Abstract
Continuous innovation of revolutionizing genome engineering technologies calls for an intensified focus on new delivery technologies that not only match the inventiveness of genome editors but also enable the combination of potent delivery and time-restricted action of genome-modifying bits and tools. We have previously demonstrated the use of lentivirus-derived nanoparticles (LNPs) as a protein delivery vehicle, incorporating and transferring DNA transposases, designer nucleases, or RNA-guided endonucleases fused to the N terminus of the Gag/GagPol polypeptide. Here, we establish LNP-directed transfer of the piggyBac DNA transposase protein by fusing the transposase to the integrase protein in the C-terminal end of GagPol. We show protein incorporation and proteolytic release of the DNA transposase within matured LNPs, resulting in high levels of DNA transposition activity in LNP-treated cells. Importantly, as opposed to conventional delivery methods based on transfection of plasmid DNA or in-vitro-transcribed mRNA, protein delivery by LNPs effectively results in time-restricted action of the protein (<24 hr) without compromising overall potency. Our findings refine LNP-directed piggyBac transposase delivery, at present the only available direct delivery strategy for this particular protein, and demonstrate a novel strategy for restricting and fine-tuning the exposure of the genome to DNA-modifying enzymes.
Keywords: DNA transposon, piggyBac, lentivirus, protein delivery, protein transduction, LNP
Introduction
For decades, the engineering of genomes has been essential in studies of cellular biology and disease modeling in cell culture and transgenic animals. With the arrival of revolutionizing genome-engineering technologies, new molecular tools, at work within the nucleus, are being implemented in tailored genetic therapies with the potential of erasing disease-causing mutations.1, 2 The rapidly expanding tool kit now includes recombinases, integrases, and transposases as well as designer nucleases and RNA-guided endonucleases (reviewed in Skipper and Mikkelsen3), some of which have already reached the clinic.4, 5, 6 Effective delivery to relevant cells and organs is a common goal for all therapeutic applications of such technologies. However, as opposed to conventional gene therapies, long-term intracellular expression and persistent activity of genome-modifying enzymes is rarely desired, and fine-tuning the cellular load of such tools remains a key challenge. Hence, safety measures and further therapeutic translation may benefit from delivery methodologies facilitating an effective but time-restricted boost of enzyme activity.
Genetic engineering protocols have traditionally relied on intracellular expression of genome-modifying proteins. Transfection, often by nucleofection, of protein-encoding plasmid DNA is used as a standard technique in many laboratories. Although effective for many in vitro applications, this approach suffers from the risks of prolonged expression and genomic insertion of the expression cassette. Delivery of the expression cassette by transduction with virus-based vectors is equally powerful and holds great potential for in vivo delivery, as was recently demonstrated by potent adeno-associated virus (AAV)-directed delivery of the RNA-guided streptococcus aureus Cas9 endonuclease to mouse liver.7 However, viral vector-based expression is potentially sustained for months or years, depending on the target organ, and viral vectors, including AAV-derived vectors,8 may potentially insert into the genome, leading to concerns related to accumulation of off-target modifications or toxicity. The use of in-vitro-transcribed mRNA as a source of genome-modifying enzymes has rapidly become the method of choice for in vitro genetic engineering of stem cells1, 2, 9 as well as many other cell types. As for conventional DNA transfection, this method facilitates heavy overexpression but nevertheless fulfills some of the criteria for optimized safety by leading to a much shorter boost of protein activity. However, nucleofection protocols are potentially hampered by cellular toxicity, and in vivo RNA delivery may be challenging despite emerging examples of successful mRNA delivery using nanoparticle carriers.10
In consideration of the safety precautions related to DNA- or RNA-directed intracellular production of genome-modifying enzymes, means of directly delivering genome-modifying proteins to cells are currently attracting increased attention. Attempts to deliver recombinant transposase proteins have not been successful,11, 12 but targeted gene knockout has been achieved by direct delivery of recombinant zinc-finger nucleases (ZFNs).13 Intriguingly, recent reports also showed potent gene disruption in cultured human cells based on the delivery on recombinant Cas9 protein.2, 9, 14, 15 For genome editing purposes, recombinant Cas9 protein can be directly complexed with a synthetic single-guide RNA (sgRNA)2, 9, 14 and cellular uptake combined with potent viral transfer of a donor sequence for repair by homologous recombination.2, 16, 17
In an attempt to develop a protein delivery strategy with potential in vivo applicability, we recently demonstrated that HIV-derived lentiviral particles are capable of ferrying foreign proteins into cells. By fusing a heterologous protein to the matrix protein in the N-terminal end of lentiviral Gag/GagPol polypeptides, we observed effective encapsidation of the protein in engineered lentivirus-derived nanoparticles (LNPs) and demonstrated protein activity in cells exposed to protein-loaded LNPs. Hence, this delivery strategy resulted in robust DNA transposition after delivery of the piggyBac transposase18 and effective genomic cleavage after delivery of zinc-finger nucleases (ZFNs) and TAL-effector nucleases (TALENs).19, 20 Despite its considerably larger size, the streptococcus pyogenes Cas9 endonuclease fused to the N terminus of Gag could also be loaded in LNPs, resulting in RNA-guided DNA cleavage in primary T cells exposed to LNPs.21 Not only is the structure of lentiviral particles sufficiently flexible to accommodate multiple copies of a foreign protein, but also cell entry and the capacity of viruses to traverse the cytoplasm and unload protein cargo in the nucleus are key features of LNP-directed protein delivery. Organized and direct delivery of the protein may explain that even relatively low intracellular levels of the transferred protein support efficient DNA transposition or DNA cleavage18, 19, 20 and thus may add to the overall safety of the procedure. Also, such LNPs can be depleted for lentiviral vector RNA and may thus serve strictly as a protein vehicle and may furthermore be delivered in a cell-targeted fashion utilizing tailored pseudotypes.22
Following this overall concept, we rethought the delivery design and engineered LNPs with the piggyBac DNA transposase fused to the integrase protein in the C terminus of GagPol. This should result in LNPs containing approximately 20 times fewer units of the protein. However, unlike LNPs carrying the DNA transposase fused directly to Gag, the protein will, together with the viral enzymes, supposedly locate to the core of the virus particle. Using this approach, we verify DNA transposition efficacy and demonstrate that LNP-directed delivery, as opposed to delivery methods based on transfection of plasmid DNA or in-vitro-transcribed mRNA, results in effective and time-restricted action of the protein without loss of potency. Our findings show a novel way of restricting and fine-tuning the exposure of the genome to DNA-modifying enzymes without compromising efficacy, and they consolidate LNP-directed protein delivery as the only currently available strategy for delivering piggyBac transposase protein directly to cells.
