Abstract
The abnormal expression of microRNAs (miRNAs) in colorectal cancer (CRC) progression has been widely investigated. It was reported that the same hairpin RNA structure could generate mature products from each strand, termed 5p and 3p, which binds different target mRNAs. Here, we explored the expression, functions, and mechanisms of miR-514b-3p and miR-514b-5p in CRC cells and tissues. We found that miR-514b-3p was significantly down-regulated in CRC samples, and the ratio of miR-514b-3p/miR-514b-5p increased from advanced CRC, early CRC to matched normal colorectal tissues. Follow-up functional experiments illustrated that miR-514b-3p and miR-514b-5p had distinct effects through interacting with different target genes: MiR-514b-3p reduced CRC cell migration, invasion and drug resistance through increasing epithelial marker and decreasing mesenchymal marker expressions, conversely, miR-514b-5p exerted its pro-metastatic properties in CRC by promoting EMT progression. MiR-514b-3p overexpressing CRC cells developed tumors more slowly in mice compared with control cells, however, miR-514b-5p accelerated tumor metastasis. Overall, our data indicated that though miR-514b-3p and miR-514b-5p were transcribed from the same RNA hairpin, each microRNA has distinct effect on CRC metastasis.
Introduction
Colorectal cancer (CRC) is the third most common and the fourth leading cause of cancer related mortality worldwide1. Due to the change in diet, the incidence of CRC in Asian countries increased remarkably during the past decades2. Although the surgical resection and postoperative chemotherapy help to get a better outcome for early-stage CRC, the clinical output for advanced CRC is still unsatisfactory, mainly due to metastasis2. Therefore, it is of critical importance to elucidate the underlying mechanism involved in metastasis process and identify biomarkers for CRC.
Epithelial mesenchymal transition (EMT) plays a pivotal role in cancer invasion and metastasis3. Within the normal intestinal epithelium, cells interact with each other to form tissue integrity. During the EMT process, the cell–cell junction disassembled and cancer cells lose their apico-basal polarity and gain the ability to migrate from the primary tumor into the surrounding tissue, which is the first step toward metastasis4,5. Besides, cells involved in EMT have many traits with stem cells in common, such as increased drug resistance, causing systemic therapy more complicated for metastatic disease6,7.
MicroRNAs (miRNAs) are small (20–25 nucleotides) non-coding RNAs. They regulate gene expression post-transcriptionally through inhibiting the translation or reducing the stability of messenger RNAs (mRNAs), directly binding to 3′ untranslated regions (3′UTRs) of target mRNA in a sequence-specific way8. Emerging evidence has demonstrated that miRNAs dysfunction is involved in various physiological or pathological process, including cell proliferation, apoptosis, drug resistance, and tumor metastasis9–13. In the miRNA biogenesis pathway, the precursor miRNA (pre-miRNA) are transcripted by RNA polymerase II (Pol II) from the DNA template and spliced into the primary miRNA (pri-miRNA) via Drosha in the nucleus. Then the pri-miRNA are transported to cytoplasm by the Exportin-5 complex and then processed to the mature miRNA via the Dicer enzyme14. Since the pri-miRNA has two arms, namely 3p and 5p arm, mature miRNAs excised from different arms of the same primary transcript have different sequences and therefore function diversely15.
It was reported that miR-514a could function as a tumor suppressor gene in testicular germ cell tumor, melanoma and renal cell carcinoma. However, there is no study investigating the function of miR-514b in CRC16–18. Here, we found that miR-514b-3p and miR-514b-5p displayed opposite functions in CRC development. The ratio of miR-514b-3p/5p is higher in normal tissue compared with paired CRC tissue and decrease with the tumor progression. These results indicate a potential role for the miR-514 family in CRC evolution and metastasis.
Materials and methods
Clinical specimen collection
In all, 62 freshly-frozen CRC tissues and adjacent non-cancerous tissues in Renji cohort 1 were obtained from patients who underwent surgery in Renji Hospital from 2010 to 2015. Similarly, we collected paraffin-fixed CRC tissues and normal colorectal tissues from 40 patients with CRC between 2014 and 2016 at Shanghai Renji Hospital (Renji cohort 2). The clinicopathological information was provided by Pathology Department of Renji Hospital and the clinicopathological traits involved gender, age, tumor size, clinical stage (AJCC), T classification, lymph node metastasis, distant metastasis, vascular invasion, and histological differentiation. None of patients had received chemotherapy or radiotherapy before surgery. Written informed consent was obtained from all patients involved in this study and ethical consent was granted from Shanghai Jiao Tong University School of Medicine, Renji Hospital.
Bioinformatics analysis
Gene set enrichment analysis (GSEA) was performed using the GO gene sets database (c2.all.v4.0.symbols.gmt) from the Molecular Signatures Database–MsigDB. Enrichment results were generated from 1000 random permutations and significance was estimated by comparing the enrichment score. FDR, 0.25 was considered to be a cut-off to identify biologically relevant genes.
Cell cultures and treatments
CRC cell lines LOVO, HCT116, SW620, SW480, SW1116, HT29, and Caco2 were purchased from ATCC (American type culture collection). All CRC cells were cultured in medium with 10% FBS (fetal bovine serum) at 37 ℃ with an atmosphere of 5% CO2.
The miRNA mimics or miRNA inhibitors were transfected into CRC cell lines using DharmaFECT 1 transfection reagent (Thermo Scientific, USA) or Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instruction. All the miRNA mimics and miRNA inhibitors were synthesized and purchased from Genepharm Technologies (China). The sequences of miRNA mimics involved in the study were listed as follows: miR-514b-3p mimics: AUUGACACCUCUGUGAGUGGA; miR-514b-5p mimics: UUCUCAAGAGGGAGGCAAUCAU; miR-514b-3p inhibitor: UCCACUCACAGAGGUGUCAAU; miR-514b-5p inhibitor: AUGAUUGCCUCCCUCUUGAGAA.
