Abstract
Coenzyme A (CoA) is an essential metabolic cofactor used by around 4% of cellular enzymes. Its role is to carry and transfer acetyl and acyl groups to other molecules. Cells can synthesize CoA de novo from vitamin B5 (pantothenate) through five consecutive enzymatic steps. Phosphopantothenoylcysteine synthetase (PPCS) catalyzes the second step of the pathway during which phosphopantothenate reacts with ATP and cysteine to form phosphopantothenoylcysteine. Inborn errors of CoA biosynthesis have been implicated in neurodegeneration with brain iron accumulation (NBIA), a group of rare neurological disorders characterized by accumulation of iron in the basal ganglia and progressive neurodegeneration. Exome sequencing in five individuals from two unrelated families presenting with dilated cardiomyopathy revealed biallelic mutations in PPCS, linking CoA synthesis with a cardiac phenotype. Studies in yeast and fruit flies confirmed the pathogenicity of identified mutations. Biochemical analysis revealed a decrease in CoA levels in fibroblasts of all affected individuals. CoA biosynthesis can occur with pantethine as a source independent from PPCS, suggesting pantethine as targeted treatment for the affected individuals still alive.
Keywords: PPCS, phospohopantothenoylcysteine synthetase, coenzyme A, pentothenate, dilated cardiomyopathy, pantethine treatment
Introduction
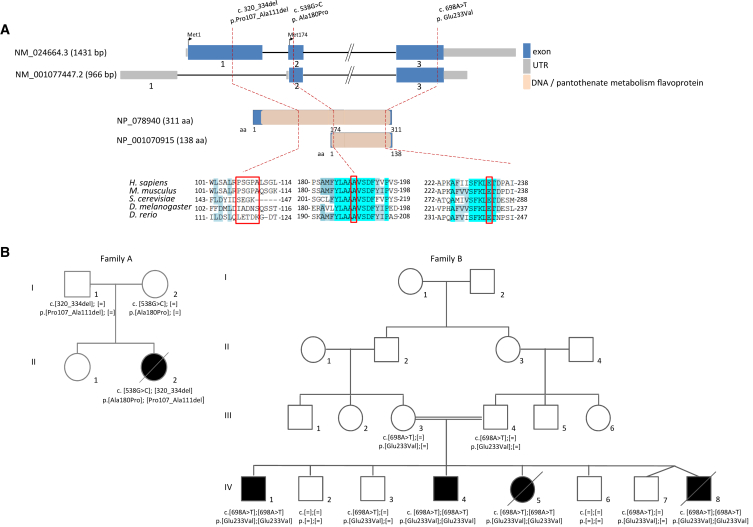
The pathway of CoA de novo biosynthesis from vitamin B5 is conserved in eukaryotes and prokaryotes and consists of five consecutive enzymatic steps. Dysfunction of two of the involved enzymes, PANK2 (MIM: 606157) and COASY (MIM: 609855), has been identified as a cause of autosomal-recessive neurodegeneration with brain iron accumulation (NBIA). Mutations in PANK2, encoding pantothenate kinase 2 necessary for the first enzymatic step of CoA biosynthesis, represent a major cause of NBIA and are detected in about half of case subjects with the eye-of-the-tiger sign, a pathognomonic finding on brain MRI.1 Mutations in COASY, encoding bifunctional CoA synthase, catalyzing the last two steps, have so far been identified in only five subjects with basal ganglia lesions.2, 3, 4, 5 To our knowledge, mutations in PPCS (MIM: 609853), encoding phosphopantothenoylcysteine synthetase, have so far not been associated with human diseases. PPCS catalyzes the second enzymatic step during which phosphopantothenate reacts with ATP (or CTP) and with cysteine to form phosphopantothenoylcysteine (Figure 1). The yeast ortholog YIL083C/CAB2 (yPPCS) is an essential gene, with haploid CAB2 mutants being non-viable even after medium supplementation with fatty acids.6 In Drosophila melanogaster, CG5629 (dPPCS) is required for tissue morphogenesis during oogenesis.7 Null mutants for dPPCS are lethal at the first instar larvae, while hypomorphic alleles exhibit locomotor defects, altered lipid homeostasis, and reduced lifespan.8 In human there are two consistently annotated isoforms of PPCS (Figure 2A): a canonical isoform (GenBank: NM_024664.3) encoding a protein of 311 amino acids and a shorter isoform (GenBank: NM_001077447.2) encoding a protein of 138 amino acids. The two isoforms share the C-terminal region. According to public databases, both isoforms are ubiquitously expressed (The Human Protein Atlas: PPCS), although with tissue-specific differences (GTEx Portal: PPCS). Here we report on the identification of biallelic mutations in PPCS in five individuals from two unrelated families presenting with severe dilated cardiomyopathy (Figure 2B). Clinical findings are summarized in Table 1.
Figure 1.
Universal Pathway for the Biosynthesis of Coenzyme A and Associated Diseases
In green are indicated genes encoding either cytosolic or mitochondrial isoforms. The mitochondrial isoforms are associated with neurodegeneration with brain iron accumulation (#234200, #615643). In gray are labeled genes predicted to have mostly a cytosolic localization (Reactome: PPCS). The asterisk (∗) indicates that the association with a disease was not reported elsewhere.
Figure 2.
Structure of PPCS and Pedigrees of Investigated Families
(A) Structure of PPCS with known conserved protein domain in the gene product and localization and conservation of amino acid residues affected by mutations identified in the two families. Intronic regions are not drawn to scale. Coloring in the sequence alignment represents the identity of amino acid residues.
(B) Pedigrees of two families with mutations in PPCS. Mutation status of affected (closed symbols) and healthy (open symbols) family members. n.d., not determined.
Table 1.
Demographic, Clinical, and Molecular Findings in Individuals with Mutations in PPCS
| Individual | FB:IV.1 | FB:IV.4 | FB:IV.6 | FB:IV.8 | FA:II.2 |
|---|---|---|---|---|---|
| Gender | M | M | F | M | F |
| Ethnic background | Arab-Muslim | Arab-Muslim | Arab-Muslim | Arab-Muslim | Central Europe |
| Consanguinity | yes | yes | yes | yes | no |
| Dysmorphic features | no | no | no | no | yes |
| Age at presentation | 2 years | 4 months | 23 months | 3 years | 2 weeks |
| Cardiac manifestations | dilated cardiomyopathy | dilated cardiomyopathy | severe dilated cardiomyopathy | severe dilated cardiomyopathy | severe dilated cardiomyopathy |
| Muscular hypotonia | no | no | N/D | N/D | severe |
| Extracardiac manifestations | none | none | none | none | yes |
| Serum lactate | normal | normal | elevated | elevated | elevated |
| Status | alive | alive | death at 23 months from DCM | death at 3 years from DCM | death at 3 months from DCM and multi organ failure |
| Brain MRI | normal | N/D | N/D | N/D | normal |
| Mutation in PPCS | c.[698A>T]; [698A>T] | c.[698A>T]; [698A>T] | c.[698A>T]; [698A>T] | c.[698A>T]; [698A>T] | c.[538G>C]; [320_334del] |
N/D, no data available
Subjects and Methods
Informed written consent to participate in the study was obtained from all individuals or their parents in the case of minors. The study was approved by the ethics committee of the Technische Universität München (Munich, Germany) and the Institutional Review Board of the Sheba Medical Center (Tel Aviv, Israel).
Subjects
In family A, individual II.2, born to healthy non-consanguineous parents of German descent, presented in the first few hours after birth with an acute life-threatening event followed by a tonic seizure. She showed minor dysmorphic features including cutis laxa, abnormally placed thumbs, abnormal dermatoglyphics, hypoplastic toe nails, and additional findings. She experienced a complicated course of illness requiring several hospitalizations. An extensive evaluation revealed severe dilated cardiomyopathy, pulmonary arterial hypertension, suspected pituitary dysfunction, and muscular hypotonia. Unfortunately, she succumbed to multi-organ failure at 3 months of age.
