Abstract
βIV spectrin links ankyrinG (AnkG) and clustered ion channels at axon initial segments (AISs) and nodes of Ranvier to the axonal cytoskeleton. Here, we report bi-allelic pathogenic SPTBN4 variants (three homozygous and two compound heterozygous) that cause a severe neurological syndrome that includes congenital hypotonia, intellectual disability, and motor axonal and auditory neuropathy. We introduced these variants into βIV spectrin, expressed these in neurons, and found that 5/7 were loss-of-function variants disrupting AIS localization or abolishing phosphoinositide binding. Nerve biopsies from an individual with a loss-of-function variant had reduced nodal Na+ channels and no nodal KCNQ2 K+ channels. Modeling the disease in mice revealed that although ankyrinR (AnkR) and βI spectrin can cluster Na+ channels and partially compensate for the loss of AnkG and βIV spectrin at nodes of Ranvier, AnkR and βI spectrin cannot cluster KCNQ2- and KCNQ3-subunit-containing K+ channels. Our findings define a class of spectrinopathies and reveal the molecular pathologies causing nervous-system dysfunction.
Keywords: cytoskeleton, axon initial segment, node of Ranvier
Introduction
Neuronal spectrins are widely expressed and function as tetramers of two αII subunits partnered with two βI, βII, βIII, or βIV spectrin subunits.1, 2 The axon’s submembranous cytoskeleton is organized into repeating circumferential actin rings, evenly spaced and connected to one another by spectrin tetramers.3 Although αII spectrin is found throughout the neuron, β spectrins are not uniformly distributed and are often found in specialized membrane domains. For example, βIV spectrin is enriched at axon initial segments (AISs) and nodes of Ranvier and forms a periodic lattice with actin.4, 5 βIV spectrin functions together with AnkG to cluster the Na+ and KCNQ2- and KCNQ3-subunit-containing K+ channels necessary for action potential initiation and propagation.6, 7, 8 Pathogenic variants in spectrins cause early infantile epileptic encephalopathy type 5 (EIEE5 [MIM: 613477]), which is characterized by seizures, hypomyelination, and brain atrophy (SPTAN1 [MIM:182810]);9, 10, 11 spinocerebellar ataxia type 5 (SCA5 [MIM: 600224]) (SPTBN2 [MIM:604985]):12 and hereditary spherocytosis (SPH2 [MIM: 616649]) (SPTB [MIM:182870]).13 A variety of “quivering” mice with spontaneous or targeted mutations in Sptbn4 have been reported and studied.14, 15, 16, 17 Human pathogenic variants in SPTBN4 (MIM:606214) were limited to a single recent case report of an individual with congenital myopathy, neuropathy, and deafness (CMND [MIM: 617519]),18 but how this variant causes disease remains unknown. We recently identified six individuals with pathogenic variants of SPTBN4 and similar neurological phenotypes. Our studies uncover common molecular pathologies among the identified variants of SPTBN4 and reveal a class of spectrinopathy that disrupts excitable domains in axons.
Methods
Research Subjects
All affected individuals presented in the neonatal period with severe hypotonia and global developmental delays. Feeding difficulties were common during infancy as a result of oromotor hypotonia, leading to placement of a gastrostomy tube in all affected individuals. Although the affected children had normal deep tendon reflexes at birth, progressive neuropathy was implied by a loss of deep tendon reflexes by early childhood. The individuals in our cohort continued to have a persistent head lag, were unable to sit unsupported, and remained non-ambulatory at the time of evaluation. Profound weakness was also evident by limited purposeful motor activities, such as reaching. All affected individuals had severe cognitive delays, with no language development, and had central-vision impairment. Epilepsy was a prominent clinical feature in three of the six individuals, and two of them had seizures refractory to medications. Specifically, the epilepsy phenotype included early-onset refractory infantile spasms and was clinically reminiscent of an early infantile epileptic encephalopathy (EIEE) phenotype, such as KCNQ2-associated epileptic encephalopathy (EIEE7 [MIM:613720]).
Clinical evaluations of individuals A.III.1 and A.III.2 were performed at the University of Florida and the University of Miami, B.II.1 and C.II.1 at Children’s Hospital of Philadelphia, D.II.1 at Kapi’olani Medical Center for Women and Children, and E.II.1 at Children’s Hospital of Buffalo. Affected individuals gave consent through the institutional review board of their respective institutions or signed a HIPAA release form to release clinical records and clinical whole-exome sequencing reports. Electromyography, nerve conduction studies (NCSs), neuroimaging, and muscle and nerve biopsies were obtained as part of the diagnostic evaluation for some affected individuals. Auditory brainstem response (ABR) studies were obtained in a few probands as a first step toward clinical functional validation of this novel disease gene because the mouse mutants are known to have auditory neuropathy.14
Exome Sequencing and Mutation Discovery
Exome sequencing of A.III.1 and A.III.2 was performed at the Oklahoma Medical Research Foundation’s Clinical Genomics Center. Prior to sequencing, genomic DNA was first quantified with a Qubit fluorometer; shearing on a Covaris S2 and sequencing library preparation with Agilent SureSelect XT2 chemistries followed as directed by the manufacturer. Exonic sequences from each sample were enriched with Agilent SureSelect v5+UTR capture baits as directed by the manufacturer, and sequencing on an Illumina Hiseq 2500 in the high-output mode with paired-end 100 bp reads followed. After sequencing, raw fastq files for each sample were aligned to the human reference genome hg37 with the program BWA followed by alignment cleanup and variant calling with the GATK suite of programs. Variant filtration, annotation, and triage were performed with the SVS program from Golden Helix. The remaining individuals had clinical whole-exome sequencing at various commercial laboratories (CHOP, GeneDx, and Ambry Genetics), which identified bi-allelic βIV spectrin variants as a candidate disease gene.
Animals
AnkG conditional knockout mice (AnkG cKO) were generated by cell-type-specific Cre-mediated deletion of Ank3. Ank3f/fmice were generated as previously described.19 Six3Cre;Ank3f/f (AnkG cKO) mice were generated as previously described.20 AnkR-null mice (Ank1pale/pale; C57BL/6) were obtained from the Jackson Laboratory (009157). βIV spectrin mutant mice (Sptbn4qv3j) were generated as described previously.14 We used both male and female mice in our studies. All animal procedures were approved by the Animal Care and Use Committee at Baylor College of Medicine and were performed in accordance with the National Institutes of Health guide for the humane care and use of animals.
DNA Constructs
The full-length βIVΣ6 spectrin with N-terminal Myc tag was kindly provided by M. Komada (Tokyo Institute of Technology). The full-length βIVΣ1 spectrin was cloned from mouse brain cDNA and inserted into plasmid pCS3+MT for expression and so that βIVΣ1 would have an N-terminal Myc tag. All variant constructs were generated with the QuikChange II XL Site-Directed Mutagenesis kit (Agilent). The pleckstrin homology (PH) domain of βIV spectrin was amplified by PCR and cloned into pGEX-5X-2 and pEGFP-C3 with N-terminal glutathione S-transferase (GST) and EGFP tags, respectively. The rat 270 kD AnkG with C-terminal GFP tag (AnkG-GFP) was a gift from V. Bennett (Duke University). αII-spectrin-EGFP was a gift from Michael Stankewich (Yale University).