Total RNA extraction and real-time PCR
Total RNA of CRC cell lines, CRC tissues and adjacent non-cancerous tissues was isolated using Trizol reagent (Takara, Japan). First-strand cDNA was synthesized using the All-in-One miRNA First-Strand cDNA Synthesis Kit (Genecopeia, USA) and real-time PCR was conducted using ABI (Thermo Fisher Scientific, USA) in the StepOne real-time PCR system. The relative mRNA expression levels were quantified by the 2−ΔΔCt method. U6 and GAPDH were used as internal controls. The gene-specific primers used in this study were listed as follows: CDH1-F: AAAGGCCCATTTCCTAAAAACCT; CDH1-R: TGCGTTCTCTATCCAGAGGCT; CLDN1-F: CCTCCTGGGAGTGATAGCAAT; CLDN1-R: GGCAACTAAAATAGCCAGACCT; FZD4-F: CCTCGGCTACAACGTGACC; FZD4-R: TGCACATTGGCACATAAACAGA; NTN1-F: GGGTGCCCTTCCAGTTCTAC; NTN1-R: GCGAGTTGTCGAAGTCGTG; GAPDH-F: GCATTGCCCTCAACGACCAC; GAPDH-R: CCACCACCCTGTTGCTGTAG; Vimentin-F: TTGACCTTGAACGCAAAGTG; Vimentin-R: TTTGGACATGCTGTTCCTGA; Fibronectin-F: GCCTTCAAGTTCCCCTGTTAC; Fibronectin-R: GACTCTCTCCGCTTGGATTCT;
Protein extraction and western blot
Protein extraction buffer (Beyotime, China), which contained a protease inhibitor mixture (protease inhibitors, phosphatase inhibitors, PMSF) was used to extract total protein. The standard curve of protein concentration was plotted using BCA Protein Assay Kit (Pierce Biotechnology). Proteins were separated by SDS-polyacrylamide gels and transferred to PVDF membranes. The membranes were incubated with second antibody accordingly overnight at 4℃ after blocked with 5% BSA (Bull Serum Albumin). The next day the membranes were incubated with species-specific secondary antibodies and visualized by the ECL detection system. Antibodies used in this study were listed as follows: Fibronectin (Abcam, UK), Vimentin (Abcam, UK), CLND1 (CST, USA), CDH1 (CST, UK) and GAPDH (KangChen, China). GAPDH was used as an internal control. The dilution of antibodies was 1:1,000.
Transwell invasion assay
LOVO and HCT116 cells were planted into 6-well plates and transfected with miRNA mimics or inhibitor accordingly. The cells were collected 24 h later and suspended in serum-free culture medium. 2 × 105 cells with 200 µl serum-free culture medium were seeded into the upper chambers which were coated with fresh matrigel and 600 µl culture medium with 20% FBS was added into the lower chambers. 48 h later, the matrigel and the cells on the upper chambers were removed gently and the cells on the lower chambers were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The invasion cells were counted under microscope.
Scratch assay
CRC cell lines were planted into 6-well plates and transfected with miRNA mimics accordingly. Scratch wounds were made by scraping the monolayer of cells when the cells were grown to almost 90% density. The wounded areas were observed at the time point of 0 h and 24 h and measured under a light microscope.
ELISA
The expression of MMP2 and MMP9 in the cell supernatants was quantified using a human MMP2 and MMP9 ELISA kit (RayBiotech, USA) according to the manufacturer’s guide.
Cell proliferation assays
HCT116 and LOVO cells were planted into 96-well plates and transfected with miRNA mimics the next day. Cells were treated with Cell Counting Kit-8 (CCK8, Dojindo, Japan) for 2 h at 37 ℃ at the time point of 24 h, 48 h, 72 h, 96 h, and 120 h after seeding. The cell proliferation curves were plotted using the absorbance, which was measured with wavelength 450 nm (450 OD). Each group was measured in 6 replicate wells and all the experiments were repeated at least 3 times.
Cell cycle analysis
CRC cells were seeded into 6-well plates and transfected with miRNA mimics accordingly. Cells were harvested 48 h after transfection and fixed with 70% pre-cooling ethanol at -20℃ overnight. The supernatant was discarded after centrifugation and the cells were washed with PBS for 2 times the next day. Then the cells were stained with 500 μL PI/RNase staining buffer (BD Pharmingen, USA) for 15 min in the dark. Finally, the samples were analyzed using a fluorescence-activated cell-sorting flow cytometer (BD Biosciences, USA).
In situ hybridization
The in situ detection of miR-514b-3p and miR-514b-5p was performed on 6-μm formalin-fixed, paraffin-embedded (FFPE) sections using DIG-labeled miRCURYTM Detection probe (Exiqon, Denmark). Briefly, the slides were hybridized with a probe (LNA-modified and DIG-labeled oligonucleotide; Exiqon) complementary to miR-514b-3p/miR-514b-5p and after incubation with anti–DIG-AP Fab fragments conjugated to alkaline phosphatase. The hybridized probes were then detected by applying nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate color substrate (Roche) to the slides. Slides were counterstained with VECTOR® nuclear fast red counterstain (VECTOR LABOTATORIES) and analyzed with a Nikon 80i microscope and Nikon NIS-Elements F 2.3 software (Nikon). The sequences of miRNA probe involved in the study were listed as follows: miR-514b-3p probe: /5DigN/TCCACTCACAGAGGTGTCAAT/3Dig_N/; miR-514b-5p probe: /5DigN/TCTACTCACAGAAGTGTCAAT/3Dig_N/.
The slides were examined independently by two investigators, who were blinded to the clinical and pathological data. MiR-514b-3p and miR-514b-5p expression were quantified using a visual grading system based on the extent of staining (percentage of positive tumor cells) and the intensity of staining (graded on a scale of 1–3: 1, weak staining; 2, moderate staining; 3, strong staining). For further analysis, the product (the corresponding score) of the extent and intensity grades was used to define the expression levels of the miRNA.
Nude mice metastatic model
HCT116 cells were infected with lenti-514b-3p and lenti-514b-5p to construct HCT116-514b-3p and HCT116-514b-5p stable expressing cell lines. Cells were collected and suspended in PBS. Male BALB/c nude mice aged 4 weeks old were purchased from Animal Center of Tongji University and maintained under SPF conditions. A total of 1 × 107 suspended cells were injected into the dorsal right flank of mice. Survival times of the mice were recorded. All the mice were euthanized 12 weeks after inoculation and lungs were obtained for fixation and HE staining. The animal experiments were approved by the Institutional Animal Care and Use Committee.
Chemotherapy resistance assays
CRC cell lines were planted into 96-well plates and transfected with miRNA mimics the next day. 6 h after transfection, the cells were treated with Irinotecan or Cisplatin with different concentrations. After 48 h of incubation, the cells were treated with Cell Counting Kit-8 (CCK8, Dojindo, Japan) for 2 h at 37 ℃. The absorbance was detected at wave length 450 nm using a microplate reader. Each group was down in six replicate wells and all the experiments were repeated at least 3 times.
In vivo chemotherapy resistance assays
HCT116 cells were infected with lenti-514b-3p, lenti-514b-5p and lenti-control virus to construct HCT116-514b-3p and HCT116-514b-5p stable expressing cell lines. Cells were collected and suspended in PBS. Male BALB/c nude mice aged 4 weeks old were purchased from Animal Center of Tongji University and maintained under SPF conditions. A total of 5 × 106 suspended cells were injected into the dorsal right flank of mice. When the volume of tumors reached 200 mm3, the mice were treated with 40 mg/kg Irinotecan, 2.5 mg/kg Cisplatin or PBS. The tumor sizes were measured every 3 days. The tumor volumes were calculated using the formula: V = (length x width2)/2, (V, volume). The animal experiments were approved by the Institutional Animal Care and Use Committee.