Family B is a multiplex consanguineous family of Arab-Muslim descent. The parents are reportedly healthy and are first-degree cousins. Four out of eight siblings exhibited dilated cardiomyopathy of varying severity, without apparent extracardiac manifestations. Among the four affected siblings, two (individuals IV.1 and IV.4) showed a milder form of dilated cardiomyopathy (Figure 3), enabling a degree of regular daily activities. The other two siblings (individuals IV.5 and IV.8) showed a severe form of dilated cardiomyopathy, fatal in early childhood (3 years of age). The four unaffected siblings are asymptomatic and healthy. The echocardiograms of the unaffected siblings and their parents were normal. Detailed clinical case reports are available in the Supplemental Note. Demographic, clinical, and molecular findings are summarized in Table 1.
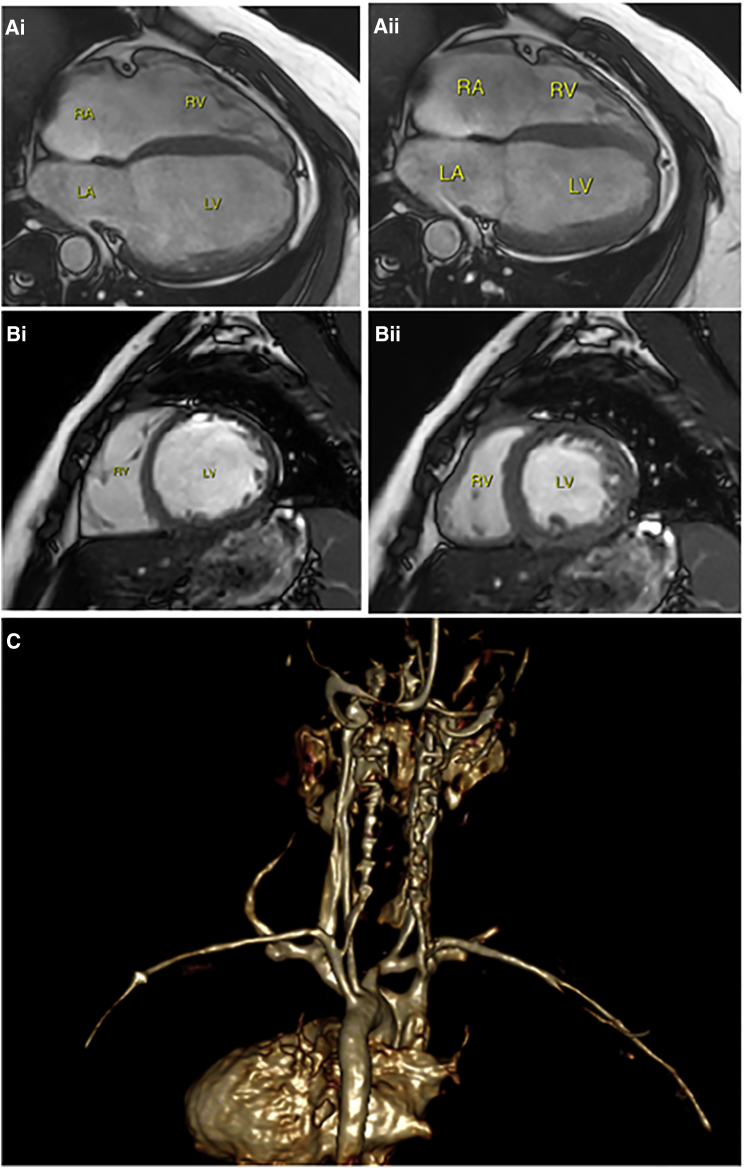
Figure 3.
Cardiac Magnetic Resonance Imaging (cMRI) Findings in Individuals with PPCS-Related Dilated Cardiomyopathy
(A) Four-chamber view of individual IV.1 (FB:IV.1 in the figure) at the age of 21 years in diastole (Ai) and systole (Aii). Note the decreased change in ventricular volume between phases. RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle.
(B) Short axis oblique of individual IV.4 (at the age of 10 years) in diastole (Bi) and systole (Bii), consistent with a low cardiac output.
(C) Volume rendering of gadolinium-enhanced angiography of the chest and head blood vessels in individual IV.4 (posterior view), demonstrating normal vasculature.
Whole-Exome Sequencing and Sanger Sequencing
We performed exome sequencing at two centers (family A in Munich, family B in Tel Aviv). In affected individuals of families A and B, coding regions were enriched with a SureSelect Human All Exon V5 Kit (Agilent) and sequenced as 100-bp paired-end runs on an Illumina HiSeq 2500. Reads were aligned to the human reference genome (UCSC Genome Browser, build hg19) with the Burrows-Wheeler Aligner (v.0.7.5a). Single-nucleotide variants and small insertions and deletions (indels) were detected by SAMtools (v.0.1.19). Sanger sequencing was used to confirm the identified mutations and test the carrier status of unaffected family members using the following primer pairs: F, 5′-TGGACAACTTCAGCAGCG-3′; R, 5′-ACAGATGCAAATAGTCCGCC-3′ and F, 5′-CCCTTACCTCCTGCTTACCC-3′; R, 5′-ACAGATGCAGAAAGGCCATC-3′ for family A, and F, 5′-TTCCATGAGATTGACCTGGTA-3′; R, 5′-CAAATGCCAAACCATATTTGC-3′ for family B.
Bioinformatic Analysis
Structural files of the human PPCS9 (PDB: 1P9O) and of the orthologous E. coli gene10 (PDB: 1U7W) were downloaded from the protein data bank (PDB).11 Sequence alignment to detect the evolutionary relation between the enzymes was done using psi-blast.12 Structural alignment between proteins was done using triangular match.13 Initial analysis of the variant location was done using G23D.14 Structural images were created using UCSF Chimera.15 Calculation of solvent-accessible area of residues at the variant positions was done using Getarea.16 A variety of programs for assessment of function and stability changes following the mutations were applied and are listed in Table S1 including the URL and reference. Conservation analysis and mapping of conservation scores on the structure were done using Consurf.17 Prediction of binding site based on structural data was done using the Coach algorithm18 in the I-Tasser website.19
RNA Extraction, RT-PCR, Cloning, and Mutagenesis of PPCS
Total RNA was isolated from fibroblasts of healthy donors (80% confluence) with the AllPrep RNA Kit (QIAGEN). RNA quantity was measured with the Nanodrop instrument (Nanodrop Technologies). For the RT-PCR, 1 μg of RNA was reverse transcribed with M-MLV reverse transcriptase (Promega, GmbH) and oligo dT. PPCS (GenBank: NM_024664.3) coding region was amplified with Thermo-Start Taq DNA Polymerase (ABgene) and primers F, 5′-CACGATGGCGGAAATGGATCCGGT-3′ and R, 5′-TCAGTTTCTGTCACCTATAAAAGCTGTG-3′. Wild-type PPCS retaining the endogenous terminal stop-codon was cloned in the vector pLenti6.3/V5-TOPO (Invitrogen).
The PPCS variants were obtained by site-directed mutagenesis of the cloned PPCS ORF using the kit QuikChange II Site-Directed Mutagenesis (Stratagene) and the primer pairs: F, 5′-TTACCTGGCTGCGCCTGTGTCAGATTTCTATG-3′; R, 5′-CATAGAAATCTGACACAGGCGCAGCCAGGTAA-3′ to introduce the change 538G>C; F, 5′-TGTCCGCTCTGCGGCTTTCGGGCTTGC-3′; R, 5′-GCAAGCCCGAAAGCCGCAGAGCGGACA-3′ for the change c.320_334del; and F, 5′-TTTCCTTTAAGTTGGTGACTGACCCCGCCATT-3′; R, 5′-AATGGCGGGGTCAGTCACCAACTTAAAGGAAA-3′ for the change 698A>T. The following plasmids were hence generated: PPCS_wild-type _pLenti6.3, PPCS _c.320_334del pLenti6.3, PPCS _c.538G>C pLenti6.3, and PPCS _c.698A>T pLenti6.3.