Results and Discussion
Functionality of LNPs Carrying PiggyBac Transposase Fused to the Integrase Protein during Particle Assembly
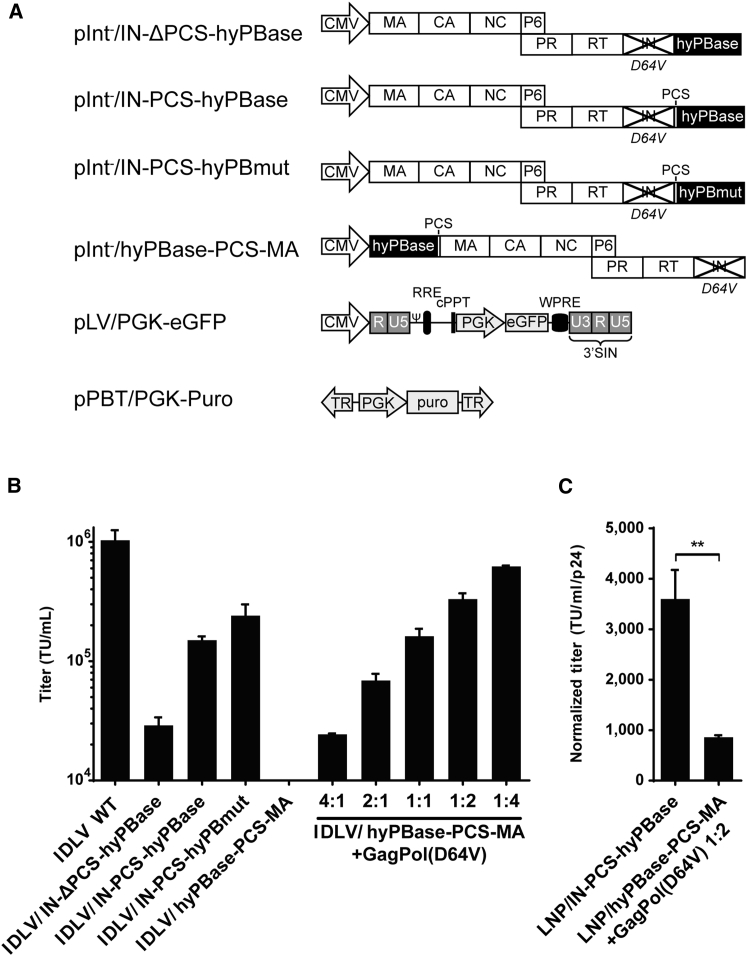
DNA transposon vectors based on the piggyBac system are mobilized from a transposon source, plasmid DNA, or viral vectors by the DNA transposase, which works in trans and is typically expressed from a co-delivered gene cassette.23, 24 We previously reported high levels of DNA transposition in cells treated with LNPs carrying the piggyBac transposase fused to the matrix (MA) protein in the N-terminal end of Gag/GagPol (reviewed in Cai and Mikkelsen25). However, we also found that particles assembled with Gag and GagPol polypeptides carrying the N-terminal fusion protein were incapable of transferring vector RNA at the same time, indicating that the added protein affected the overall structure and function of the particle. Notably, LNPs carrying fewer copies of the genome-modifying enzyme (generated by including normal unfused GagPol in the particles) were not functionally affected by the foreign cargo and still induced high levels of enzyme activity in LNP-treated cells.18, 19 Therefore, we reasoned that incorporation of the piggyBac transposase in the Pol domain and not in Gag of GagPol would allow formation of particles with a balanced content of the foreign protein, now expectedly packaged with foreign protein colocalizing with the viral enzymes within the LNP core. Inspired by work by Schenkwein and colleagues demonstrating lentiviral delivery of the I-PpoI endonuclease fused to viral integrase,26, 27, 28 we fused the hyperactive piggyBac transposase (hyPBase) to the inactive D64V integrase (IN) in the C-terminal end of the GagPol polypeptide. Using three different packaging constructs (pINT-/IN-ΔPCS-hyPBase, pINT-/IN-PCS-hyPBase, and pINT-/IN-PCS-hyPBmut; all shown in Figure 1A), we produced vesicular stomatitis virus G protein (VSV-G)-pseudotyped viral particles carrying (1) hyPBase fused directly to IN, (2) hyPBase linked to IN by a HIV protease cleavage site (PCS), or (3) a mutated variant of the transposase, hyPBmut, linked to IN via the PCS. In gene transfer experiments in HEK293 cells using a standard EGFP-encoding lentiviral vector (pLV/PGK-EGFP; Figure 1A), we measured transduction efficiencies ranging from 1.5 × 105 to 2.5 × 105 transducing units (TU)/mL for these integrase-defective lentiviral vectors (IDLVs) containing transposase variants linked to IN through the PCS, whereas the gene transfer capacity of corresponding IDLVs carrying IN-ΔPCS-hyPBase fusion protein without a protease cleavage site was reduced 10-fold (Figure 1B). The titer of control IDLVs without the hyPBase fusion was 1.0 × 106 TU/mL, showing that incorporation of hyPBase protein did to some degree affect the overall gene transfer capacity. Notably, our findings also confirmed that IDLVs carrying hyPBase fused to the Gag N terminus (LNP/hyPBase-PCS-MA) did not support any EGFP gene transfer in transduced cells (Figure 1B). Only when IDLVs carrying the hyPBase-PSC-MA fusion were produced with increasing content of normal Gag/GagPol (without the N-terminal fusion protein), the gene transfer capacity gradually increased (Figure 1B). As reduced gene transfer of protein-loaded LNPs could potentially reflect a reduction in the yield of viral particles, we also evaluated the transfer efficiency relative to amount of particles, as determined by p24 Gag ELISA. This analysis showed that each IDLV/IN-PCS-hyPBase particle was 3.5 times better at transferring the vector than particles carrying MA-fused hyPBase supplemented with wild-type Gag (Figure 1C).
Figure 1.
Titer and Transfer Efficiency of IDLVs Packaged with Integrase-Fused hyPBase
(A) Schematic representation of the constructs used in the study. (B) Evaluation of transduction titers of transposase-loaded IDLVs carrying vector RNA encoded by pLV/PGK-EGFP is shown. IDLVs with IN-fused PB transposase with or without the KARVL/AEAMS protease cleavage site (PCS) were packaged with a PGK-EGFP expression vector, and transduction titers were estimated by flow cytometry and compared to IDLVs packaged with MA-fused transposase in the presence of increasing amounts of wild-type Gag/GagPol (GagPol-D64V), as indicated by the ratios. A standard IDLV was furthermore included as a control. (C) Estimation of IDLV transfer efficiency is shown. The total amount of LNPs was estimated by p24 ELISA and used to normalize transduction titers quantified in (B) to estimate the relative transfer efficiency of IDLVs packaged with IN- or MA-fused PB transposase. cPPT, central polypurine tract; CA, capsid; IN, integrase; MA, matrix; NC, nucleocapsid; PR, protease; RT, reverse transcriptase; Ψ, psi packaging signal; RRE, rev response element; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element. Data are presented as mean ± SEM and n ≥ 3.