Luciferase reporter assays
Luciferase reporter plasmids containing CDH1, CLDN1, FZD4, or NTN1 3′ UTR miRNA target sequences (pGL3-CDH1, pGL3-CLDN1, pGL3-FZD4, pGL3-NTN1 and their respective miRNA target mutants) were generated. CRC cells were seeded into 24-well plates the day before transfection. For each well, 0.5 μg pGL3 together with 0.05 μg Renilla vector (Generay, China) were cotransfected with 3 μL miRNAs using Lipofectamine 3000 (Invitrogen, USA). 48 h later, cells were lysed with 500 μL passive lysis buffer and 10 μL supernatant was used for further detection of luciferase activity using the dual luciferase reporter assay system (Promega, USA) according to the manufacturer’s instruction. Renilla luciferase activity was used to normalize firefly luciferase activity.
Statistical analysis
All statistical analyses were carried out using the GraphPad Prism 5 and SPSS 19.0 software. Association between miR-514b-3p and miR-514b-5p expression and clinicopathologic parameters of CRC patients was analyzed using chi-square test. Overall survival of metastatic animal model was analyzed using the log-rank test and was plotted using Kaplan–Meier survival curve. All the experiments were repeated at least three times and the data were presented in the form of means ± SEM. p < 0.05 was considered to be significant.
Results
Different profiles of miR-514b-3p and miR-514b-5p in CRC tissues
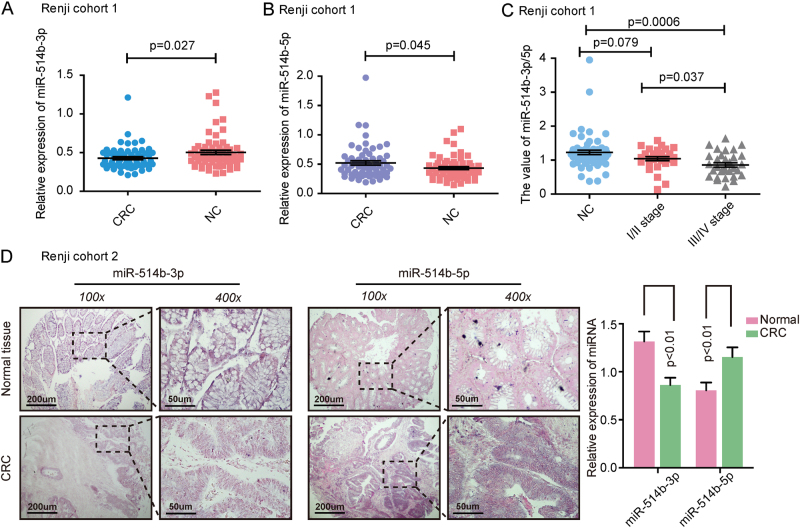
The miR-514b-3p and miR-514b-5p expression were analyzed in Renji cohort 1, consisting of 62 pairs of CRC tissues and adjacent normal tissues. The results showed that miR-514b-3p expression was significantly down-regulated in CRC samples (Fig. 1a, p < 0.05), while miR-514b-5p was significantly up-regulated in CRC samples compared with matched normal tissues (Fig. 1b, p < 0.05). Additionally, the In situ hybridization (ISH) data displayed the similar results in Renji cohort 2 patients (Fig. 1d). We further determined the ratio of miR-514b-3p/miR-514b-5p in different CRC stages in Renji cohort 1. The data indicated that the ratio of miR-514b-3p/miR-514b-5p was gradually decreased from normal colorectal tissue to stage I-II and stage III-IV of CRC tissues (Fig. 1c). We further analyzed the association between miR-514b-3p/miR-514b-5p value and clinicopathological features of CRC in Renji cohort 1. The data revealed that high miR-514b-3p/miR-514b-5p level was negatively correlated with tumor stage, histological differentiation and lymphnode invasion (Supplementary Table 1). Overall, these data indicated that miR-514b-3p and miR-514b-5p were highly associated with CRC development.
Fig. 1. The expression of miR-514b-3p and miR-514b-5p in CRC tissues.
a, b The expression of miR-514b-3p and miR-514b-5p was measured by real-time PCR in CRC tissues and matched non-tumor tissues in Renji cohort 1 (n = 62). Error bars represented SEM. c The ratio of miR-514b-3p/miR-514b-5p was analyzed in different stages of CRC progression. d The expression of miR-514b-3p and miR-514b-5p was measured by ISH (in situ hybridization) and the summarized data was presented in CRC tissues and matched non-tumor tissues in Renji cohort 2 (n = 40), the purple staining in the image indicates the specific microRNA ISH signal and red indicates the nuclear counter stain
MiR-514b-3p may inhibit cell invasion and metastasis while miR-514b-5p may promote these cell processes in CRC
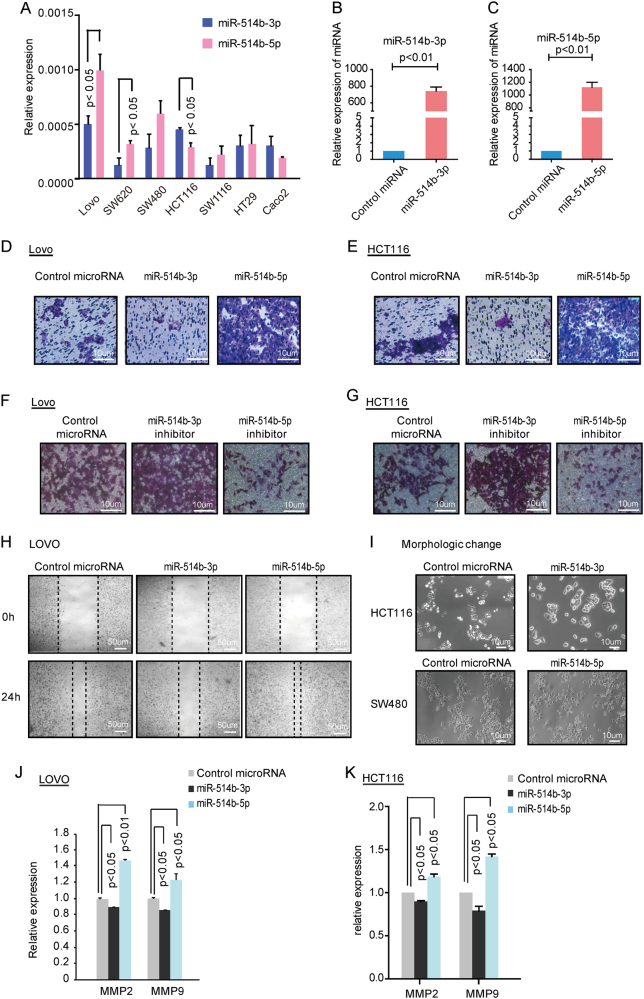
To gain a comprehensive understanding of the role of miR-514b-3p and miR-514b-5p in CRC tumorigenesis, two CRC cell line HCT116 and LOVO with low expression of miR-514b-5p or miR-514b-3p were transfected with miR-514b-3p, miR-514b-5p or control miRNA (Fig. 2a). The overexpression efficiency was confirmed by qRT-PCR (Fig. 2b, c). We first explored the miRNA-mediated cell proliferation and cell cycle arrest in HCT116 and LOVO cell lines using CCK8 assays and flow cytometry, respectively. The data showed that miR-514b-3p and miR-514b-5p had no significant difference in cell proliferation or cell cycle arrest in the two cells compared with negative control (Supplementary Fig. 1A-1C).
