Abstract
Evaluating carriage of Staphylococcus aureus, an opportunistic pathogen of humans and animals capable of causing antibiotic-resistant infections, is epidemiologically important. However, clinical and epidemiological surveillance studies of S. aureus typically rely on characterizing one isolate per individual, which may not represent the actual population diversity in a carrier. The objective of this study was to determine if one isolate is sufficient for determining carrier status of particular strains or characteristics of S. aureus in a healthy (non-hospitalized) human population. We compared spa types, genetic markers (mecA, scn), and antibiotic resistance profiles of 10 S. aureus isolates recovered from a single nasal swab for each of 19 participants (190 isolates total) selected from a cohort of industrial hog operation workers and their household members. Participants included both persistent (n = 10) and intermediate (n = 9) carriers of S. aureus. Among the participants, 17 (89%) carried a single S. aureus spa type intranasally and the other two carried dominant spa types. Less similarity was observed for genes encoded on mobile genetic elements (mecA, scn) and antibiotic resistance profiles. Statistical modeling, based on receiving operating characteristic curves, suggests that three to five isolates may be necessary to accurately assign nasal carriage status for these more variable characteristics. Variability was observed for both persistent and intermediate carriers of S. aureus. These results suggest that surveillance studies that rely on testing one S. aureus isolate are likely to identify predominant spa types but may not fully capture more variable characteristics of S. aureus, including antibiotic resistance. Surveillance studies that rely on testing one isolate may underestimate prevalence of nasal carriage of S. aureus with these more variable characteristics.
Keywords: Staphylococcus, MRSA, antibiotic resistance, opportunistic pathogen, nasal carriage, surveillance, ROC curve
Introduction
Staphylococcus aureus is an opportunistic pathogen that can colonize the nasal cavities of humans and animals and can cause superficial or life-threatening infections. Infections caused by S. aureus include skin and soft tissue infections, bacteremia, endocarditis, gastrointestinal illness, necrotizing pneumonia, post-operative infections, and toxic shock syndrome (1, 2). In the United States, annual incidence of S. aureus bacteremia is 4.3–38.2 per 100,000 person years; and the 30 day all-cause mortality rate is approximately 20% (3).
Infections caused by antibiotic-resistant strains of S. aureus including methicillin-resistant S. aureus (MRSA) and multidrug-resistant S. aureus (MDRSA) are particularly troublesome as they can be more difficult and costly to treat (4). These infections have reached epidemic proportions globally (5), and are a leading concern in the global fight against antibiotic resistance (6). MRSA first emerged in hospital settings in the 1960s and predominantly affected elderly and sick patients (2). However, MRSA is no longer confined to the hospital setting. Antibiotic resistant S. aureus strains including MRSA are increasingly detected in the community (7, 8), and among healthy individuals with livestock contact including livestock operation workers and veterinarians (9–14). Systematic surveillance is needed within and outside of hospitals and following a “One Health” approach (15) to better understand the ecology of emerging strains.
Most surveillance studies rely on characterizing a single S. aureus isolate from a nasal swab per individual (16). Exceptions include studies that test more than one anatomical site per participant or longitudinal studies that test participants over time. However, even these studies typically rely on testing a single S. aureus isolate per sample. It is not clear whether testing one isolate per sample is sufficient for surveillance of particular characteristics of S. aureus, including tests for multidrug resistance. A recent study of MRSA transmitted among staff and animal patients at a veterinary hospital demonstrated considerable within-host genetic diversity of MRSA during carriage and infection (17), and co-colonization of humans by distinct S. aureus spa types has been reported (18, 19). Surveillance studies are often reviewed for their sampling procedures including the representativeness of participating populations. However, less attention is paid to procedures applied to test for carriage in individual participants. Also, less attention tends to be paid to healthy populations though healthy individuals can act as carriers for antibiotic resistant bacteria, facilitating their spread and diversification.
The objective of this study was to determine whether analysis of a single S. aureus isolate from an individual's anterior nares is adequate to determine carrier status of a particular spa type or S. aureus characteristic. We characterized 10 rather than one S. aureus isolate for each participant and evaluated all 10 isolates for characteristics commonly tested in surveillance studies. Characteristics tested in this study include occurrence of mecA, a gene that confers resistance to β-lactam antibiotics including methicillin that is often used to indicate MRSA carriage; and occurrence of scn, a gene that serves in evasion of the human innate immune response and whose absence is sometimes used to indicate livestock association (20). All isolates were also tested for resistance to a panel of 15 antibiotics used in human and/or animal medicine (11). Among characteristics for which clonality was not observed, our secondary objective was to use mathematical models to determine the number of isolates needed to correctly characterize an individual's S. aureus nasal carrier status.
