Abstract
The transport of messenger RNAs (mRNAs) from the nucleus to cytoplasm is an essential step in the gene expression program of all eukaryotes. Recent technological advances in the areas of RNA-labeling, microscopy, and sequencing are leading to novel insights about mRNA biogenesis and export. This includes quantitative single molecule imaging (SMI) of RNA molecules in live cells, which is providing knowledge of the spatial and temporal dynamics of the export process. As this information becomes available, it leads to new questions, the reinterpretation of previous findings, and revised models of mRNA export. In this review, we will briefly highlight some of these recent findings and discuss how live cell SMI approaches may be used to further our current understanding of mRNA export and gene expression.
Keywords: mRNA export, RNA-binding protein, nuclear pore complex, in vivo single molecule imaging, MS2-MCP system, PP7-PCP system
Introduction
The efficient and timely delivery of mRNAs to the cytoplasm for translation is essential to gene expression and cellular function. This importance is reflected in the mutation of the nuclear transport machinery in human disease (e.g. cancers, developmental and neurological disorders) and targeting of this machinery by viruses to promote viral replication and survival [1–5]. Studies of mRNA export over the past several decades have provided a detailed list of components (e.g. cellular machines, proteins and small molecules) required for this process, yet it remains to be understood how these factors function in space and time to facilitate and regulate export. This is in large part due to the challenges of studying the spatial and temporal dynamics of this process. However, these types of data are essential to understanding the flux of mRNAs across the nuclear envelope to match cellular demand and how reprogramming of this process by mutation could lead to disease. Excitingly, the development of SMI strategies and powerful sequencing methodologies has now made it possible to capture the dynamics of this process and to address key questions related to mRNA biogenesis and export.
Visualization of mRNAs in vivo reveals their spatiotemporal distribution and transport kinetics
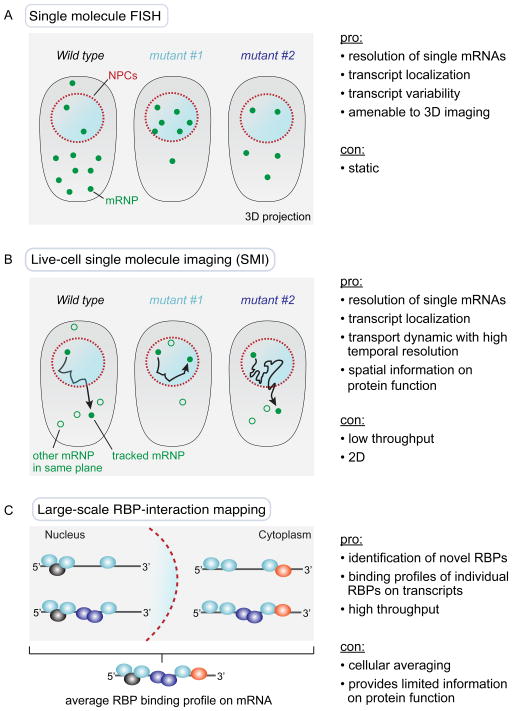
In Saccharomyces cerevisiae, it is estimated that ~60,000 mRNA molecules are present per cell [6]. Given a median transcript half-life of 11–18 min [7, 8], approximately 2,000–3,000 transcripts have to be transported out of the nucleus per minute. Taking into account a density of 60–200 nuclear pore complexes (NPCs) per yeast nucleus [9], this translates to around 10–50 mRNA export events per NPC each minute. A major bottleneck towards studying export events has been the lack of ‘sensitive’ assays to monitor where and when export occurs and to define where in the process a given export factor functions. For example, the commonly used oligo(dT) in situ hybridization assay allows bulk poly(A)+ RNA to be visualized and has led to the identification of almost all known export factors; however, all these mutants exhibit very similar phenotypes, which is the accumulation of poly(A)+ RNA in one or more compartments of the nucleus [10–14]. To step away from such bulk measurements, single transcript analyses using single molecule fluorescence in situ hybridization (smFISH) have more recently been undertaken. smFISH can reveal ‘snap-shot’-like information on transcript localization, transcript abundance and cell-to-cell variability [6, 14–16], but in vivo dynamics, such as nuclear export kinetics or transport directionality, cannot be measured (Figure 1A). To overcome these limitations, recent advancements in RNA labeling and imaging technology have allowed various groups to address the dynamics of export in vivo [17–20]. Importantly, this has provided an opportunity to spatially and temporally characterize nuclear biogenesis and export in live cells at the level of individual mRNAs (Figure 1B).
Figure 1.
Single molecule techniques can overcome limitations in understanding cellular mRNP function obtained from ensemble composition data from large-scale sequencing and proteomic approaches. A: Single molecule fluorescence in situ hybridization (smFISH) can reveal localization, abundance and cell-to-cell variability of individual mRNPs, but is a static method that does not allow visualization of in vivo dynamics, such as nuclear export kinetics or transport directionality of single transcripts. B: Single molecule imaging (SMI) enables a dynamic profiling of individual transcripts in vivo and provides spatial information on RBP function with high temporal resolution. C: Large-scale approaches analyzing RBP binding to mRNA transcripts are based on numerous data points that are compiled into an ensemble average, which may not be representative of individual mRNP composition in vivo. For example, the binding profile of an RBP may differ substantially across transcripts from different genes and/or transcripts residing in different cellular compartments.
Imaging of single RNAs in a living cell is made possible by the introduction of sequence tags into the transcript. A common strategy is based on the sequence-specific interaction of a fluorescently labeled bacteriophage coat protein (CP) with specific RNA binding sites (reviewed in [21]). This approach was initially established using protein and RNA components from the phage MS2 [22, 23]. An orthologous system has also been recently described from the phage PP7 [24, 25]. PP7 and MS2 retain binding specificity when combined in vivo, allowing even for double labeling of individual transcripts [26], which was recently employed to monitor translation in vivo [27]. Other commonly used mRNA labeling systems include lambdaN/boxB [28], and the U1A system [29,30].
To observe mRNA transport from the nucleus to the cytoplasm the MS2 or PP7 systems have been employed in Saccharomyces cerevisiae, the fly Chironomus tentans, mouse and human cells [17–20, 31, 32] (Summarized in Table 1, and reviewed in [33]). Whereas some studies observe similar mRNA behaviors (e.g. mRNA retention close to NPCs [20, 32]), the reported NPC transit times vary by several orders of magnitude. Although these numbers are likely dependent on the transcript and organism characterized, differences in image acquisition strategies and data interpretation have also been pointed out as a source of discrepancy [34, 35]. Further application of these imaging approaches is now required to clarify these differences and to address unresolved questions about directional mRNA export. In the following sections we discuss some of the major questions and highlight how SMI technologies can be used and/or adapted to answering such questions in vivo. For additional discussions of mRNA transport kinetics the reader is directed to a number of other recent reviews [33, 35–37].
Table 1.
