Abstract
Seaweeds may represent immuno-stimulants that could be used as health-promoting fish feed components. This study was performed to gain insights into the immunomodulatory effects of dietary seaweeds in Atlantic salmon. Specifically tested were 10% inclusion levels of Laminaria digitata (SW1) and a commercial blend of seaweeds (Oceanfeed®) (SW2) against a fishmeal based control diet (FMC). Differences between groups were assessed in growth, feed conversion ratio and blood parameters hematocrit and hemoglobin. After a LPS challenge of fish representing each of the three groups, RNAseq was performed on the head kidney as major immune organ to determine transcriptomic differences in response to the immune activation. Atlantic salmon fed with dietary seaweeds did not show major differences in performance in comparison with fishmeal fed fish. RNAseq resulted in ∼154 million reads which were mapped against a NCBI Salmo salar reference and against a de novo assembled S. salar reference for analyses of expression of immune genes and ontology of immune processes among the 87,600 cDNA contigs. The dietary seaweeds provoked a more efficient immune response which involved more efficient identification of the infection site, and processing and presentation of antigens. More specifically, chemotaxis and the chemokine-mediated signaling were improved and therewith the defense response to Gram-positive bacterium reduced. Specific Laminaria digitata effects included reduction of the interferon-gamma-mediated signaling. Highly upregulated and specific for this diet was the expression of major histocompatibility complex class I-related gene protein. The commercial blend of seaweeds caused more differential expression than Laminaria digitata and improved immune processes such as receptor-mediated endocytosis and cell adhesion, and increased the expression of genes involved in response to lipopolysaccharide and inflammatory response. Particularly, expression of many important immune receptors was up-regulated illustrating increased responsiveness. NF-kappa-B inhibitor alpha is an important gene that marked the difference between both seaweed diets as Laminaria digitata inhibits the expression for this cytokine while the blend of seaweeds stimulates it. It can be concluded that the inclusion of seaweeds such as Laminaria digitata can have important modulatory effects on the immune capacity of Atlantic salmon resulting in a more efficient immune response.
Keywords: Atlantic salmon Salmo salar, aquaculture, dietary seaweeds, immune response, RNAseq, head kidney transcriptome
Introduction
Seaweed additives in fish feed may have beneficial physiological effects without affecting growth performance negatively. Seaweeds may represent immuno-stimulants with anti-bacterial activity that could be used as health-promoting fish feed components thereby offering an alternative for the use of antibiotics (Bansemir et al., 2006). Although the total lipid content is generally low, seaweeds represent a good source of health promoting PUFAs as compared to other feed ingredients derived from plant and animal sources (Rajapakse and Kim, 2011). The increased immune capacity and improved disease resistance of fish by applying dietary seaweeds has been reviewed by Reverter et al. (2014) and reported by many papers ever since (e.g., Peixoto et al., 2016; Valente et al., 2016). Specifically, when we consider Laminaria digitata effects, several papers report on the beneficial immune effects of dietary Ergosan, which is based on Laminaria digitata and Ascophyllum nodosum extracts. These effects include the initial elevation in serum lysozyme and complement activity in seabass (Dicentrarchus labrax; Bagni et al., 2005) and the enhanced mucosal immune response in rainbow trout (Oncorhynchus mykiss; Sheikhzadeh et al., 2012). Oceanfeed® effects include the increase of higher total fatty acid and LC n-3 PUFA concentrations in the flesh of farm raised Atlantic salmon (Wilke et al., 2015), but specific immune effects have not yet been reported.
Immunomodulatory effects are generally studied by subjecting experimental groups to injection with an immune system activator such as lipopolysaccharide (LPS) which mimics a bacterial infection (MacKenzie et al., 2004, 2008). In contrast to humans, fish are often resistant to the endotoxic shock caused by LPS (review by Swain et al., 2008). LPS is even applied in aquaculture as an immuno-stimulative tool to promote disease and stress resistance. LPS effects in salmonids may include the polyclonal proliferation of lymphocytes, respiratory burst, phagocytic activity of macrophages, effect modification of other immune agents, increased cytokine expression and interferon induction (reviewed by Salinas et al., 2004). The endotoxic shock is absent because LPS fails to induce antiviral genes downstream of the Toll-like receptor 4 (Iliev et al., 2005).
High-throughput transcriptome sequencing (RNAseq) represents an efficacious first-step methodology to map the most important pathways in a physiological response (Palstra et al., 2013; Rurangwa et al., 2015) including the impact of nutrition on the immune system of fish (Martin and Król, 2017). Transcript mapping, expression profiling and gene ontology of immune genes in the head kidney as the most involved organ in the innate and adaptive immune response of fish (Kaattari and Irwin, 1985) should reveal the major pathways and any significant immunomodulation by experimental factors.