Complementation of PPCS-Deficient Fibroblast Cell Line
The fibroblast cell line derived from individual II.2 of family A was complemented with the wild-type copy of PPCS cloned in the pLenti6.3/V5-TOPO (Invitrogen) as described in Kremer and Prokisch.20
Yeast Strain and Plasmids
Construction of Yeast Expression Plasmids
PPCS_ wild-type pLenti6.3, PPCS_c.320_334del pLenti6.3, PPCS_c.538G>C pLenti6.3, and PPCS _c.698A>T pLenti6.3 were used as templates to amplify cDNAs of the human PPCS (hPPCS) variants. The amplification was performed with a proofreading-competent Phusion DNA polymerase (ThermoFisher) using primers PPCS-XbaIStart (5′-GATCTCTAGAAAAATGGCGGAAATGGATCCGGT-3′) and PPCS-SalIStop (5′-GATCGTCGACTCAGTTTCTGTCACCTATAAAAG-3′). PCR fragments were cleaved with XbaI and SalI and cloned into the single-copy expression yeast plasmid p415-MET25.21 p415-MET25 contains LEU2 as a selection marker and MET25 as promoter to activate heterologous genes. MET25 is a promoter of intermediary strength typical for most yeast genes with a metabolic function while extreme overexpression of heterologous genes is prevented.
We thus obtained plasmids pMET-PPCS-WT (hPPCS, wild-type), pMET-PPCS-320_334del (hPPCS deletion allele 320_334), pMET-PPCS-538G>C (hPPCS 538G>C missense variant), and pMET-PPCS-698A>T (hPPCS 698A>T missense variant).
Plasmid Shuffling
Single-copy plasmid pGE11 containing yPPCS together with URA3 as a selection marker was transformed into S. cerevisiae wild-type strain JS91.15-23 (ura3 leu2 trp1 his3 CAB2). Subsequently, the chromosomal yPPCS gene was deleted by using the gene disruption cassette from plasmid pJO9 (cab2Δ::HIS3). After selecting for His-prototrophic transformants, strain MGY9 was obtained which is viable due to the plasmid copy of yPPCS compensating for the loss of chromosomal yPPCS. The authenticity of the desired genomic alteration was verified by PCR, using gene-specific primers (not shown).
S. cerevisiae strain MGY9 was hence transformed with plasmids pMET-PPCS-WT, pMET-PPCS-320_334del, pMET-PPCS-538G>C, and pMET-PPCS-698A>T, together with the empty vector p415-MET25 (negative control) and pGE12 (authentic CAB2 as a positive control). Dilutions of Leu-prototrophic transformants were subsequently spotted on a medium containing 5-Fluoroorotic acid (5-FOA), which allows counter-selection of plasmids containing URA3 as a genetic marker. Thus, plasmid pGE11 (URA3 CAB2) is lost and growth is possible only when LEU2-containing plasmids compensate for the loss of pGE11.
Saccharomyces cerevisiae strain was MGY9 (α ura3 leu2 trp1 his3 cab2Δ::HIS3 [ARS CEN URA3 CAB2]).
Plasmids were pJO9 (cab2Δ::HIS3 deletion cassette), pGE11 (ARS CEN URA3 CAB2), pGE12 (ARS CEN LEU2 CAB2), p415-MET25 (ARS CEN LEU2 MET25Prom.), pMET-PPCS-WT (ARS CEN LEU2 MET25Prom.-hPPCS), pMET-PPCS-320_334del (ARS CEN LEU2 MET25Prom.-hPPCS-320_334del), pMET-PPCS-538G_C (ARS CEN LEU2 MET25Prom.-hPPCS 538G>C), and pMET-PPCS 698A_T (ARS CEN LEU2 MET25Prom.-hPPCS 698A>T).
Western Blotting
Analysis of PPCS protein level was performed on whole cellular lysate under denaturing conditions and proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Lonza, PAGEr EX Gels). The analysis of PPCS dimer formation was performed on whole cellular lysate under non-reducing non-denaturing conditions and separated by blue native polyacrylamide gel electrophoresis (BN-PAGE; ThermoFisher, NativePAGE Novex Bis-Tris Gel System). Denaturing and native gels were blotted onto PVDF membrane (GE-Healthcare) for subsequent incubation with primary antibodies and probing with appropriate secondary antibodies. Chemiluminescence was documented on a Fusion FX7 system (Peqlab). The following antibodies were used for western blotting: rabbit anti-PPCS (ab140626, 1:2,000) purchased from Abcam, mouse anti-Tubulin (T5168, 1:15,000) purchased from Sigma-Aldrich, and mouse anti-ATP5Α (ab14748, 1:1,000). HRP-conjugated secondary antibodies for western blot were obtained from Jackson Immunoresearch Laboratories (USA) (1:15,000).
Measurement of Cellular CoA
Total cellular CoA was measured using a fluorimetric-based kit (Abcam, ab138889) according to manufacturer instructions. Briefly, 1 × 106 cells of fibroblasts, growing in exponential phase under standard medium (Dulbecco’s Modified Eagle Medium High Glucose, 10% fetal bovine serum, 4 mM glutamine, 1% penicillin/streptomycin) were collected and the pellet was resuspended in 500 μL of lysis buffer (Abcam, ab179835). After 15 min incubation at room temperature, cell debris was spun down by centrifugation at 1,500 rpm for 5 min. For each measurement, 50 μL of clear supernatant was used. A CoA standard curve was generated by plotting relative fluorescence values (RFU) and serial dilution of CoA standard. CoA levels in test samples were calculated by using the calibration curve. Results are mean ± SD of n = 16 values. p values were calculated with an independent sample t test. All p values were two-sided with a significance level of 0.05.
Serum Pantothenic Acid, PPCS, and CoA Measurements
Serum samples were obtained from individuals IV.1 and IV.4, family B, as well as from healthy control subjects. Pantothenic acid levels were measured at Mayo Medical Laboratories (Mayo Clinic). ELISA commercial kits (MBS283149, MYBioSource) were used for measurement of PPCS levels, and Coenzyme A detection kit (ab102504, Abcam) for measurement of CoA levels in serum.
Drosophila Stocks and Cardiac Physiological Measurements
In all experiments, we used dPPCS mutant flies carrying a P element insertion in the 5′ UTR of the gene, resulting in a lower expression of dPPCS protein. This mutant, indicated as dPPCS1/TM3-GFP, was extensively described in Bosveld et al.8 All flies were maintained at 25°C on standard medium. Heterozygous and homozygous pre-pupae were selected on the basis of GFP fluorescence (homozygous pre-pupae are GFP-negative). High-speed movies of spontaneous heart wall contractions in whole pre-pupae were recorded for 3 × 10 s at a rate of 100 frames per second by using a BlueFOX3 digital camera on a Leica DM IL LED microscope with a 10× lens. Heart rate was manually analyzed with ImageJ. Kymographs were constructed using ImageJ and used to measure heart wall shortening, systolic, and diastolic length. Arrhythmia index was obtained from a 2 min/100 frames per second high-speed movie with a 20× lens of spontaneous heart wall contractions in whole pre-pupae and analyzed by via software as described before.22
Drosophila Viability and Pantethine Treatment
Heterozygous males and females dPPCS1/TM3-GFP were transferred into vials and were allowed to produce embryos on control food, food containing 12 mM vitamin B5, and food containing 8 mM pantethine, as previously described in Kiefer et al.23 After 5 days, adult flies were removed and after 12 days, the amount of homozygous and heterozygous pupae was determined.
Targeted Treatment with Pantethine in Affected Individuals
After obtaining informed consent of the affected individuals or their parents, as well as institutional review board approval as compassionate treatment, the only two living affected individuals (IV.1 and IV.4, family B) commenced oral pantethine (Now Foods) supplementation at an initial dose of 6–8 mg/kg/day (once daily), which was subsequently gradually increased to 20–24 mg/kg/day.