Materials and methods
Study participants (n = 19) were selected from a cohort of 103 industrial hog operation (IHO) workers and 80 household members who participated in a repeated-measures study to assess persistence of S. aureus nasal carriage (11). The JHSPH Institutional Review Board (IRB) approved this study (IRB00004608) and the UNC Non-Biomedical IRB approved reliance on the JHSPH IRB. In the cohort study, nasal swabs were obtained from each participant's anterior nares using a BD BBL™ CultureSwab™ once at baseline then every 2 weeks during a 4-month follow-up period. Individuals were classified as persistent carriers, intermediate carriers, or non-carriers of S. aureus. Persistent carriers had at least 80% of their nasal swabs test positive for S. aureus while intermittent carriers had <80% but more than 0% test positive for S. aureus. Non-carriers tested negative for S. aureus nasal carriage throughout the study.
For this sub-study, one nasal swab from each of 10 persistent and nine intermittent carriers, including both IHO workers (n = 11) and members of their households (n = 8; Table 1), was screened to characterize multiple isolates of S. aureus from the same source. The swabs were collected between a participant's third and eighth biweekly study visits. Ten S. aureus isolates were characterized from each of the 19 participants, and thus a total of 190 isolates were tested.
Table 1.
Spa types of S. aureus (n = 10 isolates) recovered from nasal swabs collected from each of 19 participants.
| Participant no. | IHO worker/Household member | spa types detected (# of isolates) | |
|---|---|---|---|
| Persistent carriers | 1 | IHO worker | t3446 (10) |
| 2 | IHO worker | t034 (10) | |
| 3 | IHO worker | t337 (10) | |
| 4 | IHO worker | t002 (10) | |
| 5 | IHO worker | t337 (10) | |
| 6 | Household member | t5739 (10) | |
| 7 | Household member | t5739 (10) | |
| 8 | Household member | t034 (10) | |
| 9 | Household member | t089 (10) | |
| 10* | Household member | t7226 (8), t002 (2) | |
| Intermittent carriers | 11* | IHO worker | t337 (9), t14243 (1) |
| 12 | IHO worker | t14157 (10) | |
| 13 | IHO worker | t233 (10) | |
| 14 | IHO worker | t5883 (10) | |
| 15 | IHO worker | t645 (10) | |
| 16 | IHO worker | t337 (10) | |
| 17 | Household member | t233 (10) | |
| 18 | Household member | t1937 (10) | |
| 19 | Household member | t3802 (10) |
IHO, industrial hog operation. Participants 10 and 11, marked with “*,” carried two different spa types.
To isolate S. aureus colonies, each nasal swab was clipped into a micro-centrifuge tube containing 1 mL of phosphate-buffered saline and vortexed for 1 min. From this suspension, 100 μL was spread plated onto CHROMagar™ S. aureus and incubated at 37°C for 18–24 h. Colonies with morphological characteristics of S. aureus were counted and 10 colonies that appeared easiest to isolate without touching other colonies were selected, streaked to isolation, and analyzed for genetic and antibiotic resistance characteristics.
Multiplex polymerase chain reaction (PCR) was conducted to identify the staphylococcal protein A gene (spa), mecA, and scn (11, 21). Spa is a marker for S. aureus, and the presence of both spa and mecA is indicative of MRSA. Absence of the human immune evasion cluster scn gene is an indicator of livestock association (11, 20). For each isolate, spa typing was performed as described elsewhere (22) using the Ridom Staph Type software and the Ridom SpaServer (http://spa.ridom.de/index.shtml).
The Kirby-Bauer disk diffusion method was used to assess each isolate's susceptibility to a panel of 15 antibiotics (Figure 1). Interpretations were based on Clinical and Laboratory Standards Institute (CLSI) guidelines (23). Induced clindamycin resistance was determined by the presence of a D-zone with erythromycin resistance (24). For this study, we defined isolates that demonstrated intermediate to complete resistance to an antibiotic as “resistant.”
Figure 1.