Fig. 2. The effects of miR-514b-3p and miR-514b-5p on CRC invasion and metastasis in vitro.
a The relative expression of miR-514b-3p and miR-514b-5p was quantified by real-time PCR in CRC cell lines. b, c The overexpression efficiency was confirmed in HCT116 and LOVO cells by real-time PCR. d, e Transwell Matrigel invasion assays were performed in HCT116 and LOVO cells after transfection of miR-514b-3p or miR-514b-5p mimics. f, g Transwell invasion assays were performed in HCT116 and LOVO cells after transfection of miR-514b-3p or miR-514b-5p inhibitors. h. The scratch assays were conducted in LOVO cells after control, miR-514b-3p and miR-514b-5p mimics transfection. i. The morphologic changes were observed under a light microscope in HCT116 and SW480 cells transfected with miR-514b-3p or miR-514b-5p mimics. j, k The effects of miR-514b-3p and miR-514b-5p on MMP2 and MMP9 expression were evaluated using ELISA in LOVO and HCT116 cells. All the experiments above were done for three replicates and repeated at least three times. Error bars represented SEM
Next, we analyzed the potential biological effect of miR-514b-3p and miR-514b-5p on cell migration and invasion in HCT116 and LOVO cells. Overexpression of miR-514b-3p significantly suppressed cell motility and invasiveness in both HCT116 and LOVO cells, while those cells transfected with miR-514b-5p mimics showed increased invasion ability, compared with control miRNA (Fig. 2d, e). The opposite effects were observed when cells were treated with miRNA inhibitors (Fig. 2f, g). The wound healing assay and morphological changes in cells transfected with miR-514b-3p and miR-514b-5p, further confirmed our results (Fig. 2h, i). Matrix metalloproteinase 2 (MMP2) and MMP9 possess ability to hydrolyze components of the basement membrane, and thus could promote tumor metastasis. To better understand the function of miR-514b-3p/5p in CRC metastasis, MMP2 and MMP9 were detected in cell culture supernatant by ELISA. We observed that restoration of miR-514b-3p significantly reduced MMP2 and MMP9 expression, whereas miR-514b-5p increased MMP2 and MMP9 secretion in LOVO and HCT116 cells (Fig. 2j, k).
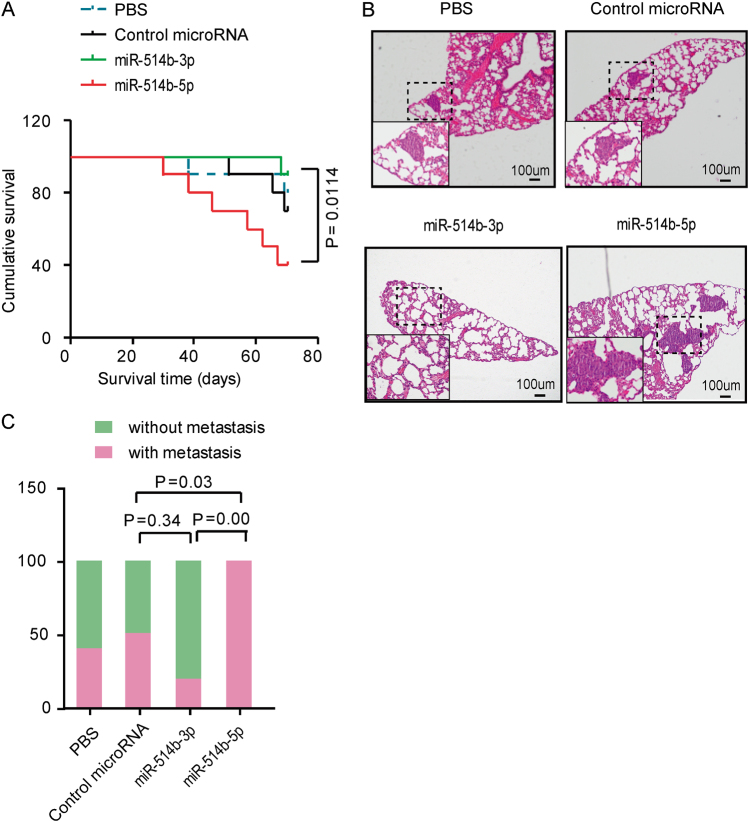
To further identify the involvement of miR-514b-3p and miR-514b-5p in vivo, we established a CRC metastasis model in nude mice. The HCT116-control miRNA cells, HCT116-miR-514b-3p and HCT116-miR-514b-5p stable expressing cells were subcutaneously injected through the right flank of nude mice, respectively. The mice inoculated with HCT116-miR-514b-3p stable expressing cells had a longer overall survival time compared with those injected with control cells (Fig. 3a). Whereas mice bearing HCT116-miR-514b-5p expressing stable cells had a significant poor survival (Fig. 3a). Hematoxylin-eosin staining showed that less metastatic CRC loci were observed in miR-514b-3p group compared to control group, while all of the mice injected with HCT116-miR-514b-5p stable expressing cells displayed lung metastases (Fig. 3b-c). Altogether, these data indicate that miR-514b-3p and miR-514b-5p may display converse function in CRC invasion and metastasis.
Fig. 3. The effects of miR-514b-3p and miR-514b-5p on CRC metastasis in vivo.
