Abstract
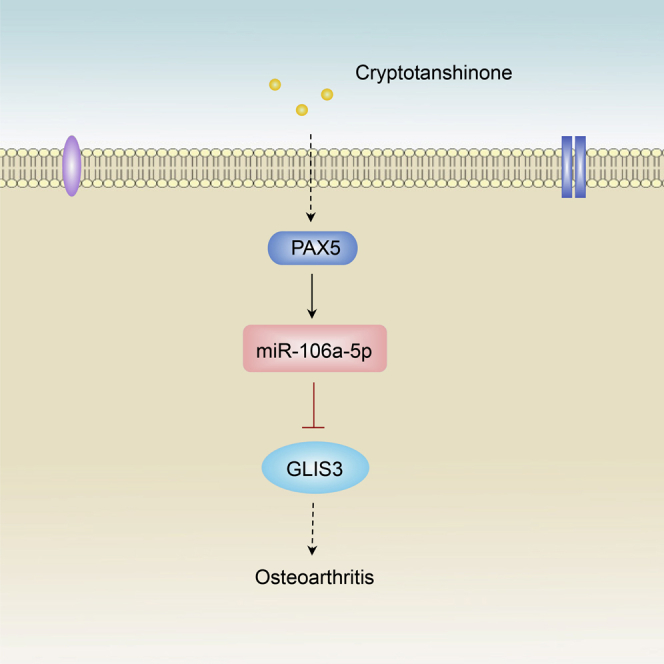
Cryptotanshinone (CTS) has emerged as an anti-inflammatory agent in osteoarthritis (OA). However, the molecular mechanism underlying its potent therapeutic effect on OA remains largely unknown. MicroRNAs (miRNAs) act as crucial regulators in maintaining cartilage homeostasis. To investigate whether CTS protects against developing OA through regulation of miRNAs, we examined the potential CTS-mediated miRNA molecules using microarray analysis. We found that CTS significantly promoted miR-106a-5p expression in chondrocytes. Using the OA mouse model created by anterior cruciate ligament transection, we revealed that intra-articular injection of miR-106a-5p agomir attenuated OA. In addition, miR-106a-5p inhibited GLI-similar 3 (GLIS3) production by directly targeting the 3′ untranslated region. CTS promoted miR-106a-5p expression through recruitment of a member of the paired box (PAX) family of transcription factors, PAX5, to the miR-106a-5p promoter. Inhibition of PAX5 mimicked the effect of miR-106a-5p and abolished the CTS ability to regulate miR-106a-5p expression. In OA patients, miR-106-5p is downregulated which is accompanied by downregulation of PAX5 and upregulation of GLIS3. Collectively, these data highlight that the PAX5/miR-106a-5p/GLIS3 axis acts as a novel pleiotropic regulator in CTS-mediated OA cartilage protection, suggesting that miR-106a-5p and PAX5 activation and GLIS3 inhibition might be useful and attractive for therapeutic strategies to treat OA patients.
Keywords: osteoarthritis, cryptotanshinone, miRNA, GLIS3, PAX5
Graphical Abstract
Introduction
Osteoarthritis (OA) is characterized by alternation of the cartilage extracellular matrix (ECM) composition and homeostasis and represents the most common form of arthritis.1, 2, 3 Articular cartilage degradation and the limited capacity to repair lead to joint integrity disruption and progressive irreversible dysfunction, resulting in disability in the worldwide population.4, 5 Consequently, therapeutic agent approaches coupled with surgical techniques have been developed to treat patients with OA. Although OA genetics have gained attention, the clinical outcomes have not yet been significantly ameliorated. Therefore, the improved identification of candidates for OA patients is urgently required, contributing to enhancing the clinical management of OA.
Cryptotanshinone (CTS), a natural constituent extracted from the Salvia miltiorrhiza Bunge root, has been identified as a powerful antioxidative and anti-inflammatory therapeutic agent.6, 7, 8 CTS suppressed p300-mediated STAT3 acetylation in rheumatoid arthritis (RA).9 In addition, CTS abolished lipopolysaccharide (LPS)-triggered nuclear factor-κB (NF-κB) activation in macrophages.10 Recently, it was revealed that CTS could protect against IL-1β-induced inflammation and ameliorate OA progression.11 However, the exact mechanisms involved in the CTS-mediated cartilage degeneration remain largely unelucidated.
MicroRNAs (miRNAs), a family of endogenous small noncoding RNAs, play an important role in regulating biogenesis, genomic organization, and other key cellular processes through preferential binding to the 3′ untranslated regions (3′-UTRs) of specific mRNA targets as posttranscriptional modulators.12, 13, 14, 15 Currently, accumulating studies have profiled miRNAs that act as regulators in the pathogenesis of OA,16, 17 thus indicating that miRNAs might contribute to the renewed understanding of therapeutic targets and might be utilized as crucial diagnostic biomarkers for OA. Abnormal expression of miRNAs may lead to impairment of normal function. Recently, several miRNAs, such as miR-29, miR-30a, miR-105, miR-221-3p, miR-145 and miR-146a, have been demonstrated to be involved in OA-like pathology and inflammation.17, 18, 19, 20, 21, 22 However, whether CTS protects cartilage against OA could be regulated through miRNAs and whether the miRNAs regulated through CTS participate in the OA pathogenesis remains obscure.
In the current study, we screened potential miRNAs using microarray analysis and found that CTS significantly upregulated miR-106a-5p expression. In addition, miR-106a-5p ameliorated OA histological signs in an OA model created by an anterior cruciate ligament transection (ACLT). We next demonstrated that GLI-similar 3 (GLIS3) was identified as a functional modulator of a novel direct target of miR-106a-5p in OA, and that CTS promoted miR-106a-5p expression though the recruitment of a member of the paired box (PAX) family of transcription factors, PAX5, to the miR-106a-5p promoter. Moreover, miR-106a-5p expression was found to be decreased in OA patients, and was positively correlated with the expression of PAX5 and negatively associated with the expression of GLIS3, which might provide insight into the mechanisms underlying CTS-modulated OA.