Antibodies
The following primary antibodies were used. Mouse anti-pan-Na+ channel monoclonal antibody (K58/35; RRID: AB_477552) was generated against a peptide containing the sequence TEEQKKYYNAMKKLGSKK, a highly conserved segment of the intracellular III–IV loop. Rabbit polycolonal anti-βIV-spectrin antibody (RRID:AB_2315634) was generated against the peptide sequence DRAEELPRRRRPERQE (in the C-terminal “specific domain” of βIV spectrin). Chicken polyclonal antibody against neurofascin (AF3235; RRID: AB_10890736) was purchased from R&D systems. Mouse monoclonal anti-Myc antibody (9E10; RRID: AB_309725) was purchased from Sigma-Aldrich. Mouse monoclonal anti-AnkG (N106/36; RRID: AB_10697718) and anti-GST (N100/13; RRID:AB_10671817) antibodies were purchased from UC Davis and the NIH NeuroMab facility. Chicken polyclonal anti-MAP2 antibody (CPCA-MAP2; RRID: AB_2138173) was purchased from EnCor Biotechnology. Rabbit polyclonal anti-GFP antibody (A11122; RRID: AB_221569) was purchased from Life Technologies. Rabbit polyclonal anti-KCNQ2 and guinea pig anti-KCNQ3 antibodies were gifts from Edward Cooper (Baylor College of Medicine). Rabbit anti-KCNQ2N antibody (RRID:AB_2314688) was generated against residues from the N-terminal region of KCNQ2 (GEKKLKVGFVGLDPGAPDSTRDC). Guinea pig anti-KCNQ3N antibody was developed against residues from the long N-terminal splice variant of KCNQ3 (AGDEERKVGLAPGDVEQVTLAL). Secondary antibodies Alexa Fluor 488 and 594 were purchased from Thermo Fisher Scientific. The validations of antibodies used against the neuronal proteins at nodes and AISs were previously described.8, 20, 21, 22
Immunofluorescence Microscopy
Cultured neurons or HEK293T cells were fixed in 4% paraformaldehyde (PFA) (pH 7.2–7.4) and immunostained as previously described.23 For immunostaining of nervous-system tissues, nerves were collected and fixed in 4% PFA on ice for 1 hr. The fixed tissues were then immersed in 20% sucrose in 0.1 M phosphate buffer overnight at 4°C. After this, tissues were embedded in optimal cutting temperature (OCT) compound (Tissue-Tek #4583) then sectioned with a cryostat (Thermo Scientific, Cryostar NX70) on glass coverslips for immunostaining. Procedures for immunofluorescence labeling were performed as previously described.24 For human biopsy samples, nerves were fixed as described above, but instead of being sectioned, nerves were teased on glass coverslips and then immunostained. Images of immunofluorescence were captured with an Axio-imager Z1 microscope (Carl Zeiss MicroImaging) or Axio-observer Z1 microscope (Carl Zeiss MicroImaging) fitted with an AxioCam digital camera (Carl Zeiss MicroImaging). Images were taken with 20× (0.8 NA), 40× (1.0 NA), 40× (0.75 NA), or 63× (1.4 NA) objective lenses. Images were then collected by Zen (Carl Zeiss MicroImaging) acquisition software.
Immunoprecipitation and Immunoblotting
COS-7 cells were transfected with AnkG-GFP or αII-spectrin-EGFP along with Myc-tagged wild-type βIV spectrin and mutants. Two days after transfection, cells were collected and lysed with 250 μL lysis buffer (20 mM Tris-HCl, pH 8.0, 10 mM EDTA [pH 8.0], and 150 mM NaCl) containing 1% Triton X-100 and protease inhibitors at 4°C for 1 hr with agitation. Cell lysates were then centrifuged at 13,000 g at 4°C for 10 minutes. The soluble supernatants were incubated overnight with GFP antibody-coupled protein A or G agarose beads (GE healthcare). The beads were washed seven times with 1 mL lysis buffer and then eluted with 20 μL 2X reducing sample buffer at 100°C for 5 min. The immunoprecipitates and the total supernatant as input were separated by SDS-PAGE, transferred to nitrocellulose membrane (GE healthcare), and detected via immunoblotting with Myc or GFP antibody.
Hippocampal Neurons
Primary cultures of hippocampal neurons were obtained from E18.5 Sprague-Dawley rat embryos. Hippocampi were dissected and dissociated. Neurons were then plated onto Poly-D-Lysine- and laminin-coated glass coverslips. Hippocampal neurons were cultured in Neurobasal medium (Life Technologies) containing 1% Glutamax (Life Technologies), 1% penicillin and streptomycin (Life Technologies), and 2% B27 supplement (Life Technologies) in an incubator with 5% CO2. Transfection of hippocampal neurons was performed as previously described.21 Neurons were then washed with 1X phosphate buffered saline (PBS) three times and then processed via the regular immunofluorescence procedures.
Phosphatidyl Inositol Phosphate (PIP) Strip Assay
The lipid-binding properties of the PH domain of βIV spectrin were measured with a PIP strip (Echelon Bioscience) assay according to the manufacturer’s instructions. In brief, PIP lipid strips are hydrophobic membranes spotted with 100 pmol of eight phosphoinositides and seven other biologically important lipids. These strips were blocked with 3% fatty-acid-free bovine serum albumin in TBS-T (0.1% v/v Tween-20) for 1 hr at room temperature and then incubated with 250 ng/mL recombinant GST-PH wild-type (WT), p.Arg2435Cys, or p.Arg2485Leufs31∗ variants or with GST alone. GST-tagged recombinant proteins were detected by mouse anti-GST antibody and HRP-conjugated goat anti-mouse secondary antibody.
Statistics
No statistical methods were used for pre-determining sample sizes, but our sample sizes are similar to those reported previously.25 All statistical analyses were performed with GraphPad Prism software or Microsoft Excel. Error bars represent SEM. An unpaired, two-tailed Student’s t test was used for statistical analyses unless otherwise specified.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author(s) upon reasonable request.