a Survival time was analyzed in nude mice bearing HCT116-control miRNA cells, HCT116-miR-514b-3p, HCT116-miR-514b-5p stable expressing cells or PBS. b Representative hematoxylin-eosin staining images of lung metastatic loci in PBS group, control-miRNA group, miR-514b-3p overexpression group and miR-514b-5p overexpression group. c The ratio of mice with metastatic loci was analyzed in each group
Distinct roles of miR-514b-3p and miR-514b-5p in chemoresistance
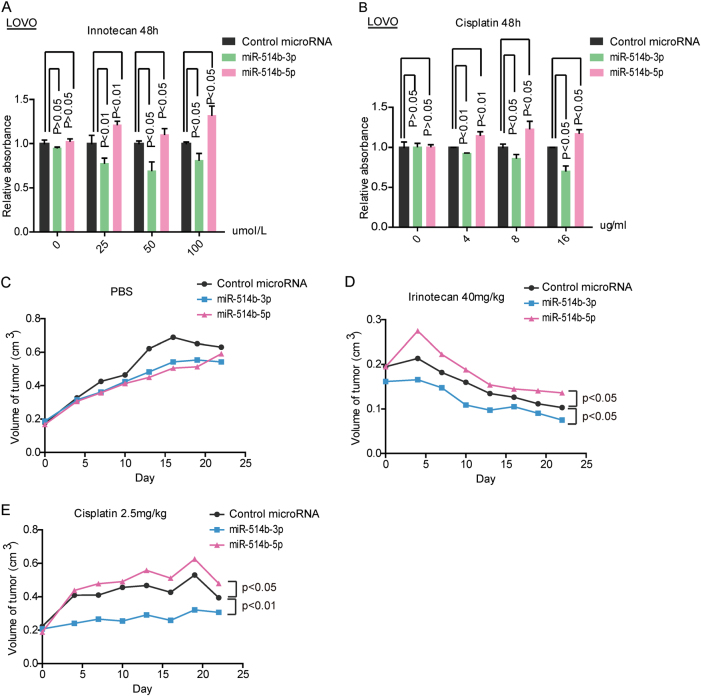
Since metastasis and chemoresistance in cancer are linked phenomenon19,20, we further assessed the potential role of miR-514b-3p and miR-514b-5p in the chemoresistance of Cisplatin and Irinotecan. The data revealed that cells with miR-514b-5p overexpression got an increased viability compared to control miRNA, and there is no significant difference in the absence of Cisplatin or Irinotecan. However, overexpression of miR-514b-3p accelerated cell death in cells treated with Cisplatin or Irinotecan compared to the control cells (Fig. 4a, b).
Fig. 4. The effects of miR-514b-3p and miR-514b-5p on drug resistance.
a, b Chemotherapy resistance assays were conducted to assess the potential role of miR-514b-3p and miR-514b-5p in the chemoresistance of Cisplatin and Irinotecan. c–e The tumor volumes were measured in mice with different treatments in the xenograft mouse model at the time point of 0, 5, 10, 15, 20, and 25 days
To better verify the exact role of miR-514b-3p and miR-514b-5p in chemoresistance in vivo, we established CRC xenograft mouse model using HCT116 cell lines. Delivery of miR-514b-3p or miR-514b-5p lentivirus had no effect on the size of tumor in the absence of Cisplatin or Irinotecan (Fig. 4c). However, when treated with Cisplatin (2.5 mg/kg) or Irinotecan (40 mg/kg), mice bearing HCT116-miR-514b-3p cells showed decreased size of tumors, whereas mice inoculated with HCT116-miR-514b-5p cells displayed an increase in tumor growth compared to control group (Fig. 4d, e). Collectively, these findings suggested that miR-514b-3p and miR-514b-5p displayed a possible role in mediating chemoresistance to Cisplatin and Irinotecan.
MiR-514b-3p and miR-514b-5p mediate CRC cell invasion via regulation of different targets
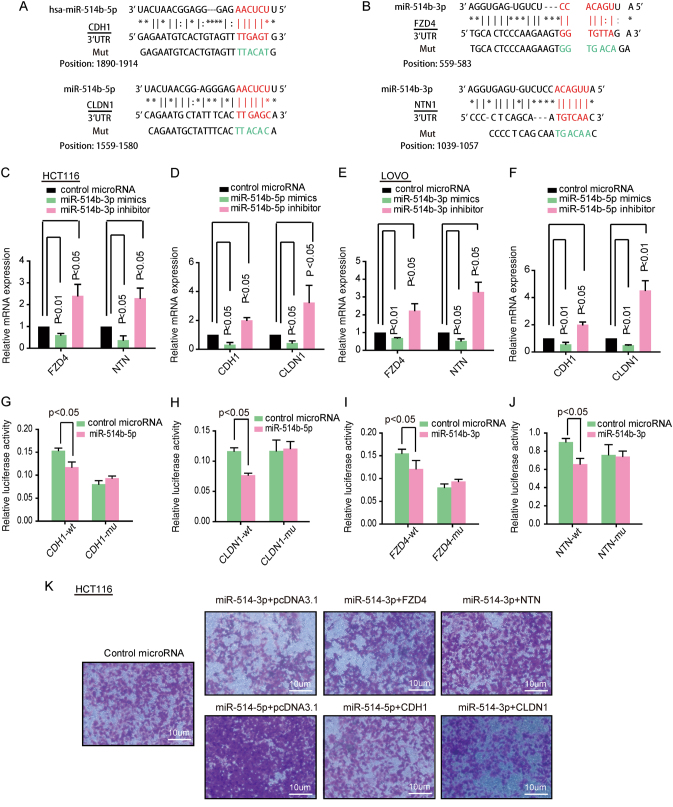
To determine the underlying mechanism of miR-514b-3p and miR-514b-5p, which is involved in CRC invasion and metastasis. The miRBase, microRNA.org and TargetScan website were used to predict target genes of miR-514b-3p and miR-514b-5p. Target prediction programs indicated that there were potential specific targets for miR-514b-3p in the seed regions of oncogenes FZD4 and NTN1, and for miR-514b-5p within the 3′UTR region of CDH1 and CLDN1 (Fig. 5a, b).
Fig. 5. The target genes of miR-514b-3p and miR-514b-5p.
a Schematic diagram of the predicted binding sites for miR-514b-5p in 3′UTR of CDH1 and CLDN1. b Schematic diagram of the predicted binding sites for miR-514b-3p in 3′UTR of FZD4 and NTN1. c–f The mRNA expression of corresponding target genes was tested by real-time PCR in HCT116 and LOVO cells transfected with miR-514b-3p and miR-514b-5p mimics or inhibitors, respectively. g–j Luciferase activities were detected in HCT116 and LOVO cells co-transfected with corresponding miRNA mimics and the indicated target gene wild-type reporter plasmids or mutant-type reporter plasmids. The experiments were done for four replicates and repeated at least three times. Error bars represented SEM. k The transwell invasion assays were performed in HCT116 after co-transfection with miR-514b-3p or miR-514b-5p mimics and their target genes overexpression plasmid. The experiments were done for three replicates and repeated at least three times
To verify the prediction, qPCR was used to test the target mRNA expression in HCT116 and LOVO cells transfected with miR-514b-3p and miR-514b-5p mimics or inhibitors, respectively. MiR-514b-3p and miR-514b-5p overexpression significantly decreased their corresponding target genes expression, and the inhibitors showed opposite effects (Fig. 5c–f). To explore whether the effect was mediated by directly binding of the miRNAs to the seed regions of their target genes, we constructed the recombined luciferase reporter plasmid, containing the predicted micoRNA binding sites of target genes. Luciferase assay data revealed that when co-transfected with wild-type reporter plasmids (CDH1 and CLDN1) and miR-514b-5p mimics, the luciferase activity was significantly decreased, whereas the mutant-type reporter plasmids made no difference (Fig. 5g, h). Also, we find that miR-514b-3p mimics significantly decreased the luciferase activity of wild-type 3′UTR (FZD4 and NTN1), but did not affect the activity of mutant-type reporter plasmids (Fig. 5i, j). These data indicated that miR-514b-5p promoted CRC progression by targeting CDH1 and CLDN1, and miR-514b-3p inhibited CRC development through direct targeting FZD-4 and NTN1.