Results
Identification of miR-106a-5p as an Important CTS-Regulated miRNA
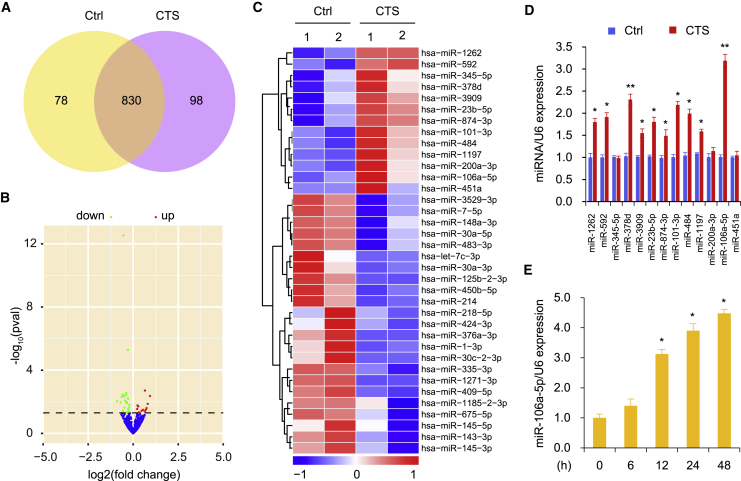
As CTS could protect against developing OA changes and ameliorate cartilage degradation, we investigated the CTS-mediated miRNA targets during the pathogenesis of OA. Microarray analysis was performed to identify the miRNA expression profiles in chondrocytes stimulated with CTS (10 μM). A total of 1,006 significantly differentially expressed miRNAs were identified in CTS-stimulated chondrocytes compared with controls (Figures 1A and 1B). The top 36 miRNAs with altered expression are shown (Figure 1C). These differentially expressed miRNAs in CTS treated cells were selected for the following criteria: |log2(foldchange)| > 1 and p < 0.05. Compared with the control, 13 miRNAs were upregulated and 23 miRNAs were downregulated (p < 0.05). To further validate the miRNA results from microarray analysis, we performed qRT-PCR. A total of 10 upregulated miRNAs (miR-23b-5p, miR-101-3p, miR-106a-5p, miR-378d, miR-484, miR-592, miR-874-3p, miR-1197, miR-1262 and miR-3909) were confirmed. We selected miR-106a-5p for further study, as this molecule was the most vigorously upregulated miRNA (Figure 1D). We next detected miR-106a-5p expression at different times in chondrocytes treated with CTS. The results revealed that CTS-mediated miR-106a-5p expression was significantly promoted after stimulation for 12 hr (Figure 1E), suggesting that CTS promoted the expression of miR-106a-5p in a time-dependent manner in human chondrocytes.
Figure 1.
Cryptotanshinone Regulates the Expression of miRNAs in Chondrocytes
(A) Venn diagram of microarray analysis comparison between control (yellow) and CTS (purple) group indicated that a total of 1,006 miRNAs were differentially expressed (brown). Ctrl, Control; CTS, Cryptotanshinone. (B) A volcano plot indicating differentially regulated miRNA expression. miRNAs upregulated and downregulated are shown in red and green, respectively. Values are presented as the log2 of tag counts. (C) Hierarchical clustering analysis of miRNAs in chondrocytes stimulated with CTS (10 μM). The level of miRNA expression is color coded: red, upregulation; blue, downregulation; white, no change. (D) qRT-PCR measurement of miRNAs upregulated in the microarray assays. (E) Cultured human chondrocytes stimulated with CTS at different times were analyzed for miR-106a-5p expression. Each bar represents the mean ± SD of at least three independent experiments performed in triplicate. *p < 0.05, **p < 0.01.
Injection of miR-106a-5p Agomir Attenuates OA
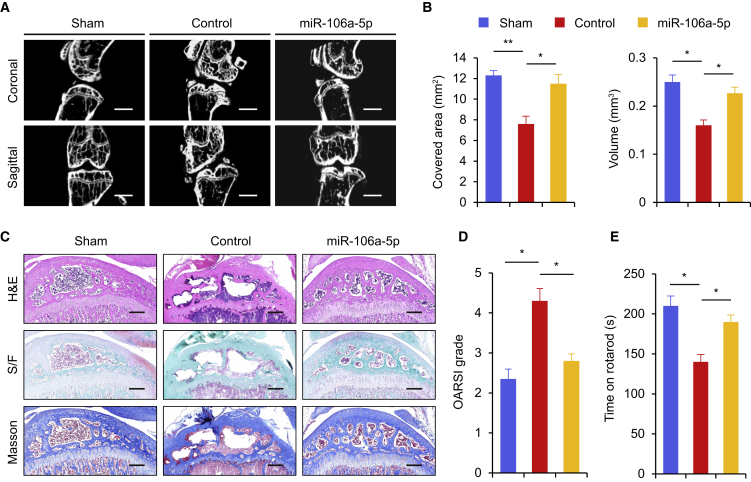
We then investigated the role of miR-106a-5p in OA treatment; 8-week-old male C57BL/6J (wild-type [WT]) mice were operated on for ACLT surgery. After 2 weeks, an intra-articular injection of miR-106a-5p agomir was performed. Cartilage volume and bone area covered by cartilage in the mice ACLT were quantified by microcomputed tomography (μCT) (Figure 2A). The mice that did not receive the injection of miR-106a-5p revealed less covered cartilage area and cartilage volume compared with the mice that received the injection (Figure 2B). Additionally, histopathologic analysis showed that the mice that received the injection of miR-106a-5p displayed less severe OA cartilage degradation (Figure 2C). Articular cartilage degeneration was graded using the Osteoarthritis Research Society International (OARSI)-modified Mankin criteria. The OARSI histologic grading scale of articular cartilage indicated that the mice that received the miR-106a-5p injection scored a lower grade on the OARSI scale than the control group (Figure 2D). Moreover, the rotarod analysis showed that the mice that did not receive the miR-106a-5p injection achieved a decreased rotarod time, while the mice that received the miR-106a-5p injection and without transection revealed no significant differences in rotarod time (Figure 2E). Taken together, these data demonstrated that miR-106a-5p protects the articular cartilage from developing OA.
Figure 2.
miR-106a-5p Protects Mice from Developing OA
(A) Representative μCT images of coronal and sagittal views of the knee joints. Scale bar, 1 μm. (B) Quantitative analysis of surface covered area and cartilage volume in mice (n = 6). (C) H&E, Safranin O/fast green staining (S/F), and Masson trichrome staining were performed to examine the proteoglycan content in the cartilage. Scale bar, 200 μm. (D) OARSI grades of the mice are shown (n = 6); yellow, sham; red, control; yellow, miR-106a-5p. (E) Time of mice on rotarod 8 weeks after an anterior cruciate ligament transection (ACLT) (n = 6); yellow, sham; red, control; yellow, miR-106a-5p. Each bar represents the mean ± SD. *p < 0.05, **p < 0.01.
miR-106a-5p Inhibits GLIS3 Expression by Directly Targeting the 3′-UTR of GLIS3
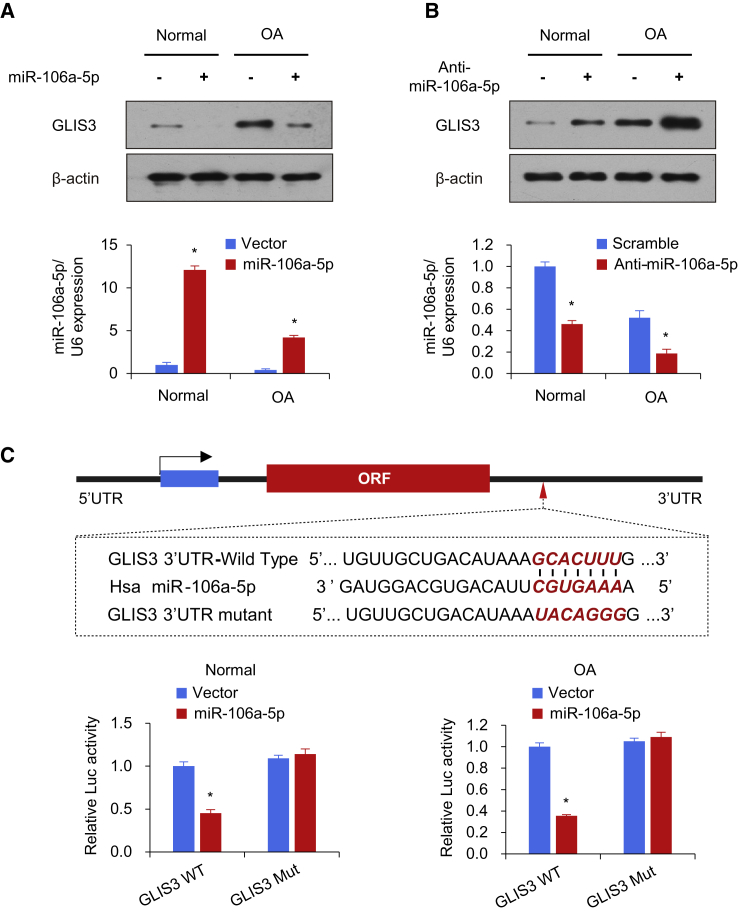
To determine the potential candidate targets of miR-106a-5p, we predicted the probability of a functional binding site of miR-106a-5p using two target prediction programs, TargetScan and miRanda. The results indicated that there were several potential miR-106a-5p-targeting genes involved in OA, including GLIS3, HMGA2, SMAD7, CCND1, and CDK6. As expected, miR-106a-5p inhibited the expression of HMGA2, as previously reported.23 Based on western blot analysis, the results revealed that GLIS3 was significantly downregulated through miR-106a-5p overexpression in human chondrocytes (Figure 3A; Figure S1). In contrast, inhibition of miR-106a-5p promoted GLIS3 expression in human chondrocytes (Figure 3B). Notably, miR-216a did not regulate the GLIS3 mRNA level, indicating that this modulation is posttranscriptional (Figure S2).