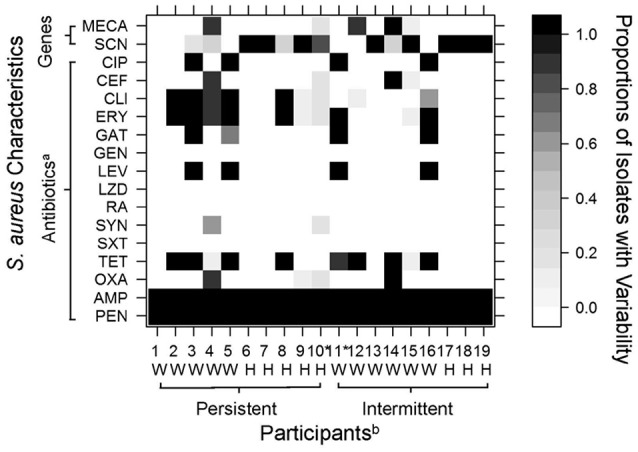
Variability among 10 S. aureus isolates recovered from nasal swabs collected from 19 industrial hog operation (IHO) workers and their household members. Variation in color indicates the observed diversity in tested characteristics, with white cells denoting that no isolates were resistant to a tested antibiotic or positive for a particular gene (i.e., mecA, scn), black cells denoting that all 10 isolates were resistant or positive for a particular gene (i.e., mecA, scn), and gray cells denoting variability among isolates as shown in the legend. aCIP, ciprofloxacin; CEF, ceftriaxone; CLI, clindamycin; ERY, erythromycin; GAT, gatifloxacin; GEN, gentamicin; LEV, levofloxacin; LZD, linezolid; RA, rifampin; SYN, quinupristin/dalfopristin; SXT, sulfamethoxazole with trimethoprim; TET, tetracycline; OXA, oxacillin; AMP, ampicillin; PEN, penicillin. mecA, methicillin resistance gene; scn, scn human immune evasion cluster gene. bW, Industrial Hog Operation (IHO) Worker; H, Household Member. *Participants 10 and 11 carried two different spa types. Other participants each carried a single spa type.
Receiver operating characteristic (ROC) curves were generated to compare the performance of different sampling scenarios needed to adequately characterize S. aureus carried intranasally. This analysis is commonly considered an effective approach for assessing the performance of diagnostic tests (here, sampling scenarios). The area under the ROC curve (AUC) is an index for measuring performance, with a larger AUC indicating a better performance (e.g., more accurate to determine the carrier status). If at least one of the tested isolates from an individual was positive for a particular gene or resistant to a particular antibiotic, we considered that person to be a true carrier of that specific characteristic. We generated three scenarios utilizing information from one, three, and five colonies, using the results from testing 10 colonies as a reference, and calculated the AUC for each sampling scenario. We considered an AUC <50% to correspond to random chance that an individual would be identified as a true carrier and an AUC >90% to indicate that the number of isolates tested was sufficiently accurate to determine the carrier status (25). ROC curves were drawn and AUCs were calculated using the “pROC” package (26) in the R statistical package, Version 3.2.1 (The R Foundation, Vienna, Austria). We were not able to generate ROC curves for the spa types because the data are not binary. Instead, we calculated positive probabilities to estimate whether testing a single isolate would have identified the dominant spa type.
Results
Ten S. aureus isolates were characterized from each of 19 participants (190 isolates total) including both persistent and intermediate carriers. For 17 of 19 (89%) participants, spa types of all 10 isolates were identical (Table 1). The remaining two participants carried two distinct spa types; but both had a predominant spa type and probability analysis suggests that testing one isolate would have been adequate in most cases. If we had tested one colony among our participants, the positive probability for identifying the dominant strain would have been 100% for 17 participants, 90% for one participant, and 80% for one participant, leading to an overall accuracy of 98%.
Variability in genes encoded on mobile genetic elements (mecA, scn) or antibiotic resistance profiles was observed among 11 (58%) of the 19 study participants (Figure 1). Four participants (21%) carried S. aureus that diverged in mecA presence. These four participants included two persistent carriers (one worker and one household member) and two intermittent carriers (one worker and one household member). Both workers had nine isolates that were positive for mecA and one isolate each that was negative. Both household members had one or two isolates positive for mecA and the remaining testing negative.
Five participants (26%) carried S. aureus that demonstrated variability in absence of scn, commonly used as a marker for livestock-association (Figure 1). These participants included four persistent carriers (two workers and two household members) and one intermittent carrier (a worker). Additionally, eight participants (42%) demonstrated variability in antibiotic resistance patterns (Figure 1). Variability of resistance patterns of intranasal S. aureus was again observed among both persistent and intermittent carriers (four participants each among persistent and intermittent carriers), and among both IHO workers and household members. Six workers and two household members showed variability in resistance patterns.