Reported mRNA dynamics in live cells.
| Cellular system | mRNA(s) | mRNA size without SL | Labeling system | Average pore transit times | Spatial precision (nm) | Temporal precision (ms) | Publications |
|---|---|---|---|---|---|---|---|
| H. sapiens | Dystrophin * | 4.8 kb | MS2-MCP | 0.5 s ** | n.d. | 1000 | [18] |
| M. musculus | β-actin | 1.9 kb | MS2-MCP | 180 ms | 26 | 20 | [17] |
| C. tentans | All hrp36-containing RNAs | various | hrp36 | 20 s | 10 | 20 | [19] |
| H. sapiens | β-actin (1), Firefly luciferase (2) | 3.3 kb (1,2) | MS2-MCP | 12 ms (1) 11 ms (2) |
8 | 2 | [31] |
| S. cerevisiae | GFA1 | 2.2 kb | PP7-PCP | 188 ms*** | 56 | 15 | [20] |
| S. cerevisiae | MDN1 (1), GLT1 (2), CLB2 (3) | 14.7 kb (1) 6.4 kb (2) 1.5 kb (3) |
PP7-PCP | n.d. | n.d. | 37 | [32] |
truncated ½-mini-Dystrophin construct
estimate, limited by temporal resolution
dwell time analysis; 215 ms with maximum likelihood estimate
n.d. not determined
Nuclear mRNP maturation: What determines the path taken?
During mRNA biogenesis, a transcript dynamically associates with various RNA-binding proteins (RBPs) to form an RNA–ribonucleoprotein particle (RNP). The repertoire of interacting proteins is responsible for directing RNA processing and maturation, which ultimately determines export competency and contributes to the cytoplasmic fate of the mRNP [38, 39]. Towards addressing questions of mRNP architecture, recent large-scale sequencing and proteomic approaches have provided important insights into mRNP composition on a global scale [40–44]. These studies identified hundreds of proteins that were not known to bind RNA, revealed many RBPs that do not have recognized RNA-binding domains, provided many examples of well-studied proteins (e.g. metabolic enzymes) binding RNA, and in some cases mapped sequence-specific binding sites [40, 43–45]. For example, Tuck and Tollervey [44] analyzed the binding patterns of individual RBPs important for mRNA export in Saccharomyces cerevisiae, which revealed protein-specific binding patterns on sets of transcripts, symmetric and asymmetric protein binding distributions along transcripts, and illustrated preferences of RBPs for spliced versus unspliced mRNAs.
While these large-scale approaches have provided important insights into RNA biology by identifying hundreds of RBPs, in most cases the transcript binding preferences, stoichiometry, and function are not known. Moreover, in the vast majority of cases these data do not capture the temporal changes an mRNP undergoes as a result of moving through different stages of biogenesis or between different subcellular compartments. As a result, the RNA-binding profile for each RBP reflects interactions occurring across multiple stages of mRNA biogenesis (i.e. an ensemble average), which may encompass multiple functions and binding interactions due to changes in mRNP architecture (Figure 1C). For example, some RBPs transiently interact with the mRNA only within the nucleus or cytoplasm, and others accompany the mRNA from the nucleus to cytoplasm [46]. Once in the cytoplasm, some RBPs are removed immediately after transition through the NPC, such as the export factor Mex67 [47], while others, like the nucleo-cytoplasmic shuttling RBP Npl3, accompany mRNA transcripts for a longer time in the cytoplasm and may be further involved in cytoplasmic events such as translation [48, 49]. Consequently, complementary approaches must now be employed to investigate how individual factors contribute to the spatial and temporal biogenesis and transport of mRNA. Live cell SMI is one such approach, with two recent publications reporting changes in the interaction of mRNPs with the nuclear basket and altered export kinetics in RBP mutants [20, 32]. This suggests that SMI approaches can identify spatial and temporal mRNA transport defects, which is a powerful means of inferring the subcellular function of RBPs in the cell on any individual mRNA (Figure 1B).
Besides the canonical mRNA export pathway discussed here, it is important to note that alternative routes for mRNP delivery to the cytoplasm have also been described. This includes, among others, eIF4E/CRM1-dependent transport of mRNA, nuclear budding of mRNA, or viral subversion of canonical protein export routes for viral RNA export. In these cases, the dynamics of export and RBP-dependencies are not well understood, but could be addressed using the technologies discussed here. We refer the reader to recent reviews on alternative RNA export mechanisms for more information [50–52].
Regulating gene expression: A role for the nuclear periphery and NPCs?
Upon maturation and becoming export competent through the acquisition of specific RBPs, an mRNP will be exported from the nucleus. Although the detailed composition of an mRNP arriving at the NPC is not clear, two factors that are thought to bind the majority of mRNAs in the nucleus prior to export are the poly(A)-binding protein Nab2 and the Mex67-Mtr2 heterodimer [42, 44, 53]. Nab2 aids nuclear export by interacting with the RNA itself, mRNA export factors and NPC components, while mRNP passage through an NPC is enabled by Mex67-Mtr2 [54–57]. Mex67-Mtr2 is recruited to fully processed (capped, spliced, polyadenylated) transcripts, possibly serving as a final ‘checkpoint’ before passage through the NPC is allowed [40, 44]. Mex67-Mtr2, together with other factors of the mRNA export machinery, have also been shown to be recruited to the site of transcription, potentially synchronizing the completion of transcription and mRNA biogenesis with the acquisition of export competency [58]. It has been observed in Saccharomyces cerevisiae and mammalian systems that when an mRNP reaches the nuclear periphery, in addition to undergoing export, it can scan along the nuclear periphery [17, 20, 32]. This scanning process remains poorly understood and has so far only been reported for a few transcripts. However, it does raise questions as to why some mRNPs are exported immediately and others are not, which may involve differential Mex67-Mtr2 binding and the acquisition of export competency. Scanning may also be indicative of other changes in mRNP composition that need to occur prior to NPC translocation and/or quality control events that have been linked to the nuclear periphery and NPCs [59–61]. By characterizing different classes of transcripts (e.g. spliced vs. non-spliced) with specific mutants (e.g. NPC components and RBPs), SMI can be employed to determine the requirements for scanning at the level of the transcript, RBPs, and processes that are supported by nuclear scanning.