Results
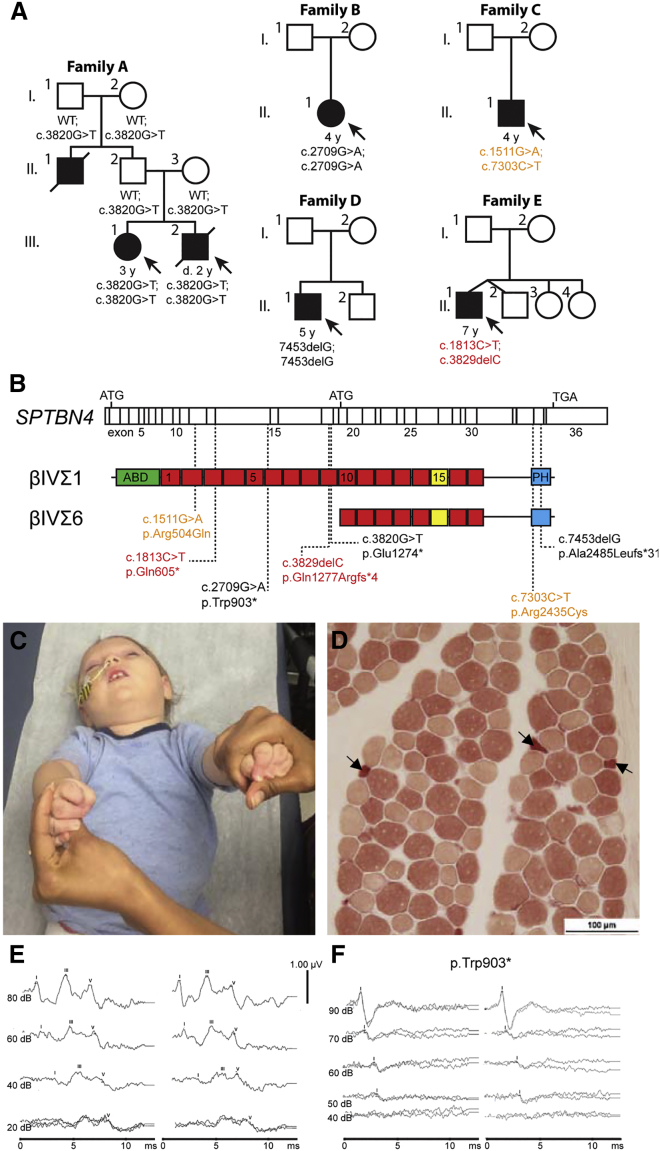
We identified six individuals with bi-allelic SPTBN4 variants (Figure 1A; the RefSeq accession number GenBank: NM_020971.2 was used to name the SPTBN4 variants). These variants distributed widely throughout βIV spectrin (Figure 1B). The Genome Aggregation Database (gnomAD) was used for variant assessment.26 None of the nonsense mutations (c.3820G>T [p.Glu1274∗], c.2709G>A [p.Trp903∗], c.7453delG [p.Ala2485Leufs∗31], c.1813C>T [p.Gln605∗], or c.3829delC [p.Gln1277Argfs∗4]) were found in this database of 123,136 exomes and 15,496 whole genomes, suggesting that these variants are exceedingly rare in the general population. The two missense mutations, c.1511G>A (p.Arg504Gln) and c.7303C>T (p.Arg2435Cys), were present in the gnomAD database but with very low allele frequencies, 6.887 × 10−5 and 8.94 × 10−5, respectively. The low frequency of these variants is consistent with the recessive model of inheritance observed in these affected individuals.
Figure 1.
Biallelic Human Pathogenic Variants in SPTBN4
(A) Pedigrees of affected individuals, with corresponding ages and genotypes. Individual A.III.2 succumbed to complications of a respiratory infection at 2 years of age. Individual A.II.1 died in infancy from a disorder associated with severe hypotonia and weakness. Both A.I.1 and A.1.2 are heterozygous for the described variant.
(B) Locations of the identified mutations in the βIVΣ1 and βIVΣ6 splice variants. ABD (actin binding domain), PH (pleckstrin homology domain), and the 17 SRs (spectrin repeats) are indicated, and the 15th SR mediating the interaction with AnkG is shown in yellow.
(C) Profound hypotonia and neuromuscular weakness (C.II.1 at 2 years of age), significant head lag and lack of truncal control.
(D) Muscle biopsy findings of neurogenic disease in individual C.II.1. Non-specific esterase stain identifies denervated fibers (darker fibers). Positively stained, atrophic muscle fibers, including angular fibers (arrows), suggesting neurogenic disease from a motor neuropathy and/or neuronopathy.
(E) Normal auditory brainstem response (ABR) tracing.
(F) ABR corresponding to individual B.II.1, showing that wave I is present but that waves III and V are absent from the affected individual.
Nodal and AIS βIV spectrins include two splice variants: a longer βIVΣ1 splice variant consisting of an N-terminal actin-binding domain (ABD), 17 spectrin repeats (SRs), and a C-terminal PH domain, and a shorter βIVΣ6 splice variant that begins in the tenth SR (SR10; Figure 1B). Five of the seven pathogenic variants identified were located N-terminal to SR10 and were predicted to only affect the βIVΣ1 splice variant (Figure 1B). The neurological features of affected individuals were remarkably similar and included congenital hypotonia, profound weakness, and areflexia (Figure 1C and Table 1). All affected individuals had profound developmental delay with no language development; three out of six had seizures, and two out of six presented clinically with severe drug-resistant epilepsy (Table 1). Other major clinical issues included feeding and respiratory difficulties (Table 1). Electromyography and nerve-conduction studies in three probands revealed evidence of motor axonopathy/neuronopathy, whereas sensory responses were normal (Table 2). Serial studies performed in one individual (B.II.1) at 6 months and 3 years of age demonstrated electrophysiological evidence for progression of motor axonopathy and neuronopathy. Serial muscle ultrasound performed on individual B.II.1 showed initially distal muscle involvement that progressed to proximal involvement (data not shown). Muscle biopsy was available in two individuals and revealed denervated and atrophic muscle fibers, consistent with a neurogenic disease (Figure 1D; individual C.II.1 is shown). Auditory brain stem responses (ABRs) were recorded from two individuals and were consistent with auditory neuropathy (compare Figures 1E and 1F; recorded from individual B.II.1).
Table 1.
Clinical Features of Individuals with Bi-allelic SPTBN4 Variants
| Individual | A.III.1 | A.III.2 | B.II.1 | C.II.1 | D.II.1 | E.II.1 |
|---|---|---|---|---|---|---|
| SPTBN4 genotype | homozygous c.3820G>T (p.Glu1274∗) |
homozygous c.3820G>T (p.Glu1274∗) |
homozygous c.2709G>A (p.Trp903∗) |
c.1511G>A (p.Arg504Gln); c.7303C>T (p.Arg2435Cys) | homozygous c.7453delG (p.Ala2485Leufs∗31) |
c.1813C>T (p.Gln605∗); c.3829delC (p.Gln1277Argfs∗4) |
| Age, Sex | 3 years, F | deceased at 2 years, M | 5 years, F | 3 years, M | 5 years, M | 7 years, M |
| Neurologic Findings | ||||||
| Congenital hypotonia | + | + | + | + | + | + |
| Profound weakness | + | + | + | + | + | + |
| Areflexia | + | + | + | + | + | + |
| Seizures | - | - | - | +, severe DRE | + | +, severe DRE |
| Severe speech delay | + | + | + | + | + | + |
| Global developmental delay | + | + | + | + | + | + |
| Cortical visual impairment | + | + | + | + | + | + |
| Diagnostic Findings | ||||||
| Auditory neuropathy/abnormal ABR | N/A | N/A | + | N/A | + | N/A |
| Axonal motor neuropathy on EMG/NCS |
+ | N/A | + | + | N/A | N/A |
| Brain MRI abnormalities | - | - | + | + | + | + |
| Systemic/Other | ||||||
| Feeding difficulties/ Gastrostomy tube |
+ | + | + | + | + | + |
| Respiratory difficulties | + | + | + | + | + | + |
Abbreviations are as follows; M = male, F = female, DRE = drug resistant epilepsy, ABR = auditory brainstem responses, and N/A = not available.