Using ROC curves, we were able to estimate the number of isolates needed to accurately determine carrier status for particular genes encoded on mobile genetic elements (mecA, scn) or phenotypic antibiotic resistance. Results suggest that testing five isolates from an individual's nasal swab would have been adequate to determine carriage of mecA, often used to define MRSA nasal carriage (Table 2). Three isolates would have been adequate to determine carriage of scn. Depending on the antibiotic assessed, different thresholds would have been needed to determine resistance (Table 2). For example, testing five isolates appeared necessary to determine clindamycin resistance, while one isolate would have been adequate to determine resistance to tetracycline, ciprofloxacin or levofloxacin. Individuals who carried isolates that diverged in resistance patterns tended to show divergence across multiple antibiotics. Participants 4, 9, 10, and 15 in particular carried isolates with variable and divergent antibiotic resistance patterns as demonstrated by the gray cells in Figure 1.
Table 2.
Area under the curve (AUC) values for Receiver Operating Characteristic (ROC) curves used to assess number of isolates needed to determine nasal carriage of S. aureus with particular characteristics.
| S. aureus characteristics | Area under the curve (%) | |||
|---|---|---|---|---|
| No. of isolates tested in sampling scenario | ||||
| 1 | 3 | 5 | ||
| Genes | mecA | 81.4 | 87.8 | 91.2* |
| scn | 88.0 | 93.4* | 96.4* | |
| Antibioticb | CIP | 100* | 100* | 100* |
| CEF | 80.5 | 91.5* | 95.8* | |
| CLI | 83.0 | 87.5 | 91.2* | |
| ERY | 86.0 | 89.5 | 92.3* | |
| GAT | 72.3 | 78.2 | 85.3 | |
| GENa | – | – | – | |
| LEV | 100* | 100* | 100* | |
| LZDa | – | – | – | |
| RAa | – | – | – | |
| SYN | 68.0 | 86.2 | 90.0* | |
| SXTa | – | – | – | |
| TET | 90.3* | 92.4* | 93.7* | |
| OXA | 69.8 | 81.2 | 84.2 | |
| AMPa | – | – | – | |
| PENa | – | – | – | |
An AUC >90% (*) indicated that the number of isolates tested was sufficient to capture the population diversity (spa types, genotypes, and drug resistance patterns) of S. aureus in tested nasal samples.
Antibiotics with a “–” denote that all of the collected isolates from each participant were the same so an AUC value could not be calculated.
Intermediate or complete resistance to: CIP, ciprofloxacin; CEF, ceftriaxone; CLI, clindamycin; ERY, erythromycin; GAT, gatifloxacin; GEN, gentamicin; LEV, levofloxacin; LZD, linezolid; RA, rifampin; SYN, quinupristin/dalfopristin; SXT, sulfamethoxazole with trimethoprim; TET, tetracycline; OXA, oxacillin; AMP, ampicillin; PEN, penicillin.
Discussion
In this study we demonstrated that genetic characteristics and antibiotic resistance patterns can vary among S. aureus isolated from a single nasal swab, although genotypes including spa types of S. aureus appear to be relatively stable per individual at a single point in time. Our results are consistent with previous studies that characterize S. aureus from human nares. In our study, two of 19 individuals (10.5%) carried two rather than one spa type of S. aureus. An earlier study of healthy individuals reported that up to 5.8% of 360 participants were co-colonized with different S. aureus spa types at any given time and that 18% were co-colonized at least once over a 2 year study period (19). Results appear similar even when other genotyping methods are employed. A study using pulsed field gel electrophoresis (PFGE) to evaluate strain diversity estimated that ~6.6% of colonized individuals would be expected to carry multiple genetic strains (18). Nasal colonization with different MRSA subtypes has also been observed, both in hospitalized human patients (27, 28), and in humans and animal patients during an outbreak that occurred as a veterinary hospital (17). This latter study concluded that multiple isolates from individuals need to be analyzed to better understand disease ecology and transmission networks. Our study further suggests that analysis of multiple isolates may be advisable to evaluate carrier status of particular characteristics of S. aureus.