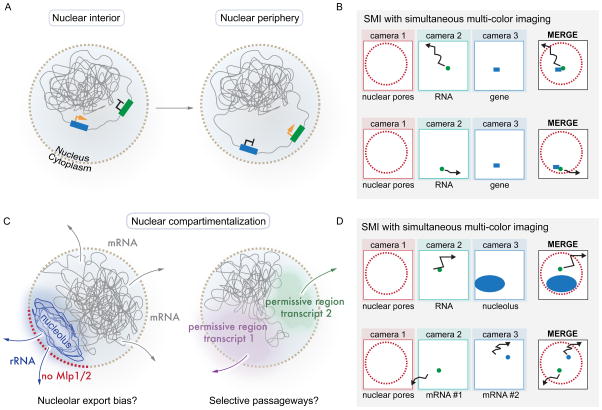
As another means to influence gene expression, genes can be positioned close to NPCs at the nuclear periphery (e.g. the GAL locus in yeast [62]) upon transcriptional activation [63–66] (Figure 2A, left panel). This has been referred to as ‘gene-gating’ and was suggested to reduce the distance between the site of transcription and the site of export, and in turn improve mRNA export efficiency [67]. If gene movement to the nuclear periphery improves mRNA export kinetics or even imposes an order of export among different classes of transcripts remains unknown. It should be pointed out that -at least in cells with smaller nuclear volumes- the process of mRNA export does not appear to be a kinetically rate-limiting step in gene expression as diffusion from the site of transcription and NPC translocation occurs in the subsecond range with an individual mRNP transiting through the pore in ~200 ms [17, 19, 20, 31]. By contrast, to produce an averaged sized protein in yeast, transcription and translation operate on the time scale of minutes. It is also debatable if gene activation through peripheral targeting is a general feature of gene expression, as other studies ascribe nuclear periphery recruitment to gene silencing [68, 69] or dampened gene expression [70]. Furthermore, during logarithmic growth few genes stably interact with the nuclear periphery, whereas changes in carbon source do not only recruit the GAL locus to the NPC but invoke global alterations in chromatin organization, which require both acetylation and deacetylation activities [71]. Consequently, it remains a challenge to ascribe changes in gene expression to any of these events. This suggests that gene positioning close to the nuclear periphery might be a mechanism to regulate and coordinate gene expression or to spatially organize the genome, rather than to enhance the kinetics of the mRNA export process. Importantly, as depicted in Figure 2A (right panel), SMI offers the ability to test these models using RNA-labeling strategies in combination with multi-camera approaches that report on the location of a gene locus (e.g. by employing the LacO/LacI system [72]).
Figure 2.
Regulating gene expression: A role for the nuclear periphery and NPCs? A: Transcriptional activation (blue with yellow arrow) and silencing (green with black arrow) have both been associated with peripheral gene positioning. The molecular basis for such chromatin rearrangements and the local environment this places a gene in to alter gene expression remains largely unknown, as does the impact gene tethering has on mRNA export. B: SMI techniques using multi-camera setups can determine the dynamic position of a transcript relative to its gene locus in different cellular conditions. C: Genome organization may also affect the transport routes taken by different classes of RNAs. rRNA export might be biased to NPCs associated with the nucleolus (blue), and mRNA export even disfavored, due to the different biophysical properties of the nucleolus or function of nearby NPCs (exemplified by the lack of Mlp1/2 in NPCs close to the nucleolus). Other genomic regions might play a similar role by facilitating local environments that select for distinct mRNPs. D: SMI can unravel such gene expression features through differential labeling of sub-nuclear structures and/or different classes of mRNAs.
Compartmentalization of the nucleus may provide a further means to influence mRNA export and regulate gene expression. As depicted in Figure 2B (left panel), NPCs adjacent to the yeast nucleolus, which is the site of rRNA transcription and pre-ribosome assembly, lack Mlp1 and Mlp2 [73], components of the NPC nuclear basket. Mlp1 has been shown to directly interact with the polyA-binding protein Nab2 [54], and absence of Mlp1 may prevent Nab2-dependent mRNA export through this subset of nucleolar-associated NPCs. In support of this, studies have shown that nucleolar regions appear to exclude mRNPs [18, 32, 74]. This local exclusion might reflect the existence of nuclear regions and specialized NPCs that favour the export of specific cargos (e.g. rRNA). In support of dedicated transport routes, mRNPs have been shown to move randomly, but discontinuously (or corralled) within the nucleus, possibly due to stalled movements in chromatin-rich regions, or due to distinct nuclear ‘paths’ that actively guide RNPs to their site of export [18, 75](Figure 2B, left panel). Such spatial preferences and their potential regulation have not yet been substantiated in vivo, but could be tested using SMI to compare export routes of rRNAs to routes taken by mRNAs (Figure 2B, right panel). Such experiments would require improved RNA labeling and imaging strategies to follow RNA particles over longer time periods and in the z-direction. Notably, recent advances in 3D image acquisition and particle tracking (see below) are making such approaches feasible [76, 77].
Directional export: Where and how does Dbp5 function?
Unlike protein transport, most mRNA transport is not directly dependent on the beta-karyopherin family or on the RanGTP gradient [78]. Instead, cytoplasmic mRNP remodeling driven by the DEAD-box ATPase Dbp5 (DDX19), together with Gle1, Nup159 (NUP214) and the small molecule co-factor inositol hexakisphosphate (InsP6), are thought to determine directional mRNA export [79–87]. This is predicted to occur due to changes in mRNP structure (i.e. remodeling) catalyzed by Dbp5 ATPase activity, which leads to loss of RBPs required for transport through an NPC (e.g. Mex67) (reviewed in [88] and [89]). Dbp5 is also highly dynamic at NPCs, binding the cytoplasmic side for ~55 ms [19], which is in the same range as mRNP residence time at the cytoplasmic side of the NPC (~80–95 ms) [17,19]. This suggests that Dbp5 cycles on and off pores to complete each mRNP remodeling event and/or travels with the mRNA. The latter possibility is supported by the fact that Dbp5 is a nuclear shuttling protein [79], interacts with transcriptional machinery [90], and is found associated with mRNPs near transcription sites and within the nucleoplasm [91].
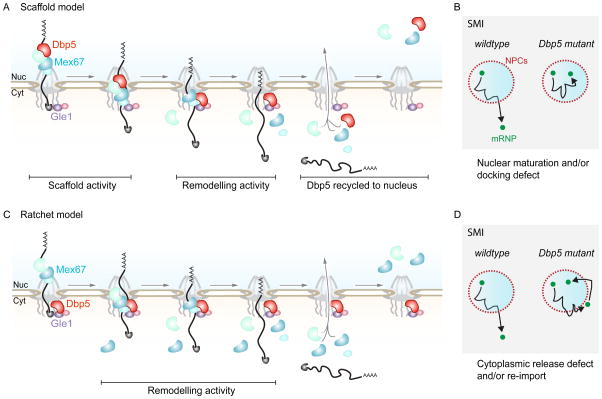
In a Dbp5 ‘scaffold’ model, as depicted in Figure 3A (left panel), nuclear shuttling would facilitate Dbp5 recruitment to the mRNA in the nucleus to generate a platform for other proteins to bind, which would persist as the mRNP travels to and through an NPC [92]. On the cytoplasmic side of the NPC, due to the presence of regulators (e.g. Nup159 and Gle1:InsP6), ATP hydrolysis and/or mRNA release would be promoted causing Dbp5 and other proteins to be displaced from the mRNA. This would lead to the removal of specific nuclear export factors from the mRNP, thereby terminating mRNA export. Alternatively, a ‘Brownian ratchet’ model has been proposed (Figure 3B, left panel), in which Dbp5 waits at the cytoplasmic site of the NPC for a translocating mRNP and continuously remodels it through regulator-stimulated ATPase activity, thereby driving directional export [93]. In the ratchet model, export factors are distributed along the transcript to allow step-wise remodeling by Dbp5 and prevent backsliding of the mRNA into the nucleus. This is supported by studies on global mRNP composition showing that export factors such Mex67 bind across the length of an mRNA with no apparent sequence specificity [40, 44]. In addition, computational modeling of mRNA export emphasized that a symmetrical distribution of export factors along the transcript strongly correlates with successful transport [34]. This model also suggested that mRNA export is very sensitive to the number of export receptors on the mRNA, and that one export factor per mRNA might not be sufficient to facilitate transport. Inherent to each of these models of Dbp5 activity are predictions about where in the cell Dbp5 binds an mRNA and what defect would result, which can be tested with SMI approaches. For example, if the scaffold model is correct, transcripts may not dock to the nuclear basket of NPCs or ever leave the nucleus due to failures in mRNP maturation (Figure 3A, right panel), whereas in a Brownian ratchet model the mRNA would dock and transit through the NPC, but show release and/or directional transport defects due to failures in mRNP remodeling (Figure 3B, right panel). Of course, it is possible that Dbp5 functions in multiple ways to facilitate export and that this may in part depend on the transcript being exported. For instance, one pathway may function preferentially on large transcripts or to bias export of specific transcripts in response to environmental stimuli (e.g. stress), which can also be tested in an SMI approach.