To further confirm our suggestion, we co-transfected HCT116 cells with miR-514b-3p mimics and their target gene. As shown in Fig. 5k, overexpression of FZD4 and NTN1 partially blocked miR-514b-3p induced anti-invasion. The same phenomenon was observed when cells were co-transfected with miR-514b-5p and its target genes’ overexpression plasmids. Collectively, the data revealed that miR-514b-3p and miR-514b-5p regulated CRC cell invasion through downregulation of different target genes.
The functional role of miR-514b-3p and miR-514b-5p in the EMT progression of CRC
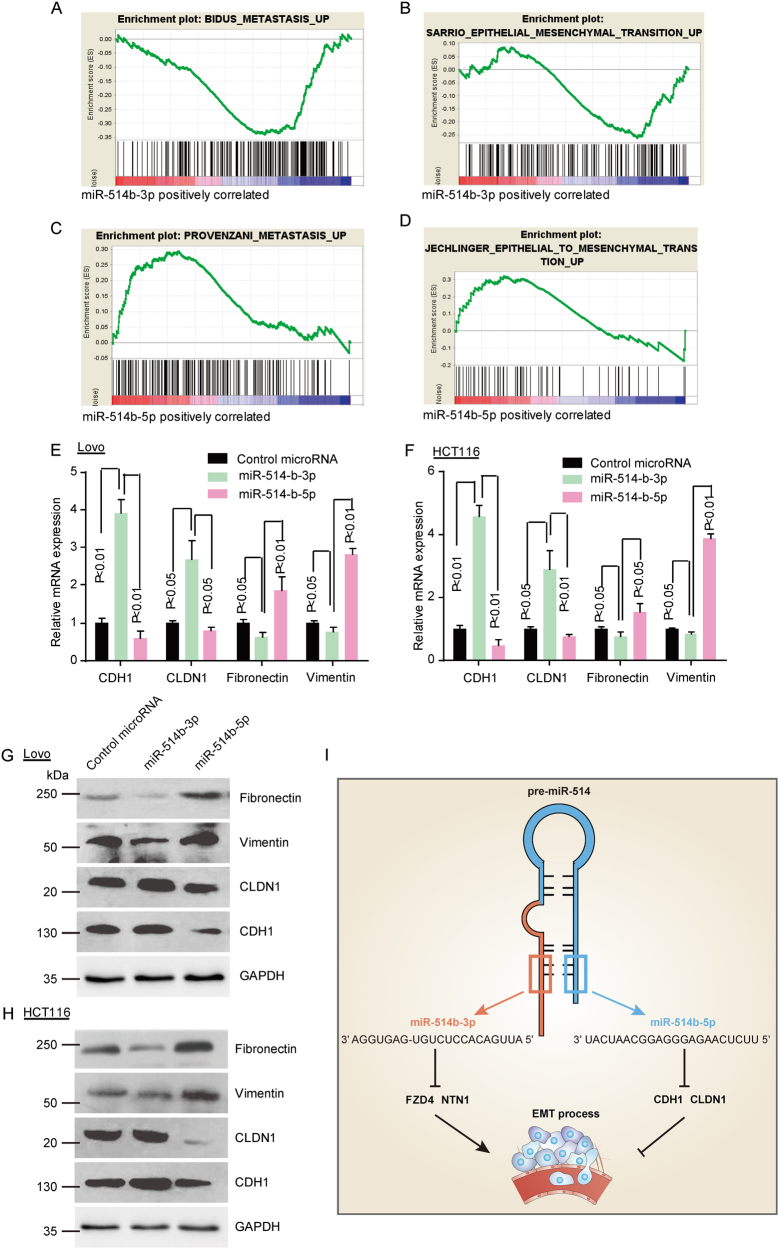
As EMT is well-known to be involved in invasion and metastasis in cancer, we are supposed to investigate whether miR-514b-3p and miR-514b-5p take part in this progression. First, RNA sequence and GSEA were analyzed in HCT116 transfected with miR-514b-3p, miR-514b-5p or control miRNAs. The enrichment plots of GSEA showed that metastasis associated gene signature and EMT pathways were enriched in cells with miR-514b-5p microRNA overexpression (Fig. 6a, b), and deficient in cells transfected with miR-514b-3p (Fig. 6c, d), but there is no difference in cells transfected with control microRNAs. These data demonstrated that miR-514b-3p and miR-514b-5p played vital roles in CRC EMT.
Fig. 6. The functional role of miR-514b-3p and miR-514b-5p in the EMT progression of CRC cell lines.
a–d GSEA was performed to identify the difference of gene profiles in control-miRNA group, miR-514b-3p overexpression group and miR-514b-5p overexpression group. e, f Real-time PCR was performed to measure the mRNA expression of EMT markers (E-cadherin, CLDN-1, fibronectin-1 and vimentin) in LOVO and HCT116 after transfection of miR-514b-3p or miR-514b-5p mimics. Error bars represented SEM. g, h Western-blot was conducted to evaluate the protein expression of EMT markers (E-cadherin, CLDN-1, fibronectin-1, and vimentin) in LOVO and HCT116 after transfection of miR-514b-3p or miR-514b-5p mimics. GAPDH was used as an internal control. i Schematic representation for the mechanism of miR-514 regulates EMT in human CRC progression
To further support our view, we tested the epithelial and mesenchymal markers level upon miR-514b-3p or miR-514b-5p overexpression. The results showed that the epithelial marker (E-cadherin and CLDN-1) increased in both mRNA and protein levels after ectopic miR-514b-3p expression, however, mesenchymal marker (fibronectin-1 and vimentin) were decreased (Fig. 6e, h). On the contrary, when cells were transfected with miR-514b-5p mimics, the expression level of E-cadherin, CLDN-1 were dramatically down-regulated, whereas mesenchymal markers were significantly up-regulated (Fig. 6e, h). These data confirmed our hypothesis that miR-514b-3p could suppress and miR-514b-5p might promote tumor metastasis via regulating EMT pathway (Fig. 6i).
Discussion
Accumulating evidence have shown that miRNA could function as important modulators in tumorigenesis21,22, emerged as oncogenes or tumor suppressors, via different target genes. Detection of specific miRNAs in urine, blood, or tissue samples could serve as novel biomarkers and contributed to tumor diagnosis and clinical outcome prediction23–26.
In the current study, we analyzed the expression of miR-514b-3p and miR-514b-5p in 2 cohorts of CRC patients in Renji hospital. It is the first time to identify miR-514b-3p and miR-514b-5p expression in CRC. We found that miR-514b-3p was significantly downregulated in CRC compared with corresponding normal tissue, and decreased along with CRC progression. On the other hand, the expression of miR-514b-5p increased in CRC patients with advanced stage compared to patients with low clinical stage. These results implied that miR-514b-3p may act as a tumor suppressor gene, yet miR-514b-5p serves as an oncogene in CRC tumorigenesis, which might provide a possibility to predict CRC progression.