Our study is the first to our knowledge to apply statistical modeling to determine the number of isolates required to determine the carrier status for particular S. aureus characteristics (i.e., genotypes, subtypes, or resistance profiles). This approach was effective in capturing the variation in S. aureus characteristics despite the limitation of a small sample size. Further, nesting of this study within a larger cohort study enabled us to choose participants based on prior knowledge of their carrier status. We were able to document that both persistent and intermediate carriers of S. aureus sometimes carry strains with different genetic characteristics or drug resistance patterns. These results can have implications from both a clinical perspective (e.g., deciding appropriate antibiotic treatment) and an epidemiological perspective (e.g., evaluating prevalence of particular strains in populations). Differences between individuals in this study were also notable, with some participants showing no variation in characteristics among tested isolates while other participants showed variation in multiple characteristics (gene markers or antibiotic resistance). Although it was beyond the scope of this study, it would be interesting to test whether these variations could be associated with host characteristics, environmental exposures, or other factors.
Our results suggest that the number of isolates necessary for surveillance studies depends on study objectives including what S. aureus characteristic is most relevant epidemiologically and/or clinically. The answer may vary depending on whether the goal is to determine spa type, MRSA carriage, livestock association, or clinical treatment based on antibiotic resistance profiles. Characterization of one isolate appears sufficient to accurately capture relatively stable characteristics of S. aureus among nasal carriers, particularly to identify the spa type of S. aureus among nasal carriage positive individuals. However, testing of additional isolates appears necessary for more plastic characteristics including the occurrence of genes encoded on mobile genetic elements (mecA, scn) routinely used to determine MRSA carriage and livestock association, respectively, and phenotypic antibiotic resistance profiles. Depending on the characteristic of interest, our analysis suggests that one to five isolates may need to be tested to accurately evaluate nasal carriage of S. aureus with these more plastic characteristics. It is concluded that by assuming S. aureus colonization by a single strain in the nares, surveillance studies that rely on testing one isolate may underestimate nasal carriage of these more variable S. aureus characteristics.
Ethics statement
The Johns Hopkins School of Public Health (JHSPH) Institutional Review Board approved this study (IRB00004608) and the University of North Carolina Non-Biomedical IRB approved reliance on the JHSPH IRB. Before participating, adults provided written informed consent. Written parental permission and informed assent were collected for participants seven to 17 years of age.
Author contributions
CH, JS, and MN contributed to the conceptualization and design of this study, and JS and CH were responsible for funding acquisition and project administration. T-TL and MN were actively involved in data collection, with JW joining them for data curation and analysis. All authors contributed to the writing and editing of this manuscript, and approved final submission.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Elizabeth Pierce and Jennifer Styles for their assistance in the laboratory, the community organizers with the Rural Empowerment Association for Community Help (REACH) who enrolled and followed the cohort, and all of the cohort participants.
Footnotes
Funding. This work was funded by NSF grant 1316318 as part of the joint NSF-NIH-USDA Ecology and Evolution of Infectious Diseases Program and by National Institute for Occupational Safety and Health (NIOSH) award K01OH010193. CH was supported by NIOSH award K01OH010193.
References
- 1.Fitzgerald JR. Livestock-associated Staphylococcus aureus: Origin, evolution and public health threat. Trends Microbiol. (2012) 20:192–8. 10.1016/j.tim.2012.01.006 [DOI] [PubMed] [Google Scholar]
- 2.Lowy F. Staphylococcus aureus infections. N Engl J Med. (1998) 339:520–32. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- 3.Holland TL, Arnold C, Fowler VG. Clinical management of Staphylococcus aureus bacteremia. JAMA (2014) 312:1330. 10.1001/jama.2014.9743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. (2004) 10:S122–9. 10.1038/nm1145 [DOI] [PubMed] [Google Scholar]
- 5.Chambers H, DeLeo F. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. (2010) 7:629–41. 10.1038/nrmicro2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization (WHO) . Antimicrobial Resistance: Global Report on Surveillance. Geneva, Switzerland (2014). Available online at: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf
- 7.Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis. (2008) 46:787–94. 10.1086/528716 [DOI] [PubMed] [Google Scholar]