Figure 3.
miR-106a-5p Inhibits GLIS3 Expression by Targeting its 3′-UTR
(A and B) Immunoblot analysis of chondrocytes transfected with miR-106a-5p (A) or anti-miR-106a-5p (B). The histograms shown under the immunoblot graphs reveal corresponding miR-106a-5p expression levels. (C) miRNA luciferase reporter assay in human chondrocytes cotransfected with wild-type or mutated GLIS3 reporter and miR-106a-5p. The top panel shows the wild-type and mutant forms of the putative miR-106a-5p target sequences in the GLIS3 3′-UTR. Bold and red italicized font indicate the putative miR-106a-5p binding sites within the human GLIS3 3′-UTR. GLIS3 WT, wild-type GLIS3 3′-UTR; GLIS3 Mut, mutated GLIS3 3′UTR. Each bar represents the mean ± SD of at least three independent experiments performed in triplicate. *p < 0.05, **p < 0.01.
Next, we examined whether GLIS3 is a direct and specific target of miR-106a-5p. Human chondrocytes were transiently co-transfected with a GLIS3 3′-UTR wild-type reporter construct or the mutated luciferase reporter and the miR-106a-5p mimics. The results showed that miR-106a-5p inhibited the 3′-UTR reporter activity of GLIS3 in chondrocytes but did not have an effect on the activity of the reporter in which the binding sites for miR-106a-5p were mutated (Figure 3C). Therefore, these findings collectively indicate that miR-106a-5p suppresses GLIS3 expression by directly targeting its 3′-UTR in human chondrocytes.
CTS Promoted the Expression of miR-106a-5p through the Recruitment of PAX5 to the Promoter of miR-106a-5p
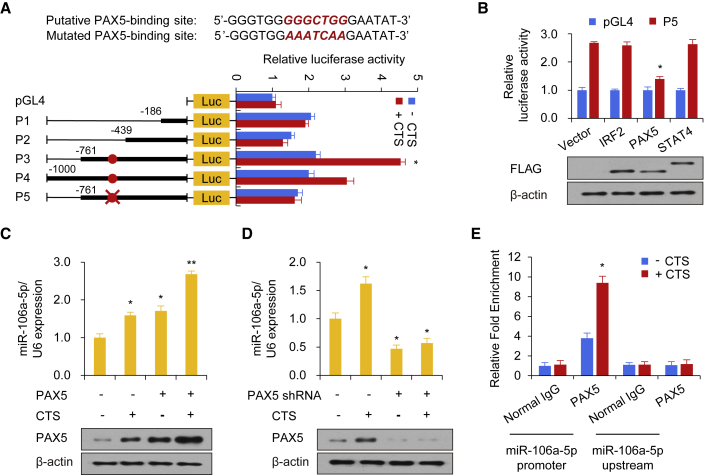
To investigate the molecular mechanism of CTS-induced miR-106a-5p expression, several regions of the miR-106a-5p promoter were generated. We then examined the regulatory regions that participated in the CTS promotion of miR-106a-5p transcription in human chondrocytes. According to the analysis of different miR-106a-5p promoter deletion reporter constructs, the promoter region from −439 to −761 base pairs (bp) contained a CTS-induced element (Figure 4A), where three putative transcriptional factors (IRF2, PAX5, and STAT4) were predicted to be capable of binding.
Figure 4.
CTS Promotes the Expression of miR-106a-5p through the Recruitment of PAX5 to the Promoter of miR-106a-5p
(A) Luciferase activity of different miR-106a-5p promoter constructs in cultured human chondrocytes with or without CTS stimulation. P1, promoter (−186 bp); P2, promoter (−439 bp); P3, promoter (−761 bp); P4, promoter (−1,000 bp); P5, mutated promoter (−761 bp). (B) Luciferase assay in cultured human chondrocytes infected with FLAG-tagged IRF2 or PAX5 or STAT4 and P5. Immunoblot shows the expression of IRF2, PAX5, and STAT4 with anti-FLAG. (C and D) Cultured human chondrocytes were infected with the PAX5 plasmid (C) and the vector control or shRNA for PAX5 (D), followed by stimulation with CTS as described in the Methods section. Immunoblot indicates the expression of PAX5. (E) Chromatin immunoprecipitation (ChIP) assay for PAX5 occupancy on the miR-106a-5p promoter or upstream of the promoter in cultured human chondrocytes. Each bar represents the mean ± SD of at least three independent experiments performed in triplicate. *p < 0.05, **p < 0.01.
We next evaluated the crucial transcriptional factors involved in the modulation of miR-106a-5p transcription. Using the reporter assay, we demonstrated that PAX5, but not IRF2 or STAT4, promoted the miR-106a-5p promoter activity described above, thus suggesting that PAX5 might be the transcriptional factor participating in the regulation of miR-106a-5p promoter reporter activity through CTS (Figure 4B). In addition, the putative PAX5-binding site mutation in this region resulted in the loss of the promotive effect of CTS on miR-106a-5p transcription (Figure 4A).
PAX5 Is Required for the CTS-Mediated Expression of miR-106a-5p
We further examined the effects of PAX5 on the expression of miR-106a-5p. PAX5 overexpression promoted miR-106a-5p production in human chondrocytes (Figure 4C). CTS stimulation increased the expression of miR-106a-5p and PAX5. Additionally, CTS cooperated with PAX5 to upregulate miR-106a-5p expression (Figure 4C). Moreover, the knockdown of endogenous PAX5 inhibited miR-106a-5p expression and abolished the ability of CTS to modulate miR-106a-5p production (Figure 4D), indicating that PAX5 is important for CTS-mediated miR-106a-5p expression.
