Abstract
There is a limited understanding about the impact of rare protein-truncating variants across multiple phenotypes. We explore the impact of this class of variants on 13 quantitative traits and 10 diseases using whole-exome sequencing data from 100,296 individuals. Protein-truncating variants in genes intolerant to this class of mutations increased risk of autism, schizophrenia, bipolar disorder, intellectual disability, and ADHD. In individuals without these disorders, there was an association with shorter height, lower education, increased hospitalization, and reduced age at enrollment. Gene sets implicated from GWASs did not show a significant protein-truncating variants burden beyond what was captured by established Mendelian genes. In conclusion, we provide a thorough investigation of the impact of rare deleterious coding variants on complex traits, suggesting widespread pleiotropic risk.
Keywords: exome sequencing, ultra-rare variants, rare variants, phewas, constraint genes, burden analysis, selection
Main Text
Protein-truncating variants (PTVs) are likely to modify gene function and have been linked to hundreds of Mendelian disorders.1, 2 However, the impact of PTVs on complex traits has been limited by the available sample size of whole-exome sequencing studies (WESs).3 Here, we assembled whole-exome sequencing data from 100,296 individuals, drawing from a combination of cohort and case/control disease studies with phenotypic information on a total of 13 quantitative traits and 10 diseases (Tables S1–S3). We used a common pipeline to process, annotate, and analyze the data (see Supplemental Material and Methods and Figure S1 for principal components plots). Ethical committees for each study approved all procedures and all subjects provided written informed consent (or legal guardian consent and subject assent).
We began by focusing our analysis on PTVs that occur in a set of 3,172 PTV-intolerant (PI) genes (see Table S4 for all gene sets used in this study). Our motivation for focusing on the PI-PTVs was two-fold. First, this gene class was identified through an unbiased approach that leveraged the observed frequency distribution in ExAC4 without relying on information from model organisms or in vitro experiments. Second, PI-PTVs have been shown to associate with early-onset neurodevelopmental and psychiatric disorders and are likely to result in reproductively disadvantageous phenotypes.5, 6, 7 To focus on those variants that are most likely to be subject to purifying selection, we considered only rare (allele frequency < 0.1%) and ultra-rare (observed in fewer than 1 in 201,176 individuals) variants (Supplemental Material and Methods).
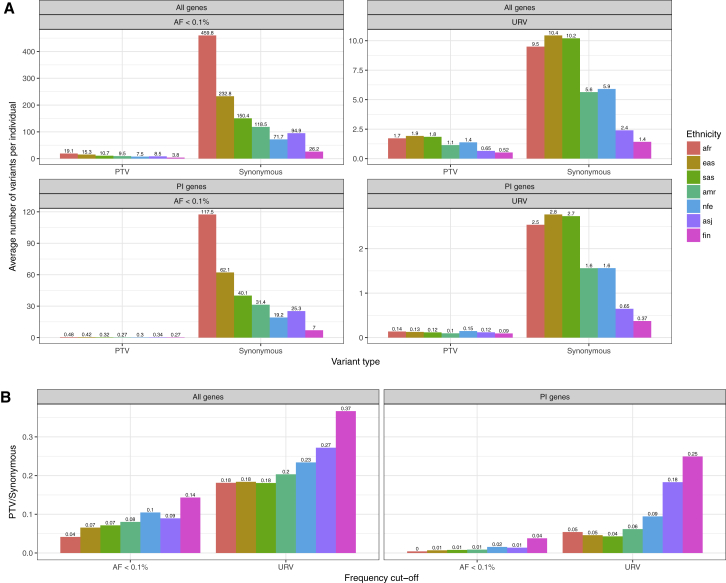
After excluding participants diagnosed with a psychiatric or neurodevelopmental disorder, we observed an average of 7.72 and 0.30 rare PTVs per individual, across all genes and in PI genes, respectively (Figure 1A); one or more ultra-rare PI-PTV was observed in 11% of the individuals. The number and frequency of rare variants differs across populations, reflecting the degree of selection compounded by recent demography, including bottlenecks, split times, and migration between populations.8 The ratio of deleterious to neutral alleles per individual increases as humans migrated out of Africa, consistent with less efficient negative selection against deleterious variants and serial founder effects that reduce the effective population size.9 Conditional on a variant being ultra-rare, we observe a higher ratio of PTVs to synonymous variants (Figure 1B); recently arisen ultra-rare variants have had less time to be purged by negative selection, which is further magnified in populations that have undergone a recent bottleneck. For example, we observed a higher ratio among Ashkenazi Jewish and Finnish populations as compared to non-Finnish Europeans, reflecting the more recent population-specific bottlenecks.10, 11
Figure 1.