This study was performed to gain insights into the immunomodulatory effects of dietary seaweeds in Atlantic salmon. Specifically tested were 10% inclusion levels of Laminaria digitata (SW1) and a commercial blend of seaweeds (Oceanfeed®) (SW2) against a fishmeal based control diet (FMC). Differences between groups were assessed in growth, feed conversion ratio (FCR) and blood parameters hematocrit (Hct) and hemoglobin (Hb). After a LPS challenge in fish representing each of the three groups, RNAseq was performed on head kidney tissue of individual fish to determine transcriptomic differences in response to the immune activation, to our knowledge for the first time in fish in this context.
Materials and Methods
Ethics Statement
The experimental protocols complied with the current laws of the Netherlands and were approved by the animal experimental committee (DEC nr. 2013113).
Experimental Diets
Laminaria digitata was made available by North Seaweed (Kapelle, Netherlands) and Oceanfeed was purchased from Ocean Harvest Technology (Milltown, Ireland). Two experimental diets were tested: a diet with 10% Laminaria digitata (SW1) and a diet with 10% of a commercial blend of seaweeds (Oceanfeed®) against a control diet based on fishmeal (FMC). In this study, seaweed is tested as an organic supplement for fish feed which, following EFSA, requires a control group that does not include the additive and a treatment group dosed at use-level with the additive. Laminaria consisted of crude protein 11.1%, ether extract 1.1%, crude fiber 5.6%, and ash 36.4%. For Oceanfeed these values were: crude protein 10.9%, ether extract 0.98%, crude fiber 8.8%, and ash 49.3%. The diets were prepared using extrusion in cooperation with Research Diet Services (RDS, Wijk bij Duurstede, Netherlands). Diets FMC, SW1 and SW2 were isonitrogenous, isoenergetic, equal in amino acid composition, calcium and phosphates levels (Tables 1–3).
Table 1.
Experimental set-up and recipes.
| Recipes | Dietary treatment |
||
|---|---|---|---|
| FMC | SW1 | SW2 | |
| Basal ingredients (%) | |||
| Fishmeala | 30.600 | 30.600 | 30.600 |
| Wheat glutenb | 15.058 | 15.058 | 15.058 |
| Wheat flourc | 17.368 | 13.500 | 13.500 |
| Fishoild | 13.500 | 13.319 | 13.319 |
| Monocalcium phosphatee | 3.071 | 3.071 | 3.071 |
| Sonac hemoglobin 92Pf | 2.841 | 2.841 | 2.841 |
| CPSP-Gg | 2.700 | 2.700 | 2.700 |
| Amino acid mix salmon1 | 2.536 | 2.536 | 2.536 |
| Premix salmon2 | 2.025 | 2.025 | 2.025 |
| Linseed oilh | 1.629 | 1.629 | 1.629 |
| Soya lecithinei | 0.999 | 0.999 | 0.999 |
| Limej | 0.901 | 0.901 | 0.901 |
| Soya oilk | 0.470 | 0.470 | 0.470 |
| Cholesteroll | 0.297 | 0.297 | 0.297 |
| Carophyll Pinkm | 0.054 | 0.054 | 0.054 |
| 90% | 90% | 90% | |
| Test ingredients (%) | |||
| Wheat glutenb | 0.212 | 0.000 | 0.000 |
| Wheat flourc | 3.868 | 0.000 | 0.000 |
| CMCn | 0.690 | 0.000 | 0.000 |
| Diamolm | 4.473 | 0.000 | 0.000 |
| Soy protein concentrateo | 0.515 | 0.000 | 0.000 |
| Corn gluten mealp | 0.242 | 0.000 | 0.000 |
| Semolinaq | 0.001 | 0.000 | 0.000 |
| Laminaria digitatar | 0.000 | 10.000 | 0.000 |
| Ocean feeds | 0 | 0.000 | 10.000 |
| 10% | 10% | 10% | |
| Check | 100 | 100 | 100 |
aDanish LT fishmeal. Type LT (Triple Nine Fish Protein Esbjerg, Denmark), bWheat gluten, Gluvital 21000 Cargill, Netherlands, cWheat flour, cWheatflour (Meneba Weert, Netherlands), dCoppens International, Netherlands, eTessenderlo Chemie, Belgium, fHemoglobin powder (Sonac, Bad Bramstedt, D), gSopropeche Boulogne sur Mer, hLinagro, Lichtervelde, Belgium, iNutripur G Cargill, Hamburg, Germany, jInducal 250 van Sibelco/Ankerpoort, Maastricht, kSoya oil refined, (Smilde foods, Heerenveen), lCholesterol SF van Dishman Netherlands B.V., Veenendaal, mCAROPHYLL PINK, F. Hoffmann-La Roche Ltd., nCarboxy methyl cellulose, fDamolin A/S, Hamburg, 0Soycomil ADM Eurpoort B.V., Netherlands, pCargill, Sas van Gent, Netherlands, qFa Gebr van Eck, Wijk bij Duurstede, Netherlands, rNorth SeaWeed, Netherlands, sOcean Harvest, Ireland. 1Amino acid mix: lysine 5 g kg-1, threonine 2 g kg-1, methionine 3 g kg-1, arginine 5 g kg-1, histidine 5 g.kg-1. 2Premix: vitamins (mg or IU kg-1 diet): vitamin A (retinyl acetate), 15000 IU; D3 (cholecalciferol), 3000 IU; K3 (menadione), 8 mg; B12 (cyanocobalamin), 0.10 mg; B1 (thiamine), 10 mg; B2 (riboflavin), 15 mg; B6 (pyridoxine hydrochloride), 15 mg; folic acid, 10 mg; biotin, 0.5 mg; inositol, 600 mg; niacin, 50 mg; pantothenic acid, 50 mg, choline chloride, 2000 mg; vitamin C, 300 mg; vitamin E, 500 mg; minerals (g or mg kg-1 diet): Mn (manganese sulfate), 30 mg; I (potassium iodide), 5 mg; Cu (copper sulfate), 5 mg; Co (cobalt sulfate), 2 mg; Cr (chromium sulfate), 1 mg; Mg (magnesium sulfate), 300 mg; K (potassium chloride), 2600 mg; Zn (zinc sulfate), 100 mg; Se (sodium selenite), 1 mg; Fe (iron sulfate), 60 mg. BHT (E300-321), 100 mg; calcium propionate, 1000 mg.