In addition, the chromatin immunoprecipitation (ChIP) assay revealed that PAX5 was recruited to the promoter of miR-106a-5p but not to the region approximately 1.0-kb upstream of the miR-106a-5p promoter (Figure 4E), and this recruitment was enhanced after treatment with CTS. Taken together, these findings reveal that CTS promotes miR-106a-5p transcription through the enhanced recruitment of PAX5 to the promoter of miR-106a-5p.
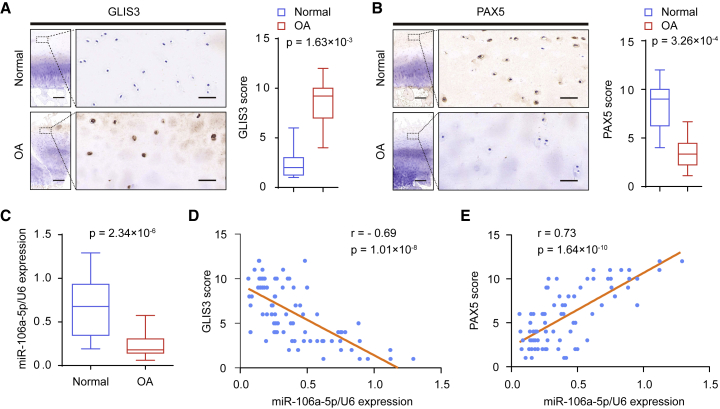
miR-106a-5p Is Downregulated in OA Patients and Is Positively Correlated with PAX5 Expression and Negatively Correlated with GLIS3 Expression
To investigate the potential role of GLIS3, PAX5, and miR-106a-5p in the pathogenesis of OA, we detected the expression of GLIS3 and PAX5 using immunohistochemistry, and assessed the levels of miR-106a-5p using qRT-PCR in 40 pairs of OA cartilage tissues and corresponding normal cartilage samples (Figures 5A–5C). In OA patients, miR-106-5p is downregulated which is accompanied by downregulation of PAX5 and upregulation of GLIS3 (Figures 5A–5C).
Figure 5.
Expression of GLIS3, PAX5, and miR-106a-5p in OA Patients
(A) Representative immunohistochemistry assay of GLIS3 in normal and OA cartilage tissues. Scale bar, left, 500 μm; right, 50 μm. The scores of GLIS3 in normal and OA cartilage tissues based on the immunohistochemistry assay are shown. (B) Representative immunohistochemistry assay of PAX5 in normal and OA cartilage tissues. Scale bar, left, 500 μm; right, 50 μm. The scores of PAX5 in normal and OA cartilage tissues based on the immunohistochemistry assay. (C) qRT-PCR measurement of miR-106a-5p in normal and OA cartilage tissues. Data are expressed as the mean ± SD. (D and E) The relationship among GLIS3 (D), PAX5 (E), and miR-106a-5p expression was determined through Pearson’s χ2 analysis.
We next performed correlation analysis. The results demonstrated a significant negative relationship between miR-106a-5p and GLIS3 in OA cartilage tissues, which was consistent with the miR-106a-5p-mediated inhibition of GLIS3 expression in human chondrocytes (Figure 5D). Moreover, the expression of miR-106a-5p was positively correlated with that of PAX5 in OA cartilage tissues (Figure 5E). Taken together, these data suggest crucial pathological roles for miR-106a-5p, PAX5, and GLIS3 in OA.
Discussion
Currently, accumulating studies have indicated miRNAs as crucial modulators in regulating various biological processes and cellular functions.24, 25, 26, 27 Aberrant miRNA expression is closely associated with cartilage degeneration, degradation, and regeneration in the pathogenesis of OA.28, 29, 30 Hence, a better understanding of the miRNA-mediated networks might open novel gates for the mechanism of cartilage degradation and facilitate therapeutic treatment for patients with OA. Studies have implicated that miR-106a-5p participated in the proliferation, invasion, metastasis, and progression of various types of cancers, including osteosarcoma,23 renal cell carcinoma,31 hepatocellular carcinoma,32, 33 and colon cancer.34 However, the exact role and function of miR-106a-5p in OA is still unknown. In the present study, we found that miR-106a-5p protected against developing OA cartilage degradation and ameliorated cartilage damage by directly targeting GLIS3. Moreover, miR-106a-5p expression was significantly decreased in patients with OA, suggesting that miR-106a-5p exerts a protective role in OA.
CTS is an active compound extracted from the root of Salvia miltiorrhiza Bunge. Studies have demonstrated that CTS exerts its therapeutic potential roles on diverse diseases, such as malignant glioma, prostate cancer, rheumatoid arthritis, and cardiac fibrosis, among others.7, 9, 35, 36, 37, 38, 39 A recent report revealed that CTS could exert effects as an anti-inflammatory agent in OA.11 However, the mechanism of CTS-mediated OA therapy has not been fully elucidated. In addition, whether CTS could protect cartilage against developing OA perhaps regulated through miRNAs and whether the miRNAs regulated through CTS participate in the OA pathogenesis remain obscure. In this study, we found that CTS promoted the expression of miR-106a-5p through the recruitment of the transcription factor PAX5 to the promoter of miR-106a-5p in human chondrocytes, providing a novel mechanism for CTS-mediated OA cartilage amelioration in human OA.
GLIS3, a member of the Glis subfamily of Krüppel-like zinc-finger transcription factors, was characterized as modulating a set of essential cellular functions, such as insulin production, β-cell maintenance and maturation, stem and progenitor cell differentiation, and development.40, 41 Although studies converge on GLIS3 functioning in cancer, diabetes, hypothyroidism, and polycystic kidney disease,40, 42, 43, 44 the exact role of GLIS3 in human OA is unknown, and the miRNA regulating the expression of GLIS3 remains unclear. In this study, we demonstrated that GLIS3 was a direct target of miR-106a-5p. We also identified that the high expression levels of GLIS3 were more frequent in OA cartilage tissues than the normal tissues. Conceivably, GLIS3 downregulation could be a promising molecular strategy for OA therapy. Additionally, GLIS3 was negatively correlated with miR-106a-5p expression, revealing that the miR-106a-5p/GLIS3 axis might be a novel target for the prevention of OA.
PAX5 acts as a potent transcription factor that plays an important role in cancerous processes and development.45 Emerging information on PAX5 indicates that abnormal functions of PAX5 protein contribute to breast cancer, bladder cancer, and HIV.46, 47, 48 GWAS studies have revealed that in B-cell precursor acute lymphoblastic leukemia (B-ALL), leukemic cells from all affected individuals in both families exhibited 9p deletion, with loss of heterozygosity and retention of the mutant PAX5 allele at 9p13, leading to the reduced function of transcriptional activity.49, 50, 51 However, the exact role of PAX5 in human OA remains unelucidated, and the relationship between PAX5, CTS, and miRNAs is unclear. In this study, we demonstrated that CTS promoted the expression of miR-106a-5p through the recruitment of PAX5 to the promoter of miR-106a-5p. Furthermore, introduction of PAX5 in CTS-stimulated chondrocytes promoted the protective ability of CTS in the pathogenesis of OA. Moreover, we found decreased expression levels of PAX5 in OA cartilage tissues, and PAX5 was positively correlated with miR-106a-5p expression, suggesting that PAX5 and miR-106a-5p act in the same pathway. Downregulation of miR-106a-5p in OA patients is also associated with downregulation of PAX5, since PAX5 regulates miR-106a-5p, the effect of miR-106a-5p might be a consequence of the reduction in PAX5 expression. Thus, PAX5 might be the primary effect in these patients, and that the CTS/PAX5/miR-106a-5p axis might be a promising therapeutic intervention for OA.