Variants Frequency Distribution across Different Ethnic Group and Gene Sets
(A) Average number of variants per individual in n = 83,439 participants without neurodevelopmental/psychiatric disorders. We report the results separately for each ethnic group.
(B) Ratio between PTV/Synonymous for each ethnic group.
Abbreviations: Afr, African American; Eas, East Asian; Sas, South Asian; Amr, Latinos; Nfe, non-Finnish European; Asj, Ashkenazi Jewish; Fin, Finnish.
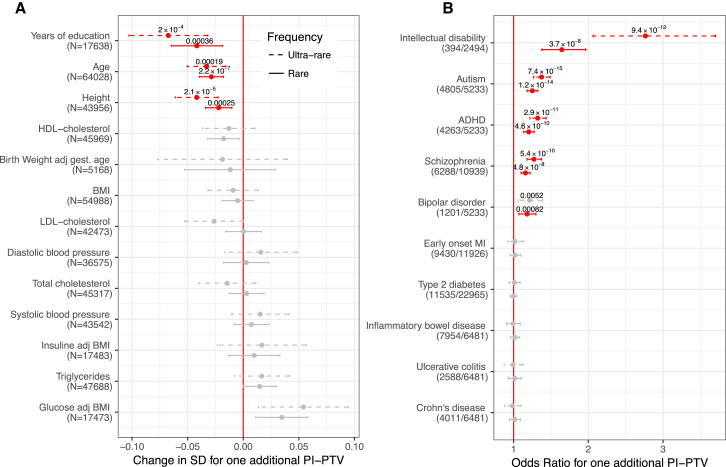
We tested the association between a burden of PI-PTVs and the 13 traits and 10 disease diagnoses (Figure 2) by performing study-specific and ethnicity-specific linear or logistic regression analysis adjusting for potential confounders such as overall mutation rate (Table S5). The results of these separate analyses were then meta-analyzed (Supplemental Material and Methods). We used an experiment-wise p value threshold of 2 × 10−3 to account for multiple testing (0.05/23 traits tested). Among the quantitative traits, we found that carriers of at least one rare PI-PTV had fewer years of education (−2.2 months, p = 4 × 10−4), as we have previously reported,12 were shorter (−0.2 cm, p = 3 × 10−4), and were younger (−3.7 months, p = 2 × 10−7).
Figure 2.
Association Analysis for Rare and Ultra-rare Variant Burden
(A) Association between PI-PTV burden and continuous traits. We reported the association in standard deviations (SD) to allow for comparison across traits. In parentheses, we reported the number of individual included in the analysis for each trait. The p values are reported only for experiment-wise significant results (p < 2 × 10−3), highlighted in red. Bars indicate 95% confidence intervals. All the results are obtained from meta-analyzing study and ethnicity-specific associations.
(B) Odds ratio for association between PI-PTV burden and dichotomous traits. In brackets, we reported the number of case and control subjects.