Results
Molecular Diagnosis by Next Generation Sequencing
In family A, a search for homozygous or potentially compound heterozygous non-synonymous rare variants with a minor allele frequency (MAF) <0.1% prioritized in the Munich in-house database, containing 11,525 control exomes, rare variants in two genes, PPCS and C4orf21. Neither of these genes have so far been associated with human disease. Due to the function of the encoded protein and in silico prediction, we considered PPCS (GenBank: NM_024664.3) as the top candidate gene. In family B, a search for recessive inheritance resulted in 26 genes harboring rare (MAF < 0.01%) non-synonymous variants, for which the proband was homozygous and the mother heterozygous (Table S2). Further analysis excluded 21 variants (benign, reported in unaffected individuals, genes unrelated to the phenotype), resulting in 5 candidate genes. As 3 of these variants were located within introns, the remaining 2 (in PPCS and PPID) were further evaluated for segregation in the family. Finally, PPCS, which was not previously associated with a human phenotype, however, is known for its role in the CoA biosynthesis pathway, remained as the most promising candidate for further analysis.
Exome sequencing in individual II.2 of family A led to the identification of the heterozygous changes c.320_334del (p.Pro107_Ala111del) and c.538G>C (p.Ala180Pro) in PPCS (Figure 2B). Carrier testing confirmed that the variant c.320_334del was located on the paternal allele and the variant c.538G>C on the maternal allele.
In family B, WES identified the homozygous c.698A>T mutation (p.Glu233Val) in exon 3 of the PPCS gene of individual IV.8 (Figure 2B). Segregation analysis in all available family members confirmed full segregation, with all four affected siblings found to harbor the c.178A>T mutation in homozygous form, both parents and two of the four unaffected siblings (subjects IV.3 and IV.7) found to be heterozygous for the mutation, and the remaining two unaffected siblings (subjects IV.2 and IV.6) found to be wild-type.
None of the PPCS missense variants (c.698A>T, c.538G>C) are listed in the gnomAD (Genome Aggregation Database) browser (12/2017). The in-frame deletion c.320_334del is listed 10× only in a heterozygous state in 271,726 alleles of the genomAD browser. No additional rare biallelic PPCS variants were observed in the Munich in-house database containing exome dataset from more than 11,000 individuals with unrelated phenotypes.
Bioinformatic Analysis of the Variants Found in PPCS
The two SNV mutations p.Glu233Val and p.Ala180Pro (canonical isoform) are categorized as highly deleterious by almost all prediction tools (Table S1), including PolyPhen, Sift, LRT, MutationTaster, MutationAssessor, and Provean. These tools base their predictions on different considerations but heavily rely on evolutionary conservation. Indeed, the mutated amino acids and their vicinity are conserved throughout evolution (Figure 2A). Mapping the conservation scores to the protein structure using Consurf also reveals conserved structural cluster in the general region of the mutations (Figure S1A). These mutations are not likely to change drastically the protein stability as they are partially exposed to the surface and not integral part of the protein.
The two positions share several topological and structural commonalities. Residues Glu233 and Ala180 of PPCS have their side chain partially exposed to the surface (33% and 20% relative accessibility, respectively). Both are located within the core Coa-B like domain. The two positions are also spatially close and separated by only about 10 Å as indicated by the structure of the human protein, which was resolved to 2.3 Å resolution (PDB: 1P9O). Unfortunately, PDB file 1P9O does not contain ligands or cofactors, complicating the analysis of possible involvement of Ala180 and Glu233 in molecular binding. We thus attempted to examine additional related structures which include cofactors. A sensitive psi-blast sequence search identified the phosphopantothenoylcysteine synthetase protein from E. coli, as a remote homolog of PPCS (20% identity). This protein has been crystalized (PDB: 1U7W) with a CTP nucleotide cofactor.10 Structural alignment of PDB: 1P9O and 1U7W revealed a highly similar fold (Figure S1C) with the best alignment encompasses 159 residues with RMSD of 0.85 Å (p value for structure similarity < E−15). Glu233 was found to be located in close vicinity to the phosphate recognition region of the nucleotide binding site. This residue is conserved in the bacterial protein (residue Phe330 therein) in spite of low overall sequence similarity between the proteins. Phe330 creates contact with the CTP phosphate group in the bacterial protein. It is not entirely clear how the negatively charged residue creates favorable contacts with the negative phosphate. One option is that the recognition is mediated by a divalent cation. Indeed, the binding of the phosphate in the other side of the binding cleft is mediated by a positive calcium ion (Figure S1D) which is coordinated to a negatively charged aspartate (Asp276). It can well be that Phe330 also coordinates a mediator ion that was not resolved by the X-ray crystallography. The other option is that an electrostatic repulsive force helps to orientate the nucleotide in the binding site.
Ala180 is also located in the nucleotide binding cleft, but in a closer vicinity to the sugar moiety. The equivalent residue in the bacterial protein is Ala276, suggesting the need for a small residue at this spot. The side chain in both the bacterial and human proteins point to the opposite side of the nucleotide.
Although the nucleotide involved in the human protein reaction is ATP and not CTP, nevertheless the phosphate and sugar binding mode are likely to be conserved as these parts are identical.
In conclusion, there is a consensus agreement regarding the deleterious effect of both p.Glu233Val and p.Ala180Pro on the protein function, which reflects the high evolutionary conservation of both sites. The changes are likely to affect the binding to the ATP co-factor of PPCS.
The mutation c.320_334del affects a region which is not conserved in evolution (Figure 2A). However, prediction programs such as MutationTaster predict the change to be deleterious since the missing amino acids may be relevant to keep the conformation of the protein (Figure S1F). The human PPCS forms a dimer that is biologically relevant for function.9, 10 The dimer has two dimerization regions including a non-conserved sequence between helices α4 and α5 (residues 107–111). In the crystal structure of human PPCS (PDB: 1P9O),9 these residues are intertwined in the other monomer, although for part of them there is no electron density for residues 107–111 (marked in red in Figure S1G [zoom]).
Deletion of residues 107–111 could result in a destabilization of this dimerization region. In particular, it is possible that the deletion leads to a shortening of the strand β5 as seen in Figures S1F and S1G of the SWISS-MODEL model of the deletion mutant.24, 25, 26, 27 Additionally, the deletion could lead to rearrangement of helices α4 and α5 and dynamics of the protein and eventually to change the substrate binding region.
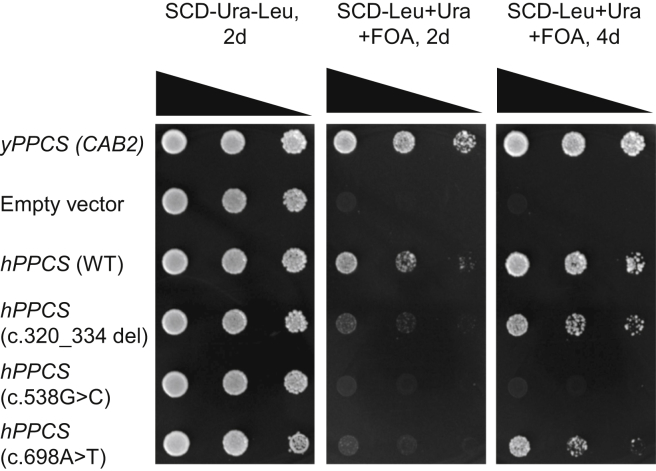
Validation of Pathogenicity of Mutations in Yeast
yPPCS is a gene essential for yeast viability.6 In order to evaluate the functional relevance of identified PPCS variants, we performed a functional complementation of the yPPCS null background with the human wild-type PPCS and its mutant variants using the method of plasmid shuffling.28 The genuine yPPCS gene was used as positive control, while an empty vector was used as negative control.