Table 3.
Calculated amino acid composition.
| Code | Unit | FMC | SW1 | SW2 |
|---|---|---|---|---|
| Calculated amino acids | ||||
| Lysinea | g.kg-1 | 29.4 | 29.7 | 29.3 |
| Methioninea | g.kg-1 | 12.8 | 12.8 | 12.7 |
| Cysteineb | g.kg-1 | 5.4 | 5.3 | 5.4 |
| Threoninea | g.kg-1 | 16.9 | 17.1 | 17.0 |
| Tryptophana | g.kg-1 | 4.3 | 4.2 | 4.3 |
| Isoleucinea | g.kg-1 | 15.3 | 15.5 | 15.3 |
| Argininea | g.kg-1 | 27.0 | 32.9 | 32.9 |
| Phenylalaninea | g.kg-1 | 28.4 | 27.0 | 26.9 |
| Histidinea | g.kg-1 | 17.4 | 17.4 | 17.2 |
| Leucinea | g.kg-1 | 30.8 | 30.9 | 30.5 |
| Tyrosineb | g.kg-1 | 12.5 | 12.7 | 12.5 |
| Valinea | g.kg-1 | 19.8 | 20.1 | 19.8 |
| Alanine | g.kg-1 | 21.3 | 22.2 | 24.9 |
| Asparagine | g.kg-1 | 30.6 | 30.9 | 30.8 |
| Glutamate | g.kg-1 | 80.4 | 79.0 | 80.4 |
| Glycine | g.kg-1 | 22.4 | 22.6 | 22.4 |
| Proline | g.kg-1 | 28.3 | 28.0 | 27.9 |
| Serine | g.kg-1 | 17.6 | 17.5 | 17.6 |
aEssential, bconditionally essential.
Table 2.
Calculated and analyzed proximate composition of diets.
| Code | Unit | FMC | SW1 | SW2 |
|---|---|---|---|---|
| Calculated (macro) nutritional composition | ||||
| DM | (g.kg-1) | 911 | 912 | 909 |
| Ash | (g.kg-1) | 128 | 121 | 134 |
| CP | (g.kg-1) | 434 | 435 | 434 |
| EE | (g.kg-1) | 213 | 213 | 212 |
| CF | (g.kg-1) | 8.5 | 6.9 | 10.1 |
| NFE | (g.kg-1) | 126 | 136 | 117 |
| P | (g.kg-1) | 14.1 | 14.0 | 14.0 |
| Ca | (g.kg-1) | 16.6 | 16.6 | 16.6 |
| GE | (MJ.kg-1) | 21.1 | 21.2 | 20.9 |
| CP/GE | – | 20.6 | 20.5 | 20.8 |
| DM | (g.kg-1) | 926 | 947 | 965 |
| Ash | (g.kg-1) | 136 | 126 | 139 |
| CP | (g.kg-1) | 469 | 467 | 469 |
| EE | (g.kg-1) | 134 | 126 | 118 |
| CF | (g.kg-1) | 3 | 6 | 6 |
| NFE | (g.kg-1) | 184 | 223 | 232 |
| GE | (MJ.kg-1) | 16.7 | 16.4 | 16.2 |
| CP/GE | – | 28.0 | 28.4 | 29.0 |
Experimental Animals and Procedures
Atlantic salmon (378 ± 57 g) were purchased as juveniles from Meridian Salmon (Furnace, United Kingdom) and transported by Solway (Dumfriesshire, United Kingdom) to the experimental aquaculture facilities of Wageningen Marine Research (WMR) in Yerseke (Netherlands). Fish were accommodated in nine tanks (800 l each) which were integrated in a single recirculation system. The experiment consisted of a 15-day acclimatization followed by a 42-day experimental period. Diets were tested in triplicate with 20 fish per tank, adding up to 180 fish in total.