Table 2.
Electrophysiologic Data in Individuals with Biallelic SPTBN4 Mutations
| Individual | Age |
SNAP (uV) |
CMAP (mV) |
Motor NCV (m/s) |
EMG |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | M | U | P | T | M | U | P | T | M | U | Fibs | Dur | Amp | Recruit | ||
| A.III.1 | 2 yr, 7 mo | 8 | 68 | - | - | 7.6 | 3.3 | - | - | 40 | 40 | - | 0 | ↑ | ↑ | Discret |
| B.II.1 | 6 m | 6.8 | 9.2 | 3.8 | 1.2 | 8.0 | 3.5 | 2.5 | 28 | 26 | 35 | 44 | 0 | ↑ | ↑ | ↓ |
| 3 yr, 9 mo | 32.7 | - | - | 0.4 | 14.1 | 8.4 | - | 38 | 33 | 44 | - | 2+ | ↑ | ↑ | Discret | |
| C.II.1 | 1 yr, 10 mo | 9.2 | 13.2 | 10.8 | 1.7 | 13.2 | 5.2 | - | 43 | 40 | 55 | - | 0 | ↑ | ↑ | ↓ |
Abbreviations are as follows: SNAP = sensory nerve action potential, CMAP = compound motor action potential, NCV = nerve conduction velocity, S = sural or superficial peroneal, M = median, U = ulnar, p = peroneal, T = tibial, Fibs = fibrillations, Dur = motor unit duration, Amp = motor unit amplitude, Recruit = motor unit recruitment pattern, yr = year, mo = month, - = not done, and Discret = single motor unit firing rapidly.
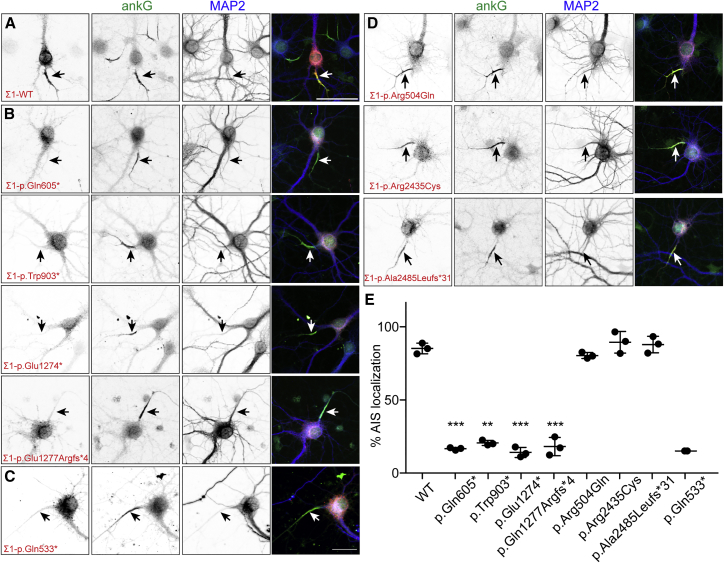
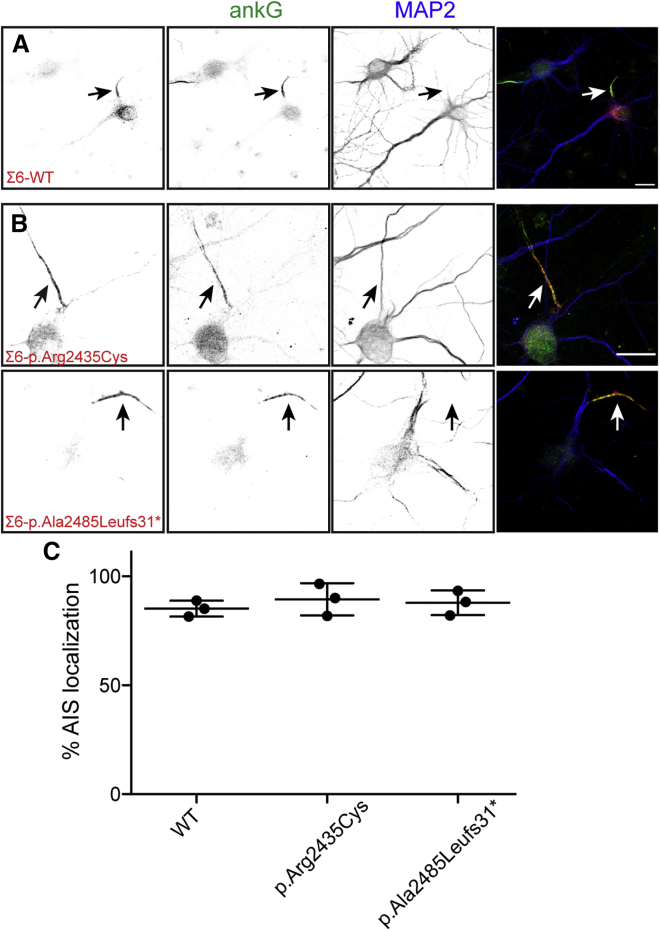
Experiments in quivering mice previously showed that neurons with a variety of βIV spectrin loss-of-function variants still express βIV spectrin mRNA,14 and protein in these mice can still be detected at nodes and AISs.16 Thus, human variants with premature termination codons might not undergo nonsense-mediated decay. Knowing that it would be difficult to obtain neuronal cells from individuals affected with βIV-spectrin-related neuropathy, we opted instead to express the pathogenic human βIV-spectrin variants in neurons to determine how each affected βIV-spectrin functions. We introduced the equivalent human variants in Myc-tagged mouse βIVΣ1 and βIVΣ6, then expressed the proteins in cultured rat hippocampal neurons; sequence alignment of human and mouse βIV spectrin indicated that each of the mutated amino acids is conserved (Figure S1). Transfected wild-type βIVΣ1 was highly enriched at AISs, where it co-localized with AnkG (Figure 2A). In contrast, variants p.Gln605∗, p.Trp903∗, p.Glu1274∗, and p.Gln1277Argfs∗4 failed to localize at the AIS (Figures 2B and 2E). The previously reported truncation variant, p.Gln533∗,18 also failed to be clustered at the AIS (Figure 2C). Variants p.Arg504Gln, p.Arg2435Cys, and p.Ala2485Leufs∗31 were still properly targeted to the AIS (Figures 2D and 2E). Similarly, transfected wild-type βIVΣ6 was highly enriched at the AIS (Figure 3A), and variants p.Arg2435Cys and p.Ala2485Leufs∗31, predicted to also affect βIVΣ6 (Figure 1B), were also properly localized to the AIS (Figures 3B and 3C).
Figure 2.
The Localization of βIVΣ1 Spectrin Variants in Neurons
(A) Transfection of cultured rat hippocampal neurons with Myc-tagged βIVΣ1 spectrin. The AIS is labeled with antibodies against AnkG (green) and is indicated with an arrow. The somatodendritic domain is labeled with antibodies against MAP2 (blue). The scale bar represents 50 μm.
(B–D) Transfection of hippocampal neurons with Myc-tagged βIVΣ1 spectrin plasmids containing the indicated variants. AISs are labeled with antibodies against AnkG (green) and are indicated with an arrow. The somatodendritic domain is labeled with antibodies against MAP2 (blue). The scale bar represents 50 μm.