Figure 3.
Models of Dbp5-dependent directional mRNA export. A: Schematic of a ‘scaffold’ model of Dbp5 dependent export. Dbp5 is recruited to the mRNA in the nucleus and incorporated into the mRNP to generate a platform for other proteins to bind. Dbp5 travels through the pore with the mRNP and upon reaching the cytoplasmic side of the NPC remodels the mRNP though its ATPase activity due to the presence of regulators (e.g. Gle1:InsP6 and Nup159). Dbp5 is then recycled back to the nucleus. B: Using SMI techniques, individual mRNPs would be expected to show nuclear maturation and/or docking defects in the ‘scaffold’ scenario. C: Schematic of a ‘Brownian ratchet’ model of Dbp5 dependent export. Dbp5 waits at the cytoplasmic side of the NPC for a translocating mRNP with regulators (e.g. Gle1:InsP6 and Nup159) and continuously remodels the mRNP through its ATPase activity. D: In contrast to the ‘scaffold’ model, individual mRNPs in the ‘ratchet’ scenario would be expected to show cytoplasmic release defects and/or mRNP re-import using SMI.
Beyond Dbp5, post-translational modifications (PTMs) could also contribute to directional mRNA export. PTMs provide the advantage of a fast response to environmental changes and can rapidly modulate protein-protein and protein-RNA interactions in a reversible manner (reviewed in [94] and [95]). The phosphorylation-dephosphorylation cycle of the export factor Npl3 was one of the first PTMs associated with the establishment of transport directionality [96, 97]. Another prevalent modification found in many RNA binding proteins, including Npl3, is arginine methylation by type I methyltransferases [98]. These modifications have been implicated in regulating transport by weakening contacts between proteins, thereby contributing to mRNP remodeling and potentially imposing directionality [94]. Alternatively, PTMs might not be essential for mRNP formation and export, but rather serve to differentiate the free RBPs from mRNA-bound RBPs [99]. The application of sequencing technologies to the identification of RNA modifications has also lead to the cataloguing of abundant PTMs on mRNA, including N6-methyladenosine (m6A), 5-methylcytosine (m5C), and pseudouridine (Ψ) [100–102]. In these cases, SMI could be employed to address how mutations in protein or RNA modification sites alter mRNA transport kinetics.
mRNA transit through NPCs: Is there extensive mRNP remodeling?
Transit through the central channel of the NPC (~50 nm in length) has been suggested to require mRNP remodeling. This notion stems from observations in Chironomus tentans salivary glands of the ~40 kb Balbiani Ring (BR) mRNA undergoing export [103]. By EM, large BR mRNPs (~25 nm wide and ~135 nm long) are observed to unfold into elongated fibrils to transit through an NPC. A similar unfolding/remodeling process has also been proposed for the large human Dystrophin mRNP [18]. In contrast, Saccharomyces cerevisiae mRNPs have been described as being much smaller (5 nm wide, 20–30 nm long) [53] and most mRNPs might therefore fit through an NPC without extensive remodeling. Yeast mRNPs with an average mass of 2 MDa [104–106] are similar in size to eukaryotic ribosomal subunits, for which no remodeling within NPCs has been described [107]. It also remains to be tested if transcript identity defines the “degree” to which mRNPs are remodeled, with longer mRNA substrates requiring more remodeling and longer transport times, as suggested by a mathematical model of mRNA transport [34]. Analysis of NPC transport times for individual β-actin transcripts in mouse cells suggested that transit through the channel is not the rate-limiting step. Instead, events close to the nucleo- and cytoplasmic face of the NPC were observed to contribute the most to overall export times [17]. If this is a general feature of mRNA export or changes in relation to mRNP size remains to be tested, but this could be addressed by selecting transcripts of various sizes for SMI imaging of export dynamics.
Besides mRNAs, other RNA and protein cargos undergo transport through NPCs and it remains poorly understood how these concurrent processes are spatially and temporally coordinated. For example, it is possible that mRNAs share transport routes and compete with other cargos. Alternatively, cargo-specific paths within NPCs and/or specialized NPCs may exist to facilitate transport of select cargos. However, work to date has only studied a few cargos in vivo and the technical challenges of imaging these fast processes with high spatial precision requires much more work to be done [35]. However, we expect that multi-color SMI of different cargo types, in concert with transport receptors or NPC components, will help elucidate how various transport processes are coordinated within and across NPCs.
mRNP export and translation: Are they coupled?
Where and when the first round of translation occurs in relation to NPCs and the completion of export is not well understood. Towards addressing this question, an RNA biosensor was recently developed that could visualize the pioneer round of translation in live Drosophila oocytes, showing that the translation event first occurs within minutes of export for the transcript tested [27]. Moreover, the authors showed that a subpopulation of their reporter construct diffused several micrometers away from the nucleus before being translated, indicating that, at least in Drosophila oocytes and for this transcript, the first round of translation does not need to be spatially confined to NPCs or the immediate vicinity. However, EM images have shown that BR mRNAs in the process of export are often oriented to export 5′ cap first [108] and the cytoplasmic portion of the exporting mRNP can be associated with ribosomes [103], which suggests that the mRNA can associate with translation machinery during the export process. Whether translation plays a role in export and mRNP remodeling of this extremely large mRNP or whether it is a result of slow transport times is not known. In yeast, export factors are not asymmetrically distributed over mRNA transcripts in a manner that would explain a mechanism for asymmetric export [40, 44]. Furthermore, during the pioneer round of translation, mRNAs are subject to quality control to ensure upstream biogenesis events have successfully completed [109]. Key to the surveillance process is the presence of factors in the mRNP that were loaded in the nucleus (e.g. exon-junction complex), which interact with cytoplasmic components to provide an opportunity for crosstalk between nuclear and cytoplasmic processes for the purposes of quality control. Whether certain aspects of quality control are linked to NPCs and export is an open question. Consequently, SMI approaches could be applied to transcripts of different types, including those subject to quality control (e.g. non-sense mediated decay), in different mutant backgrounds to understand the links between export, translation, and RNA quality control activities.
mRNP visualization: What improvements are needed?