Fish were fed using feeding belts and feeding level was restricted. Tanks were checked frequently to verify if all feed was consumed. Dry matter content of diets was analyzed at the start of the experiment to be able to determine equal feeding levels on dry matter for all diets. Proximate compositions were analyzed by Nutrilab B.V. (Rijswijk, Netherlands).
Husbandry conditions during the experimental period were: photoperiod 16L:8D, temperature 13.8 ± 1.6°C, oxygen 8.0 ± 0.5 mg.l-1, pH 7.51 ± 0.04, TAN (total ammonia nitrogen) 0.05 ± 0.09 mg.l-1, NO2-, 0.07 ± 0.05 mg.l-1, salinity 25–30 ppt and flow 39.5 ± 2.3 l.min-1, which stayed within pre-set limits. Temperature and oxygen were measured daily. Flow, pH, TAN, NO2- were measured weekly.
At the start and at the end of the experimental period all fish per tank (N = 20) were anesthetized using phenoxy ethanol (2 ml l-1) and weighed. Fish were not fed 1 day prior to weighing. N = 18 fish at the start and N = 10 fish per tank at the end of the experiment were sampled for blood to determine Hct and Hb. Blood was obtained by caudal venous puncture, using a heparinized syringe (0.6 mm/60 mm needle). The samples were transferred to Eppendorf tubes, stored on ice and processed within 15 min. Hct was determined by centrifuging blood samples for 5 min at 11,000 rpm (SpinCrit micro hematocrit centrifuge, Indianapolis, IN, United States). Hb content was determined using the method described by van Kampen and Zijlstra (1961).
At the end, N = 1 fish per tank (or N = 3 fish per diet) were subjected to a LPS challenge (LPSs from Escherichia coli; 6 mg.kg-1 in 0.2 ml physiological salt solution injected intraperitoneally) and between 68 and 70 h after injection, fish were sacrificed and dissected for head kidney tissue that was stored in RNAlater (Ambion). Sham injections were not applied as their effects would be filtered out by the comparisons that were made in our experimental design, e.g., comparing each of the experimental groups vs. the control. The potential effect of the intraperitoneal injection itself was not expected to be any different between the individuals of the three experimental groups, and certainly not for sequencing RNA expression in the head kidney.
Growth
From the individual weight data, average body weight at the start (BW0) and at the end (BWt) was calculated per tank as experimental unit. Feed intake (FI) and FCR were calculated as well as growth rates in %.day-1 and g.kg-0.8.day-1. The specific growth rate (SGR) and growth expressed in metabolic body weight (GMBW) during the growth period were calculated with the formulas:
| (1) |
with t as the duration of the growth period and LN the natural logarithm,
and
| (2) |
Statistics of Growth and Blood Data
Data were analyzed using ANOVA to test for diet effects. Homogeneity of variance was tested using Levene’s test. When necessary, data were transformed or tested using the Kruskal–Wallis test. For all tests a probability p < 0.05 was considered significant. When significant, depending on the hypothesis; equal, higher- or lower, mean values were compared using the one sided or two sided Fisher’s LSD multiple comparisons post hoc test.
RNAseq
RNA was isolated from homogenized head kidney samples (TissueRuptor, Qiagen, Venlo, Netherlands) of the three individual fish per diet using the miRNAeasy mini kit (Qiagen). RNA concentrations were between 158 and 1,200 ng μl-1 and RIN values ∼10. Illumina multiplexed RNAseq libraries were prepared from 2 μg total RNA using the Illumina TruSeq RNA Sample Prep Kit v2 according to the manufacturer’s instructions (Illumina Inc.). All nine RNAseq libraries (3 individual fish × 3 diets = 9 samples) were sequenced on an Illumina HiSeq 2500 sequencer as 1 × 50 nucleotides paired-end (PE50) reads according to the manufacturer’s protocol. Image analysis and base calling were done by the Illumina pipeline. A total of ∼154 million single-read 1 × 50 nt reads (∼8 Gb data) were derived from all nine RNAseq libraries. Two strategies were used for quantitative analysis of the head kidney RNAseq data sets: referred to as mapping reads against NCBI Salmo salar reference (strategy 1) and mapping reads against de novo assembled Salmo salar reference (strategy 2). Strategy 1 was more appropriate for salmon and particularly performed for analyses on gene level while strategy 2 was performed for more extensive and comprehensive data analyses on biological process level and detection of new immune salmon genes.