To ensure the robustness of the age (at enrollment) result, we performed a series of quality control analyses to guard against the impact of technical confounders or specific study designs that might bias the results. We first confirm that the signal was not observed among PTVs in non-PI genes and synonymous variants in PI genes, our negative controls (Figure S2). We further found that the effect was consistent across ethnicities and study cohorts (Figure S3), when INDELs and SNPs were considered separately, when mutations possibly caused by cytosine deamination (C>T or G>A) were excluded, and when a highly stringent QC was used (Figure S4). Similarly, the association did not change after adjusting for eight QC metrics capturing most of the sample properties (Figure S5). Although we could not exclude the impact of unmeasured confounder, we find this result consistent with reduced survival, detrimental health, or decreased study participation over time among PI-PTVs carriers. If reduced survival or detrimental health effects drive this association, we see it as a signature of viability selection, overall in the population. Analyses with time at death as the outcome will be needed to confirm the interpretation of this finding.
We then focused on dichotomous traits (Figure 2). We observed significant associations with all psychiatric disorders that were tested: intellectual disability (ID) (odds ratio [OR] = 1.7, p = 4 × 10−8), autism (OR = 1.3, p = 1 × 10−14), schizophrenia (OR = 1.2, p = 5 × 10−8), ADHD (OR = 1.2, p = 5 × 10−10), and bipolar disorder (OR = 1.2, p = 8 × 10−4). We did not, however, find PI-PTV burden to be associated with later-onset, non-brain-related diseases such as type 2 diabetes, early-onset myocardial infarction, inflammatory bowel disease, ulcerative colitis, or Crohn disease. Across all significantly associated phenotypes, the effect size was stronger among the subset of ultra-rare PI-PTV carriers, confirming that rarer PTVs are, on average, more deleterious.
The association with these five neurodevelopmental/psychiatric disorders and three quantitative traits was observed only for PI-PTVs and not for PTVs in non-PI genes nor for synonymous variants in PI genes. These results suggest that the association to PI-PTVs is not driven by population stratification or technical bias (Figure S6).
Our approach so far focuses on assuming that all PI-PTVs act on the phenotype in the same direction, that is, they are all either protective or risk conferring. We relaxed this hypothesis, allowing rare PI-PTVs to have different directions as well as different magnitudes of effects, and repeated these tests using SKAT.13 We did not identify any additional associations (Figure S7), suggesting that PI genes do not account for a substantial fraction of variability in the traits for which no PTV burden was identified. Further, the observed burden of PI-PTVs for neurodevelopmental/psychiatric disorders, height, educational attainment, and age suggests that the majority of those PI genes that have an effect, do so in the same direction. Although case ascertainment bias reduces the power of detecting protective variants, this is not the case for continuous traits like height.
We also evaluated whether damaging missense variants, which are on average more common and less severely deleterious than PTVs, showed a similar signal. Damaging missense variants have been associated with complex disorders such as coronary heart disease and inflammatory bowel disease.14, 15 We found an independent signal for damaging missense variants in PI genes for all disorders and traits that were also associated with PI-PTVs. Furthermore, the strength of the association increased as a function of the number of prediction algorithms that confidently classified a missense variant as “damaging” (Figure S8), suggesting that these missense mutations are similar to PTVs in biological effect, potentially abrogating gene function. We note that this effect was particularly strong for ultra-rare variants, reinforcing the observation that variant frequency is a marker of selection and aids in the identification of pathogenic damaging missense variation.16, 17
Given the high degree of shared comorbidities across neurodevelopmental/psychiatric disorders, we leveraged information from the Danish National Psychiatric registry to evaluate whether the signal was driven by a specific disorder or shared across multiple disorders. Individuals with multiple neurodevelopmental/psychiatric disorders, and especially those with ID, showed a stronger enrichment of PI-PTVs (Figure S9). Nevertheless, among those without comorbidities, the signal remained significant and remarkably similar across disorders (OR = 1.12, 1.15, 1.21, 1.18 for schizophrenia, bipolar, autism, and ADHD, respectively; Cochran’s Q test for heterogeneity p = 0.282). We further found that carriers of ultra-rare PI-PTVs had earlier onset of ADHD (−4.0 months, p = 0.008; Table S6). However, this was partially explained by the fact that individuals with earlier diagnosis of ADHD were also more likely to be diagnosed with ID (14.7 versus 15.5 years for individuals with ADHD with and without ID, t test p value = 0.009). Indeed, when we considered ADHD-affected case subjects without major comorbidities, the effect was attenuated (−2.9 months, p = 0.12). Finally, in control subjects with none of these psychiatric diagnoses, we still observed a significant association with the broader ICD-10 category of mental, behavioral, and neurodevelopmental disorders, suggesting that PI-PTVs influence the broader cognitive spectrum (Table S7).