Together, our findings suggested that the vector transfer efficiency of IDLV/IN-PCS-hyPBase was in the same range as IDLVs containing fused and wild-type Gag/GagPol in a 1:1 ratio, indicating overall functionality of viral particles ferrying protein fused to the integrase in the C-terminal end of GagPol. This led us to examine whether the transposon donor sequence could be co-delivered with IN-fused hyPBase in IDLVs (Figure S1A). We produced IDLVs carrying vector RNA containing a PGK-puro cassette embedded in a piggyBac transposon sequence, which is converted to an active DNA transposon upon reverse transcription in transduced cells. Robust DNA transposition was observed in IDLV-treated HEK293 cells, resulting in 3.5 × 103 puromycin-resistant colonies after transduction of 4 × 105 cells (Figure S1C). In comparison, 200 colonies appeared as a result of random integration after treatment of the cells with the corresponding IDLVs loaded with the inactive hyPBmut transposase. Southern blot analysis of genomic DNA isolated from 12 expanded clones unveiled a single insertion in each of the clones (Figure S1D). Mapping of the insertion site in each clone (except clone 7, for which mapping was not successful) verified hyPBase-directed gene insertion (Figure S1E). Four of the ten mapped insertions were located in intergenic regions, whereas the remaining six were mapped to genes, the five of them to introns.
Potent DNA Transposition after LNP-Directed Transposase Protein Delivery
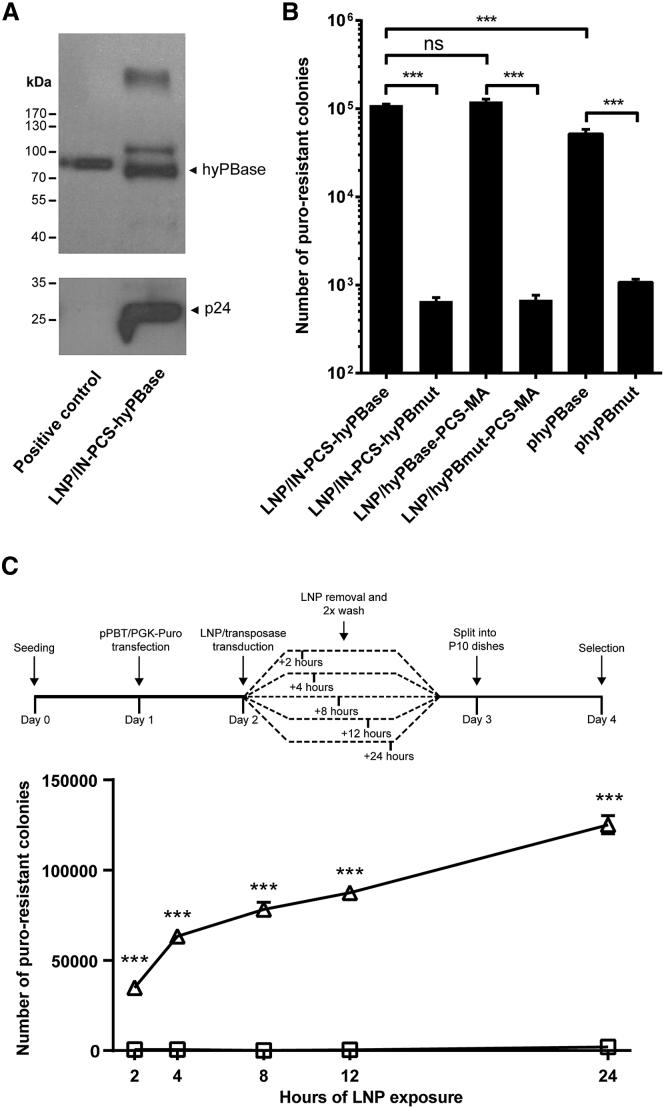
We were primarily interested in studying the protein delivery capacity of viral particles that do not carry genetic information (we refer to such particles as LNPs) and produced, thus, vector-void LNPs carrying a hemagglutinin (HA)-tagged variant of the hyPBase fused to IN with an intervening PCS. Western blot analysis of the particle content confirmed incorporation of hyPBase into the particles and showed that the transposase was released from IN during virus maturation (Figure 2A). We also detected an additional and considerably weaker band (evident in both Figures 2A and S2) indicative of the presence of IN-hyPBase fusion protein in the population of maturing LNPs. However, neither p24 nor hyPBase was detectable in LNP samples isolated from producer cells treated with the protease inhibitor saquinavir (SQV) (Figure S2), suggesting that transposase release was dependent on protease activity during particle maturation.
Figure 2.
Efficient and Rapid Cellular Uptake of hyPBase-Loaded LNPs
(A) Evidence of LNP encapsidation of IN-fused hyPBase and subsequent proteolytic release of hyPBase. Western blot analysis on a LNP lysate incorporating an HA-tagged version of the hyPBase transposase is shown. Cell lysates from HEK293T cells transfected with 1 μg HA-hyPBase expressing plasmid served as positive control. (B) Colony formation after transposase delivery by hyPBase-loaded LNPs is shown. Initially, the PB transposon donor pPBT/PGK-Puro was delivered to HeLa cells by plasmid DNA transfection. The cells were subsequently treated with LNPs loaded with either IN- or MA-fused PB transposase, and transposase activity was estimated by quantification of puromycin-resistant colonies. (C) Limited exposure demonstrates rapid cellular uptake of LNPs. HeLa cells were exposed to PB-loaded LNPs for 2, 4, 8, 12, or 24 hr after delivery of the pPBT/PGK-Puro PB transposon donor. Transposase activity was subsequently estimated as described in (B). Triangles, LNP/hyPBase; squares, LNP/hyPBmut. Data are presented as mean ± SEM and n ≥ 3.
To investigate the activity of LNP-delivered hyPBase in DNA transposition assays, we produced LNPs containing the two different fusion variants (IN-PCS-hyPBase and hyPBase-PCS-MA) and measured formation of puromycin-resistant colonies in HeLa cells transfected with donor plasmid carrying a PGK-puro expression cassette in the context of a DNA transposon (Figure 1A). Both fusion variants resulted in more than 105 colonies after exposure of 4.4 × 105 cells to LNPs, demonstrating that they equally well supported potent mobilization of the transposon from plasmid to the genome (Figure 2B). In both cases, the number of colonies in cells treated with hyPBase-loaded LNPs was approximately 150-fold higher than in cells treated with LNPs carrying the hyPBmut transposase. Interestingly, as we have seen previously,18 a control experiment using our standard conditions for co-transfection of hyPBase-encoding plasmid and the transposon donor plasmid resulted in significantly fewer puromycin-resistant colonies (Figure 2B). Also, levels of DNA transposition was considerably higher than we observed after IDLV-directed delivery of the transposon in HEK293 cells (Figure S1B), which most likely indicates that transposition from transfected plasmid DNA is more efficient than transposition from an IDLV-delivered substrate, which is supposedly less abundant. As opposed to delivery methods based on intracellular transposase production, protein delivery is expected to facilitate immediate activity in cells exposed transiently to LNPs. To test this, we transfected HeLa cells with transposon donor plasmid (pPBT/PGK-puro) and on the following day exposed the cells to LNPs for a duration of 2–24 hr followed by a thorough wash. Most transposition events leading to the largest number of colonies was detected after 24 hr of exposure, but robust levels of transposition were measured also in cells exposed to the LNPs for only 2 hr (Figure 2C). Although the average number of integrations per cell was not measured and quantification of colony formation therefore did not represent a strict measurement of protein activity, formation of transposon-containing colonies did indeed provide a good estimation of efficacy after LNP-mediated protein delivery. Hence, throughout the study, colony formation was used as a measure of successful transposase delivery, resulting in intracellular activity of the transposase. Collectively, our findings indicate that LNP-directed protein uptake is rapid and leads to high enzymatic activity, supporting the use of protein-loaded LNPs for delivery of genomic tools.