Mapping Reads Against NCBI Salmo salar Reference
In strategy 1, reads were aligned to 48,223 Salmo salar cDNA sequences (downloaded from NCBI) using TopHat (version 2.0.5) (Trapnell et al., 2009) and about 15% of the RNAseq reads could be mapped. Reference alignment was done and the resulting files were filtered using SAMtools (version 0.1.18) (Li et al., 2009) to exclude secondary alignment of reads. For statistical comparison of gene expression levels between groups, aligned fragments per predicted gene were counted from SAM alignment files using the Python package HTSeq (version 0.5.3p9) (Anders et al., 2014). In order to make comparisons across samples possible, these fragment counts were corrected for the total amount of sequencing performed for each sample. As a correction scaling factor, we employed library size estimates determined using the R/Bioconductor (release 2.11) package DESeq (Anders and Huber, 2010). Read counts were normalized by dividing the raw counts obtained from HTSeq by its scale factor. Correction for false positives is included in the statistical analysis of gene expression through DESeq.
Mapping Reads Against de Novo Assembled Salmo salar Reference
In strategy 2, CLC bio’s de novo assembler was used to generate cDNA contigs from the total of ∼154 million reads. This resulted in 87,600 cDNA contigs corresponding to mRNAs that were expressed in the head kidneys and ranging in size from 200 to 14,086 nt. The de novo cDNA contigs were annotated to known genes via custom BLAST searches against four reference databases: (a) a UNIPROT protein database (36.3% hits), (b) a Teleost Refseq mRNA database (26.5% hits), (c) a Teleost “all mRNA” database (80.1% hits) and (d) the Danio rerio Zv9 genome (6.3% hits). RNAseq reads from the nine individual samples were then aligned to this de novo assembled cDNA reference database. About 60% of the RNAseq reads could be mapped against the de novo assembled reference database. Statistical comparison between groups of fish SW1 vs. FMC and SW2 vs. FMC was done using HTseq and DESeq as described above under “strategy 1.” The set of differentially expressed contigs at P < 0.01 was cleaned by removing undefined mRNA sequences: 792 Oncorhynchus mykiss sequences; 40 Plecoglossus altivelis sequences; 10 Ictalurus punctatus sequences; 202 Salmo salar Sasaskin sequences; 11 Gadus morhua strain sequences; 5 Fundulus grandis transcripts; 2 Anoplopoma fimbria sequences; 2 Scophthalmus maximus sequences, and 391 blanks.
Gene Ontology
From the cleaned set of differentially expressed contigs at P < 0.01 of strategy 2, contigs were selected that were associated with genes with apparent immune response function as determined by manually scanning gene ontology (GO) with Uniprot, QuickGO, Genecards, NCBI, Wikigene, and Wikipedia. These genes were identified in Uniprot and complete GO annotation on biological process level was analyzed in QuickGO. GO terms that reflected apparent immune response processes were collected per comparison SW1 vs. FMC and SW2 vs. FMC. GO differences were analyzed and significant differences identified applying Mann–Whitney U tests.
Availability of Data and Material
The datasets (raw RNAseq reads) supporting the conclusions of this article are available in the NCBI GEO repository, accession numbers GSM2705950–GSM27059581.
Results
Growth and Blood
Average feed intake was 5.97–5.98 g.dm.fish-1.d-1 and similar for all treatments (Figure 1A). Feed conversion on dry matter basis (FCRdm) between diets tended to differ but not significantly (P = 0.075; Figure 1B).
FIGURE 1.
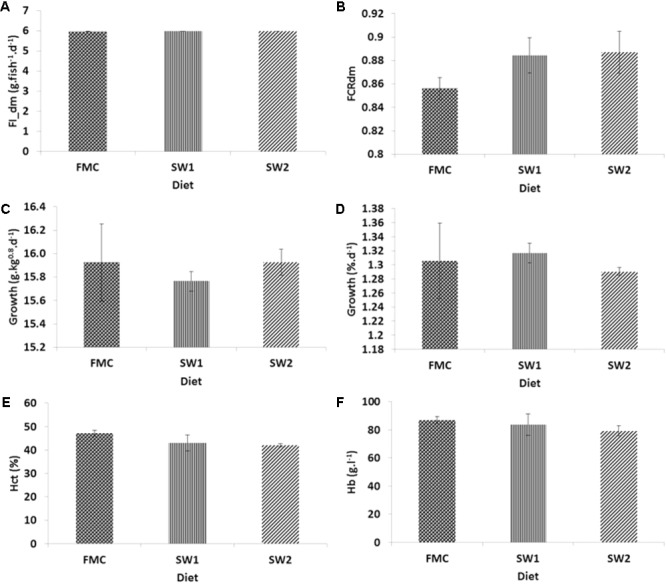
Atlantic salmon performance when fed with the FMC, SW1 and SW2 diets. (A) Feed intake in g.fish-1.d-1 on dry matter basis. (B) Feed conversion ratio (FCR) on dry matter basis. (C) Growth in metabolic body weight (g.kg0.8.d-1). (D) Specific growth rate (%.d-1). (E) Hct in %. (F) Hemoglobin (g.l-1). Fish fed with seaweed diets tended to have higher FCRdm values (P = 0.075) and less individual variation in growth.
No differences in growth performance were detected between FMC, SW1, and SW2 fish (Figures 1C,D). Interestingly, fish fed with seaweed diets showed much less individual variation in growth performance than the fish fed the FMC control diet.