Since previous studies have shown a higher rate of PI de novo PTVs in autism-affected females as compared to males,18, 19 we wondered whether sex played a role here. In this study, however, we did not have parent-offspring subjects needed to distinguish de novo variants from those that have recently arisen in the population, the latter being the majority of observed rare variants. This would potentially dilute the sex-specific effect if it is in fact a property of de novo variants but not of rare variants more generally. We found both weak and insignificant differences between males and females in the effect of PI-PTVs on four neurodevelopmental/psychiatric disorders (Table S8). Interestingly, we did not observe differences in ADHD-affected males and females, in contrast with the hypothesis that affected females might be enriched for rare deleterious variants.20 We cannot exclude that differences in the diagnostic criteria used in these European studies compared to those of previous studies, which were mostly conducted in the U.S., might explain these results.
We also assessed whether the observed burden of PTVs was specific to PI genes or whether such a burden could be identified for other gene sets that are likely to contain functionally relevant genes. First, we examined other experimental and literature-based gene sets linked to severe phenotypes. Specifically, we considered all genes (1) reported in ClinVar,2 (2) that resulted in lethal or subviable phenotypes in mice,21 (3) that were required for proliferation and survival in a human cancer cell line,22 and (4) were categorized as haploinsufficient by ClinGen (Table S4 and Supplemental Material and Methods). Except for the haploinsufficient genes, which showed a significantly stronger association with autism alone, none of the other gene sets tested showed the PTV burden that was captured by PI genes (Figure S10). This suggests that the degree of natural selection against PTVs in a gene is indeed an important indicator of whether such PTVs are likely to be implicated as strong effects for neurodevelopmental/psychiatric disorders, height, educational attainment, and age. It also highlights that observed associations are not simply reflecting an aggregate signal from known Mendelian disorders. Indeed, we could not detect significant association for any of the traits considered in this analysis when focusing on just ClinVar genes.
We reasoned that a single variant approach, rather than a gene-based test, might provide increased resolution. We considered all high-quality ClinVar variants (0.76 on average per individual) and a set of variants deemed to be recessive lethal (0.03) (Supplemental Material and Methods). Carriers of these variants were not enriched in any of the disorders or traits examined here (Table S9).
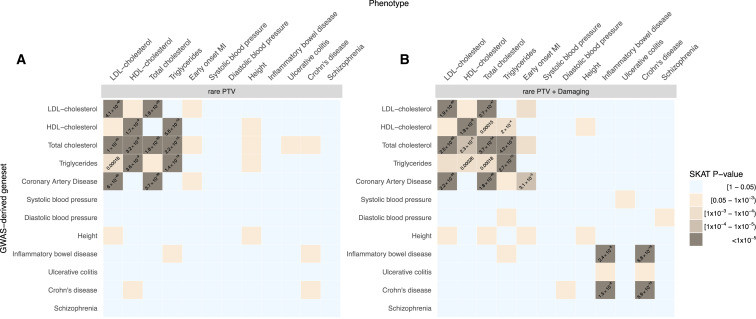
Second, we examined whether results from GWASs conducted on the same phenotypes as those included in this study could implicate genes containing an aggregate PTV burden. We used DEPICT23 to link genome-wide significant hits to candidate genes (Table S10 and Supplemental Material and Methods) and, within each GWAS-derived gene set, we studied the association between rare PTVs and the phenotypes using the SKAT test. GWAS-derived gene-sets captured associations between rare PTVs and different classes of lipids (Figure 3 and Table S11). For example, the association between rare PTVs and HDL cholesterol was captured by gene sets derived from GWASs of HDL (p = 2 × 10−9), total cholesterol (p = 3 × 10−8), and triglycerides (p = 4 × 10−9), but not by those of coronary heart diseases (p = 0.25), consistent with previous observations about non-causality of HDL cholesterol on coronary heart diseases.24 The inclusion of both rare damaging missense and PTVs resulted in additional signal co-localization between inflammatory bowel disease, early-onset myocardial infarction, and the corresponding GWAS-derived gene sets. However, it appeared that all these signals were being driven by well-known