Effective Time-Restricted Transposition Activity after LNP-Directed Protein Delivery
For every 20 Gag molecules incorporated in lentiviral particles, one molecule contains an additional Pol domain encompassing the enzymatic proteins. This means that every LNP unit contains in the order of 250 GagPol molecules,29 allowing packaging of a similar and relatively low number of proteins fused to the C terminus of GagPol. This may partially explain why IN-fused proteins are better tolerated than MA-fused proteins, as evaluated by the capacity of the viral particles to successfully transfer vector RNA. Notably, however, protein activity after LNP delivery is high despite the reduced amount of protein delivered by each virus particle. Intuitively, the safety of genome engineering will increase with methods that allow genomic intervention after delivery of a limited number of effector molecules. Furthermore, time restriction of the activity of these tools will add to the safety. To investigate the temporal pattern of protein activity after exposure of cells to LNPs and compare to other conventional delivery methods, we took advantage of DNA transposition as a model system. For three distinct transposase delivery methodologies, (1) cationic polymer-based DNA transfection, (2) liposome-assisted transfection of in-vitro-transcribed mRNA, and (3) protein transduction using VSV-G-pseudotyped LNPs, we initially established experimental conditions that resulted in comparable numbers of puromycin-resistant colonies (approximately 15,000 colonies) after co-delivery of plasmid DNA carrying the PBT/PGK-puro transposon.
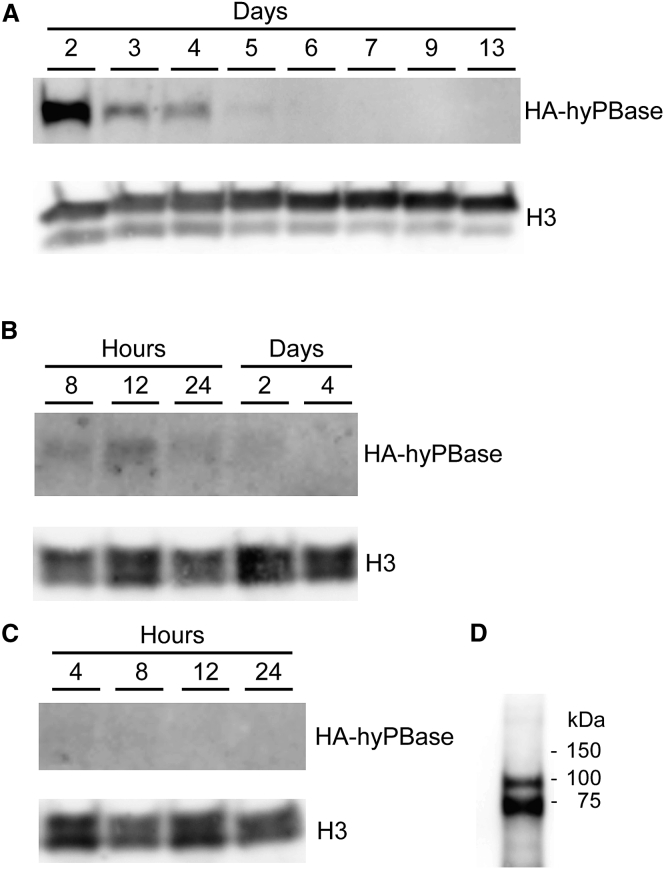
Using DNA-, RNA-, and protein-based delivery strategies resulting in similar levels of DNA transposition, we investigated the time span of transposase activity. First, the cellular content of HA-tagged hyPBase protein over time after delivery was examined by western blot analyses. The level of hyPBase protein peaked at day 2 after plasmid DNA transfection and the protein was detectable for up to 5 days after transfection (Figure 3A). These findings reproduce previous observations by Saha and co-workers, showing detection of a piggyBac transposase up to 7 days after transfection.30 After RNA transfection, the transposase protein was detectable at reduced levels compared to plasmid transfection and for a duration of only 2 days with peak amounts observed at 12 hr after RNA delivery (Figure 3B). In sharp contrast to intracellular production-based methods, LNP-directed protein delivery did not result in levels of protein that were detectable by western blot (Figure 3C). Therefore, we analyzed the transposase protein content in purified LNPs and verified a high content of protein in the particles (Figure 3D). These data reproduce our previous observations showing the lack of detection of hyPBase (by western blot analysis) in cells treated with LNPs carrying Gag-fused hyPBase.18 Together, our findings suggested that cells treated with hyPBase-loaded LNPs contained only relatively low levels of transposase protein but that LNP-delivered protein nevertheless supported high levels of DNA transposition. This lends support to the notion that short-term activity of small amounts of LNP-delivered protein is sufficient to facilitate robust enzymatic activity in cells.
Figure 3.
Western Blot Analysis of hyPBase Protein Levels over Time
(A–C) HeLa cell lysates were analyzed by western blotting at the indicated time points after delivery of HA-tagged versions of the hyPBase transposase by either plasmid DNA transfection of 100 ng of a hyPBase expression vector (A), mRNA transfection of 150 ng in-vitro-transcribed hyPBase mRNA (B), or protein transduction of 400 ng P24 hyPBase-loaded LNPs (C). (D) A LNP lysate was furthermore included to verify the presence of hyPBase in the LNPs. For all blots, H3 was used as a loading control.
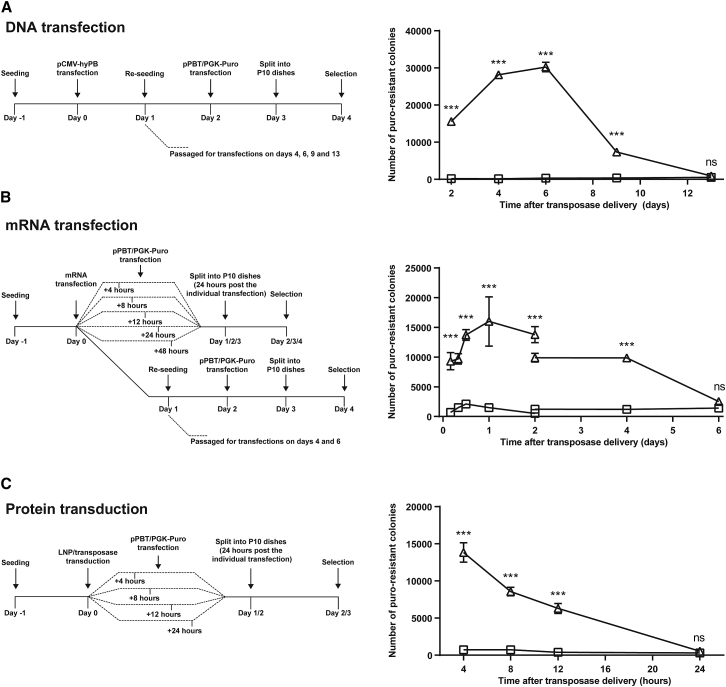
To examine the temporal properties of LNP-directed protein delivery in more detail, we used the three above-mentioned methods to deliver the transposase to HeLa cells and subsequently at different time points delivered the transposon donor plasmid by DNA transfection. For each time point, the number of resistant colonies after puromycin selection was determined, providing an indication of the presence of active hyPBase transposase at the given time point. Time lines for each of the three delivery strategies are provided in the left panels of Figure 4.