(E) Quantification of the percentage of neurons with the indicated βIV spectrin construct found at the AIS. n = 3 independent experiments for each construct, except for p.Gln533∗, where n = 2. The mean ± SEM is shown. ∗∗p < 0.001; ∗∗∗p < 0.0001.
Figure 3.
βIVΣ6 Spectrin Variants Are Localized to the AIS
(A and B) Transfection of cultured rat hippocampal neurons with plasmids for Myc-tagged βIVΣ6 (Σ6-WT) or two variants of βIVΣ6, p.Arg2435Cys and p.Ala2485Leufs∗31. DIV 7 hippocampal neurons were transfected with the indicated βIV spectrin constructs, then immunolabeled with antibodies against Myc (red), AnkG (green), and MAP2 (blue). AISs are indicated by an arrow. The scale bar represents 50 μm.
(C) Quantification of the percentage of transfected cells that had clustered Myc-tagged βIV spectrin at their AIS. n = 3 independent experiments for each construct. The mean ± SEM is shown.
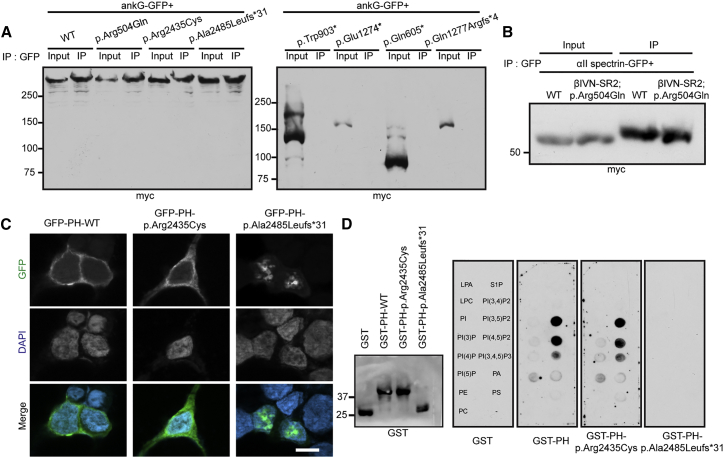
Our previous studies suggested that AnkG interactions through spectrin repeat 15 drive the recruitment of βIV spectrin to the AIS and nodes of Ranvier.7 Therefore, to confirm that the variants with disrupted localization to the AIS also affect AnkG binding, we co-expressed AnkG-GFP with the different βIVΣ1 variants, then performed immunoprecipitation by using GFP antibodies. These experiments showed that all AIS-targeted constructs (p.Arg504Gln, p.Arg2435Cys, and p.Ala2485Leufs∗31) interacted with AnkG-GFP, whereas those that were not found at the AIS (p.Gln605∗, p.Trp903∗, p.Glu1274∗, and p.Gln1277Argfs∗4) failed to interact with AnkG-GFP (Figure 4A). Thus, p.Gln605∗, p.Trp903∗, p.Glu1274∗, and p.Gln1277Argfs∗4 pathogenic variants disrupt βIVΣ1 function because they cannot bind AnkG.
Figure 4.
βIV Spectrin Protein and Lipid Interactions
(A) Cotransfections and immunoprecipitations between AnkG-GFP and the indicated βIV spectrin constructs. Immunoprecipitation reactions were performed with anti-GFP antibodies, and immunoblots were performed wih anti-Myc antibodies. Both input and immunoprecipitates (IPs) are shown.
(B) Cotransfection and immunoprecipitation of αII spectrin and βIV spectrin’s N terminus, including spectrin repeat 2 with and without the p.Arg504Gln variant. IPs were performed with anti-GFP antibodies, and immunoblots were performed with anti-Myc antibodies.
(C) Immunostaining of HEK cells transfected with the indicated βIV spectrin PH domain constructs. The scale bar represents 10 μm.
(D) Immunoblot of GST and βIV spectrin PH-domain-containing fusion proteins and PIP strip blots showing that the same fusion proteins bind to phosphoinositides. GST fusion proteins were detected with anti-GST antibodies.
How does the p.Arg504Gln variant disrupt βIVΣ1 function even though it still interacts with AnkG and is localized to the AIS (Figure 2B)? Arg504 is located in SR2 (Figure 1B), and SR2 is reported to underlie α and β subunit heterodimer interactions.27 Therefore, we co-expressed αII spectrin-GFP together with an N-terminal fragment of wild-type βIVΣ1 or the p.Arg504Gln variant in the SR2 domain (βIVN-SR2). Although this minimal segment of βIV spectrin interacts with αII spectrin, the p.Arg504Gln variant did not disrupt the interaction (Figure 4B).
How do the p.Arg2435Cys and p.Ala2485Leufs∗31 variants disrupt βIV spectrin function? Both variants are found in the C-terminal PH domain of βIV spectrin (Figure 1B). PH domains are found in many cytoskeletal proteins and participate in their recruitment or retention at specific membrane domains, sometimes through high-affinity binding to phosphoinositides.28 To determine whether βIV spectrin’s PH domain function is compromised by the p.Arg2435Cys or p.Ala2485Leufs∗31 variants, we expressed GFP-tagged PH domains in HEK293T cells to determine their subcellular localizations. We found that both WT and p.Arg2435Cys-containing PH domain constructs were diffusely distributed throughout cells (Figure 4C). However, the p.Ala2485Leufs∗31-containing PH domain was found in small intracellular puncta, suggesting that disruption of the PH domain alters the distribution of βIV spectrin in membrane compartments (Figure 4C). To directly determine the phosphoinositide binding capacity of βIV spectrin’s PH domain, we constructed GST fusion proteins containing WT or βIV spectrin PH domain variants, then tested their ability to bind to phosphoinositides immobilized on PIP strips (Figure 4D). Whereas both WT and p.Arg2435Cys-containing PH domain fusion proteins bound strongly to PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3, the variant p.Ala2485Leufs∗31 failed to bind to any phosphoinositides (Figure 4D). Together, these results show that the pathogenic variants p.Trp903∗, p.Glu1274∗, p.Gln605∗, p.Gln1277Argfs∗4, and p.Ala2485Leufs∗31 result in a loss of function through disrupted AnkG or phosphoinositide binding, whereas the mechanisms of pathology due to p.Arg504Gln and p.Arg2435Cys variants (found in individual C.II.1) remain unknown.