genes, involved in rare familial forms of these diseases. Specifically, when Mendelian lipid genes and NOD2 were removed from the cardiovascular and inflammatory bowel disease-related GWAS gene sets, respectively, no signal remained (Figure S11). This might reflect a lack of power (despite this being the largest WES study for the majority of the traits), inaccurate links between genome-wide significant hits and the corresponding candidate genes or PTVs, and common variants acting on partially distinct pathways. Nevertheless, we observed similar results when including SNPs below genome-wide significance to increase power and when using different methods to link SNPs with corresponding candidate genes to increase precision, including gene-based testing25 and eQTL mapping (Figure S11 and Supplemental Material and Methods).
Figure 3.
Signal Overlap between Rare Variants and GWAS-Derived Gene Sets
(A) Association (SKAT test p value) in GWAS-derived gene sets (y axis) between rare PTVs and the phenotypes reported on the x axis. Each gene set is obtained using DEPICT to link SNPs derived from GWAS with p value < 5 × 10−8 and a candidate gene. In brackets we report the number of genes with at least one PTV in our dataset. p values are reported only for experiment-wise associations (p < 0.0003).
(B) Association (SKAT test p value) in GWAS-derived gene sets (y axis) between rare PTVs + damaging missense and the phenotypes reported on the x axis.
The choice of the 10 diseases and 13 quantitative traits included in the main analysis was driven by data availability and power considerations but was not truly unbiased. Therefore, we leveraged national population health registries to increase the scope of disorders we could examine. These well-studied and validated registries26, 27 include diagnostic codes from 14,117 individuals (n = 8,493 from Finland and 5,624 from Sweden), recorded between 1968 and 2015 (Table S12). Individuals with psychiatric disorders were excluded from our analyses. To maximize the validity of the diagnoses, we used a curated list of disease definitions aggregating related ICD codes (Table S13). We studied the association between rare PI-PTVs and 101 diseases with at least 50 case subjects, using a survival analysis model. We identified an association (multi-testing significance threshold = 0.05/101; 5 × 10−4) with chronic kidney failure (hazard ratio = 1.9, p = 3 × 10−6; number of case subjects = 120; Figure S12). The association was strong among the Finnish data and only significant when considering ultra-rare PI-PTVs in the Swedish data (Table S14).
We speculated that this association might reflect a burden of underlying comorbidities that were too rare to be included in this analysis. To evaluate epidemiological associations, we extended our analysis to 28,709 Finnish individuals that were not exome sequenced but were linked to the registries. We found that individuals with chronic kidney failure also have a higher rate of cardiovascular-related comorbidities, as well as skin infections, kidney cancer, and other abnormalities of the renal system (Table S15). Therefore, it is challenging to determine whether it is the chronic kidney failure or some more rare comorbid condition that drives the association with PI-PTVs. Nevertheless, five PI-PTVs in Finnish individuals with chronic kidney failure were in genes involved in Mendelian-type disorders characterized by renal or endocrine abnormalities (ARNT2, COL4A1, DMXL2, FBN1, and NNT; Supplemental Material and Methods).
We also examined whether the association between PI-PTVs and diminished cognition and detrimental health would result in a higher number of hospital visits, counting the number of in-patient visits associated with a unique ICD codes. In both the Swedish and Finnish datasets, we observed a significant increase in the rate of hospital visits with a greater burden of PI-PTVs (+7.6% per additional PI-PTV, p = 0.0002). We used different strategies to model the outcome and observed similar results (Table S16 and Figure S13).