Fish fed FMC had average hematocrit (Hct) and hemoglobin (Hb) levels of 28.9 ± 9.9% and 62.9 ± 18.5 g.l-1, respectively, at the start of the experiment. Start values of both Hct and Hb were lower than values at the end of the experiment (P < 0.05), yet Hct and Hb did not differ between treatments at the end (P > 0.05) (Figures 1E,F).
RNAseq Differential Expression After Mapping Reads Against NCBI Salmo salar Reference
The comparison SW1 vs. FMC yielded 28,312 expressed contigs associated with S. salar genes: 59% of the NCBI sequences. 144 of these contigs were differentially expressed: 74 up-regulated (51%) and 70 down-regulated (49%). Twenty contigs were considered as direct immune response genes of which nine genes (five up-regulated, four down-regulated) were specific for SW1 vs. FMC (Table 4).
Table 4.
Differentially expressed immune response genes.
| Specific for SW1 | fc | |
|---|---|---|
| MHC class I (UBA) mRNA, UBA∗0901 allele | 362 | |
| T-cell receptor alpha chain V region 2B4 precursor putative mRNA | 7.19 | |
| MHC class I antigen (UBA) mRNA, UBA∗4001 allele | 6.61 | |
| Macrophage migration inhibitory factor putative mRNA | Inf | |
| MHC class I antigen (Sasa-UBA) mRNA, Sasa-UBA∗0902 allele | Inf | |
| BOLA class I histocompatibility antigen, alpha chain BL3-7 precursor putative mRNA | 0.154 | |
| MHC class I (UBA) mRNA, UBA∗0501 allele | 0.131 | |
| MHC class I mRNA | 0.0467 | |
| MHC class I (UBA) mRNA, UBA∗0701 allele | 0.00130 | |
| Specific for SW2 | fc | |
| MHC class I antigen (Sasa-UBA) mRNA, Sasa-UBA∗3701 allele | 288 | |
| H-2 class II histocompatibility antigen gamma chain putative mRNA | 137 | |
| MHC class II antigen alpha chain (Sasa-DAA) mRNA | 39.7 | |
| MHC class II alpha mRNA | 18.6 | |
| Tumor necrosis factor receptor superfamily member 11B precursor putative mRNA | 4.14 | |
| CD209 antigen-like protein E putative mRNA | 3.21 | |
| Ig kappa chain V-IV region Len putative mRNA | 2.86 | |
| partial mRNA for MHC class II antigen beta chain (DAB gene) | Inf | |
| MHC class I antigen (Sasa-UBA) mRNA, Sasa-UBA∗1402 allele | Inf | |
| Class I histocompatibility antigen, F10 alpha chain precursor putative mRNA | 0.254 | |
| MHC class I (UBA) mRNA, UBA∗0501 allele | 0.131 | |
| MHC class I antigen (Sasa-UBA) mRNA, Sasa-UBA∗3501 allele | 0.125 | |
| Proliferating cell nuclear antigen putative mRNA | 0.113 | |
| MHC class I (UBA) mRNA, UBA∗1501 allele | 0.00567 | |
| Anamorsin putative mRNA | 0 | |
| Common SW1-SW2 | SW1 (fc) | SW2 (fc) |
| MHC class II antigen beta chain (DAB gene) | 309 | 171 |
| MHC-Sasa class II (clone c144) | 54.7 | 7.76 |
| MHC class I (UBA) mRNA, UBA∗0201 allele | 36.8 | 35.0 |
| MHC class I antigen (Sasa-UBA) mRNA | Inf | Inf |
| MHC class I antigen (Sasa-UBA) mRNA, Sasa-UBA∗0202 allele | Inf | Inf |
| MHC class II antigen alpha chain (Sasa-DAA) mRNA | Inf | Inf |
| HLA class II histocompatibility antigen, DQW1.1 beta chain precursor putative mRNA | Inf | Inf |
| CD209 antigen-like protein E putative mRNA | Inf | Inf |
| MHC class I (UBA) mRNA, UBA∗1401 allele | 0.350 | 0.351 |
| MHC-Sasa class II B (clone c22) | 0.276 | 0.003 |
| MHC class I antigen (Sasa-UBA) mRNA, Sasa-UBA∗3901 allele | 0.00349 | 0 |
Differentially expressed immune response genes at P < 0.05, specific for the comparison SW1 vs. FMC, SW2 vs. FMC and those in common as a result of strategy 1; Mapping reads against NCBI Salmo salar reference. Expression is given by fold change (fc) and Inf indicates that expression was unique for the particular seaweed.
The comparison SW2 vs. FMC yielded 28,189 expressed contigs associated with S. salar genes: 58% of the NCBI sequences. Two hundred and forty-six of these contigs were differentially expressed: 136 up-regulated (55%) and 110 down-regulated (45%). Twenty-six contigs were considered as direct immune response genes of which 15 genes (nine up-regulated, six down-regulated) were specific for SW2 vs. FMC (Table 4).