Tumor invasion and metastasis is the leading cause of cancer-related death worldwide. Numerus studies indicated that miRNA dysfunction was involved in tumor migration and invasion27,28. Here, we first evaluated the correlation between miR-514b-3p/5p expression and clinicopathological parameters in Renji cohort 1. Intriguingly, we found that the ratio of miR-514b-3p/5p expression was negatively correlated with lymph node invasion, AJCC stage and histological differentiation. Consistent with the finding, transfection of miR-514b-3p mimics damaged cell invasion ability and decreased metastatic CRC loci in the lung of nude mice. In contrast, overexpression of miR-514b-5p showed the opposite tendency. These data suggested that miR-514b-3p and miR-514b-5p involved in metastasis process in CRC development.
EMT is a morphogenic process which occurs during embryonic development, tissue repair, organ fibrosis and tumor invasion and metastasis29,30. During EMT, epithelial cells acquire fibroblast-like properties, exhibit reduced intercellular adhesion and show increased invasion ability. The activation of the epithelial-mesenchymal transition (EMT) program is generally considered as a major driver of tumor progression from initiation to metastasis. Given the fact that the role of EMT in promoting metastasis has recently been challenged, in particular concerning effects of the EMT transcription factors (EMT-TFs) Snail and Twist in pancreatic cancer31. Although this study showed that pancreatic cancer may metastasize without activating of EMT programs, the author’s conclusion can not be sustained by their data, because of the technical defect32. In addition, Krebs et al. showed that the EMT-TF Zeb1 is a key factor for the formation of precursor lesions, invasion and notably metastasis in the same pancreatic cancer model as well33. Furthermore, major studies remain support that EMT is crucial for tumor invasion and metastasis in majority type of cancers, including breast cancer, hepatocellular carcinoma, CRC, and our research model34–38. Our previous data demonstrated that miR-514b-3p and miR-514b-5p could regulate CRC invasion and metastasis, we next attempt to figure out whether this modulation was mediated by EMT. To confirm our assumption, we performed the GSEA analysis and detected the epithelial and mesenchymal markers in CRC cells transfected with miR-514b-3p or miR-514b-5p mimics. The results showed that miR-514b-3p and miR-514b-5p induced CRC invasion and metastasis was mediated by EMT. It was observed that patients with metastasis are prone to be more resistant to chemotherapy39. In addition, increasing evidences have demonstrated that chemoresistance is included in the EMT progression, which is the pivotal step in tumor invasion and metastasis40. Besides, Zheng et al further identified that EMT program induces chemoresistance in pancreatic cancer31. In this study, since miR-514 could influence EMT progress through target genes, we are supposed to further explore its function on chemoresistance. Interestingly, we found that miR-514b-3p, which suppresses EMT process, could undermine chemo-resistance in vitro or in vivo, in accordance with the fact that EMT induces stem cell properties, prevents apoptosis and contributes to immunosuppression41.
The specific mechanism of miR-514b-3p and miR-514b-5p involved in CRC is still unknown. To elucidate the underlying mechanism, we searched for three microRNA databases. E-cadherin and Claudin-1, translated from CDH1 and CLDN1, are well-known epithelial markers responsible for maintaining cell polarity and cell-cell adhesion42,43. Loss of them was considered as hallmarks of EMT process44. Our data showed that miR-514b-5p targets CDH1 and CLDN1, which was consistent with its function. In addition, we found that miR-514b-3p directly binds to 3′UTR of FZD4 and NTN1. It was documented that NTN1 could increase invasiveness and metastasis by promoting EMT process in hepatocellular and prostate carcinoma cancer cells45,46. FZD4 silencing induced E-cadherin expression, and thus restrained EMT initiation in human prostate cancer47. Consistent with previous studies, our results indicated that FZD4 and NTN1 were activated in CRC EMT process and could be blocked by miR-514b-3p overexpression.
There were still some deficiencies in our study. Further investigations were needed to reveal the underlying regulatory mechanisms which mediated the change in expression from precursor of miR-514 to mature miR-514b-3p and miR-514b-5p.
To sum up, the present study supplies the evidence of miR-514b-3p and miR-514b-5p involvement in CRC, for the first time. Functional studies demonstrated that miR-514b-3p and miR-514b-5p emerged to play distinct roles, both in vitro or in vivo, repressing or promoting tumor metastasis and chemoresistance, respectively. In line with previous studies, our discovery of the important role which miR-514 family plays in the CRC metastasis made it possible for miR514b-3p/5p used as a biomarker in tumor diagnosis and prognosis. Therefore, the detection and disruption of miR-514b-3p and miR-514b-5p could monitor and suppress CRC development, providing a new potential therapeutic strategy for human CRC.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the National Natural Science Foundation (81320108024, 81421001, 81530072, 81522008, 81790632, 31371420, 31371273, 81372267, and 81402347); the Shanghai Natural Science Foundation (Grant No. 20144Y0253); the Program for Professor of Special Appointment (Eastern Scholar No. 201268 and 2015 Youth Eastern Scholar NO. QD2015003) at Shanghai Institutions of Higher Learning;Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support (No.20152512, 20161309) and Chenxing Project of Shanghai Jiao-Tong University to H.Y.C. and J.H.
Author contributions
L.L.R., T.T.Y., C.Q.S., Y.C.W., and X.K. performed the experiments and analyzed data. J.Y.T., J.C., Q.L., and M.Z. provided colon cancer specimens and clinical and pathological information. J.H. and H.Y.C. conceived, wrote the manuscript, and supervised the study. J.Y.F. designed or/and supervised this project and revised the manuscript.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Lin-Lin Ren, Ting-Ting Yan, Chao-Qin Shen
Edited by E. Candi
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41419-018-0732-5).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ming Zhong, Email: drzhongming@hotmail.com.
Hao-Yan Chen, Email: haoyanchen@sjtu.edu.cn.
Jie Hong, Email: jiehong97@sjtu.edu.cn.
Jing-Yuan Fang, Email: jingyuanfang@sjtu.edu.cn.