By aggregating WES data on more than 100,000 individuals for 23 different traits and disorders, we have gained insight into the role of PTVs in conferring risk for these conditions. First, PTVs occurring in PI genes had a remarkably similar effect on autism, schizophrenia, bipolar disorder, and ADHD. The majority of this signal was driven by ultra-rare PI-PTVs and we observed only a marginal additional contribution of non-ultra-rare PTVs with allele frequency < 0.1% (Figure S14). The observed effects of PI-PTVs on psychiatric disorders were not driven by major underlying comorbidities. This suggests that these PI-PTVs as a whole are likely to be pleiotropic, influencing some core intermediate phenotypes that relate to risk across many psychiatric disorders. Nevertheless, we could consider only “bulk” pleiotropy, which is the combined impact of PTVs in PI genes, and we are not powered to detect whether single variants have disease-specific effects. Further, this burden suggests that individual PI genes will be eventually discovered conclusively for each of these disorders, not just autism, but that such associations will need to be interpreted in the light of this shared effect across disorders. The strong enrichment of PI-PTVs in individuals with neurodevelopmental/psychiatric disorders does not exclude the existence of non-PI genes involved in the etiology of these disorders. These genes, however, are more likely to have weaker and, possibly, trait-specific effect.
Second, we detected a significant association between PI-PTVs and decreased human height. In contrast to this, a recent large-scale study using the exome chip has shown a similar numbers of height-increasing and height-decreasing rare variants.28 This discrepancy could be because, by using a more stringent frequency cut-off and focusing on a subset of genes likely to cause early-onset severe disease, we effectively considered variants related to a burden of (incompletely) penetrant Mendelian-type disorders, often characterized by reduced growth. Such an interpretation is consistent with a tighter link to directional selection on stronger impact mutations for human height.
Third, we systematically compared the co-localization of signal between GWAS-candidate genes and rare PTVs. We found few overlaps (cardiovascular-related traits, inflammatory bowel disease) which, we revealed, were entirely driven by a few genes previously identified by both GWASs and WES studies. Other traits did not show any overlap. Schizophrenia, for example, which is highly enriched for PI-PTVs, did not show overlap with GWAS candidate genes. Even among traits where genes with low-frequency coding variants have been previously identified by exome-chip-based studies, such as height and systolic blood pressure, we found no substantial rare PTVs enrichment. These results suggest that the relationship between GWAS signal and rare coding variants is not always straightforward, and that, when interpreting WES data, other complementary approaches such as those that integrate population genetic models and large sample resources might be more suitable to nominate gene sets of interest. The degree of overlap, and therefore the most effective strategy to identify pathogenic variants, is likely to depend on the selective pressure shaping the genetic architecture of the trait under investigation. Moreover, it cannot be overlooked that individuals carrying rare PTVs in genes implicated by common variant-based approaches might present phenotypic outcomes that deviate from those under investigation. Finally, it is interesting to notice that, while PTVs tend to have a consistent directional effect within a PI gene, this is not the case for GWAS-derived gene sets, where most of signals could be captured only by assuming heterogeneity in effect direction (Supplemental Material and Methods).
In conclusion, in this large WES study, we showed that PI genes are well suited to capture the impact of rare to ultra-rare PTVs on the cognitive, behavioral, and developmental spectra. This is less the case for major later-onset complex traits with modest effect on reproductive fitness. Strategies to prioritize gene sets relevant for these traits would need to consider the role that relaxed selective pressure has been playing in shaping the frequency distribution of disease-causing PTVs.
Declaration of Interests
Benjamin M. Neale is a member of the scientific advisory board at Deep Genomics and a paid consultant at Camp4 Therapeutics Corporation, Merck & Co., and Avanir Pharmaceuticals.