Collectively, our findings demonstrated that CTS protected cartilage against developing OA through the PAX5/miR-106a-5p/GLIS3 axis. miR-106a-5p can suppress articular cartilage degradation by targeting GLIS3 expression. miR-106a-5p was downregulated in OA patients and was positively correlated with the expression of PAX5 and negatively associated with the expression of GLIS3, thus suggesting that the CTS/PAX5/miR-106a-5p/GLIS3 axis may be an attractive therapeutic strategy for treating OA patients. Furthermore, GWAS studies of single nucleotide polymorphisms in PAX5 or miR-106a-5p associated with risk of OA need to be investigated in the following future study, contributing to provide with new promising options for the treatment of OA.
Materials and Methods
Patients and Specimens
Articular cartilage samples were collected from 40 patients with knee OA undergoing knee arthroplasty surgery. OA was macroscopically diagnosed according to the Modified Outerbridge Classification. Specimens that included all cartilage layers and subchondral bone were separately harvested from sites of the tibial plateau. Clinical information was collected from patient records. All human studies were conducted with informed consent of the patients and were approved through the institutional ethics review board (No. 20090611-3) of the General Hospital of the People’s Liberation Army (Beijing, China). linical and demographic characteristics of the study population are shown in Table S1.
Cell Isolation and Culture
Chondrocytes were isolated as described previously.19 Briefly, normal chondrocytes were obtained from the knees of patients who had died of diseases unrelated to arthritis. Arthritic chondrocytes were obtained from patients undergoing elective total knee arthroplasty for end-stage OA according to a modified Outerbridge scale (Grade III: maximal fibrillation). The chondrocytes were incubated in high glucose DMEM with 10% fetal calf serum (FCS), 100 IU/mL penicillin and 100 μg/mL streptomycin at 37°C in an atmosphere of 5% CO2. The characterization of the chondrocyte phenotype was established through the analysis of COL2/COL1 gene expression. First-passage chondrocytes at 85% confluence were used for all experiments.
Treatment with CTS
The chondrocytes (serum-starved overnight) were maintained at 2.1 × 105 cells per well in 250 μL of medium in 12-well plates, stimulated with CTS (10 μM) for the indicated time, and were subsequently used for further analysis.
RNA Extraction and qRT-PCR
Total RNA from tissues or cultured chondrocyte samples containing miRNA was extracted and reverse-transcribed into cDNA using a RNeasy Mini kit (QIAGEN, Valencia, CA, USA), according to the manufacturer’s instructions. The expression of mRNAs and mature miRNAs was determined using SYBR Premix Ex Taq Master Mix (2×) (Takara, Mountain View, CA, USA), as previously described.22 The relative expression level of the target was calculated using the comparative cycle threshold (Ct) method. RNU6B or β-actin was used as an internal control to normalize sample differences. The sequences of the primers used for qRT-PCR analysis are presented in Table S2.
Microarray Analysis
For miRNA expression analysis, total RNA, including miRNA, was isolated from chondrocytes stimulated with CTS (10 μM) according to the manufacturer’s instructions and analyzed using Affymetrix GeneChip miRNA 2.0 arrays (Affymetrix, Santa Clara, CA, USA) containing 4,560 probe sets for human small RNAs. All of the steps of the procedure were performed according to the standardized protocol for Affymetrix miRNA 2.0 arrays. Briefly, 1 μg of total RNA of each sample was subjected to a tailing reaction labeled with the Flashtag RNA labeling kit (Genisphere, Hatfield, PA) followed by ligation of the biotinylated signal molecule to the RNA sample according to the manufacturer’s instructions. Each sample was then hybridized to a GeneChip 2.0 miRNA Array at 48°C for 16 hr and then washed and stained on a Fluidics Station 450. After staining, the arrays were scanned on an Affymetrix 3000 GeneScanner. The intensity values for miRNA transcripts were calculated using Affymetrix GeneChip Command Console 3.2. The quality control for the microarray was performed with the Affymetrix miRNA QC Tool. The miRNA expression microarray data were deposited according to minimum information about a microarray experiment (MIAME) guidelines to the Gene Expression Omnibus database. Statistical comparisons were performed using ANOVA.
ACLT Surgery
Eight-week-old male mice underwent ACL surgical transection of the right knee to induce mechanical instability and create an experimental OA model. Sham operations were performed on independent mice. For the mice, we analyzed them 8 weeks after surgery. Investigators were blinded to the genotype of the mice when the surgical transection was performed. The OARSI system (grades 0–6) was used to quantify OA severity, which was also evaluated by observers who were blinded to the experimental group.
Intra-articular Administration
Mice were anesthetized with 3% isoflurane, and then the articular joint was shaved. Mice were treated with miRNA control (5 nmol) and miRNA-106a-5p agomir (5 nmol) via injection with 33-gauge needles (Hamilton Company) and 25 μL CASTIGHT syringes (Hamilton Company).
μCT Analysis
Mouse knee joints were harvested, and soft tissues were dissected. The remaining tissues were stored in 70% ethanol. CT scanning was performed using high-resolution μCT (Skyscan 1172). Images were analyzed and reconstructed with CTAn v1.9 and Nrecon v1.6. Three-dimensional model visualization software (CTVol v2.0) was also used. A voltage of 50 kVp, a resolution of 5.7 μm per pixel, and a current of 200 μA were set for the scanner. The transverse, coronal and sagittal images of the knee joints were used for analyses. The region of interest covering the surface area of the tibia and cartilage volume was collected. After scanning, each sample was assigned a random number for ensuring blinded assessment and then was processed for image analysis.
Rotarod Analysis
Mice were put onto an accelerating rotarod (Ugo Basile). Briefly, the first round time was recognized as the duration of staying atop the rod until the first failure. Mice at different time points after surgery were randomly assigned to different groups. Each trial had a maximum time of 5 min. Mice were given an inter-trial rest interval of 30 min.
Plasmids, Lentiviruses, and Reagents
Wild-type or mutant promoter luciferase reporters were generated through the insertion of PCR-amplified promoter fragments from genomic DNA into the pGL4-Basic vector (Promega, Madison, WI, USA). Lentiviral vectors overexpressing PAX5 tagged with FLAG were obtained through lentiviral transduction using the pCDH plasmid (System Biosciences). Lentiviral shRNA vectors were generated after cloning short hairpin RNA (shRNA) fragments into the lentiviral vector pSIH-H1-Puro (System Biosciences, Mountain View, CA, USA). The lentiviruses were produced through the co-transfection of HEK293T cells with recombinant lentivirus vectors and pPACK Packaging Plasmid Mix (System Biosciences, Mountain View, CA, USA) using Megatran reagent (Origene, Rockville, MD, USA). Lentiviruses were collected at 48 hr after transfection and added to the medium of target cells with 8 μg/mL Polybrene (Sigma-Aldrich, St. Louis, MO, USA).