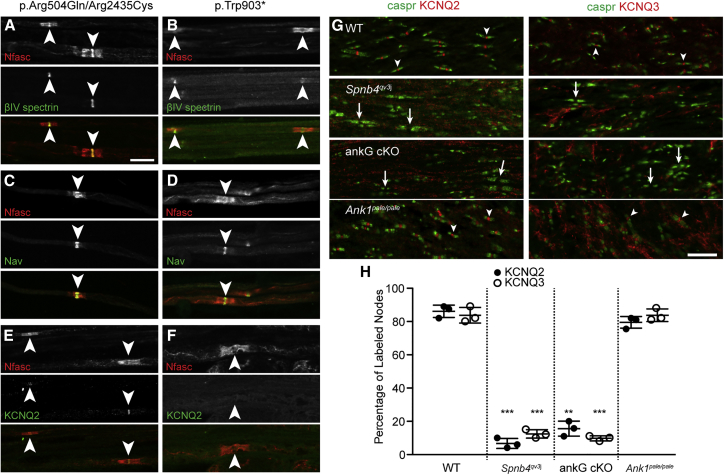
To further determine the consequence of human βIV spectrin variants on the structure and function of axons, we compared the molecular organization of nodes of Ranvier in sural nerve biopsies from individual C.II.1, who harbors the p.Arg504Gln; p.Arg2435Cys variants, the molecular pathology of which remains unclear, and the p.Trp903∗ variant, which did not localize to the AIS. Immunostaining with a pan-neurofascin (Nfasc) antibody that recognizes both axonal NF186 and glial NF155 showed normal paranodal NF155 and highly clustered nodal NF186 in p.Arg504Gln; p.Arg2435Cys nerves (Figure 5A) but significantly reduced nodal NF186 in p.Trp903∗ nerves (Figure 5B, arrowheads). Labeling of the same nodes with antibodies that recognize both βIVΣ1 and βIVΣ6 showed strong nodal enrichment in the p.Arg504Gln; p.Arg2435Cys nerve (Figure 5A) but nearly undetectable nodal βIVΣ6 immunoreactivity in the p.Trp903∗ nerve (Figure 5B). Immunostaining with a pan-Na+ channel antibody (Nav) showed strong nodal enrichment in p.Arg504Gln; p.Arg2435Cys nerves but weak nodal labeling in p.Trp903∗ nerves (Figures 5C and 5D). KCNQ2- and KCNQ3-subunit-containing K+ channels are also clustered at nodes of Ranvier.29 Because loss-of-function variants in KCNQ2 and KCNQ3 cause epilepsy and infantile spasms, both of which were present in some βIV spectrin probands (Table 1), we immunostained the p.Arg504Gln; p.Arg2435Cys and p.Trp903∗ nerves by using antibodies against KCNQ2 K+ channels. In p.Arg504Gln; p.Arg2435Cys nerves we found weak nodal KCNQ2 immunoreactivity (Figure 5E, arrowheads). In p.Trp903∗ nerves we could not detect any KCNQ2 K+ channels (Figure 5F, arrowhead). Together, these findings indicate that in the p.Trp903∗ variant, the shorter and unaffected βIVΣ6 splice variant (Figure 1B) is not sufficient to rescue nodal properties. This finding is consistent with the neurological phenotypes of mice that lack βIVΣ1 but retain βIVΣ6.17 Because the p.Arg504Gln; p.Arg2435Cys proband has mostly normal nodes, with only a possible reduction in KCNQ2 K+ channel clustering, the molecular pathology of these variants remains unclear. It is possible that the variants are synergistic and that defects in βIV-spectrin function are only observed upon tetramerization. In contrast, because all truncating variants of βIV spectrin were unable to interact with AnkG or cluster at the AIS, we speculate that pathological and molecular features of the p.Trp903∗ variant might be generalizable to all variants and that this might result in premature truncation.
Figure 5.
Human βIV Spectrin Variants Disrupt Nodal Ion Channel Clustering
(A–F) Immunostaining of human sural nerve biopsies from individuals with the p.Arg504Gln; p.Arg2435Cys and p.Trp903∗ variants was performed with antibodies against neurofascin (Nfasc, red) and βIV spectrin (green) (A and B). Immunostaining of human sural nerve biopsies from individuals with p.Arg504Gln; p.Arg2435Cys and p.Trp903∗ variants was performed with antibodies against neurofascin (Nfasc, red) and Na+ channels (Nav, green) (C and D). Immunostaining of human sural nerve biopsies from individuals with p.Arg504Gln; p.Arg2435Cys and p.Trp903∗ variants was performed with antibodies against neurofascin (Nfasc, red) and KCNQ2 K+ channels (green) (E and F). Nodes are indicated by arrowheads. NB: the exposure times for immunostaining of βIV spectrin, Na+ channels, and KCNQ2 in the p.Trp903∗ variant are much longer than the exposure times in the p.Arg504Gln; p.Arg2435Cys variant to allow detection of these proteins. This is evident from the high background seen along the axons. The scalebar represents 10 μm and applies to all panels (A–F).
(G) Immunostaining of the optic nerve from the indicated genotypes with antibodies against Caspr (green), KCNQ2 (red), or KCNQ3 (red). Arrowheads indicate nodes with KCNQ2 or KCNQ3 immunostaining, whereas arrows indicate nodes without any nodal labeling for these channels. The scale bar represents 10 μm.
(H) Quantification of the percent of nodes labeled for KNCQ2 or KCNQ3 in the indicated mice. n = 3 mice for each genotype. The mean ± SEM is shown. ∗∗p < 0.001; ∗∗∗p < 0.0001.
The lack of nodal KCNQ2 K+ channel immunoreactivity in p.Trp903∗ axons is surprising because both Na+ channels and KCNQ2 K+ channels are recruited to the AIS and nodes of Ranvier through a similar AnkG-binding motif.8 Furthermore, we previously showed that after loss of βIV spectrin or AnkG, a secondary mechanism based on AnkR/βI spectrin can compensate for nodal (but not AIS) Na+ channel clustering.20 Why does this secondary clustering mechanism not rescue nodal KCNQ2 K+ channels in p.Trp903∗ myelinated axons? To answer this question we first modeled loss of βIV spectrin by using optic nerves from quivering3j (Sptbn4qv3j) and Six3Cre;Ank3f/f (AnkG cKO) mice, which have dramatically reduced amounts of nodal βIV spectrin.20 Sptbn4qv3j mice have a point mutation that is proximal to the PH domain and results in a frameshift mutation, a novel 49 amino acid extension, and deletion of the PH domain;14 this mutation is similar to p.Ala2485Leufs∗31. Sptbn4qv3j mice are ataxic, deaf, and die at about 6 months of age. The AnkG cKO mice are viable and have no apparent behavioral abnormalities because the recombination is restricted to the retina and optic nerves, and AnkR compensates for the loss of AnkG from nodes in these mice.20 As controls, in both wild-type and AnkR-deficient mice (Ank1pale/pale), we found KCNQ2- and KCNQ3-subunit-containing K+ channels clustered at nodes of Ranvier flanked by Caspr-labeled paranodal junctions (Figure 5G, arrowhead); the AnkR-deficient mice have severe hemolytic anemia due to spherocytosis. But since AnkG is expressed normally, nodes of Ranvier are unaffected (Figures 5G and 5H). In contrast to findings in these controls, we did not detect any KCNQ2 or KCNQ3 K+ channel subunits at nodes in Sptbn4qv3j mice (Figure 5G, arrows, and Figure 5H). This observation replicates the loss of nodal KCNQ2 and KCNQ3 immunoreactivity previously reported in these mice.30 Importantly, AnkG cKO mice also lacked clustered KCNQ2- and KCNQ3-subunit-containing K+ channels (Figure 5G, arrows, and Figure 5H), indicating that although AnkR and βI spectrin can rescue nodal Na+ channel clustering,20 this complex cannot rescue clustering of nodal KCNQ2- and KCNQ3-subunit-containing K+ channels.