SMI requires a sufficient signal to noise ratio (SNR) to both detect and track particles [110]. Current mRNA labeling approaches suffer from low SNR when used to follow dynamic processes due to the requirement of short exposure times (~10–20 ms) and photobleaching over an imaging series. Consequently, 6 – 24 repeats of the MS2- or PP7-specific RNA hairpins are commonly introduced into an mRNA to achieve the required SNR. Both MS2 and PP7 bind as dimers, therefore up to 48 FP-CP can bind to an array of 24 RNA hairpins, adding significant protein mass to the labeled mRNP. This must be taken into consideration, in addition to the location of the hairpins in the transcript, to ensure that the mRNP retains its functionality. With such proper characterization, transcripts have been identified that when tagged show no differences in abundance or functionality [20, 26]. However, recent publications have reported that the presence of multiple MS2 hairpins can lead to the stabilization of MS2-containing decay fragments [111–113], which may act as false-positive signals in imaging experiments.
To increase SNR and spatial detection precision, and in turn reduce the number of hairpins needed to visualize the transcript, fluorophores with improved brightness and photostability are an attractive labeling alternative. Examples include fluorescent proteins such as mNeonGreen [114] or dye-based labeling systems [115–118]. However, these optimized probes are still fluorescent even if not bound to the RNA hairpins, which can lead to substantial background fluorescence. Labeling strategies that bypass undesired background fluorescence include dye-binding aptamers, which only become brightly fluorescent when the dye is in complex with RNA. A range of dye-aptamer combinations has been developed in recent years (Spinach [119], Spinach2 [120], Broccoli/dBroccoli [121], and Mango [122]. So far only the Spinach/Spinach2 system has been applied to eukaryotic RNA imaging, although it did not allow for single molecule detection [120, 123]. Fluorescence complementation systems such as bimolecular fluorescence complementation (BiFC) or dimerization-dependent FPs provide another mRNA labeling alternative that strongly reduce fluorescent background signal [124, 125]. However, such complementation systems exhibit unfavorable dissociation constants (KD >10 μM), as well as slow folding and maturation rates, which restricts mRNA visualization to the cytoplasm. In addition, BiFC fragment fusion is irreversible, with fusion products potentially exceeding mRNA transcript lifetime and therefore complicate single mRNA particle tracking and introduce background fluorescence. A recent development in mRNA labeling involves the CRISPR/Cas9-dependent tagging of RNA [126]. In this approach, the RNA itself does not harbor a genetically encoded tag, but a nuclease-inactive fluorescently labeled Cas9 can bind to the RNA in a sequence-specific manner and thus allow endogenous RNA labeling. This approach has the advantage that the RNA does not require modification and would allow visualization of endogenous transcripts, although this technique has so far only been applied to bulk mRNA localization studies and needs further development in order to track single mRNPs.
In addition to RNA labeling strategies, advancements in imaging technology have been instrumental to the imaging of mRNPs in vivo with high spatial precision and temporal resolution. These developments were critical since discrete steps within many RNA-related processes take place in the millisecond range, but the complete event can last seconds or more. Therefore, single particle imaging systems have to balance imaging speed, sufficient SNR to reliably detect particles over background, and illumination intensities that minimize photobleaching and phototoxic effects over time. Specialized single particle microscope systems that were developed to meet these criteria include widefield [17, 20, 31] and light sheet microscopy solutions [19]. The latter shows improved SNR, reduced phototoxicity and increased 2D and 3D resolution compared to conventional widefield or confocal microscopy [127]. Temporal resolution with these systems ranged from 2 ms [31] to 15 ms [20] or 20 ms [17, 19] using EMCCD based systems (Table 1). New camera developments, such as high-sensitivity, low-noise sCMOS cameras, allow for detection speeds below 1 ms, but whether they will permit sufficient SNR for single particle detection in vivo remains to be seen.
The required temporal resolution also depends on the biological question to be addressed. To visualize discrete steps of mRNA remodeling within and around the NPC, (sub-)millisecond resolution and high SNR are critical, but the higher laser powers that are currently required for this can lead to photodamage and phototoxicity. Conversely, to follow an mRNA throughout its lifetime, minimizing photo-bleaching becomes more important than high-speed imaging, as transcription, translation and decay take minutes in Saccharomyces cerevisiae and are even slower in mammalian cells (reviewed in [128]). In addition, extracting positional information of single particles relative to distinct subcellular or sub-organelle structures requires both high spatial precision as well as multi-color imaging setups. Several recently reported single mRNA particle studies employ two-camera microscopy systems to simultaneously detect the transcript relative to fluorescently labeled NPCs, with comparable co-localization precision (8nm [31], 10 nm [19], 26 nm [17]; 56 nm [20]). A recent study, adapting the imaging setup by Grunwald et al. (2010), attempted to visualized mRNPs with individual ribosomes or polysomes by using a total internal reflection fluorescence (TIRF) setup to excite only a subset of fluorescently-labeled ribosomes, but imaging was limited to a few frames due to photobleaching [129]. Taking it one step further, several recent studies visualized translation in vivo by direct labeling of the nascent protein that emerges from ribosomes translating an MS2-labeled reporter mRNA [118, 130–132].
Finally, prolonged RNA tracking will require imaging of larger cellular volumes, which demands high-speed 3D imaging modalities. Light sheet microscopy, in its various forms [133], allows for long-term imaging and high 3D resolution via fast sectioning along the optical z axis. Other novel approaches that do not rely on z-sectioning are able to image 3D volumes simultaneously through multi-focus imaging [76] or using novel 3D point spread functions [134–136]. Further development and application of these approaches will be central to in vivo RNA imaging and addressing the many questions raised in this review.
Conclusions
In summary, recent work by multiple groups has begun to address the spatial and temporal dynamics of directional mRNA transport in live cells using SMI approaches. However, we are far from understanding nuclear and cytoplasmic mRNA processing at the level of a single transcript. Improved SMI labeling strategies, 3D imaging, and multi-camera setups that extend beyond two-colors will allow for further investigation of functional relationships between transcripts, RBPs, and cellular structures that include the gene locus, NPCs, nucleoli, and cytoplasmic processing bodies. This will undoubtedly provide information on mRNA export dynamics and the spatial and temporal control of the eukaryotic gene expression program.