Cryptotanshinone (CTS) (C5624) and antibodies to Flag (A8592) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Antibodies to SMAD7 (42-0400), GLIS3 (PA5-41677), CCND1 (AHF0082), IRF2 (700226), and PAX5 (PA1-109) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Antibodies to HMGA2 (ab97276), CDK6 (ab124821), STAT4 (ab215428) and β-actin (ab8226) were purchased from Abcam (Cambridge, MA, USA). Chromatin immunoprecipitation kits were purchased from Merck Millipore (Billerica, MA, USA).
Transfection
miRNA mimics were transfected into human normal and OA chondrocytes (passage 1) using FuGENE HD (Promega, Madison, WI, USA), according to the manufacturer’s instructions. miRNA inhibitors (Ambion, Grand Island, NY, USA) were transfected at a concentration of 50 nM.
Luciferase Assay
The chondrocytes were harvested and examined for β-galactosidase and luciferase activities as previously described.22 Cells seeded into 24-well plates were cotransfected with miR-106a-5p and luciferase reporter constructs containing wild-type or mutated GLIS3 3′UTR. The chondrocytes were harvested 48h after transfection and washed with PBS. The cell extracts were assayed for luciferase activity using the luciferase assay system (Promega) and β-galactosidase activity was used as an internal control for transfection efficiency. At least three independent experiments were performed with triplicate results.
Western Blotting
Total protein extracts from tissues or cultured chondrocytes were prepared for western blot analysis as previously described.22 The membranes were incubated with antibodies to GLIS3 (1:500 dilution), PAX5 (1:500 dilution), IRF2 (1:100 dilution), STAT4 (1:1,000 dilution), HMGA2 (1:500 dilution), SMAD7 (1:100 dilution), CCND1 (1:1,000 dilution) and CDK6 (1:5,000 dilution), FLAG-HRP (1:3,000 dilution), and β-actin (1:500 dilution). The immunocomplexes were visualized through chemiluminescence using an ECL kit (Amersham Biosciences, Piscataway, NJ, USA).
ChIP
A ChIP assay was performed according to the manufacturer’s instructions. Briefly, chondrocytes were fixed in 37% formaldehyde, pelleted, and resuspended in lysis buffer. The cells were sonicated and centrifuged to remove insoluble material. The supernatants were collected, and Pellet Protein G magnetic beads were added and incubated for 1 hr at 4°C with antibodies against PAX5 (1:50 dilution). The chromatin was collected, purified, and de-crosslinked at 62°C for 2 hr, followed by incubation at 95°C for 10 min. The precipitated DNA fragments were quantified through RT-PCR analysis.
Histology and Immunohistochemistry
Human cartilage tissues were fixed in 4% buffered paraformaldehyde for 48 hr and subsequently decalcified with buffered EDTA (20% EDTA, pH 7.4). The tissues were embedded in paraffin, sectioned and stained with hematoxylin-eosin (HE), Safranin O/Fast Green and Masson’s trichrome. The histopathological changes were blindly scored using the modified Mankin grading system.
For the IHC assay, briefly, the articular cartilage sections were pre-treated for 10 min with trypsin (0.05%) prior to treatment with 3% (vol/vol) H2O2 for 15 min. Subsequently, the sections were blocked with 10% goat serum for 1 hr at room temperature. After washing with PBS, antibodies to GLIS3 (1:10 dilution) and PAX5 (1:50 dilution) were applied to the sections and incubated overnight at 4°C. The sections were subsequently washed with PBS and incubated for 15 min with biotinylated secondary antibodies, followed by incubation for 30 min using the Histostain Plus kit (Invitrogen, Carlsbad, CA, USA). Lastly, the sections were washed and incubated for 2 min with 3,3′-diaminobenzidine (DAB) substrate. Using light microscopy, IHC staining was detected by two experienced pathologists blindly reviewing the stained tissue sections. The widely accepted German semiquantitative scoring system, considering the staining intensity and area extent, was used: 0, no staining; 1, weak staining; 2, moderate staining; and 3, strong staining. In addition, the percentage of staining was given a score of 0 (< 5%), 1 (5%–25%), 2 (25%–50%), 3 (51%–75%), or 4 (>75%). These two scores were multiplied as the final score. For IHC staining of GLIS3 or PAX5, we defined a 0 score as negative and scores of 1-12 as positive.
Statistical Analysis
The miRNA statistical analysis was examined using the GeneSifter v. 4.0 (VizX Labs, Seattle, WA). A two-tail t test with Benjamini and Hochberg correction was used. All data were log transformed. miRNA clusters were identified by web-based programs MirClust (http://fgfr.ibms.sinica.edu.tw/MetaMirClust/MetaMirClustSearch.php) and miRGen Cluster (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=mirgenv3%2Findex). The evaluation of qRT-PCR data was performed using one-way ANOVA with Tukey’s post hoc test. The correlation of the expression of miR-106a-5p, GLIS3 and PAX5 was examined through Pearson’s χ2 analysis using GraphPad PRISM 6 (GraphPad, San Diego, CA, USA). All statistical tests were two-sided. Statistical calculations were performed using SPSS 17.0. The data are presented as mean ± SD. P < 0.05 was considered statistically significant.
Author Contributions
Y.W. conceived the project and designed the experiments; Q.J., X.X., Y.X., Q.S., and Z.F. performed experiments; Q.J. and Y.W. wrote the manuscript; all authors analyzed data; and W.M., S.G., and Q.Y. supervised the project.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
This study was financially supported through grants from the National Natural Science Foundation (81672195, 81472589, and 81371976) and the National Programs for High Technology Research and Development (2015AA033701).
Footnotes
Supplemental Information includes two figures and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.02.001.
Contributor Information
William J. Maloney, Email: wmaloney@stanford.edu.
Yan Wang, Email: yanwang301@126.com.