Aligning vs. NCBI S. salar sequences and comparing SW1 vs. FMC and SW2 vs. FMC revealed 54 common differentially expressed contigs. Expression of all these contigs was in the same direction, either up- or down-regulated. Eleven contigs (eight up-regulated, three down-regulated) were considered as direct immune response genes representing the common genes in the immunomodulatory effects of the tested dietary seaweeds (Table 4).
RNAseq Differential Expression and Gene Ontology After Mapping Reads Against de Novo Assembled Salmo salar Reference
The comparison SW1 vs. FMC yielded 1,951 differentially expressed contigs: 2.23% of the total of 87,600 contigs. Expression of 1,092 contigs (56%) was up-regulated, of 859 contigs (44%) down-regulated. On basis of more the stringent criterion of P < 0.01, 736 contigs were selected for further analyses.
The comparison SW2 vs. FMC yielded 4,350 differentially expressed contigs: 4.97% of the total of 87,600 contigs. Expression of 1,485 contigs (34%) was up-regulated, of 2,865 contigs (66%) down-regulated. On basis of the more stringent criterion of P < 0.01, 1,703 contigs were selected for further analyses.
BLAST searching against four reference databases and comparing SW1 vs. FMC and SW2 vs. FMC revealed 224 common differentially expressed contigs. Expression of these contigs was in the same direction, except for five: Glutamate dehydrogenase 1, Serine incorporator 1 and two Oncorhynchus mykiss contig sequences, of which expression was in opposite direction, as well as for NF-kappa-B inhibitor alpha (S. salar).
Interestingly, contig 86142 had by far the highest fold change (fc) for both SW1 vs. FMC and SW2 vs. FMC comparisons with fc 2,550 and 2,968, respectively. This contig was mapped against the Oncorhynchus mykiss mRNA sequence with gene identifier 299677870 that resulted from a characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches by Salem et al. (2010). With a BLASTn search, a 98% homology was found aligning this sequence with PREDICTED: Salmo salar integrin alpha-2-like (LOC106561807).
For the comparison SW1 vs. FMC, 67 contigs were related to immune response genes. Specific for this comparison were 43 immune response genes: 32 up-regulated, 11 down-regulated (Supplementary material). Twenty-four genes were shared with the comparison SW2 vs. FMC: 12 up-regulated, 12 down-regulated (Supplementary material). In total 39 genes contributed to the pool of 168 GO terms for this comparison (84 specific and 84 in common with comparison SW2 vs. FMC).
For the comparison SW2 vs. FMC, 156 contigs were related to immune response genes. Specific for this comparison were 132 genes: 67 up-regulated, 65 down-regulated (Supplementary material). Of course, 24 genes were shared with the comparison SW1 vs. FMC, but now with 11 up-regulated and 13 down-regulated because NF-kappa-B inhibitor alpha (S. salar) was down-regulated and not up-regulated like for SW1 vs. FMC (Supplementary material). In total 82 genes contributed to the pool of 256 GO terms for this comparison (172 specific).
The number of GO terms was narrowed down removing those with less than five scores, which left us with the 15 most important GO terms (Figure 2). Six of these GO terms did not show differences between comparisons. More regular GO terms such as immune system process (GO:0002376), immune response (GO:0006955), and viral process (GO:0016032) had as many scores for SW1 vs. FMC as for SW2 vs. FMC for both up- and down-regulated genes. Chemotaxis (GO:0006935) and chemokine-mediated signaling pathway (GO:0070098) only contained up-regulated gene expression for both comparisons, expression of genes involved in the defense response to Gram-positive bacterium (GO:0050830) was down-regulated for both comparisons. Other GO terms were more abundant or even specific for either the SW1 vs. FMC or SW2 vs. FMC comparison. Abundant or specific for the SW1 vs. FMC comparison were cytokine-mediated signaling pathway (GO:0019221) and the interferon-gamma-mediated signaling pathway (GO:0060333). More terms were abundant or specific for the SW2 vs. FMC comparison: receptor-mediated endocytosis (GO:0006898), inflammatory response (GO:0006954), cell adhesion (GO:0007155), and response to lipopolysaccharide (GO:0032496) all up-regulated; innate immune response (GO:0045087) up- and down-regulated; NIK/NF-kappaB signaling (GO:0038061) and defense response to virus (GO:0051607) down-regulated.
FIGURE 2.
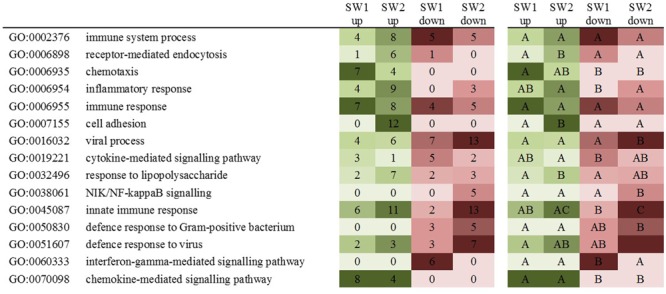
Heat map of the 15 main biological processes indicating the differences between diet comparisons. Shown are GO terms and descriptions; seaweed (SW) diets 1 and 2 scores of up- and down-regulated genes. Also indicated the significant differences (P < 0.05).