Acknowledgments
A.G. is supported by the Knut and Alice Wallenberg Foundation (2015.0327) and the Swedish Research Council (2016-00250). This study was supported by grants from the National Human Genome Research Institute (U54 HG003067, R01 HG006855), the National Institute of Mental Health (1U01MH105666-01, 1R01MH101244-02, R01 MH077139, and RC2 MH089905), the National Institute of Diabetes and Digestive and Kidney Disease (1U54DK105566-02), the Stanley Center for Psychiatric Research, the Alexander and Margaret Stewart Trust, and the Sylvan C. Herman Foundation. V.S. was supported by the Finnish Foundation for Cardiovascular Research.
Published: May 31, 2018
Footnotes
Supplemental Data include 14 figures, 16 tables, and Supplemental Material and Methods and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.05.002.
Contributor Information
Andrea Ganna, Email: aganna@broadinstitute.org.
Benjamin M. Neale, Email: bneale@broadinstitute.org.
Accession Numbers
We have made the code and variants used in the paper available at https://github.com/andgan/ultra_rare_pheno_spectrum.
We are extremely committed to data sharing. However, we recognize that due to national regulation, not all individual-level data and phenotypes can be shared using dbGap. Specifically, access to the Danish and Finnish phenotypic and genetic data can be obtained only by Danish and Finnish national institutions. Information about getting access to the Danish data can be obtained at http://ipsych.au.dk/about-ipsych/. Access to the Finnish data can be obtained at https://www.thl.fi/en/web/thl-biobank/for-researchers. We are happy to help other researchers with information to simplify the application process to obtain these datasets. All the remaining datasets are available via dbGap, as follows. Migen: phs000814.v1.p1, phs000902.v1.p1, phs000917.v1.p1, phs000883.v1.p1, phs001058.v1.p1, phs000806.v1.p1, phs001000.v1.p1, phs000990.v1.p1, phs000916.v1.p1, phs001101.v1.p1; T2D-GENES/GoT2D/SIGMA: phs001099, phs001098, phs000849, phs001097, phs001096, phs001095, phs001093, phs001100, phs001102, phs000840; Swedish Schizophrenia: phs000473.v2.p2; IBD: phs001076.v1.p1.
Web Resources
Codes and variants, https://github.com/andgan/ultra_rare_pheno_spectrum
iPSYCH, http://ipsych.au.dk/about-ipsych
THL Biobank, https://www.thl.fi/en/web/thl-biobank/for-researchers
Supplemental Data
References
- 1.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuk O., Schaffner S.F., Samocha K., Do R., Hechter E., Kathiresan S., Daly M.J., Neale B.M., Sunyaev S.R., Lander E.S. Searching for missing heritability: designing rare variant association studies. Proc. Natl. Acad. Sci. USA. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genovese G., Fromer M., Stahl E.A., Ruderfer D.M., Chambert K., Landén M., Moran J.L., Purcell S.M., Sklar P., Sullivan P.F. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat. Neurosci. 2016;19:1433–1441. doi: 10.1038/nn.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson E.B., St Pourcain B., Anttila V., Kosmicki J.A., Bulik-Sullivan B., Grove J., Maller J., Samocha K.E., Sanders S.J., Ripke S., iPSYCH-SSI-Broad Autism Group Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat. Genet. 2016;48:552–555. doi: 10.1038/ng.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathieson I., McVean G. Demography and the age of rare variants. PLoS Genet. 2014;10:e1004528. doi: 10.1371/journal.pgen.1004528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henn B.M., Botigué L.R., Peischl S., Dupanloup I., Lipatov M., Maples B.K., Martin A.R., Musharoff S., Cann H., Snyder M.P. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc. Natl. Acad. Sci. USA. 2016;113:E440–E449. doi: 10.1073/pnas.1510805112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostrer H., Skorecki K. The population genetics of the Jewish people. Hum. Genet. 