Discussion
Our report expands the clinical phenotype of human disease associated with SPTBN4 variants via the largest cohort reported to date. We provide functional validation to help establish the mechanisms of disease. Our results suggest a mechanism for ion-channel dysfunction based not on mutations in the channels themselves, but rather on disruption of the cytoskeletal machinery controlling proper localization of channels and function of axonal domains where axonal ion channels are normally clustered in high density (i.e., AIS and nodes of Ranvier).
Structural and biochemical studies of ankyrin in complex with the Nav1.2 or KCNQ2 ion channels show that the affinity of Nav1.2 for ankyrin is ∼100-fold that of KCNQ2.31 Furthermore, AnkR also contains an auto-inhibitory segment that binds to the same region on AnkR as Nav1.2 and KCNQ2, but with an affinity close to that for Nav1.2. Thus, KCNQ2 K+ channels have 100-fold reduced affinity for AnkR and must compete for binding with AnkR’s higher affinity auto-inhibitory segment.31 We speculate that together these properties of AnkR disrupt clustering of nodal KCNQ2- and KCNQ3-subunit-containing K+ channels when βIV spectrin variants result in loss of function.
Our sural nerve biopsy studies with human teased fibers only examined the consequence of disrupted βIV spectrin at nodes and not at the AIS, where βIV spectrin forms a complex with AnkG to cluster Na+ channels. The AIS functions not only to initiate action potentials but also to maintain neuronal polarity and regulate the proper trafficking of somatodendritic and axonal cargos.23, 32 We speculate that the extensive neurologic involvement with central and peripheral features associated with pathogenic human βIV spectrin variants reflect both disruptions to nodes of Ranvier and the function of the AIS. Indeed, with the exception of neurons lacking an AIS (e.g., peripheral sensory neurons32), it is not possible to uncouple the consequences of loss of βIV spectrin at nodes from the loss at the AIS. Nevertheless, King et al.33 showed that loss of KCNQ2 from sensory neurons had no effect on the structural organization of nodes but did result in increased thermal sensitivity, allodynia, and increased excitability.
Our work significantly expands on the neurologic phenotype as well as the underlying mechanism of βIV spectrinopathies. A previous single case report18 coincides with our findings in describing congenital hypotonia and weakness in an individual with the p.Gln533∗ variant. Our clinical phenotyping and neurophysiological studies in multiple affected individuals with a variety of pathogenic SPTBN4 variants suggests that weakness is primarily due to severe motor axonal neuropathy and neuronopathy as opposed to a myopathy. Serial electrophysiological studies available in one individual (proband B.II.1) suggest a progressive nature of the neuropathy. Consistent with our findings, the affected person reported by Knierim et al. (2017)18 showed evidence of an axonal motor neuropathy as well. Nevertheless, we did not find evidence of a primary congenital myopathy or a demyelinating neuropathy. Instead, the muscle biopsies in our cohort were suggestive of a neurogenic process with evidence of denervation. It is therefore likely that muscle involvement is secondary to denervation, which could confound the etiology of the neuromuscular weakness.
In summary, we identified pathogenic SPTBN4 variants that define a class of spectrinopathy. Most variants affect the molecular organization and function of excitable domains by disrupting βIV spectrin’s capacity to interact with AnkG or phosphoinositides. Furthermore, these variants in SPTBN4 cause profound neurological dysfunction, probably by reducing or disrupting the clustering of Na+ and KCNQ2- and KCNQ3-subunit-containing K+ channels. Although hypotonia and global developmental delays are clinically non-specific, we propose that the clinical scenario of progressive axonal neuropathy and neuronopathy along with severe congenital hypotonia and intellectual disability should raise consideration to include this disorder in the differential diagnosis. Because the neuropathy might not always be ascertained clinically, particularly during infancy, genetic assessment of βIV spectrin should be considered in children presenting with the spectrum of symptoms overlapping severe psychomotor delays, hypotonia, and epilepsy.
Acknowledgments
Supported by grants NS044916 (M.N.R.), NS069688 (M.N.R.), AR056360 (P.M.G.), AR063124 (P.M.G.), AI082714 (P.M.G.), GM110766 (P.M.G), and NS049453 (X.O.G.) from the National Institutes of Health and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (M.N.R.). We thank Ed Cooper for helpful discussions of AnkR and KCNQ2 and KCNQ3 interactions, and Peter Kang and Stephanie Sacharow for the evaluations of individuals A.III.1 and A.III.2. We thank Brian Harding (CHOP) for the muscle pathology photomicrographs. We thank Jun-Xian Zhang and JIan Li (University of Pennsylvania) for preparation of teased fibers from the human sural nerve biopsy. We thank Avni Santani and Addie Nesbitt at the CHOP Division of Genomics Diagnostics and Katherine Helbig at Ambry Genetics for clinical exome interpretation and referrals. X.O.G. is supported by the Robert Wood Johnson Harold Amos Faculty Development Award.
Published: May 31, 2018
Footnotes
Supplemental Data include one figure and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.04.012.
Contributor Information
Klaas J. Wierenga, Email: klaas-wierenga@ouhsc.edu.
Matthew N. Rasband, Email: rasband@bcm.edu.