Figure 4.
Determination of the Temporal Pattern of Protein Activity after Transposase Delivery by LNPs Compared to Standard Delivery Methods
HeLa cells were transfected with 900 ng of the pPBT/PGK-Puro transposon donor, and subsequently, PB transposase was delivered to the cells by either plasmid DNA transfection (A), transfection of in-vitro-transcribed mRNA (B), or transduction of transposase-loaded LNPs (C) at the indicated time points. Detailed delivery schemes can be seen in the left panels of (A), (B), and (C). Transposase activity was subsequently estimated as described earlier. Triangles, hyPBase; squares, hyPBmut. Data are presented as mean ± SEM and n ≥ 3.
For delivery of the transposase by plasmid DNA transfection, we initially transfected the cells with pCMV-hyPBase and subsequently, at different time points, transfected the cells with pPBT/PGK-puro (Figure 4A, left panel). With this setup, we observed transposition at its peak 6 days after transfection of transposase-encoding plasmid (Figure 4A, right panel). At day 9, the level of transposition was reduced to 20% of the peak level. For studies of transposition after delivery of in-vitro-transcribed RNA encoding the transposase, we wanted to determine the activity shortly after mRNA transfection and therefore re-transfected the cells with the transposon donor plasmid 4, 8, 12, 24, and 48 hr after RNA delivery (Figure 4B, left panel). For analysis of persistence of activity, we also re-seeded mRNA-transfected cells after one day, allowing for subsequent pPBT/PGK-puro transfections. With these two setups, we produced two complimentary sets of data overlapping at the day 2 time point. After transfection of hyPBase-encoding mRNA, transposase activity peaked already 1 day after mRNA transfection and gradually declined over the following days (Figure 4B, right panel). Robust DNA transposition was evident 4 days after mRNA transfection, whereas the activity was almost gone at day 6. Notably, transposase activity after both DNA- and RNA-based transposase delivery was initially significantly reduced compared to later time points, despite the fact that the levels of transposase protein were highest at these early time points. For example, levels of transposase peaked at day 2 (Figure 3A), whereas activity peaked with a delay on day 6 after transfection (Figure 4A), suggesting that DNA transposition was transiently suppressed by high piggyBac expression levels, potentially by mechanisms of “overproduction inhibition”. These observations mimic previous studies suggesting that piggyBac transposition in DNA transfection assays is negatively affected by transposase overproduction.31, 32 Cells treated with transposase-loaded LNPs were transfected with the transposon donor plasmid after LNP exposure times of 4, 8, 12, and 24 hr, following a thorough wash to remove free LNPs (Figure 4C, left panel). With this delivery strategy, the maximum number of transposition events was observed in cells transfected with the DNA transposon 4 hr after LNP treatment (Figure 4C, right panel). The level of DNA transposition then gradually declined, reaching 50% of the peak level already 8 or 9 hr after the initial exposure to LNPs. At 24 hr after LNP addition, active DNA transposition could not be detected in the cells, indicating that the hyPBase had been degraded and/or diluted during this time span. Interestingly, the decay rate of LNP-delivered transposase, measured by activity after LNP delivery (Figure 4C), was within the same range as the decay rate of transposase protein after DNA transfection, measured by western blotting (Figure 3A). Although the production of transposase was maintained for some time after DNA transfection and this assay therefore did not strictly illustrate protein decay, the observation of comparable decay rates served as an internal validation of the two assays. In summary, our findings demonstrate that LNP-based delivery of genome-modifying enzymes results in a pattern of activity, which is markedly time-restricted relative to delivery methods based on intracellular protein production using transfected DNA or mRNA sources. Given the fact that these time courses were established under conditions showing similar initial levels of hyPBase activity, these observations also stress that a short-lived and effective boost of enzymatic activity—an attractive criterion for safe delivery of tools to the genome—could be established only by direct delivery of the protein.
So far, attempts to produce and deliver recombinant piggyBac transposase have not been successful,11 and LNP-directed protein delivery currently represents the only available option for direct delivery of the piggyBac transposase protein, allowing time-restricted activity within cells. This capacity to deliver the protein using virus transduction may be relevant for effective footprint-free removal of the piggyBac DNA transposon during production of induced pluripotent stem cells (iPSCs)33, 34, 35 or in genome editing strategies that require genomic excision of a selective reporter gene.36, 37, 38, 39 It may also allow effective DNA transposition in primary cells or hard-to-transfect cells or tissues.18 For the CRISPR/Cas9 system, in contrast, production of recombinant Cas9 is now well established,14, 15 and direct delivery of ribonucleoprotein (RNP) complexes consisting of Cas9 and sgRNA by nucleofection results in robust DNA cleavage.9 Alternatively, a cell-penetrating peptide fused to Cas9 can facilitate cellular uptake.15 Other endonucleases, like recombinant ZFNs, seem to have intrinsic cell-penetrating properties that facilitate direct uptake in cells in vitro.13 Although virus- and RNP-based delivery strategies each offer advantages, both allow the establishment of a short-lived boost of activity. However, whereas RNP delivery may grow to become the most popular delivery method for ex vivo modification of patient cells, incorporation of nucleases in LNPs may be compatible with criteria related to efficiency and safety of performing genome editing in vivo for future genetic therapies. We have previously established LNP-based delivery of ZFN and TALEN proteins19, 20 as well as of Cas9 protein,21 and we now know that this delivery route facilitates only short-term and therefore likely safer activity of genome-modifying tools. But how can the relatively low protein content delivered by LNP-based protein vehicles be sufficient to reach protein activity levels competing with those of other delivery systems? This is yet unclear. However, although we still lack formal evidence that LNPs, due to viral origin and their capacity to traverse cytoplasm and membranes, drop foreign protein cargo directly at the chromatin, it is tempting to speculate that such mechanisms make virus-derived systems optimal for protein delivery. In the future, such delivery may even be targeted to a specific cell type or organ.