Abbreviations
- NPC
nuclear pore complex
- RBP
RNA-binding protein
- RNP
RNA–ribonucleoprotein particle
- SMI
single molecule imaging
References
- 1.Capelson M, Hetzer MW. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 2009;10:697–705. doi: 10.1038/embor.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hurt JA, Silver PA. mRNA nuclear export and human disease. Dis Model Mech. 2008;1:103–8. doi: 10.1242/dmm.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon DN, Rout MP. Cancer and the nuclear pore complex. Advances in Exp Med Biol. 2014;773:285–307. doi: 10.1007/978-1-4899-8032-8_13. [DOI] [PubMed] [Google Scholar]
- 4.Yarbrough ML, Mata MA, Sakthivel R, Fontoura BM. Viral subversion of nucleocytoplasmic trafficking. Traffic. 2014;15:127–40. doi: 10.1111/tra.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tran DD, Koch A, Tamura T. THOC5, a member of the mRNA export complex: a novel link between mRNA export machinery and signal transduction pathways in cell proliferation and differentiation. Cell Commun Signal. 2014;12:3. doi: 10.1186/1478-811X-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–71. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munchel SE, Shultzaberger RK, Takizawa N, Weis K. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol Biol Cell. 2011;22:2787–95. doi: 10.1091/mbc.E11-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller C, Schwalb B, Maier K, Schulz D, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol. 2011;7:458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winey M, Yarar D, Giddings TH, Jr, Mastronarde DN. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol Biol Cell. 1997;8:2119–32. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadowaki T, Chen S, Hitomi M, Jacobs E, et al. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126:649–59. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Segref A, Sharma K, Doye V, Hellwig A, et al. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–71. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–76. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hieronymus H, Silver PA. Genome-wide analysis of RNA–protein interactions illustrates specificity of the mRNA export machinery. Nat Genet. 2003;33:155–61. doi: 10.1038/ng1080. [DOI] [PubMed] [Google Scholar]
- 14.Paul B, Montpetit B. Altered RNA processing and export lead to retention of mRNAs near transcription sites and nuclear pore complexes or within the nucleolus. Mol Biol Cell. 2016;27:2742–56. doi: 10.1091/mbc.E16-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raj A, Peskin CS, Tranchina D, Vargas DY, et al. Stochastic mRNA synthesis in mammalian cells. PLOS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–90. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 17.Grünwald D, Singer RH. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–7. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mor A, Suliman S, Ben-Yishay R, Yunger S, et al. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12:543–52. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- 19.Siebrasse JP, Kaminski T, Kubitscheck U. Nuclear export of single native mRNA molecules observed by light sheet fluorescence microscopy. Proc Nat Acad Sci USA. 2012;109:9426–31. doi: 10.1073/pnas.1201781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith C, Lari A, Derrer CP, Ouwehand A, et al. In vivo single-particle imaging of nuclear mRNA export in budding yeast demonstrates an essential role for Mex67p. J Cell Biol. 2015;211:1121–30. doi: 10.1083/jcb.201503135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urbanek MO, Galka-Marciniak P, Olejniczak M, Krzyzosiak WJ. RNA imaging in living cells – methods and applications. RNA Biol. 2014;11:1083–95. doi: 10.4161/rna.35506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol. 1999;9:569–S1. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- 23.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, et al. Localization of ASH1 mRNA Particles in Living Yeast. Mol Cell. 1998;2:437–45. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 24.Chao JA, Patskovsky Y, Almo SC, Singer RH. Structural basis for the coevolution of a viral RNA–protein complex. Nat Struct Mol Biol. 2008;15:103–5. doi: 10.1038/nsmb1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larson DR, Zenklusen D, Wu B, Chao JA, et al. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332:475–8. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hocine S, Raymond P, Zenklusen D, Chao JA, et al. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat Methods. 2013;10:119–21. doi: 10.1038/nmeth.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halstead JM, Lionnet T, Wilbertz JH, Wippich F, et al. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science. 2015;347:1367–671. doi: 10.1126/science.aaa3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daigle N, Ellenberg J. λN-GFP: an RNA reporter system for live-cell imaging. Nat Methods. 2007;4:633–6. doi: 10.1038/nmeth1065. [DOI] [PubMed] [Google Scholar]
- 29.Brodsky AS, Silver PA. Pre-mRNA processing factors are required for nuclear export. RNA. 2000;6:1737–49. doi: 10.1017/s1355838200001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takizawa PA, Vale RD. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc Nat Acad Sci USA. 2000;97:5273–8. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma J, Liu Z, Michelotti N, Pitchiaya S, et al. High-resolution three-dimensional mapping of mRNA export through the nuclear pore. Nat Commun. 2013;4:2414. doi: 10.1038/ncomms3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saroufim M-A, Bensidoun P, Raymond P, Rahman S, et al. The nuclear basket mediates perinuclear mRNA scanning in budding yeast. J Cell Biol. 2015;211:1131–40. doi: 10.1083/jcb.201503070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palazzo AF, Truong M. Single particle imaging of mRNAs crossing the nuclear pore: Surfing on the edge. BioEssays. 2016;38:744–50. doi: 10.1002/bies.201600038. [DOI] [PubMed] [Google Scholar]
- 34.Azimi M, Bulat E, Weis K, Mofrad MRK. An agent-based model for mRNA export through the nuclear pore complex. Mol Biol Cell. 2014;25:3643–53. doi: 10.1091/mbc.E14-06-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musser SM, Grünwald D. Deciphering the structure and function of nuclear pores using single molecule fluorescence approaches. J Mol Biol. 2016;428:2091–119. doi: 10.1016/j.jmb.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ben-Yishay R, Ashkenazy AJ, Shav-Tal Y. Dynamic encounters of genes and transcripts with the nuclear pore. Trends Genet. 2016;32:419–31. doi: 10.1016/j.tig.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Buxbaum AR, Yoon YJ, Singer RH, Park HY. Single-molecule insights into mRNA dynamics in neurons. Trends Cell Biol. 2015;25:468–75. doi: 10.1016/j.tcb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh G, Pratt G, Yeo GW, Moore MJ. The clothes make the mRNA: Past and present trends in mRNP fashion. Annu Rev Biochem. 2015;84:325–54. doi: 10.1146/annurev-biochem-080111-092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet. 2013;14:275–87. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 40.Baejen C, Torkler P, Gressel S, Essig K, et al. Transcriptome maps of mRNP biogenesis factors define pre-mRNA recognition. Mol Cell. 2014;55:745–57. doi: 10.1016/j.molcel.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 41.Baltz AG, Munschauer M, Schwanhausser B, Vasile A, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–90. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 42.Castello A, Fischer B, Eichelbaum K, Horos R, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 43.Ray D, Kazan H, Cook KB, Weirauch MT, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–7. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuck Alex C, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154:996–1009. doi: 10.1016/j.cell.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13:508–14. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Köhler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–73. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 47.Lund MK, Guthrie C. The DEAD-Box Protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;20:645–51. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 48.Estrella LA, Wilkinson MF, González CI. The shuttling protein Npl3 promotes translation termination accuracy in Saccharomyces cerevisiae. J Mol Biol. 2009;394:410–22. doi: 10.1016/j.jmb.2009.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, et al. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol Cell Biol. 2004;24:10479–91. doi: 10.1128/MCB.24.23.10479-10491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delaleau M, Borden KLB. Multiple export mechanisms for mRNAs. Cells. 2015;4:452–73. doi: 10.3390/cells4030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montpetit B, Weis K. Cell biology. An alternative route for nuclear mRNP export by membrane budding. Science. 2012;336:809–10. doi: 10.1126/science.1222243. [DOI] [PubMed] [Google Scholar]