As expected, transformants containing pGE12 (LEU2 CAB2) are able to grow while empty vector transformants fail to grow (Figure 4, lines 1 and 2, respectively). Importantly, expression of the cDNA of wild-type hPPCS in yeast restores growth even in the absence of yPPCS, although slightly less efficient than authentic yPPCS (line 3). In contrast, transformants with deletion or missense variants of hPPCS were unable to grow on 5-FOA-containing medium after 2 days of cultivation (lines 4–6). However, after 4 days, mutant variants hPPCS 320_334del (line 4) and hPPCS 698A>T (line 6) could support a minimal growth while variant hPPCS 538G>C (line 5) turned out to be completely non-functional even when the time of cultivation was extended.
Figure 4.
Functional Complementation of a Yeast cab2 Deletion Mutation by Human PPCS Gene Variants
Strain MGY9, used as a recipient for transformation, contains a chromosomal cab2 null mutation which was complemented by the single-copy URA3 CAB2 plasmid pGE11. Single-copy LEU2 plasmids used for complementation studies contain yeast CAB2 (positive control) and human PPCS alleles (wild-type and variants c.320_334del, c.538G>C, c.698A>T). Transformants containing two autonomously replicating plasmids are shown (left, SCD-Ura-Leu. incubation for 2 days). To counter-select URA3 plasmid pGE11, transformants were transferred to a medium supplemented with 5-FOA and incubated for 2 or 4 days (middle, right).
Stability of PPCS Protein in Cells
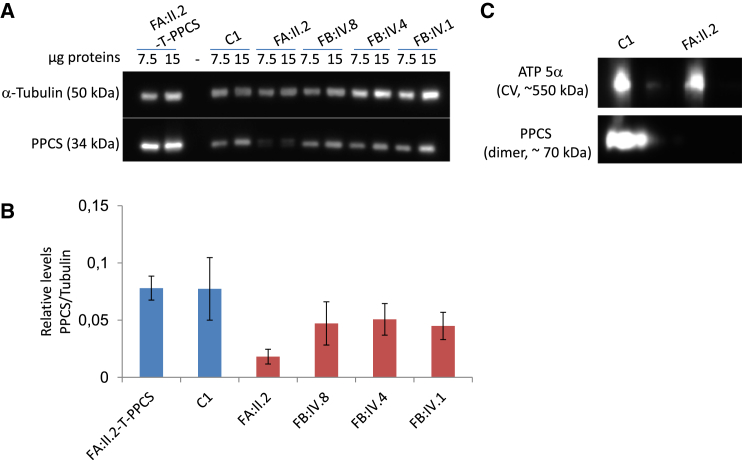
In order to gain insight on the effects of identified mutations on PPCS stability, we performed an immunoblotting analysis on fibroblasts extracts from the four affected individuals, three unrelated control subjects, and one affected individual’s cell transduced with the wild-type PPCS cDNA (Figures 5A and S3). Under denaturing conditions, the monoclonal PPCS antibody, directed against the C-terminal region present in both PPCS isoforms, detected both in control and affected individuals a main band of 34 kDa, corresponding to the canonical isoform. In subject II.2, family A (FA:II.2 in Figures 5A and S3), there is a second band corresponding to the deletion-carrying allele, in addition to the 34 kDa band, corresponding to the missense-carrying allele. The normalization of the signal of PPCS to the loading control α-tubulin revealed a marked reduction in PPCS levels in all affected individuals, with the strongest reduction observed in individual II.2 (Figures 5B and S3). The expression of the wild-type PPCS in the fibroblasts of individual II.2 (FA:II.2-T-PPCS in Figure 5A) normalized the PPCS protein levels (Figure 5B).
Figure 5.
Western Blot Analysis of PPCS Protein in PPCS Mutant Fibroblasts
(A) Western blot analysis of fibroblasts from the four affected individuals (FA:II.2, FB:IV.8, FB:IV.4, FB:IV.1), one healthy control subject (C1), and individual II.2, family A complemented by lentiviral transduction with wild-type PPCS. 7.5 and 15 μg proteins were loaded. Tubulin was used as a loading control.
(B) Densitometric analysis of this western blot. Error bars indicate the standard error of the mean.
(C) Blue native gel electrophoresis of total cellular extract from individual II.2, family A, and control C1 solubilized by 1% laurylmaltoside. ATP5α, sub-unit of the respiratory chain complex V, was used as a loading control
As the c.320_314del allele carried by individual II.2, family A was predicted to be associated with a dimerization defect, we performed an immunoblotting analysis under native conditions on fibroblasts extracted from this subject to verify whether at least the missense allele could form a dimer (Figure 5C). The PPCS antibody detected one band of about 70 kDa in the cell extract from a healthy control (C1 in Figure 5C), corresponding to the expected size of a PPCS dimer (about 68 kDa). No signal was instead present in the cell extract of the affected individual (Figure 5C). This pointed out that either the proteins encoded by the deletion and missense alleles are extremely unstable and do not form a dimer, or if a dimer is made by the missense carrying protein, that the level is so low that the antibody cannot detect it. We looked also for the monomeric PPCS in control and individual II.2, but in none of them was the monomeric PPCS detected, indicating that in physiological conditions PPCS exists mainly as a dimer.
Intracellular CoA Levels in the Presence of PPCS Mutations
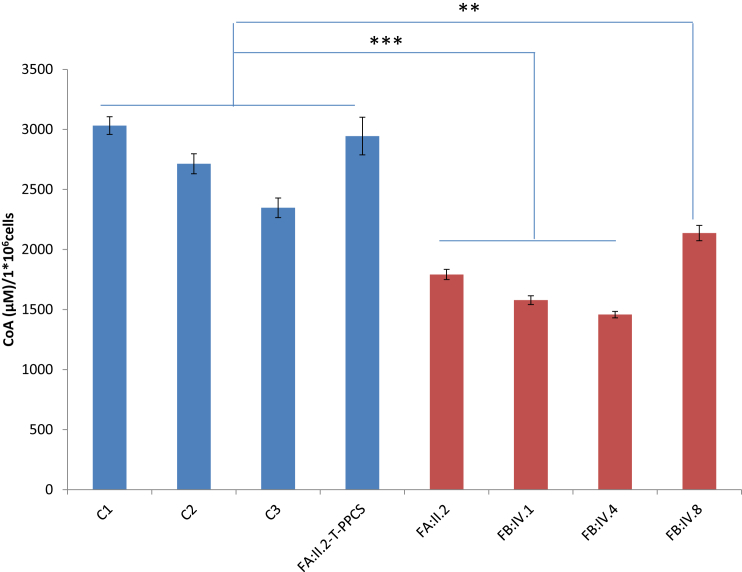
To gain further insight into the impact of PPCS variants on the steady-state level of CoA, we measured total cellular CoA levels in fibroblasts from all four PPCS-deficient individuals and compared this to the levels of CoA in fibroblasts from a commercially available control subject (NDHFneo, C1) and two healthy control subjects (C2 and C3) (Figure 6). The investigation revealed a significant reduction of CoA in all four affected individuals, which was rescued by reintroducing the wild-type copy of the gene (Figure 6).
Figure 6.
Total Cellular CoA in Fibroblasts
Fibroblasts growing in exponential phase under standard medium were collected, homogenized, and evaluated for CoA content. Relative fluorescence values (RFU) of serial dilution of CoA standard were measured to generate a calibration curve. CoA levels in fibroblasts were calculated by making use of the calibration curve. Fibroblasts from three unrelated controls (C1, C2, C3 in the figure) and from an affected individual transduced with wild-type PPCS (FA:II.2-T-PPCS) are labeled in blue; affected individuals (FA:II.2, FB:IV.1, FB:IV.4, FB:IV.8) are labeled in red. Results are mean ± SD of n = 16 values. p values were calculated with an independent sample t test. All p values were two-sided with a significance level of 0.05.
Metabolic Investigation in Blood and Serum of PPCS-Deficient Individuals
Acylcarnitine profiles in dried blood spots of the probands revealed normal levels of free carnitine (C0) but relatively low concentrations of long chain acylcarnitines C16 and C18, leading to an elevated ratio C0/(C16+C18) and reduced ratio (C16+C18:1)/C0 (Table 2).