Supplemental Information
References
- 1.Glyn-Jones S., Palmer A.J., Agricola R., Price A.J., Vincent T.L., Weinans H., Carr A.J. Osteoarthritis. Lancet. 2015;386:376–387. doi: 10.1016/S0140-6736(14)60802-3. [DOI] [PubMed] [Google Scholar]
- 2.Goldring S.R., Goldring M.B. Changes in the osteochondral unit during osteoarthritis: structure, function and cartilage-bone crosstalk. Nat. Rev. Rheumatol. 2016;12:632–644. doi: 10.1038/nrrheum.2016.148. [DOI] [PubMed] [Google Scholar]
- 3.Mobasheri A., Rayman M.P., Gualillo O., Sellam J., van der Kraan P., Fearon U. The role of metabolism in the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2017;13:302–311. doi: 10.1038/nrrheum.2017.50. [DOI] [PubMed] [Google Scholar]
- 4.Loeser R.F., Collins J.A., Diekman B.O. Ageing and the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2016;12:412–420. doi: 10.1038/nrrheum.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malfait A.M. Osteoarthritis year in review 2015: biology. Osteoarthritis Cartilage. 2016;24:21–26. doi: 10.1016/j.joca.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi J., Wang H., Guan H., Shi S., Li Y., Wu X., Li N., Yang C., Bai X., Cai W. IL10 inhibits starvation-induced autophagy in hypertrophic scar fibroblasts via cross talk between the IL10-IL10R-STAT3 and IL10-AKT-mTOR pathways. Cell Death Dis. 2016;7:e2133. doi: 10.1038/cddis.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu C.F., Klauck S.M., Efferth T. Anticancer activity of cryptotanshinone on acute lymphoblastic leukemia cells. Arch. Toxicol. 2016;90:2275–2286. doi: 10.1007/s00204-015-1616-4. [DOI] [PubMed] [Google Scholar]
- 8.Liu Z., Xu S., Huang X., Wang J., Gao S., Li H., Zhou C., Ye J., Chen S., Jin Z.G., Liu P. Cryptotanshinone, an orally bioactive herbal compound from Danshen, attenuates atherosclerosis in apolipoprotein E-deficient mice: role of lectin-like oxidized LDL receptor-1 (LOX-1) Br. J. Pharmacol. 2015;172:5661–5675. doi: 10.1111/bph.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Zhou C., Gao H., Li C., Li D., Liu P., Huang M., Shen X., Liu L. Therapeutic effect of Cryptotanshinone on experimental rheumatoid arthritis through downregulating p300 mediated-STAT3 acetylation. Biochem. Pharmacol. 2017;138:119–129. doi: 10.1016/j.bcp.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Tang S., Shen X.Y., Huang H.Q., Xu S.W., Yu Y., Zhou C.H., Chen S.R., Le K., Wang Y.H., Liu P.Q. Cryptotanshinone suppressed inflammatory cytokines secretion in RAW264.7 macrophages through inhibition of the NF-κB and MAPK signaling pathways. Inflammation. 2011;34:111–118. doi: 10.1007/s10753-010-9214-3. [DOI] [PubMed] [Google Scholar]
- 11.Feng Z., Zheng W., Li X., Lin J., Xie C., Li H., Cheng L., Wu A., Ni W. Cryptotanshinone protects against IL-1β-induced inflammation in human osteoarthritis chondrocytes and ameliorates the progression of osteoarthritis in mice. Int. Immunopharmacol. 2017;50:161–167. doi: 10.1016/j.intimp.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 12.Hamam R., Hamam D., Alsaleh K.A., Kassem M., Zaher W., Alfayez M., Aldahmash A., Alajez N.M. Circulating microRNAs in breast cancer: novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017;8:e3045. doi: 10.1038/cddis.2017.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skommer J., Rana I., Marques F.Z., Zhu W., Du Z., Charchar F.J. Small molecules, big effects: the role of microRNAs in regulation of cardiomyocyte death. Cell Death Dis. 2014;5:e1325. doi: 10.1038/cddis.2014.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDermott R., Gabikian P., Sarvaiya P., Ulasov I., Lesniak M.S. MicroRNAs in brain metastases: big things come in small packages. J. Mol. Med. (Berl.) 2013;91:5–13. doi: 10.1007/s00109-012-0971-3. [DOI] [PubMed] [Google Scholar]
- 15.Bak R.O., Hollensen A.K., Mikkelsen J.G. Managing microRNAs with vector-encoded decoy-type inhibitors. Mol. Ther. 2013;21:1478–1485. doi: 10.1038/mt.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J.F., Zhang S.J., Zhao C., Qiu B.S., Gu H.F., Hong J.F., Cao L., Chen Y., Xia B., Bi Q., Wang Y.P. Altered microRNA expression profile in synovial fluid from patients with knee osteoarthritis with treatment of hyaluronic acid. Mol. Diagn. Ther. 2015;19:299–308. doi: 10.1007/s40291-015-0155-2. [DOI] [PubMed] [Google Scholar]
- 17.Le L.T., Swingler T.E., Crowe N., Vincent T.L., Barter M.J., Donell S.T., Delany A.M., Dalmay T., Young D.A., Clark I.M. The microRNA-29 family in cartilage homeostasis and osteoarthritis. J. Mol. Med. (Berl.) 2016;94:583–596. doi: 10.1007/s00109-015-1374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng X., Zhao F.C., Pang Y., Li D.Y., Yao S.C., Sun S.S., Guo K.J. Downregulation of miR-221-3p contributes to IL-1β-induced cartilage degradation by directly targeting the SDF1/CXCR4 signaling pathway. J. Mol. Med. (Berl.) 2017;95:615–627. doi: 10.1007/s00109-017-1516-6. [DOI] [PubMed] [Google Scholar]
- 19.Ji Q., Xu X., Xu Y., Fan Z., Kang L., Li L., Liang Y., Guo J., Hong T., Li Z. miR-105/Runx2 axis mediates FGF2-induced ADAMTS expression in osteoarthritis cartilage. J. Mol. Med. (Berl.) 2016;94:681–694. doi: 10.1007/s00109-016-1380-9. [DOI] [PubMed] [Google Scholar]
- 20.Hu G., Zhao X., Wang C., Geng Y., Zhao J., Xu J., Zuo B., Zhao C., Wang C., Zhang X. MicroRNA-145 attenuates TNF-α-driven cartilage matrix degradation in osteoarthritis via direct suppression of MKK4. Cell Death Dis. 2017;8:e3140. doi: 10.1038/cddis.2017.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X., Wang C., Zhao J., Xu J., Geng Y., Dai L., Huang Y., Fu S.C., Dai K., Zhang X. miR-146a facilitates osteoarthritis by regulating cartilage homeostasis via targeting Camk2d and Ppp3r2. Cell Death Dis. 2017;8:e2734. doi: 10.1038/cddis.2017.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji Q., Xu X., Zhang Q., Kang L., Xu Y., Zhang K., Li L., Liang Y., Hong T., Ye Q., Wang Y. The IL-1β/AP-1/miR-30a/ADAMTS-5 axis regulates cartilage matrix degradation in human osteoarthritis. J. Mol. Med. (Berl.) 2016;94:771–785. doi: 10.1007/s00109-016-1418-z. [DOI] [PubMed] [Google Scholar]
- 23.He Q.Y., Wang G.C., Zhang H., Tong D.K., Ding C., Liu K., Ji F., Zhu X., Yang S. miR-106a-5p suppresses the proliferation, migration, and invasion of osteosarcoma cells by targeting HMGA2. DNA Cell Biol. 2016;35:506–520. doi: 10.1089/dna.2015.3121. [DOI] [PubMed] [Google Scholar]
- 24.Schwarzenbach H., Nishida N., Calin G.A., Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen L.H., Diao H.J., Chew S.Y. MicroRNAs and their potential therapeutic applications in neural tissue engineering. Adv. Drug Deliv. Rev. 2015;88:53–66. doi: 10.1016/j.addr.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 26.McManus D.D., Freedman J.E. MicroRNAs in platelet function and cardiovascular disease. Nat. Rev. Cardiol. 2015;12:711–717. doi: 10.1038/nrcardio.2015.101. [DOI] [PubMed] [Google Scholar]