Materials and Methods
Plasmid Cloning
The pMDLg/p-RRE(D64V) plasmid encoding GagPol with the D64V mutation in Integrase was used to generate Integrase fusions with either hyPBmut or hyPBase, the latter with and without a C-terminal HA tag. pInt-/IN-PCS-hyPBase, pInt-/IN-ΔPCS-hyPBase, and pInt-/IN-PCS-hyPBmut were created by fusing the transposase expression cassette to the viral integrase with and without the codon-optimized PCS KARVL/AEAMS, which separates capsid and SP1 in wild-type HIV-1 by overlap extension PCR. The resulting PCR products were subsequently cloned into AflII- and BspEI-digested pMDLg/p-RRE(D64V). phyPBase-gagpol-D64V or phyPBmut-gagpol-D64V (described in Cai et al.18) was used as template for amplification of the hyPBase and hyPBmut sequences, respectively. The piggyBac transposon plasmid pPBT/PGK-puro containing an expression cassette for the PAC gene conferring puromycin resistance, the HA-tagged hyPBase-expressing plasmid pCMV-HA_hyPBase, and pLV/PGK-EGFP were previously described in Cai et al.,18 Sharma et al.,40 and Jakobsen et al.,41 respectively. Vectors for in vitro transcription of hyPBase and hyPBmut were generated by PCR amplification and subsequent insertion into BglII-digested pT3TS/SB1142 by Gibson Assembly (New England Biolabs, Ipswich, MA) according to manufacturer’s instructions. The lentiviral hybrid vector containing the PBT/PGK-Puro transposon placed in a reverse orientation in the lentiviral transfer vector (pLV/puro-PGK-PBT) was generated in Cai et al.18
IDLV and LNP Production
Protein-transducing LNPs devoid of vector RNA were generated by standard calcium phosphate transfection of lentiviral packaging plasmids into HEK293T cells seeded the day before in 10-cm dishes at 4 × 106 cells per dish. The amounts of plasmids used for production of particles in 10-cm dishes were pRSV-Rev: 3 μg, pMD2.G: 3.75 μg, and GagPol-encoding plasmid: 26 μg. Protein-transducing IDLVs carrying vector RNA were generated in a similar manner but with the following amounts of plasmids: pRSV-Rev: 3 μg; pMD2.G: 3.75 μg; GagPol-encoding plasmid: 13 μg; and lentiviral transfer vector: 13 μg. One day after transfection, medium was replaced, and two days after transfection, supernatant was harvested by filtration through a 0.45-μm filter (Sarstedt, Nümbrecht, Germany) and stored in aliquots at −70°C. When necessary, concentrated virus preparations were produced by scaling up the production to 15-cm dishes and ultracentrifugation of viral supernatant through a 4-mL 20% sucrose cushion at 25,000 rpm at 4°C for 2 hr followed by resuspension of the pelleted virus in DPBS−/−. The yield of each vector preparation was determined by p24 ELISA using kits provided either by Zeptometrix (Buffalo, NY) or XpressBio (Thurmont, MD) following manufacturers’ protocols.
Cell Culturing
HEK293, HEK293T, and HeLa cells were cultured at standard conditions at 37°C in 5% CO2. They were cultured in DMEM with L-glutamine (Lonza, Basel, Switzerland) supplemented with 5% fetal calf serum, penicillin (100 U/mL), and streptomycin (0.1 mg/mL).
Titer Assays
Functional titer assays were performed by the limiting dilution method with lentiviral vectors encoding EGFP. HEK293T cells, seeded the day before in 6-well plates at 50,000 cells per well, were transduced with serially diluted vectors supplemented with 8 μg/mL polybrene (Sigma-Aldrich, St. Louis, MO). Four days post-transduction, cells were fixed in 2% buffered formalin (Lillie’s fixative) and analyzed for EGFP fluorescence by flow cytometry using a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ). Diluted viral preparations giving rise to 1%–20% EGFP+ cells were used to calculate the biological titer (TU/mL) using the dilution factor and the total cell number counted on the day of transduction.
Southern Blotting
HEK293 cells were transduced with LNP/IN-PCS-hyPBase and subjected to puromycin selection. Single colonies were isolated and expanded, after which DNA was extracted using a standard NaCl/EtOH precipitation protocol. Fifteen micrograms of genomic DNA (gDNA) from each clone was digested overnight with DraI before gel electrophoresis and vacuum blotting. A PCR-amplified puromycin probe was randomly labeled using the Prime-It random primer labeling kit (Agilent Technologies) according to the manufacturer’s instructions.
LDI-PCR
Genomic DNA was extracted by a standard NaCl/EtOH precipitation protocol. 3 μg genomic DNA was digested with Fastdigest NheI and Fastdigest XbaI (Thermo Fisher Scientific) for six hours and column purified with E.Z.N.A. Gel Extraction Kit (Omega Bio-tek, Norcross, GA). Purified DNA was ligated overnight with T4 DNA ligase (New England Biolabs) in a total volume of 500 μL. 2 μL ligated DNA was used as template for the first PCR using DreamTaq DNA polymerase (Thermo Fisher Scientific) following manufacturer’s instructions (57°C annealing temperature; 2 min extension time; 50 μL total volume) with a piggyBac-specific sense primer and a PGK-specific antisense primer. 2 μL of the first round of PCR was used as a template for a nested PCR reaction employing identical settings as the first-round PCR. The products of the nested PCR were separated by gel electrophoresis, and discrete bands were excised and column purified (Gel Extraction Kit; Omega Bio-tek). Purified products were sequenced (GATC Biotech, Cologne, Germany) using the sense primer from the nested PCR. Resulting sequences were mapped to the human reference assembly (GRCh37/hg19) using Blat.
In Vitro Transcription
For generating transposase transcripts, hyPBase and hyPBmut were PCR amplified from pT3TS/hyPBase or pT3TS/hyPBmut, respectively, using a T3 and T7 primer set. Purified PCR products (Figure S3B, left panel) were then used as templates for in vitro transcription. Capped RNA transcripts were generated using the Ambion mMessage Machine T3 transcription kit (Thermo Fisher Scientific, Waltham, MA) according to manufacturer’s instructions. RNAs were purified by lithium chloride precipitation and visualized by denaturing agarose gel electrophoresis in a 1% formaldehyde gel in 3-morpholinopropane-1-sulfonic acid (MOPS) buffer (Figure S3B, right panel).
Colony-Forming Assays
To investigate ability of LNP-delivered, IN-fused transposase to mobilize an episomal transposon donor, we performed a series of colony-forming assays. HeLa cells seeded in 6-well plates the day before at a density of 2.5 × 105 cells/well were transfected with 900 ng pPBT/PGK-Puro transposon donor together with 100 ng pCDNA3.1-PL4 using Turbofect (Thermo Fisher Scientific). 24 hr after transfection, the cells were transduced using 400 ng p24 of LNP/IN-PCS-hyPBase, LNP/IN-PCS-hyPBmut, LNP/hyPBase-PCS-MA, or LNP/hyPBmut-PCS-MA in the presence of polybrene (8 μg/mL). The day after transduction, cells were split into P10 dishes in appropriate dilutions, and subsequently, the following day, medium was changed to medium supplemented with 1 μg/mL puromycin (Sigma-Aldrich). Puromycin-resistant colonies were stained with 0.6% methylene blue (Sigma-Aldrich) and counted. For the plasmid DNA transfection, control cells were co-transfected with pPBT/PGK-Puro and either pCMV-hyPBase or pCMV-hyPBmut in a 9:1 ratio with 1 μg total DNA and subsequently split into P10 dishes the day after.