Table 2.
Acylcarnitine Profiles in Individuals with Mutations in PPCS before and during Panthetine Supplementation
| Individual | Controls | FA:II.2 | FB:IV.1 | FB:IV.4 | FB:III.3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | c.[538G>C];[320_334del] | c.[698A>T];[698A>T] | c.[698A>T];[698A>T] | c.[698A>T];[ = ] | |||||||
| Pantethine supplementation | no | no | 600 mg/d | 1800 mg/d | 1800 mg/d | no | 300 mg/d | 900 mg/d | 900 mg/d | no | |
| C0 | 6–65 | 30 | 42 | 45 | 44.5 | 38.5 | 46 | 39 | 44 | 42 | 22.5 |
| C16 | 0.79–5.72 | 0.67 | 1.21 | 1.85 | 1.17 | 1.56 | 1.33 | 0.95 | 1.1 | 1.12 | 0.95 |
| C18 | 0.38–1.78 | 0.33 | 0.71 | 0.68 | 0.43 | 0.59 | 0.6 | 0.46 | 0.46 | 0.56 | 0.47 |
| C18:2 | 0–0.49 | 0.11 | 0.46 | 0.65 | 0.63 | 0.56 | 0.54 | 0.49 | 0.39 | 0.47 | 0.43 |
| C0/(C16+C18) | 2.30–18 | 30 | 21.88 | 17.79 | 27.81 | 17.91 | 23.83 | 27.66 | 28.21 | 25.00 | 15.85 |
| (C16+C18:1)/C0 | 0.08–0.42 | 0.04 | 0.05 | 0.07 | 0.05 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 |
Pantothenic acid (vitamin B5) in the serum of available affected individuals (IV.1 and IV.4, family B) was within normal limits for both affected siblings (74.95 μg/L and 73.84 μg/L, respectively; normal range for >10 years 37–147 μg/L).
Unfortunately, the levels of PPCS and CoA were consistently undetectable in both the affected and control samples.
Functional Metabolic Tests during Fasting
Following the identification of pathogenic variants in PPCS, known to be involved in the CoA synthesis pathway, we sought to evaluate the respiratory quotient (RQ) and basal metabolic rate (BMR) for the two live affected siblings (subjects IV.1 and IV.4, family B). This investigation (performed during fasting, at 21 years of age for individual IV.1 and at 9.5 years of age for individual IV.4) showed the RQ to be 0.773 and 0.796, respectively, both consistent with normal fat oxidation. BMR was found to be elevated for both siblings, 2,771 kcal/day for individual IV.1 and 2,091 kcal/day for individual IV.4, consistent with 142% and 160% of expected for their body weight and composition, respectively. This elevation was to be expected and indeed was attributable to their current state of heart failure.
Homozygous dPPCS1 Flies Show Altered Physiological Cardiac Parameters
To examine the effect of the dPPCS1 mutation on cardiac function, we determined heart rate, arrhythmia index, heart wall shortening, and systolic and diastolic length in heterozygous and homozygous dPPCS1 Drosophila melanogaster. In comparison with heterozygous dPPCS1 flies, homozygous dPPCS1 flies showed a significant increase in heart rate, heart wall shortening, and arrhythmia index and a decrease in systolic length (Figure 7). These findings indicate that homozygous variants in dPPCS are associated with cardiac dysfunction in Drosophila.
Figure 7.
Physiological Cardiac Parameters Are Changed in Homozygous dPPCS1Drosophila melanogaster
Parameters: (A) heart rate in beats per minute, (B) arrhythmia index, (C) systolic and (D) diastolic length in seconds, (E) heart wall shortening, and (F) representative kymographs (10 s). ∗∗p ≤ 0.01 and ∗∗∗p ≤ 0.001. N = 50 for heterozygous and N = 36 for homozygous for (A) and (B); N = 18 for heterozygous and N = 15 for homozygous for (C), (D), and (E).
Pantethine Supplementation in Drosophila with dPPCS Deficiency Improves Viability
Based on Mendelian inheritance, the genotype of the F1 embryonal progeny of crossing dPPCS1/TM3-GFP adults is the following: 50% will be dPPCS1/TM3-GFP, 25% will be dPPCS1/dPPCS1, and 25% will be TM3-GFP/TM3-GFP. The genotype TM3-GFP/TM3-GFP (homozygous for the balancer chromosome) induces a fully penetrant early developmental lethality and this genotype is not present in the F1 adult progeny, which therefore based on Mendelian inheritance consists of 66.6% dPPCS1/TM3-GFP and 33.3% dPPCS1/dPPCS1 flies only. dPPCS1/dPPCS1 homozygous adults are viable.7, 8 The exact percentages of the two viable genotypes of the F1 are not only determined by Mendelian inheritance but also can be influenced by the impact of the PPCS1 allele on viability. In the case a homozygous mutation in dPPCS negatively influencing viability, less than 33.3% of the F1 progeny will consist of dPPCS1/dPPCS1 homozygous mutant flies. In our crosses, the percentage of homozygous dPPCS1 pupae ranged from 9% to 22%, suggesting a reduced viability in homozygous mutants. Pantethine can serve as a source for CoA de novo biosynthesis, and since pantothenate kinase is the first enzyme of the canonical CoA biosynthesis pathway, it can phosphorylate pantethine to form 4-phosphopantetheine.29, 30 This route therefore does not require PPCS. Addition of pantethine to the fly food indeed rescued the percentage of homozygous survivors to levels that can be expected based on Mendelian inheritance. Vitamin B5 was used as a control. Experiments were repeated twice. In both experiments, a rescue of 10% was observed (Figure S2).
Effects of Pantethine Supplementation in Individuals with PPCS Deficiency
The two living affected individuals (IV.1 and IV.4, family B) commenced oral pantethine supplementation, initially at a dose of 6–8 mg/kg/day (once daily), which was subsequently gradually increased over a period of 2 months, reaching 20–24 mg/kg/day. At 2-month intervals, serial clinical evaluations, including pediatric cardiology visits and echocardiograms and serum acylcarnitine profiles using dried blood spots were obtained (Table 2). Compared to their baseline evaluations, both clinical and echocardiographic evaluations had shown individual IV.1 to experience a mild improvement in exertional dyspnea and an increase of ejection fraction (EF, using the Simpson mode) from 36% (at baseline) to 48% (at a recent visit); subject IV.4 remained stable, with symptoms of heart failure upon exertion and an EF of 45%.
Discussion
Here, we report on five individuals from unrelated families in whom biallelic mutations in PPCS lead to a cardiac phenotype. Variants identified in PPCS-deficient individuals are extremely rare. In our in-house database containing >10,000 WES datasets, we could not identify additional individuals carrying other rare biallelic PPCS variants. Moreover, in the gnomAD browser, neither of the two missense variants are present either as homozygous or heterozygous variants. The in-frame deletion is instead present only as a heterozygous variant with a MAF of 0.0001.
In addition to frequency considerations, identified variants are also predicted to be deleterious according to structural assumptions. In fact, mutations change either amino acids involved in the binding of ATP to PPCS or amino acids relevant in humans for the dimerization of PPCS.
In order to verify the pathogenicity of identified variants, we utilized a yeast null mutant for yPPCS. It is known that ablation of yPPCS is lethal, so we investigated whether transforming the yPPCS-null mutant with the wild-type PPCS or variant-carrying PPCS constructs could rescue the lethal phenotype. For validation experiments, we considered the long canonical isoform GenBank: NM_024664.3 as the isoform associated with the pathology since the mutation p.Pro107_Ala111del carried by subject II.2 of family A affects only this isoform.
While expression of the wild-type PPCS complemented the lethal phenotype in the yPPCS deletion strain, a clear growth defect was observed for all strains transformed with the identified human variants, thus confirming their deleterious nature. c.538G>C in particular was completely inactive, indicating that this variant impairs PPCS function more dramatically than the other variants.