- 27.Nassar F.J., Nasr R., Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol. Ther. 2017;172:34–49. doi: 10.1016/j.pharmthera.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Vicente R., Noël D., Pers Y.M., Apparailly F., Jorgensen C. Deregulation and therapeutic potential of microRNAs in arthritic diseases. Nat. Rev. Rheumatol. 2016;12:211–220. doi: 10.1038/nrrheum.2015.162. [DOI] [PubMed] [Google Scholar]
- 29.Miyaki S., Asahara H. Macro view of microRNA function in osteoarthritis. Nat. Rev. Rheumatol. 2012;8:543–552. doi: 10.1038/nrrheum.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beyer C., Zampetaki A., Lin N.Y., Kleyer A., Perricone C., Iagnocco A., Distler A., Langley S.R., Gelse K., Sesselmann S. Signature of circulating microRNAs in osteoarthritis. Ann. Rheum. Dis. 2015;74:e18. doi: 10.1136/annrheumdis-2013-204698. [DOI] [PubMed] [Google Scholar]
- 31.Pan Y.J., Wei L.L., Wu X.J., Huo F.C., Mou J., Pei D.S. MiR-106a-5p inhibits the cell migration and invasion of renal cell carcinoma through targeting PAK5. Cell Death Dis. 2017;8:e3155. doi: 10.1038/cddis.2017.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J., Huang J., Wang W., Xu J., Yin M., Cheng N., Yin J. Long non-coding RNA Fer-1-like protein 4 acts as a tumor suppressor via miR-106a-5p and predicts good prognosis in hepatocellular carcinoma. Cancer Biomark. 2017;20:55–65. doi: 10.3233/CBM-170090. [DOI] [PubMed] [Google Scholar]
- 33.Hu B., Cai H., Zheng R., Yang S., Zhou Z., Tu J. Long non-coding RNA 657 suppresses hepatocellular carcinoma cell growth by acting as a molecular sponge of miR-106a-5p to regulate PTEN expression. Int. J. Biochem. Cell Biol. 2017;92:34–42. doi: 10.1016/j.biocel.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Yue B., Sun B., Liu C., Zhao S., Zhang D., Yu F., Yan D. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 2015;106:1323–1332. doi: 10.1111/cas.12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu L., Zhang S., Li C., Zhou C., Li D., Liu P., Huang M., Shen X. Cryptotanshinone inhibits human glioma cell proliferation in vitro and in vivo through SHP-2-dependent inhibition of STAT3 activation. Cell Death Dis. 2017;8:e2767. doi: 10.1038/cddis.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park I.J., Kim M.J., Park O.J., Park M.G., Choe W., Kang I., Kim S.S., Ha J. Cryptotanshinone sensitizes DU145 prostate cancer cells to Fas(APO1/CD95)-mediated apoptosis through Bcl-2 and MAPK regulation. Cancer Lett. 2010;298:88–98. doi: 10.1016/j.canlet.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Liu W., Yu B., Xu G., Xu W.R., Loh M.L., Tang L.D., Qu C.K. Identification of cryptotanshinone as an inhibitor of oncogenic protein tyrosine phosphatase SHP2 (PTPN11) J. Med. Chem. 2013;56:7212–7221. doi: 10.1021/jm400474r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z., Zhu R., Zheng J., Chen C., Huang C., Ma J., Xu C., Zhai W., Zheng J. Cryptotanshinone inhibits proliferation yet induces apoptosis by suppressing STAT3 signals in renal cell carcinoma. Oncotarget. 2017;8:50023–50033. doi: 10.18632/oncotarget.18483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lo S.H., Hsu C.T., Niu H.S., Niu C.S., Cheng J.T., Chen Z.C. Cryptotanshinone inhibits STAT3 signaling to alleviate cardiac fibrosis in type 1-like diabetic rats. Phytother. Res. 2017;31:638–646. doi: 10.1002/ptr.5777. [DOI] [PubMed] [Google Scholar]
- 40.Chou C.K., Tang C.J., Chou H.K., Liu C.Y., Ng M.C., Chang Y.T., Yuan S.F., Tsai E.M., Chiu C.C. The potential role of Kruppel-Like zinc-finger protein Glis3 in genetic diseases and cancers. Arch Immunol Ther Exp (Warsz) 2017;65:381–389. doi: 10.1007/s00005-017-0470-x. [DOI] [PubMed] [Google Scholar]
- 41.Scoville D.W., Kang H.S., Jetten A.M. GLIS1-3: emerging roles in reprogramming, stem and progenitor cell differentiation and maintenance. Stem Cell Investig. 2017;4:80. doi: 10.21037/sci.2017.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen X., Yang Y. Emerging roles of GLIS3 in neonatal diabetes, type 1 and type 2 diabetes. J. Mol. Endocrinol. 2017;58:R73–R85. doi: 10.1530/JME-16-0232. [DOI] [PubMed] [Google Scholar]
- 43.Fu C., Luo S., Long X., Li Y., She S., Hu X., Mo M., Wang Z., Chen Y., He C. Mutation screening of the GLIS3 gene in a cohort of 592 Chinese patients with congenital hypothyroidism. Clin. Chim. Acta. 2018;476:38–43. doi: 10.1016/j.cca.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y., Bush S.P., Wen X., Cao W., Chan L. Differential gene dosage effects of diabetes-associated gene GLIS3 in pancreatic β cell differentiation and function. Endocrinology. 2017;158:9–20. doi: 10.1210/en.2016-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Brien P., Morin P., Jr., Ouellette R.J., Robichaud G.A. The Pax-5 gene: a pluripotent regulator of B-cell differentiation and cancer disease. Cancer Res. 2011;71:7345–7350. doi: 10.1158/0008-5472.CAN-11-1874. [DOI] [PubMed] [Google Scholar]
- 46.Shukla S., Awasthi N.P., Singh P., Husain N. CD20 negative primary diffuse large B cell lymphoma of breast: Role of Pax-5. J. Cancer Res. Ther. 2015;11:658. doi: 10.4103/0973-1482.140792. [DOI] [PubMed] [Google Scholar]
- 47.Feng C., Jin J., Zou Q., Chen X., Zhou C., Wu B., Weiner D.B., Wang B. Interleukin-21 inhibits humoral response to an HIV DNA vaccine by enhancing Bcl-6 and Pax-5 expression. Viral Immunol. 2012;25:131–140. doi: 10.1089/vim.2011.0073. [DOI] [PubMed] [Google Scholar]
- 48.Denzinger S., Burger M., Hammerschmied C.G., Wieland W.F., Hartmann A., Obermann E.C., Stoehr R. Pax-5 protein expression in bladder cancer: a preliminary study that shows no correlation to grade, stage or clinical outcome. Pathology. 2008;40:465–469. doi: 10.1080/00313020802197871. [DOI] [PubMed] [Google Scholar]
- 49.Shah S., Schrader K.A., Waanders E., Timms A.E., Vijai J., Miething C., Wechsler J., Yang J., Hayes J., Klein R.J. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat. Genet. 2013;45:1226–1231. doi: 10.1038/ng.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iacobucci I., Lonetti A., Paoloni F., Papayannidis C., Ferrari A., Storlazzi C.T., Vignetti M., Cilloni D., Messa F., Guadagnuolo V. The PAX5 gene is frequently rearranged in BCR-ABL1-positive acute lymphoblastic leukemia but is not associated with outcome. A report on behalf of the GIMEMA Acute Leukemia Working Party. Haematologica. 2010;95:1683–1690. doi: 10.3324/haematol.2009.020792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawamata N., Ogawa S., Zimmermann M., Niebuhr B., Stocking C., Sanada M., Hemminki K., Yamatomo G., Nannya Y., Koehler R. Cloning of genes involved in chromosomal translocations by high-resolution single nucleotide polymorphism genomic microarray. Proc. Natl. Acad. Sci. USA. 2008;105:11921–11926. doi: 10.1073/pnas.0711039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.