- 52.Natalizio BJ, Wente SR. Postage for the messenger: Designating routes for Nuclear mRNA Export. Trends Cell Biol. 2013;23:365–73. doi: 10.1016/j.tcb.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Batisse J, Batisse C, Budd A, Böttcher B, et al. Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J Biol Chem. 2009;284:34911–7. doi: 10.1074/jbc.M109.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fasken MB, Stewart M, Corbett AH. Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export. J Biol Chem. 2008;283:27130–43. doi: 10.1074/jbc.M803649200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hector RE, Nykamp KR, Dheur S, Anderson JT, et al. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 2002;21:1800–10. doi: 10.1093/emboj/21.7.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, et al. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010;24:1927–38. doi: 10.1101/gad.583310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sträβer K, Baβler J, Hurt E. Binding of the Mex67p/Mtr2p heterodimer to Fxfg, Glfg, and Fg repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gwizdek C, Iglesias N, Rodriguez MS, Ossareh-Nazari B, et al. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Nat Acad Sci USA. 2006;103:16376–81. doi: 10.1073/pnas.0607941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonnet A, Palancade B. Regulation of mRNA trafficking by nuclear pore complexes. Genes. 2014;5:767–91. doi: 10.3390/genes5030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sood V, Brickner JH. Nuclear pore interactions with the genome. Curr Opin Genet Dev. 2014;25:43–9. doi: 10.1016/j.gde.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stancheva I, Schirmer EC. Nuclear Envelope: Connecting Structural Genome Organization to Regulation of Gene Expression. In: Schirmer EC, Heras JIdl, editors. Cancer Biology and the Nuclear Envelope. Springer; New York: 2014. pp. 209–44. [DOI] [PubMed] [Google Scholar]
- 62.Casolari JM, Brown CR, Komili S, West J, et al. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–39. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 63.Ahmed S, Brickner DG, Light WH, Cajigas I, et al. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol. 2010;12:111–8. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol. 2006;26:7858–70. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taddei A, Van Houwe G, Hediger F, Kalck V, et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–8. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 67.Blobel G. Gene gating: a hypothesis. Proc Nat Acad Sci USA. 1985;82:8527–9. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–5. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- 69.Gartenberg MR, Neumann FR, Laroche T, Blaszczyk M, et al. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell. 2004;119:955–67. doi: 10.1016/j.cell.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 70.Green EM, Jiang Y, Joyner R, Weis K. A negative feedback loop at the nuclear periphery regulates GAL gene expression. Mol Biol Cell. 2012;23:1367–75. doi: 10.1091/mbc.E11-06-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dultz E, Tjong H, Weider E, Herzog M, et al. Global reorganization of budding yeast chromosome conformation in different physiological conditions. J Cell Biol. 2016;212:321–34. doi: 10.1083/jcb.201507069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 73.Galy V, Gadal O, Fromont-Racine M, Romano A, et al. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- 74.Vargas DY, Raj A, Marras SA, Kramer FR, et al. Mechanism of mRNA transport in the nucleus. Proc Nat Acad Sci USA. 2005;102:17008–13. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siebrasse JP, Veith R, Dobay A, Leonhardt H, et al. Discontinuous movement of mRNP particles in nucleoplasmic regions devoid of chromatin. Proc Nat Acad Sci USA. 2008;105:20291–6. doi: 10.1073/pnas.0810692105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith CS, Preibisch S, Joseph A, Abrahamsson S, et al. Nuclear accessibility of β-actin mRNA is measured by 3D single-molecule real-time tracking. J Cell Biol. 2015;209:609–19. doi: 10.1083/jcb.201411032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith CS, Stallinga S, Lidke KA, Rieger B, et al. Probability-based particle detection that enables threshold-free and robust in vivo single-molecule tracking. Mol Biol Cell. 2015;26:4057–62. doi: 10.1091/mbc.E15-06-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Madrid AS, Weis K. Nuclear transport is becoming crystal clear. Chromosoma. 2006;115:98–109. doi: 10.1007/s00412-005-0043-3. [DOI] [PubMed] [Google Scholar]
- 79.Hodge CA, Colot HV, Stafford P, Cole CN. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1–1 cells. EMBO J. 1999;18:5778–88. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, et al. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 2011;25:1052–64. doi: 10.1101/gad.2041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, et al. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–42. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Murphy R, Watkins JL, Wente SR. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell. 1996;7:1921–37. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schmitt C, von Kobbe C, Bachi A, Pante N, et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999;18:4332–47. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Strahm Y, Fahrenkrog B, Zenklusen D, Rychner E, et al. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr 255p. EMBO J. 1999;18:5761–77. doi: 10.1093/emboj/18.20.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weirich CS, Erzberger JP, Flick JS, Berger JM, et al. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–76. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 86.Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–6. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 87.Noble KN, Tran EJ, Alcazar-Roman AR, Hodge CA, et al. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 2011;25:1065–77. doi: 10.1101/gad.2040611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ledoux S, Guthrie C. Regulation of the Dbp5 ATPase cycle in mRNP remodeling at the nuclear pore: a lively new paradigm for DEAD-box proteins. Genes Dev. 2011;25:1109–14. doi: 10.1101/gad.2062611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Folkmann AW, Noble KN, Cole CN, Wente SR. Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus. 2011;2:540–8. doi: 10.4161/nucl.2.6.17881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Estruch F, Cole CN. An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Mol Biol Cell. 2003;14:1664–76. doi: 10.1091/mbc.E02-09-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao J, Jin SB, Bjorkroth B, Wieslander L, et al. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 2002;21:1177–87. doi: 10.1093/emboj/21.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–54. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 93.Stewart M. Ratcheting mRNA out of the nucleus. Mol Cell. 2007;25:327–30. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 94.Tutucci E, Stutz F. Keeping mRNPs in check during assembly and nuclear export. Nat Rev Mol Cell Biol. 2011;12:377–84. doi: 10.1038/nrm3119. [DOI] [PubMed] [Google Scholar]