It is likely that the decreased enzymatic activity of PPCS is due to decreased protein stability, as indicated by the strong reduction of PPCS signal in all PPCS-deficient individuals in the western blotting experiment or by the inability to assemble into a functional PPCS dimer. Of note, the monoclonal PPCS antibody detected only one band of 34 kDa under denaturing conditions, despite the antibody’s epitope residing within the C terminus region, common to the two PPCS isoforms. The presence of a single band corresponding to the canonical isoform suggests that this is likely the only splice variant translated into a protein.
In contrast to the neurodegenerative NBIA-related phenotypes associated with defects in PANK2 and COASY, PPCS-deficient individuals present with a clear dilated cardiomyopathy with a variable degree of severity and no neurodegeneration. Only in one individual (II.2, family A) were extra-cardiac features present in addition to severe dilated cardiomyopathy. Presumably, the presence of a completely inactive PPCS allele determines a more severe phenotype in this individual, as expected from the lack of rescue in yeast. Nonetheless, no neurodegeneration was present.
In fruit flies, impairment of dPPCS is associated with neurological impairment. In view of the fact that PPCS-deficient individuals have dilated cardiomyopathy, we reinvestigated dPPCS mutants with a focus on cardiac function. With deeper inspection, dPPCS mutants presented with reduced viability, and moreover showed increased heart rate, increased arrhythmia index, reduced systolic function, and increased heart wall shortening, all manifestations indicative of dilate cardiomyopathy in fruit flies.31
We therefore conclude that the cardiac phenotype of dPPCS flies recapitulates the pathophysiological alteration seen in PPCS-affected individuals, suggesting that alteration in PPCS can cause dilated cardiomyopathy.
In zebrafish, for instance, the downregulation of pank2 leads to perturbation in the vasculature development, reduced heart beat, and slower blood flux in addition to neurodegenerative signs,32 while the downregulation of coasy leads to edema at a cardiac level,33 suggesting already a link between CoA biosynthesis and cardiac function. With this knowledge in mind, it would be interesting to also investigate whether fruit flies with dPANK and dCOASY impairment have a subtle cardiac dysfunction, accompanying the most obvious neurodegeneration.
PPCS is the second enzyme of the CoA de novo biosynthesis pathway. Mutations in enzymes taking part in the route are expected to result in reduced CoA biosynthesis. Indeed, in PPCS-deficient individuals, the level of free CoA is significantly lower than in healthy control subjects. By reintroducing the wild-type copy of the PPCS in fibroblasts of an affected individual, the level of CoA is restored to normal values.
In contrast to PPCS-affected individuals, subjects with PKAN (pantothenate kinase-associated neurodegeneration) and COPAN (COASY protein-associated neurodegeneration) do not have detectable decrease of CoA levels in fibroblasts, despite the fact that most of the mutations in PANK2 and COASY impair the enzymatic activity of the recombinant enzymes, respectively.2, 34
Explanations of why mutations in PPCS do lead to decreased CoA levels in fibroblasts and mutations in PANK2 and COASY do not show a decrease in CoA levels and how this is possibly linked to a cardiac disease versus a neurodegenerative disease are currently not present. In order to understand these differences, CoA should be measured in relevant tissue of all conditions, meaning in the globus pallidus of PKAN and COPAN models and in cardiac tissue of PPCS-deficient models. Analysis of the abundancy of PANK2, PPCS, and COASY in relevant tissue, the identification of exact intracellular localizations of these three de novo CoA biosynthesis enzymes in disease-affected tissues, elucidation of possible redundancies in their roles in CoA synthesis, and identification of potential moonlighting functions are required to obtain answers to these intriguing questions.
Reduced cytosolic CoA concentration in PPCS-affected individuals may lead to a reduced synthesis of activated long-chain fatty acids in PPCS-affected individuals. Therefore, there is only limited substrate for carnitine palmitoyltransferase I (CPT1), resulting in reduced long-chain acylcarnitine concentrations, similar to individuals with carnitine palmitoyltransferase I (CPT1) deficiency. Indeed, this pattern is present in all individuals with PPCS deficiency, but not in the asymptomatic carrier (III.3), and has also been reported in two subjects with COPAN.5
Since CoA has a pivotal role in heart metabolism, not only as a source of energy but also as a modulator of fatty acid and glucose oxidation,35 it is expected that alteration in CoA metabolism impair cardiac function.
We cannot exclude that cardiomyopathy in PPCS-deficient individuals is also due to the generation of toxic metabolites. For instance, L-cysteine normally funneled in the synthesis of phosphopantothenoylcysteine could accumulate and eventually exceed the cytotoxicity threshold leading to detrimental effects in tissues, especially in the heart.36 Consistent with this hypothesis, dPPCS mutants have been reported to be hypersensitive to cysteine.8
However, considering the ubiquitous presence of the enzymatic PPCS activity, it remains unexplained why the brain and in general the other organs are not affected in PPCS-deficient individuals.
Aside from this open question which future studies will address, the identification of PPCS variant as the causative mutation in families A and B suggested a therapeutic trial of oral pantethine supplementation as targeted treatment aimed to bypass the deficient enzyme in the CoA synthesis pathway.37 We treated dPPCS mutants with pantethine, and the compound successfully rescued the viability in homozygous mutants. These results are also consistent with studies performed in PPCS mutants in bacteria.30 Following the promising results from the Drosophila model, empirical targeted treatment with pantethine was introduced for the two living affected siblings of family B. Our results thus far had not shown significant clinical improvement. This can be explained by the late introduction of the treatment in their clinical course, after cardiac damage had already occurred. Future studies, following early detection and initiation of treatment in additional affected families, may shed light on whether this therapeutic intervention can indeed bring more substantial clinical benefit, as demonstrated in the Drosophila model.
In summary, although the precise mechanism by which decreased CoA leads to cardiac rather than neurological problems remains to be fully elucidated, we demonstrate that pathogenic changes in PPCS lead to decreased CoA concentrations and cause cardiomyopathy with variable progression.
Acknowledgments
This study was supported by the German BMBF and Horizon2020 through E-Rare project GENOMIT (01GM1603 and 01GM1207 to H.P.); Horizon2020 Project SOUND (633974 to HP); German Network for Mitochondrial Disorders (mitoNET 01GM1113C to H.P. and T.M.); and the German Center for Heart Research (Z76010017300 and Z56010015300 to T.M.). We thank Dr. Angelika Seitz (Department of Neuroradiology, University Hospital Heidelberg) for the interpretation of the cranial MRI of individual II.2, family A, and Dr. Yussef Shahin who participates in the cardiology care of family B. S.A. is an MD PhD student at the Sackler Faculty of Medicine, Tel-Aviv University, and a candidate for the Graduate Partnership Program of the National Institutes of Health (NIH). The authors wish to thank the affected persons and their families for their continued support and collaboration.
Published: May 10, 2018
Footnotes
Supplemental Data include three figures, two tables, Supplemental Note, and one video and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.022.
Contributor Information
Dorothea Haas, Email: dorothea.haas@med.uni-heidelberg.de.
Yair Anikster, Email: yair.anikster@sheba.health.gov.il.