Detection of hyPBase Protein in LNPs and Transduced Cells by Western Blotting
For detection of hyPBase protein in HeLa cells, 2 × 105 cells/well were seeded in 6-well plates. The day after, for plasmid DNA transfection, cells were transfected with 100 ng pCMV-hyPBase together with 900 ng pCDNA3.1-PL4 as stuffer using Turbofect. For RNA-based delivery, cells were transfected with 150 ng of rT3TS/hyPBase using Lipofectamine 2000 (Thermo Fisher Scientific), and for LNP-mediated transduction, 400 ng p24 was used as described above. Cell lysates for each time point were prepared with cells from three separate transfections/transductions, which were pooled and lysed in Pierce RIPA buffer (Thermo Fisher Scientific) supplemented with 10 mM NaF and 1× complete protease inhibitor cocktail (Roche, Basel, Switzerland). For western blots on LNPs containing piggyBac transposase, ultracentrifuged LNPs were lysed in Pierce RIPA buffer as described above. Proteins were separated by SDS-PAGE and blotted in to a polyvinylidene fluoride (PVDF) membrane. Membranes were blocked in 5% skim-milk dissolved in TBS supplemented with 0.05% Tween-20 (Sigma-Aldrich) and then incubated overnight with either an HIV-1 p24 polyclonal antibody (Thermo Fisher Scientific), an HA monoclonal antibody (Covance, Princeton, NJ), or an H3 monoclonal antibody (Abcam, Cambridge, Great Britain). The membranes were then washed and subsequently incubated either with horseradish peroxidase (HRP)-conjugated anti-rabbit (Agilent Technologies, Glostrup, Denmark) or anti-mouse (Agilent Technologies) secondary antibodies and visualized by chemiluminescence using a horseradish peroxidase substrate (Thermo Fisher Scientific).
Quantification of PB Transposase Potency and Activity
To test the potency of the LNPs, HeLa cells were seeded in 6-well plates (2 × 105 cells/well) and, the day after, transfected with 900 ng pPBT/PGK-Puro together with 100 ng pCDNA3.1-PL4 as stuffer. 24 hr after transfection, the cells were transduced with 400 ng p24 of either LNP/IN-PCS-hyPBase or LNP/IN-PCS-hyPBmut. After 2, 4, 8, 12, or 24 hr incubation, the virus-containing medium was removed from the transduced cells and the cells were subsequently washed twice in DPBS−/− before addition of standard medium. On day 3, the cells were split into P10 dishes and allowed to form colonies under puromycin selection. For analyses of persistence of hyPBase activity following delivery by either plasmid DNA or in-vitro-transcribed mRNA, HeLa cells were seeded in 6-well plates (2 × 105 cells/well). The day after, for plasmid DNA transfection, cells were transfected with 100 ng pCMV-hyPBase or pCMV-hyPBmut together with 900 ng pCDNA3.1-PL4 as stuffer using Turbofect. For RNA-based delivery, cells were transfected with 150 ng of either rT3TS/hyPBase or rT3TS/hyPBmut using Lipofectamine 2000 according to manufacturer’s instructions. Each replicate constituted of three pooled transfections bringing the total number of transfections per construct up to nine. To quantify colony formation on days 2, 4, 6, 9, and 13, colony-forming assays was carried out as described above. Cells were seeded at a density of 2 × 105 cells/well. On the day of transfection, 900 ng pPBT/PGK-Puro was co-transfected together with 100 ng pCDNA3.1-PL4. To determine short-term activity following either transduction of transposase-loaded LNPs or transfection of transposase mRNA, HeLa cells were seeded in 6-well plates (2 × 105 cells/well). The day after, cells were either transduced with 400 ng p24 of LNP/IN-PCS-hyPBase or LNP/IN-PCS-hyPBmut or transfected with 150 ng rT3TS/hyPBase or rT3TS/hyPBmut. The cells were then incubated for 4, 8, 12, 24, or 48 hr before transfection with 900 ng pPBT/PGK-Puro together with 100 ng pCDNA3.1-PL4. The cells were then incubated for another 24 hr before being split into P10 dishes. For the LNP-transduced cells, two washing steps with DPBS−/− were introduced four hours after transduction to limit exposure of the cells to LNPs.
Statistical Analysis
Two-tailed Student’s t tests were performed to compare differences between two groups. The assumption of equal variances was tested by the F test. Viral transductional titers were compared by statistical analysis on log-transformed data. To compare colony formation of LNPs loaded with IN- or MA-fused PB transposase, a one-way ANOVA with Turkey’s multiple comparisons test was used. In the investigation of LNP-delivered transposase potency and activity over time, a two-way ANOVA was utilized to determine how time and the transposase used affected the results. Bonferroni post-tests were subsequently used to compare hyPBase and hyPBmut at each time point. The results were displayed in the figures with the following annotations: ns, p > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
Author Contributions
K.A.S., R.O.B., and J.G.M. conceived the project and designed the experiments. K.A.S., M.G.N., S.A., L.B.R., and R.O.B performed the experiments. K.A.S. and J.G.M. wrote the manuscript and assembled the figures.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was made possible through support of the Danish Council for Independent Research/Medical Sciences (grant DFF-4004-00220), The Lundbeck Foundation (grant R126-2012-12456), the Hørslev Foundation, Aase og Ejnar Danielsens Fond, Grosserer L. F. Foghts Fond, Agnes og Poul Friis Fond, Oda og Hans Svenningsens Fond, Snedkermester Sophus Jacobsen and Hustru Astrid Jacobsens Fond, and Familien Hede Nielsens Fond. J.G.M. is head of Gene Therapy Initiative Aarhus (GTI-Aarhus) funded by the Lundbeck Foundation and a member of the Aarhus Research Center for Innate Immunology (ARCII) established through funding from the AU-Ideas program at Aarhus University.
Footnotes
Supplemental Information includes three figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.02.006.