- 95.Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17:666–72. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 96.Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell. 2004;13:201–12. doi: 10.1016/s1097-2765(04)00030-9. [DOI] [PubMed] [Google Scholar]
- 97.Gilbert W, Siebel CW, Guthrie C. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA. 2001;7:302–13. doi: 10.1017/s1355838201002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McBride AE, Cook JT, Stemmler EA, Rutledge KL, et al. Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J Biol Chem. 2005;280:30888–98. doi: 10.1074/jbc.M505831200. [DOI] [PubMed] [Google Scholar]
- 99.Lukong KE, Richard S. Arginine methylation signals mRNA export. Nat Struct Mol Biol. 2004;11:914–5. doi: 10.1038/nsmb1004-914. [DOI] [PubMed] [Google Scholar]
- 100.Bodi Z, Bottley A, Archer N, May ST, et al. Yeast m6A Methylated mRNAs Are Enriched on Translating Ribosomes during Meiosis, and under Rapamycin Treatment. PLoS One. 2015;10:e0132090. doi: 10.1371/journal.pone.0132090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–6. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Squires JE, Patel HR, Nousch M, Sibbritt T, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–33. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mehlin H, Daneholt B, Skoglund U. Structural interaction between the nuclear pore complex and a specific translocating RNP particle. J Cell Biol. 1995;129:1205–16. doi: 10.1083/jcb.129.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Carroll JS, Munchel SE, Weis K. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J Cell Biol. 2011;194:527–37. doi: 10.1083/jcb.201007151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Futcher B, Latter GI, Monardo P, McLaughlin CS, et al. A sampling of the yeast proteome. Mol Cell Biol. 1999;19:7357–68. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miura F, Kawaguchi N, Yoshida M, Uematsu C, et al. Absolute quantification of the budding yeast transcriptome by means of competitive PCR between genomic and complementary DNAs. BMC Genomics. 2008;9:574. doi: 10.1186/1471-2164-9-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, et al. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 2012;19:560–7. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- 108.Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maquat LE, Tarn W-Y, Isken O. The pioneer round of translation: Features and functions. Cell. 2010;142:368–74. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Waters JC. Accuracy and precision in quantitative fluorescence microscopy. J Cell Biol. 2009;185:1135–48. doi: 10.1083/jcb.200903097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Garcia JF, Parker R. MS2 coat protein bound to yeast mRNAs block 5′ to 3′degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system. RNA. 2015;21:1393–5. doi: 10.1261/rna.051797.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Garcia JF, Parker R. Ubiquitous accumulation of 3′mRNA decay fragments in Saccharomyces cerevisiae mRNAs with chromosomally integrated MS2 arrays. RNA. 2016;22:657–9. doi: 10.1261/rna.056325.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haimovich G, Zabezhinsky D, Haas B, Slobodin B, et al. Use of the MS2 aptamer and coat protein for RNA localization in yeast: A response to “MS2 coat proteins bound to yeast mRNAs block 5′ to 3′ degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system”. RNA. 2016;22:660–6. doi: 10.1261/rna.055095.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shaner NC, Lambert GG, Chammas A, Ni Y, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods. 2013;10:407–9. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Grimm JB, English BP, Chen J, Slaughter JP, et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods. 2015;12:244–50. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Keppler A, Pick H, Arrivoli C, Vogel H, et al. Labeling of fusion proteins with synthetic fluorophores in live cells. Proc Nat Acad Sci USA. 2004;101:9955–9. doi: 10.1073/pnas.0401923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Los GV, Encell LP, McDougall MG, Hartzell DD, et al. HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem Biol. 2008;3:373–82. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 118.Morisaki T, Lyon K, DeLuca KF, DeLuca JG, et al. Real-time quantification of single RNA translation dynamics in living cells. Science. 2016;352:1425–9. doi: 10.1126/science.aaf0899. [DOI] [PubMed] [Google Scholar]
- 119.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–6. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Strack RL, Disney MD, Jaffrey SR. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat Methods. 2013;10:1219–24. doi: 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Filonov GS, Moon JD, Svensen N, Jaffrey SR. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc. 2014;136:16299–308. doi: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dolgosheina EV, Jeng SCY, Panchapakesan SSS, Cojocaru R, et al. RNA Mango Aptamer-Fluorophore: A Bright, High-Affinity Complex for RNA Labeling and Tracking. ACS Chem Biol. 2014;9:2412–20. doi: 10.1021/cb500499x. [DOI] [PubMed] [Google Scholar]
- 123.Guet D, Burns LT, Maji S, Boulanger J, et al. Combining Spinach-tagged RNA and gene localization to image gene expression in live yeast. Nat Commun. 2015;6:8882. doi: 10.1038/ncomms9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alford SC, Ding Y, Simmen T, Campbell RE. Dimerization-dependent green and yellow fluorescent proteins. ACS Synth Biol. 2012;1:569–75. doi: 10.1021/sb300050j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu B, Chen J, Singer RH. Background free imaging of single mRNAs in live cells using split fluorescent proteins. Sci Rep. 2014;4:3615. doi: 10.1038/srep03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nelles DA, Fang MY, O’Connell MR, Xu JL, et al. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell. 2016;165:488–96. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stelzer EHK. Light-sheet fluorescence microscopy for quantitative biology. Nat Methods. 2015;12:23–6. doi: 10.1038/nmeth.3219. [DOI] [PubMed] [Google Scholar]
- 128.Oeffinger M, Zenklusen D. To the pore and through the pore: A story of mRNA export kinetics. BBA - Gene Regul Mech. 2012;1819:494–506. doi: 10.1016/j.bbagrm.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Katz ZB, English BP, Lionnet T, Yoon YJ, et al. Mapping translation ‘hot-spots’ in live cells by tracking single molecules of mRNA and ribosomes. eLife. 2016;5:e10415. doi: 10.7554/eLife.10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang C, Han B, Zhou R, Zhuang X. Real-time imaging of translation on single mRNA transcripts in live cells. Cell. 2016;165:990–1001. doi: 10.1016/j.cell.2016.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yan X, Hoek TA, Vale RD, Tanenbaum ME. Dynamics of translation of single mRNA molecules in vivo. Cell. 2016;165:976–89. doi: 10.1016/j.cell.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wu B, Eliscovich C, Yoon YJ, Singer RH. Translation dynamics of single mRNAs in live cells and neurons. Science. 2016;352:1430–5. doi: 10.1126/science.aaf1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Follain G, Mercier L, Osmani N, Harlepp S, et al. Seeing is believing: multi-scale spatio-temporal imaging towards in vivo cell biology. J Cell Sci. 2016 doi: 10.1242/jcs.189001. jcs.189001. [DOI] [PubMed] [Google Scholar]
- 134.Pavani SRP, Thompson MA, Biteen JS, Lord SJ, et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Nat Acad Sci USA. 2009;106:2995–9. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Backlund MP, Joyner R, Weis K, Moerner WE. Correlations of three-dimensional motion of chromosomal loci in yeast revealed by the double-helix point spread function microscope. Mol Biol Cell. 2014;25:3619–29. doi: 10.1091/mbc.E14-06-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shechtman Y, Weiss LE, Backer AS, Lee MY, et al. Multicolour localization microscopy by point-spread-function engineering. Nat Photon. 2016;10:590–4. doi: 10.1038/nphoton.2016.137. [DOI] [PMC free article] [PubMed] [Google Scholar]