2013;132:119–127. doi: 10.1007/s00439-012-1235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim E.T., Würtz P., Havulinna A.S., Palta P., Tukiainen T., Rehnström K., Esko T., Mägi R., Inouye M., Lappalainen T., Sequencing Initiative Suomi (SISu) Project Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganna A., Genovese G., Howrigan D.P., Byrnes A., Kurki M., Zekavat S.M., Whelan C.W., Kals M., Nivard M.G., Bloemendal A. Ultra-rare disruptive and damaging mutations influence educational attainment in the general population. Nat. Neurosci. 2016;19:1563–1565. doi: 10.1038/nn.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stitziel N.O., Stirrups K.E., Masca N.G., Erdmann J., Ferrario P.G., König I.R., Weeke P.E., Webb T.R., Auer P.L., Schick U.M., Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N. Engl. J. Med. 2016;374:1134–1144. doi: 10.1056/NEJMoa1507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo Y., de Lange K.M., Jostins L., Moutsianas L., Randall J., Kennedy N.A., Lamb C.A., McCarthy S., Ahmad T., Edwards C. Exploring the genetic architecture of inflammatory bowel disease by whole-genome sequencing identifies association at ADCY7. Nat. Genet. 2017;49:186–192. doi: 10.1038/ng.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavalli-Sforza L.L. Population structure and human evolution. Proc. R. Soc. Lond. B Biol. Sci. 1966;164:362–379. doi: 10.1098/rspb.1966.0038. [DOI] [PubMed] [Google Scholar]
- 17.Price G.R. Selection and covariance. Nature. 1970;227:520–521. doi: 10.1038/227520a0. [DOI] [PubMed] [Google Scholar]
- 18.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., Raja A., Coe B.P., Stessman H.A., He Z.X. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosmicki J.A., Samocha K.E., Howrigan D.P., Sanders S.J., Slowikowski K., Lek M., Karczewski K.J., Cutler D.J., Devlin B., Roeder K. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat. Genet. 2017;49:504–510. doi: 10.1038/ng.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor M.J., Lichtenstein P., Larsson H., Anckarsater H., Greven C.U., Ronald A. Is There a Female Protective Effect Against Attention-Deficit/Hyperactivity Disorder? Evidence From Two Representative Twin Samples. J Am Acad Child Adolesc Psychiatry. 2016;55:504–512 e502. doi: 10.1016/j.jaac.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickinson M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., Meehan T.F., Weninger W.J., Westerberg H., Adissu H., International Mouse Phenotyping Consortium. Jackson Laboratory. Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS) Charles River Laboratories. MRC Harwell. Toronto Centre for Phenogenomics. Wellcome Trust Sanger Institute. RIKEN BioResource Center High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., Sabatini D.M. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pers T.H., Karjalainen J.M., Chan Y., Westra H.J., Wood A.R., Yang J., Lui J.C., Vedantam S., Gustafsson S., Esko T., Genetic Investigation of ANthropometric Traits (GIANT) Consortium Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voight B.F., Peloso G.M., Orho-Melander M., Frikke-Schmidt R., Barbalic M., Jensen M.K., Hindy G., Hólm H., Ding E.L., Johnson T. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludvigsson J.F., Andersson E., Ekbom A., Feychting M., Kim J.L., Reuterwall C., Heurgren M., Olausson P.O. External review and validation of the Swedish national inpatient register. BMC Public Health. 2011;11:450. doi: 10.1186/1471-2458-11-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sund R. Quality of the Finnish Hospital Discharge Register: a systematic review. Scand. J. Public Health. 2012;40:505–515. doi: 10.1177/1403494812456637. [DOI] [PubMed] [Google Scholar]
- 28.Marouli E., Graff M., Medina-Gomez C., Lo K.S., Wood A.R., Kjaer T.R., Fine R.S., Lu Y., Schurmann C., Highland H.M., EPIC-InterAct Consortium. CHD Exome+ Consortium. ExomeBP Consortium. T2D-Genes Consortium. GoT2D Genes Consortium. Global Lipids Genetics Consortium. ReproGen Consortium. MAGIC Investigators Rare and low-frequency coding variants alter human adult height. Nature. 2017;542:186–190. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.