Figure 4. Effect of Nup116 GLFG-repeats on cell fitness and the connectivity of Nup116 with scaffold Nups. See also Figures S3–S4.
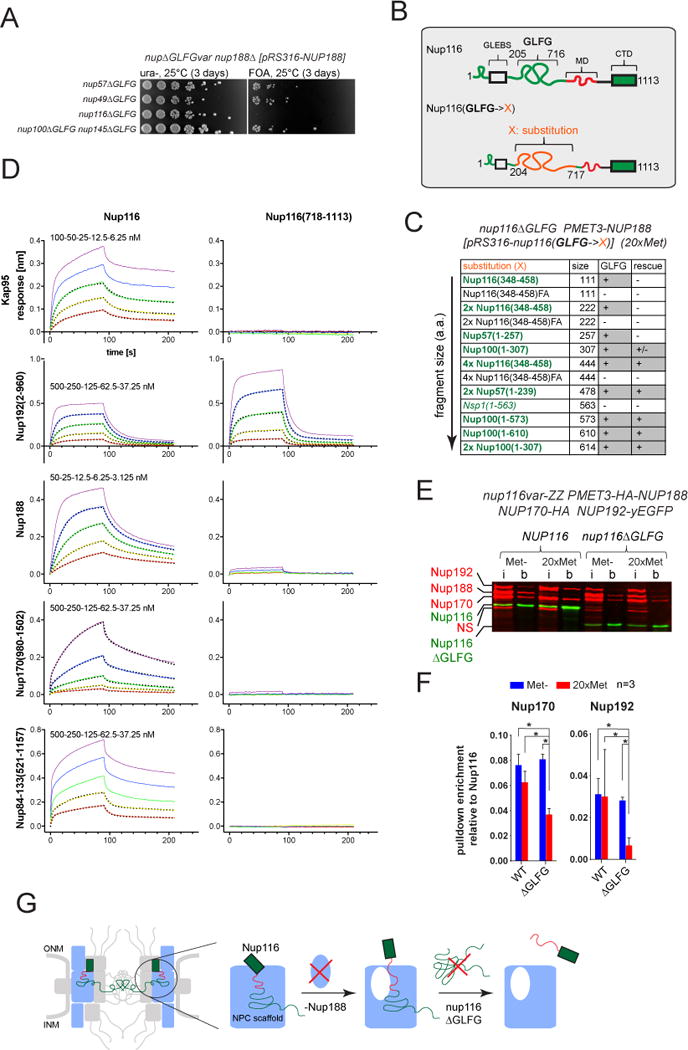
(A) Deletion of NUP188 is synthetically lethal in combination with the deletion of Nup116 GLFG-repeats (nup116ΔGLFG) but not with any other GLFG-repeats (nup57ΔGLFG, nup49ΔGLFG or nup100ΔGLFG nup145NΔGLFG).
(B) Outline of GLFG-repeat segment substitutions in Nup116.
(C) Growth rescue of PMET3-NUP188 nup116ΔGLFG strain by ectopically expressing Nup116 GLFG-repeat substitutions upon repression of Nup188. “+”, “+/−” and “−” - complete rescue, partial rescue and no rescue of cell growth, respectively. See also Figure S3B.
(D) Representative BLI curves showing association and dissociation kinetics for various scaffold Nups and Kap95 with the respective immobilized Nup116 variants. Curves were corrected for buffer background. Five two-fold dilution series of analyte were used for each experiment (purple, blue, green, yellow, red). The highest concentration (purple) was 50 nM (Nup188), 100 nM (Kap95) and 500 nM (Nup84-133, Nup170, Nup192). Dotted lines show fitted curves after global fitting analysis. See also Figures S4A–B.
(E and F) Effects of GLFG-repeats and Nup188 depletion on the connection of Nup116 with scaffold Nups. PMET3-NUP188 strains expressing tagged scaffold Nups (NUP170-HA and NUP192-yEGFP) and either of the ZZ-tagged Nup116 variants (NUP116-ZZ or nup116ΔGLFG-ZZ) were incubated for 12h in Met- or 20xMet media prior to processing for affinity pulldowns with IgG-Dynabeads. (E) Representative Western blot image used to quantify protein amounts in the input (i) and IgG-Dynabeads bound (b) fractions. (NS) – a protein cross-reacting with anti-HA-tag antibody. (F) Plot showing Nup116 co-purification efficiencies of Nup170 and Nup192 based on the corresponding band intensities as (b/i ratio of prey)/(b/i ratio of Nup116). Mean +/− SD from three independent experiments. Asterisks (*) – significant differences, Student t-test p-values (< 0.01). See also Figures S4C–D.
(G) Model describing the connectivity function of Nup116 GLFG-repeats in the absence of Nup188.