Web Resources
gnomAD Browser, http://gnomad.broadinstitute.org/
GTEx Portal, https://www.gtexportal.org/home/
OMIM, http://www.omim.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Reactome, https://reactome.org/
The Human Protein Atlas, https://www.proteinatlas.org
Supplemental Data
References
- 1.Hartig M.B., Hörtnagel K., Garavaglia B., Zorzi G., Kmiec T., Klopstock T., Rostasy K., Svetel M., Kostic V.S., Schuelke M. Genotypic and phenotypic spectrum of PANK2 mutations in patients with neurodegeneration with brain iron accumulation. Ann. Neurol. 2006;59:248–256. doi: 10.1002/ana.20771. [DOI] [PubMed] [Google Scholar]
- 2.Dusi S., Valletta L., Haack T.B., Tsuchiya Y., Venco P., Pasqualato S., Goffrini P., Tigano M., Demchenko N., Wieland T. Exome sequence reveals mutations in CoA synthase as a cause of neurodegeneration with brain iron accumulation. Am. J. Hum. Genet. 2014;94:11–22. doi: 10.1016/j.ajhg.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogarth P. Neurodegeneration with brain iron accumulation: diagnosis and management. J. Mov. Disord. 2015;8:1–13. doi: 10.14802/jmd.14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annesi G., Gagliardi M., Iannello G., Quattrone A., Iannello G., Quattrone A. Mutational analysis of COASY in an Italian patient with NBIA. Parkinsonism Relat. Disord. 2016;28:150–151. doi: 10.1016/j.parkreldis.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Evers C., Seitz A., Assmann B., Opladen T., Karch S., Hinderhofer K., Granzow M., Paramasivam N., Eils R., Diessl N. Diagnosis of CoPAN by whole exome sequencing: Waking up a sleeping tiger’s eye. Am. J. Med. Genet. A. 2017;173:1878–1886. doi: 10.1002/ajmg.a.38252. [DOI] [PubMed] [Google Scholar]
- 6.Olzhausen J., Schübbe S., Schüller H.J. Genetic analysis of coenzyme A biosynthesis in the yeast Saccharomyces cerevisiae: identification of a conditional mutation in the pantothenate kinase gene CAB1. Curr. Genet. 2009;55:163–173. doi: 10.1007/s00294-009-0234-1. [DOI] [PubMed] [Google Scholar]
- 7.Bosveld F., Rana A., Lemstra W., Kampinga H.H., Sibon O.C. Drosophila phosphopantothenoylcysteine synthetase is required for tissue morphogenesis during oogenesis. BMC Res. Notes. 2008;1:75. doi: 10.1186/1756-0500-1-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosveld F., Rana A., van der Wouden P.E., Lemstra W., Ritsema M., Kampinga H.H., Sibon O.C. De novo CoA biosynthesis is required to maintain DNA integrity during development of the Drosophila nervous system. Hum. Mol. Genet. 2008;17:2058–2069. doi: 10.1093/hmg/ddn105. [DOI] [PubMed] [Google Scholar]
- 9.Manoj N., Strauss E., Begley T.P., Ealick S.E. Structure of human phosphopantothenoylcysteine synthetase at 2.3 A resolution. Structure. 2003;11:927–936. doi: 10.1016/s0969-2126(03)00146-1. [DOI] [PubMed] [Google Scholar]
- 10.Stanitzek S., Augustin M.A., Huber R., Kupke T., Steinbacher S. Structural basis of CTP-dependent peptide bond formation in coenzyme A biosynthesis catalyzed by Escherichia coli PPC synthetase. Structure. 2004;12:1977–1988. doi: 10.1016/j.str.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Berman H.M., Westbrook J.D., Gabanyi M.J., Tao W., Shah R., Kouranov A., Schwede T., Arnold K., Kiefer F., Bordoli L. The protein structure initiative structural genomics knowledgebase. Nucleic Acids Res. 2009;37:D365–D368. doi: 10.1093/nar/gkn790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nussinov R., Wolfson H.J. Efficient detection of three-dimensional structural motifs in biological macromolecules by computer vision techniques. Proc. Natl. Acad. Sci. USA. 1991;88:10495–10499. doi: 10.1073/pnas.88.23.10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solomon O., Kunik V., Simon A., Kol N., Barel O., Lev A., Amariglio N., Somech R., Rechavi G., Eyal E. G23D: Online tool for mapping and visualization of genomic variants on 3D protein structures. BMC Genomics. 2016;17:681. doi: 10.1186/s12864-016-3028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 16.Fraczkiewicz R., Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 1998;19:319–333. [Google Scholar]
- 17.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44(W1) doi: 10.1093/nar/gkw408. W344-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., Roy A., Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29:2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kremer L.S., Prokisch H. Identification of disease-causing mutations by functional complementation of patient-derived fibroblast cell lines. Methods Mol. Biol. 2017;1567:391–406. doi: 10.1007/978-1-4939-6824-4_24. [DOI] [PubMed] [Google Scholar]
- 21.Mumberg D., Müller R., Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nolte I.M., Munoz M.L., Tragante V., Amare A.T., Jansen R., Vaez A., von der Heyde B., Avery C.L., Bis J.C., Dierckx B. Erratum: Genetic loci associated with heart rate variability and their effects on cardiac disease risk. Nat. Commun. 2017;8:16140. doi: 10.1038/ncomms16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rana A., Seinen E., Siudeja K., Muntendam R., Srinivasan B., van der Want J.J., Hayflick S., Reijngoud D.J., Kayser O., Sibon O.C. Pantethine rescues a Drosophila model for pantothenate kinase-associated neurodegeneration. Proc. Natl. Acad. Sci. USA. 2010;107:6988–6993. doi: 10.1073/pnas.0912105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku340. W252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 27.Guex N., Peitsch M.C., Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(Suppl 1):S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 28.Sikorski R.S., Boeke J.D. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 29.Sibon O.C., Strauss E. Coenzyme A: to make it or uptake it? Nat. Rev. Mol. Cell Biol. 2016;17:605–606. doi: 10.1038/nrm.2016.110. [DOI] [PubMed] [Google Scholar]
- 30.Balibar C.J., Hollis-Symynkywicz M.F., Tao J. Pantethine rescues phosphopantothenoylcysteine synthetase and phosphopantothenoylcysteine decarboxylase deficiency in Escherichia coli but not in Pseudomonas aeruginosa. J. Bacteriol. 2011;193:3304–3312. doi: 10.1128/JB.00334-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taghli-Lamallem O., Akasaka T., Hogg G., Nudel U., Yaffe D., Chamberlain J.S., Ocorr K., Bodmer R. Dystrophin deficiency in Drosophila reduces lifespan and causes a dilated cardiomyopathy phenotype. Aging Cell. 2008;7:237–249. doi: 10.1111/j.1474-9726.2008.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zizioli D., Tiso N., Guglielmi A., Saraceno C., Busolin G., Giuliani R., Khatri D., Monti E., Borsani G., Argenton F., Finazzi D. Knock-down of pantothenate kinase 2 severely affects the development of the nervous and vascular system in zebrafish, providing new insights into PKAN disease. Neurobiol. Dis. 2016;85:35–48. doi: 10.1016/j.nbd.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khatri D., Zizioli D., Tiso N., Facchinello N., Vezzoli S., Gianoncelli A., Memo M., Monti E., Borsani G., Finazzi D. Down-regulation of coasy, the gene associated with NBIA-VI, reduces Bmp signaling, perturbs dorso-ventral patterning and alters neuronal development in zebrafish. Sci. Rep. 2016;6:37660. doi: 10.1038/srep37660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotzbauer P.T., Truax A.C., Trojanowski J.Q., Lee V.M. Altered neuronal mitochondrial coenzyme A synthesis in neurodegeneration with brain iron accumulation caused by abnormal processing, stability, and catalytic activity of mutant pantothenate kinase 2. J. Neurosci. 2005;25:689–698. doi: 10.1523/JNEUROSCI.4265-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abo Alrob O., Lopaschuk G.D. Role of CoA and acetyl-CoA in regulating cardiac fatty acid and glucose oxidation. Biochem. Soc. Trans. 2014;42:1043–1051. doi: 10.1042/BST20140094. [DOI] [PubMed] [Google Scholar]
- 36.Stipanuk M.H., Dominy J.E., Jr., Lee J.I., Coloso R.M. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J. Nutr. 2006;136(6, Suppl):1652S–1659S. doi: 10.1093/jn/136.6.1652S. [DOI] [PubMed] [Google Scholar]
- 37.Srinivasan B., Baratashvili M., van der Zwaag M., Kanon B., Colombelli C., Lambrechts R.A., Schaap O., Nollen E.A., Podgoršek A., Kosec G. Extracellular 4′-phosphopantetheine is a source for intracellular coenzyme A synthesis. Nat. Chem. Biol. 2015;11:784–792. doi: 10.1038/nchembio.1906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.