc.M113.453464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou Y., Cox T.C. A likely role for a novel PH-domain containing protein, PEPP2/PLEKHA5, at the membrane-microtubule cytoskeleton interface. Biocell. 2013;37:55–61. [Google Scholar]
- 45.Paschoud S., Jond L., Guerrera D., Citi S. PLEKHA7 modulates epithelial tight junction barrier function. Tissue Barriers. 2014;2:e28755. doi: 10.4161/tisb.28755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynolds A.B., Carnahan R.H. Regulation of cadherin stability and turnover by p120ctn: implications in disease and cancer. Semin. Cell Dev. Biol. 2004;15:657–663. doi: 10.1016/j.semcdb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Warzecha C.C., Shen S., Xing Y., Carstens R.P. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009;6:546–562. doi: 10.4161/rna.6.5.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrova Y.I., Schecterson L., Gumbiner B.M. Roles for E-cadherin cell surface regulation in cancer. Mol. Biol. Cell. 2016;27:3233–3244. doi: 10.1091/mbc.E16-01-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boggon T.J., Murray J., Chappuis-Flament S., Wong E., Gumbiner B.M., Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 50.Prakasam A., Chien Y.H., Maruthamuthu V., Leckband D.E. Calcium site mutations in cadherin: impact on adhesion and evidence of cooperativity. Biochemistry. 2006;45:6930–6939. doi: 10.1021/bi060213m. [DOI] [PubMed] [Google Scholar]
- 51.Sotomayor M., Schulten K. The allosteric role of the Ca2+ switch in adhesion and elasticity of C-cadherin. Biophys. J. 2008;94:4621–4633. doi: 10.1529/biophysj.107.125591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.