References
- 1.Song M, Garrett WS, Chan AT. Nutrients, foods, and colorectal cancer prevention. Gastroenterology. 2015;148:1244–1260. doi: 10.1053/j.gastro.2014.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, et al. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Cano A, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 4.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J. Mammary Gland Biol. Neoplasia. 2010;15:117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Bu P, et al. A miR-34a-numb feedforward loop triggered by inflammation regulates asymmetric stem cell division in intestine and colon cancer. Cell Stem Cell. 2016;18:189–202. doi: 10.1016/j.stem.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehta A, et al. The MicroRNA-132 and MicroRNA-212 cluster regulates hematopoietic stem cell maintenance and survival with age by buffering FOXO3 expression. Immunity. 2015;42:1021–1032. doi: 10.1016/j.immuni.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-Barrueco R, et al. miR-424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev. 2017;31:553–566. doi: 10.1101/gad.292318.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valeri N, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25:469–483. doi: 10.1016/j.ccr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandbothe M, et al. The microRNA-449 family inhibits TGF-beta-mediated liver cancer cell migration by targeting SOX4. J. Hepatol. 2017;66:1012–1021. doi: 10.1016/j.jhep.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 15.Mitra R, Sun J, Zhao Z. microRNA regulation in cancer: one arm or two arms? Int J. Cancer. 2015;137:1516–1518. doi: 10.1002/ijc.29512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozata DM, et al. Loss of miR-514a-3p regulation of PEG3 activates the NF-kappa B pathway in human testicular germ cell tumors. Cell Death Dis. 2017;8:e2759. doi: 10.1038/cddis.2016.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wotschofsky Z, et al. Diagnostic and prognostic potential of differentially expressed miRNAs between metastatic and non-metastatic renal cell carcinoma at the time of nephrectomy. Clin. Chim. Acta. 2013;416:5–10. doi: 10.1016/j.cca.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Stark M. S. et al. miR-514a regulates the tumour suppressor NF1 and modulates BRAFi sensitivity in melanoma. Oncotarget6,17753–17763 (2015). [DOI] [PMC free article] [PubMed]
- 19.Chen D, et al. Targeting BMI1 + cancer stem cells overcomes chemoresistance and inhibits metastases in squamous cell carcinoma. Cell Stem Cell. 2017;20:621–634. doi: 10.1016/j.stem.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acharyya S, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, et al. MicroRNA-191 acts as a tumor promoter by modulating the TET1-p53 pathway in intrahepatic cholangiocarcinoma. Hepatology. 2017;66:136–151. doi: 10.1002/hep.29116. [DOI] [PubMed] [Google Scholar]
- 22.Ou C, et al. MiR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer. Cancer Lett. 2017;399:53–63. doi: 10.1016/j.canlet.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Kakkassery V, et al. Vitreous microRNA levels as diagnostic biomarkers for vitreoretinal lymphoma. Blood. 2017;129:3130–3133. doi: 10.1182/blood-2017-01-765180. [DOI] [PubMed] [Google Scholar]
- 24.Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol. Ther. 2017;172:34–49. doi: 10.1016/j.pharmthera.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Torres-Ferreira J, et al. MiR-193b promoter methylation accurately detects prostate cancer in urine sediments and miR-34b/c or miR-129-2 promoter methylation define subsets of clinically aggressive tumors. Mol. Cancer. 2017;16:26. doi: 10.1186/s12943-017-0604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karakas M, et al. Circulating microRNAs strongly predict cardiovascular death in patients with coronary artery disease-results from the large AtheroGene study. Eur. Heart J. 2017;38:516–523. doi: 10.1093/eurheartj/ehw250. [DOI] [PubMed] [Google Scholar]
- 27.Zhou W, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang B, et al. MicroRNA-1301 suppresses tumor cell migration and invasion by targeting the p53/UBE4B pathway in multiple human cancer cells. Cancer Lett. 2017;401:20–32. doi: 10.1016/j.canlet.2017.04.038. [DOI] [PubMed] [Google Scholar]
- 29.Lin Y, et al. Snail1-dependent control of embryonic stem cell pluripotency and lineage commitment. Nat. Commun. 2014;5:3070. doi: 10.1038/ncomms4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murdoch CE, et al. Endothelial NADPH oxidase-2 promotes interstitial cardiac fibrosis and diastolic dysfunction through proinflammatory effects and endothelial-mesenchymal transition. J. Am. Coll. Cardiol. 2014;63:2734–2741. doi: 10.1016/j.jacc.2014.02.572. [DOI] [PubMed] [Google Scholar]
- 31.Zheng X, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aiello NM, et al. Upholding a role for EMT in pancreatic cancer metastasis. Nature. 2017;547:E7–E8. doi: 10.1038/nature22963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krebs AM, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017;19:518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 34.Gujral TS, et al. A noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell. 2014;159:844–856. doi: 10.1016/j.cell.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng JM, et al. Actin cytoskeleton remodeling drives epithelial-mesenchymal transition for hepatoma invasion and metastasis. Hepatology. 2017;67:2226–2243. doi: 10.1002/hep.29678. [DOI] [PubMed] [Google Scholar]
- 36.Lee JH, et al. A20 promotes metastasis of aggressive basal-like breast cancers through multi-monoubiquitylation of Snail1. Nat. Cell Biol. 2017;19:1260–1273. doi: 10.1038/ncb3609. [DOI] [PubMed] [Google Scholar]
- 37.Sikandar SS, et al. Role of epithelial to mesenchymal transition associated genes in mammary gland regeneration and breast tumorigenesis. Nat. Commun. 2017;8:1669. doi: 10.1038/s41467-017-01666-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan TT, et al. miR-508 defines the stem-like/mesenchymal subtype in colorectal cancer. Cancer Res. 2018;78:1751–1765. doi: 10.1158/0008-5472.CAN-17-2101. [DOI] [PubMed] [Google Scholar]
- 39.Lim E, et al. Preoperative versus postoperative chemotherapy in patients with resectable non-small cell lung cancer: systematic review and indirect comparison meta-analysis of randomized trials. J. Thorac. Oncol. 2009;4:1380–1388. doi: 10.1097/JTO.0b013e3181b9ecca. [DOI] [PubMed] [Google Scholar]
- 40.Lee JW, et al. miR-708-3p mediates metastasis and chemoresistance through inhibition of epithelial-to-mesenchymal transition in breast cancer. Cancer Sci. 2018;109:1404–1413. doi: 10.1111/cas.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Toh YC, Xing J, Yu H. Modulation of integrin and E-cadherin-mediated adhesions to spatially control heterogeneity in human pluripotent stem cell differentiation. Biomaterials. 2015;50:87–97. doi: 10.1016/j.biomaterials.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Stache C, et al. Tight junction protein claudin-1 is differentially expressed in craniopharyngioma subtypes and indicates invasive tumor growth. Neuro Oncol. 2014;16:256–264. doi: 10.1093/neuonc/not195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah PP, et al. Ubiquilin1 represses migration and epithelial-to-mesenchymal transition of human non-small cell lung cancer cells. Oncogene. 2015;34:1709–1717. doi: 10.1038/onc.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H, Chen Q, Luo Q. Expression of netrin-1 by hypoxia contributes to the invasion and migration of prostate carcinoma cells by regulating YAP activity. Exp. Cell Res. 2016;349:302–309. doi: 10.1016/j.yexcr.2016.10.023. [DOI] [PubMed] [Google Scholar]
- 46.Yan W, et al. Netrin-1 induces epithelial-mesenchymal transition and promotes hepatocellular carcinoma invasiveness. Dig. Dis. Sci. 2014;59:1213–1221. doi: 10.1007/s10620-013-3016-z. [DOI] [PubMed] [Google Scholar]
- 47.Gupta S, et al. FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2010;70:6735–6745. doi: 10.1158/0008-5472.CAN-10-0244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.