- 8.Deleo FR, Chambers HF. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest. (2009) 119:2464–74. 10.1172/JCI38226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis. (2005) 11:1965–6. 10.3201/eid1112.050428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N, Beaujean D, et al. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis. (2007) 13:1834–9. 10.3201/eid1312.070384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nadimpalli M, Stewart JR, Pierce E, Pisanic N, Love DC, Hall D, et al. Livestock-associated, antibiotic-resistant Staphylococcus aureus nasal carriage and recent skin and soft tissue infection among industrial HOG operation workers. PLoS ONE (2016) 11:e165713. 10.1371/journal.pone.0165713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatcher SM, Rhodes SM, Stewart JR, Silbergeld E, Pisanic N, Larsen J, et al. The prevalence of antibiotic-resistant Staphylococcus aureus nasal carriage among industrial hog operation workers, community residents, and children living in their households: North Carolina, USA. Environ Health Perspect. (2017) 125:560–9. 10.1289/EHP35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun J, Yang M, Sreevatsan S, Bender JB, Singer RS, Knutson TP, et al. Longitudinal study of Staphylococcus aureus colonization and infection in a cohort of swine veterinarians in the United States. BMC Infect Dis. (2017) 17:1. 10.1186/s12879-017-2802-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahibzada S, Hernandez-Jover M, Jordan D, Thomson PC, Heller J. Emergence of highly prevalent CA-MRSA ST93 as an occupational risk in people working on a pig farm in Australia. PLoS ONE (2018) 13:e0195510. 10.1371/journal.pone.0195510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis MF, Rankin SC, Schurer JM, Cole S, Conti L, Rabinowitz P, et al. Checklist for One Health Epidemiological Reporting of Evidence (COHERE). One Health (2017) 4:14–21. 10.1016/j.onehlt.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sollid JUE, Furberg AS, Hanssen AM, Johannessen M. Staphylococcus aureus: determinants of human carriage. Infect Genet Evol. (2014) 21:531–41. 10.1016/j.meegid.2013.03.020 [DOI] [PubMed] [Google Scholar]
- 17.Paterson GK, Harrison EM, Murray GGR, Welch JJ, Warland JH, Holden MTG, et al. Capturing the cloud of diversity reveals complexity and heterogeneity of MRSA carriage, infection and transmission. Nat Commun. (2015) 6:1–10. 10.1038/ncomms7560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cespedes C, Said-Salim B, Miller M, Lo SH, Kreiswirth BN, Gordon RJ, et al. The clonality of Staphylococcus aureus nasal carriage. J Infect Dis. (2005) 191:444–52. 10.1086/427240 [DOI] [PubMed] [Google Scholar]
- 19.Votintseva AA, Miller RR, Fung R, Knox K, Godwin H, Peto TEA, et al. Multiple-strain colonization in nasal carriers of Staphylococcus aureus. J Clin Microbiol. (2014) 52:1192–200. 10.1128/JCM.03254-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stegger M, Liu CM, Larsen J, Soldanova K, Aziz M, Contente-Cuomo T, et al. Rapid differentiation between livestock-associated and livestock- independent Staphylococcus aureus CC398 clades. PLoS ONE (2013) 8:e79645. 10.1371/journal.pone.0079645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stegger M, Andersen PS, Kearns A, Pichon B, Holmes MA, Edwards G, et al. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin Microbiol Infect. (2012) 18:395–400. 10.1111/j.1469-0691.2011.03715.x [DOI] [PubMed] [Google Scholar]
- 22.European Union Reference Laboratory for Antimicrobial Resistance Protocol for spa typing. (2009). Available online at: http://www.crl-ar.eu/data/images/tc_april-2009/7-protocols for spa typing.pdf
- 23.CLSI (Clinical and Laboratory Standards Institute) M100-S23: Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. Wayne, PA (2013). Available online at: http://www.facm.ucl.ac.be/intranet/CLSI/CLSI-M100S23-susceptibility-testing-2013-no-protection.pdf
- 24.Steward CD, Raney PM, Morrell AK, Williams PP, McDougal LK, Jevitt L, et al. Testing for induction of clindamycin resistance in erythromycin-resistant isolates of Staphylococcus aureus. J Clin Microbiol. (2005) 43:1716–21. 10.1128/JCM.43.4.1716-1721.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swets JA. Measuring the accuracy of diagnostic systems. Science (1988) 240:1285–93. 10.1126/science.3287615 [DOI] [PubMed] [Google Scholar]
- 26.Robin X, Turck N, Hainard A, Tiberti N, Frederique L, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics (2011) 12:77. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lima DF, Cohen RWF, Rocha GA, Albano RM, Marques EA, Leão RS. Genomic information on multidrug-resistant livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolated from a Brazilian patient with cystic fibrosis. Mem Inst Oswaldo Cruz (2017) 112:79–80. 10.1590/0074-02760160342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lauderdale TLY, Wang JT, Lee WS, Huang JH, McDonald LC, Huang IW, et al. Carriage rates of methicillin-resistant Staphylococcus aureus (MRSA) depend on anatomic location, the number of sites cultured, culture methods, and the distribution of clonotypes. Eur J Clin Microbiol Infect Dis. (2010) 29:1553–9. 10.1007/s10096-010-1042-8 [DOI] [PubMed] [Google Scholar]