Supplemental Information
References
- 1.De Ravin S.S., Reik A., Liu P.Q., Li L., Wu X., Su L., Raley C., Theobald N., Choi U., Song A.H. Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat. Biotechnol. 2016;34:424–429. doi: 10.1038/nbt.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skipper K.A., Mikkelsen J.G. Delivering the goods for genome engineering and editing. Hum. Gene Ther. 2015;26:486–497. doi: 10.1089/hum.2015.063. [DOI] [PubMed] [Google Scholar]
- 4.Cyranoski D. Chinese scientists to pioneer first human CRISPR trial. Nature. 2016;535:476–477. doi: 10.1038/nature.2016.20302. [DOI] [PubMed] [Google Scholar]
- 5.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kebriaei P., Singh H., Huls M.H., Figliola M.J., Bassett R., Olivares S., Jena B., Dawson M.J., Kumaresan P.R., Su S. Phase I trials using Sleeping Beauty to generate CD19-specific CAR T cells. J. Clin. Invest. 2016;126:3363–3376. doi: 10.1172/JCI86721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gil-Farina I., Fronza R., Kaeppel C., Lopez-Franco E., Ferreira V., D’Avola D., Benito A., Prieto J., Petry H., Gonzalez-Aseguinolaza G., Schmidt M. Recombinant AAV integration is not associated with hepatic genotoxicity in nonhuman primates and patients. Mol. Ther. 2016;24:1100–1105. doi: 10.1038/mt.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C.Y., Li J.F., Liou J.S., Charng Y.C., Huang Y.W., Lee H.J. A gene delivery system for human cells mediated by both a cell-penetrating peptide and a piggyBac transposase. Biomaterials. 2011;32:6264–6276. doi: 10.1016/j.biomaterials.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 12.Järver P., Fernaeus S., El-Andaloussi S., Tjörnhammer M.-L., Langel Ü. Co-transduction of sleeping beauty transposase and donor plasmid via a cell-penetrating peptide: a simple one step method. Int. J. Pept. Res. Ther. 2008;14:58–63. [Google Scholar]
- 13.Gaj T., Guo J., Kato Y., Sirk S.J., Barbas C.F., 3rd Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat. Methods. 2012;9:805–807. doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramakrishna S., Kwaku Dad A.B., Beloor J., Gopalappa R., Lee S.K., Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J., Exline C.M., DeClercq J.J., Llewellyn G.N., Hayward S.B., Li P.W., Shivak D.A., Surosky R.T., Gregory P.D., Holmes M.C., Cannon P.M. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling C., Bhukhai K., Yin Z., Tan M., Yoder M.C., Leboulch P., Payen E., Srivastava A. High-efficiency transduction of primary human hematopoietic stem/progenitor cells by AAV6 vectors: strategies for overcoming donor-variation and implications in genome editing. Sci. Rep. 2016;6:35495. doi: 10.1038/srep35495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai Y., Bak R.O., Krogh L.B., Staunstrup N.H., Moldt B., Corydon T.J., Schrøder L.D., Mikkelsen J.G. DNA transposition by protein transduction of the piggyBac transposase from lentiviral Gag precursors. Nucleic Acids Res. 2014;42:e28. doi: 10.1093/nar/gkt1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Y., Bak R.O., Mikkelsen J.G. Targeted genome editing by lentiviral protein transduction of zinc-finger and TAL-effector nucleases. eLife. 2014;3:e01911. doi: 10.7554/eLife.01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai Y., Laustsen A., Zhou Y., Sun C., Anderson M.V., Li S., Uldbjerg N., Luo Y., Jakobsen M.R., Mikkelsen J.G. Targeted, homology-driven gene insertion in stem cells by ZFN-loaded ‘all-in-one’ lentiviral vectors. eLife. 2016;5:e12213. doi: 10.7554/eLife.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi J.G., Dang Y., Abraham S., Ma H., Zhang J., Guo H., Cai Y., Mikkelsen J.G., Wu H., Shankar P., Manjunath N. Lentivirus pre-packed with Cas9 protein for safer gene editing. Gene Ther. 2016;23:627–633. doi: 10.1038/gt.2016.27. [DOI] [PubMed] [Google Scholar]
- 22.Lévy C., Verhoeyen E., Cosset F.L. Surface engineering of lentiviral vectors for gene transfer into gene therapy target cells. Curr. Opin. Pharmacol. 2015;24:79–85. doi: 10.1016/j.coph.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Ding S., Wu X., Li G., Han M., Zhuang Y., Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Wilson M.H., Coates C.J., George A.L., Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 25.Cai Y., Mikkelsen J.G. Driving DNA transposition by lentiviral protein transduction. Mob. Genet. Elements. 2014;4:e29591. doi: 10.4161/mge.29591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schenkwein D., Turkki V., Ahlroth M.K., Timonen O., Airenne K.J., Ylä-Herttuala S. rDNA-directed integration by an HIV-1 integrase--I-PpoI fusion protein. Nucleic Acids Res. 2013;41:e61. doi: 10.1093/nar/gks1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schenkwein D., Turkki V., Kärkkäinen H.R., Airenne K., Ylä-Herttuala S. Production of HIV-1 integrase fusion protein-carrying lentiviral vectors for gene therapy and protein transduction. Hum. Gene Ther. 2010;21:589–602. doi: 10.1089/hum.2009.051. [DOI] [PubMed] [Google Scholar]
- 28.Turkki V., Schenkwein D., Timonen O., Husso T., Lesch H.P., Ylä-Herttuala S. Lentiviral protein transduction with genome-modifying HIV-1 integrase-I-PpoI fusion proteins: studies on specificity and cytotoxicity. BioMed Res. Int. 2014;2014:379340. doi: 10.1155/2014/379340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swanson C.M., Malim M.H. SnapShot: HIV-1 proteins. Cell. 2008;133:742. doi: 10.1016/j.cell.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Saha S., Woodard L.E., Charron E.M., Welch R.C., Rooney C.M., Wilson M.H. Evaluating the potential for undesired genomic effects of the piggyBac transposon system in human cells. Nucleic Acids Res. 2015;43:1770–1782. doi: 10.1093/nar/gkv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grabundzija I., Irgang M., Mátés L., Belay E., Matrai J., Gogol-Döring A., Kawakami K., Chen W., Ruiz P., Chuah M.K. Comparative analysis of transposable element vector systems in human cells. Mol. Ther. 2010;18:1200–1209. doi: 10.1038/mt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woodard L.E., Downes L.M., Lee Y.C., Kaja A., Terefe E.S., Wilson M.H. Temporal self-regulation of transposition through host-independent transposase rodlet formation. Nucleic Acids Res. 2017;45:353–366. doi: 10.1093/nar/gkw1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woltjen K., Hämäläinen R., Kibschull M., Mileikovsky M., Nagy A. Transgene-free production of pluripotent stem cells using piggyBac transposons. Methods Mol. Biol. 2011;767:87–103. doi: 10.1007/978-1-61779-201-4_7. [DOI] [PubMed] [Google Scholar]
- 34.Woltjen K., Michael I.P., Mohseni P., Desai R., Mileikovsky M., Hämäläinen R., Cowling R., Wang W., Liu P., Gertsenstein M. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaji K., Norrby K., Paca A., Mileikovsky M., Mohseni P., Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osborn M., Lonetree C.L., Webber B.R., Patel D., Dunmire S., McElroy A.N., DeFeo A.P., MacMillan M.L., Wagner J., Balzar B.R. CRISPR/Cas9 targeted gene editing and cellular engineering in Fanconi anemia. Stem Cells Dev. 2016 doi: 10.1089/scd.2016.0149. Published online August 18, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie F., Ye L., Chang J.C., Beyer A.I., Wang J., Muench M.O., Kan Y.W. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24:1526–1533. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yusa K. Seamless genome editing in human pluripotent stem cells using custom endonuclease-based gene targeting and the piggyBac transposon. Nat. Protoc. 2013;8:2061–2078. doi: 10.1038/nprot.2013.126. [DOI] [PubMed] [Google Scholar]
- 39.Ye L., Wang J., Beyer A.I., Teque F., Cradick T.J., Qi Z., Chang J.C., Bao G., Muench M.O., Yu J. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. USA. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma N., Moldt B., Dalsgaard T., Jensen T.G., Mikkelsen J.G. Regulated gene insertion by steroid-induced PhiC31 integrase. Nucleic Acids Res. 2008;36:e67. doi: 10.1093/nar/gkn298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jakobsen M., Stenderup K., Rosada C., Moldt B., Kamp S., Dam T.N., Jensen T.G., Mikkelsen J.G. Amelioration of psoriasis by anti-TNF-alpha RNAi in the xenograft transplantation model. Mol. Ther. 2009;17:1743–1753. doi: 10.1038/mt.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilber A., Wangensteen K.J., Chen Y., Zhuo L., Frandsen J.L., Bell J.B., Chen Z.J., Ekker S.C., McIvor R.S., Wang X. Messenger RNA as a source of transposase for sleeping beauty transposon-mediated correction of hereditary tyrosinemia type I. Mol. Ther. 2007;15:1280–1287. doi: 10.1038/sj.mt.6300160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.