Web Resources
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Bennett V., Baines A.J. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol. Rev. 2001;81:1353–1392. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu K., Zhong G., Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leterrier C., Potier J., Caillol G., Debarnot C., Rueda Boroni F., Dargent B. Nanoscale architecture of the axon initial segment reveals an organized and robust scaffold. Cell Rep. 2015;13:2781–2793. doi: 10.1016/j.celrep.2015.11.051. [DOI] [PubMed] [Google Scholar]
- 5.D’Este E., Kamin D., Balzarotti F., Hell S.W. Ultrastructural anatomy of nodes of Ranvier in the peripheral nervous system as revealed by STED microscopy. Proc. Natl. Acad. Sci. USA. 2017;114:E191–E199. doi: 10.1073/pnas.1619553114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berghs S., Aggujaro D., Dirkx R., Jr., Maksimova E., Stabach P., Hermel J.M., Zhang J.P., Philbrick W., Slepnev V., Ort T., Solimena M. betaIV spectrin, a new spectrin localized at axon initial segments and nodes of ranvier in the central and peripheral nervous system. J. Cell Biol. 2000;151:985–1002. doi: 10.1083/jcb.151.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y., Ogawa Y., Hedstrom K.L., Rasband M.N. betaIV spectrin is recruited to axon initial segments and nodes of Ranvier by ankyrinG. J. Cell Biol. 2007;176:509–519. doi: 10.1083/jcb.200610128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Z., Kao T., Horvath Z., Lemos J., Sul J.-Y., Cranstoun S.D., Bennett V., Scherer S.S., Cooper E.C. A common ankyrin-G-based mechanism retains KCNQ and NaV channels at electrically active domains of the axon. J. Neurosci. 2006;26:2599–2613. doi: 10.1523/JNEUROSCI.4314-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Writzl K., Primec Z.R., Stražišar B.G., Osredkar D., Pečarič-Meglič N., Kranjc B.S., Nishiyama K., Matsumoto N., Saitsu H. Early onset West syndrome with severe hypomyelination and coloboma-like optic discs in a girl with SPTAN1 mutation. Epilepsia. 2012;53:e106–e110. doi: 10.1111/j.1528-1167.2012.03437.x. [DOI] [PubMed] [Google Scholar]
- 10.Saitsu H., Tohyama J., Kumada T., Egawa K., Hamada K., Okada I., Mizuguchi T., Osaka H., Miyata R., Furukawa T. Dominant-negative mutations in alpha-II spectrin cause West syndrome with severe cerebral hypomyelination, spastic quadriplegia, and developmental delay. Am. J. Hum. Genet. 2010;86:881–891. doi: 10.1016/j.ajhg.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tohyama J., Nakashima M., Nabatame S., Gaik-Siew C., Miyata R., Rener-Primec Z., Kato M., Matsumoto N., Saitsu H. SPTAN1 encephalopathy: distinct phenotypes and genotypes. J. Hum. Genet. 2015;60:167–173. doi: 10.1038/jhg.2015.5. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda Y., Dick K.A., Weatherspoon M.R., Gincel D., Armbrust K.R., Dalton J.C., Stevanin G., Dürr A., Zühlke C., Bürk K. Spectrin mutations cause spinocerebellar ataxia type 5. Nat. Genet. 2006;38:184–190. doi: 10.1038/ng1728. [DOI] [PubMed] [Google Scholar]
- 13.Delaunay J. The molecular basis of hereditary red cell membrane disorders. Blood Rev. 2007;21:1–20. doi: 10.1016/j.blre.2006.03.005. Published online May 30, 2006. [DOI] [PubMed] [Google Scholar]
- 14.Parkinson N.J., Olsson C.L., Hallows J.L., McKee-Johnson J., Keogh B.P., Noben-Trauth K., Kujawa S.G., Tempel B.L. Mutant beta-spectrin 4 causes auditory and motor neuropathies in quivering mice. Nat. Genet. 2001;29:61–65. doi: 10.1038/ng710. [DOI] [PubMed] [Google Scholar]
- 15.Komada M., Soriano P. [Beta]IV-spectrin regulates sodium channel clustering through ankyrin-G at axon initial segments and nodes of Ranvier. J. Cell Biol. 2002;156:337–348. doi: 10.1083/jcb.200110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y., Lacas-Gervais S., Morest D.K., Solimena M., Rasband M.N. BetaIV spectrins are essential for membrane stability and the molecular organization of nodes of Ranvier. J. Neurosci. 2004;24:7230–7240. doi: 10.1523/JNEUROSCI.2125-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lacas-Gervais S., Guo J., Strenzke N., Scarfone E., Kolpe M., Jahkel M., De Camilli P., Moser T., Rasband M.N., Solimena M. BetaIVSigma1 spectrin stabilizes the nodes of Ranvier and axon initial segments. J. Cell Biol. 2004;166:983–990. doi: 10.1083/jcb.200408007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knierim E., Gill E., Seifert F., Morales-Gonzalez S., Unudurthi S.D., Hund T.J., Stenzel W., Schuelke M. A recessive mutation in beta-IV-spectrin (SPTBN4) associates with congenital myopathy, neuropathy, and central deafness. Hum. Genet. 2017;136:903–910. doi: 10.1007/s00439-017-1814-7. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins P.M., Vasavda C., Hostettler J., Davis J.Q., Abdi K., Bennett V. E-cadherin polarity is determined by a multifunction motif mediating lateral membrane retention through ankyrin-G and apical-lateral transcytosis through clathrin. J. Biol. Chem. 2013;288:14018–14031. doi: 10.1074/jbc.M113.454439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho T.S., Zollinger D.R., Chang K.J., Xu M., Cooper E.C., Stankewich M.C., Bennett V., Rasband M.N. A hierarchy of ankyrin-spectrin complexes clusters sodium channels at nodes of Ranvier. Nat. Neurosci. 2014;17:1664–1672. doi: 10.1038/nn.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedstrom K.L., Xu X., Ogawa Y., Frischknecht R., Seidenbecher C.I., Shrager P., Rasband M.N. Neurofascin assembles a specialized extracellular matrix at the axon initial segment. J. Cell Biol. 2007;178:875–886. doi: 10.1083/jcb.200705119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amor V., Zhang C., Vainshtein A., Zhang A., Zollinger D.R., Eshed-Eisenbach Y., Brophy P.J., Rasband M.N., Peles E. The paranodal cytoskeleton clusters Na+ channels at nodes of Ranvier. eLife. 2017;6 doi: 10.7554/eLife.21392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hedstrom K.L., Ogawa Y., Rasband M.N. AnkyrinG is required for maintenance of the axon initial segment and neuronal polarity. J. Cell Biol. 2008;183:635–640. doi: 10.1083/jcb.200806112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogawa Y., Schafer D.P., Horresh I., Bar V., Hales K., Yang Y., Susuki K., Peles E., Stankewich M.C., Rasband M.N. Spectrins and ankyrinB constitute a specialized paranodal cytoskeleton. J. Neurosci. 2006;26:5230–5239. doi: 10.1523/JNEUROSCI.0425-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Susuki K., Chang K.J., Zollinger D.R., Liu Y., Ogawa Y., Eshed-Eisenbach Y., Dours-Zimmermann M.T., Oses-Prieto J.A., Burlingame A.L., Seidenbecher C.I. Three mechanisms assemble central nervous system nodes of Ranvier. Neuron. 2013;78:469–482. doi: 10.1016/j.neuron.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Speicher D.W., Weglarz L., DeSilva T.M. Properties of human red cell spectrin heterodimer (side-to-side) assembly and identification of an essential nucleation site. J. Biol. Chem. 1992;267:14775–14782. [PubMed] [Google Scholar]
- 28.Lemmon M.A. Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 2007;74:81–93. doi: 10.1042/BSS0740081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devaux J.J., Kleopa K.A., Cooper E.C., Scherer S.S. KCNQ2 is a nodal K+ channel. J. Neurosci. 2004;24:1236–1244. doi: 10.1523/JNEUROSCI.4512-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devaux J.J. The C-terminal domain of ßIV-spectrin is crucial for KCNQ2 aggregation and excitability at nodes of Ranvier. J. Physiol. 2010;588:4719–4730. doi: 10.1113/jphysiol.2010.196022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C., Wei Z., Chen K., Ye F., Yu C., Bennett V., Zhang M. Structural basis of diverse membrane target recognitions by ankyrins. eLife. 2014;3 doi: 10.7554/eLife.04353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuijpers M., van de Willige D., Freal A., Chazeau A., Franker M.A., Hofenk J., Rodrigues R.J., Kapitein L.C., Akhmanova A., Jaarsma D., Hoogenraad C.C. Dynein Regulator NDEL1 Controls Polarized Cargo Transport at the Axon Initial Segment. Neuron. 2016;89:461–471. doi: 10.1016/j.neuron.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 33.King C.H., Lancaster E., Salomon D., Peles E., Scherer S.S. Kv7.2 regulates the function of peripheral sensory neurons. J. Comp. Neurol. 2014;522:3262–3280. doi: 10.1002/cne.23595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author(s) upon reasonable request.