Discussion
Seaweeds may represent immuno-stimulants that could be used as health-promoting fish feed components thereby offering an alternative for the use of antibiotics. This study was performed to gain insights into the immunomodulatory effects of dietary seaweeds in Atlantic salmon. Specifically tested were 10% inclusion levels of Laminaria digitata (SW1) and a commercial blend of seaweeds (Oceanfeed®) (SW2) against a fishmeal based control diet (FMC). By RNAseq of the head kidney transcriptome of LPS challenged fish representing all three experimental groups, we could determine the common immunomodulatory effects of dietary seaweeds, but also the specific immunomodulatory effects of Laminaria digitata and of the commercial blend of seaweeds.
No Major Difference in Performance
From our results it can be concluded that the inclusion of dietary seaweeds did not lead to major differences in performance of Atlantic salmon: growth and blood Hct and Hb were similar between experimental groups. The FCR of the fish fed with dietary seaweeds was not significantly different. Growth variation in the fish fed with dietary seaweeds, however, was lower which would lead to a more uniform market size. Similarly as in previously performed studies, dietary seaweeds did not affect growth performance of rainbow trout nor seabass at inclusion levels of 10% (Oncorhynchus mykiss: Soler-Vila et al., 2009, Dicentrarchus labrax; Valente et al., 2006; Peixoto et al., 2016). Several other studies did report on growth improvement by dietary seaweed supplementation, but the variety of both fish and seaweed species, and their inclusion levels, is extensive which makes it complex to generalize conclusions. Anyway, the absence of major difference in the performance of Atlantic salmon would pave the way for dietary seaweed application if clear health-promoting effects would exist.
Nutrigenomic Approach Investigating Dietary Immunomodulation
Recent studies have provided more insights into the dietary immunomodulation of gene expressions since the review by Tacchi et al. (2011). Several studies have a similar nutrigenomic approach as our study and support immune enhancement on basis of up-regulated gene expressions in Atlantic salmon. Martinez-Rubio et al. (2012, 2014) reported on the dietary immunomodulation of gene expressions in the heart in response to immune challenging. Núñez-Acuña et al. (2015) investigated the effects of plant-derived additives on the skin and heart kidney transcriptome in sea lice-infested Atlantic salmon. Eslamloo et al. (2017) investigated whether vegetable and fish oils can alter antiviral responses of salmon macrophage-like cells by transcriptomic profiling but found similar activation of the immune-related pathways and functions between experimental groups. Caballero-Solares et al. (2017) concluded that plant-based diets may enhance the immune response on basis of increased expression of transcripts involved in the synthesis of pro-inflammatory eicosanoids and chemotaxis. Our study supports the general immune enhancement of dietary seaweeds but also shows that the seaweed specific effects are important and may vary significantly.
Common Immunomodulatory Effects of Dietary Seaweeds
From the de novo assembled reference we learned that GO terms that represented common immunomodulatory effects were referring to quite general processes (immune system process, immune response, and viral process) but also more specifically to chemotaxis, chemokine-mediated signaling pathway and defense response to Gram-positive bacterium. From the direction of regulation of the genes that were representing these GO terms, we can conclude that the common immunomodulatory effects of the dietary seaweeds in our study improved chemotaxis and the chemokine-mediated signaling pathway, and reduced the defense response to Gram-positive bacterium. Thus, based on the regulation of these immune genes, fish fed with seaweeds have a more efficient immune response. The infection site is more efficiently identified and antigens are more efficiently processed and presented.
The contig 86142 with fc values of 2,550 and 2,968 for SW1 vs. FMC and SW2 vs. FMC comparisons, respectively, that had a 98% homology with the predicted Salmo salar integrin alpha-2-like, may certainly represent a key marker gene of the LPS immune response in salmonids. This gene is predicted to express a subunit of the heterodimeric integral membrane glycoprotein integrin which is involved in cell adhesion and cell-surface mediated signaling of T cells (the NKT cells), NK cells, fibroblasts and platelets. Integrin alpha-2 is known to show up-regulated expression at the attachment site of salmon louse in order to regulate a cell proliferation response (Robledo et al., unpublished) but our results show that its up-regulated expression is not restricted to response to a skin wound.
Read annotation to S. salar NCBI sequences revealed predominantly MHC class I and II and other genes involved in antigen processing and presentation. Common for both SW1 vs. FMC and SW2 vs. FMC was also the expression of CD209 antigen-like protein E which is a putative pathogen-recognition receptor that may mediate the endocytosis of pathogens (Dettleff et al., 2017; Rozas-Serri et al., 2018). The particular transcript that was common for both groups was expressed only in the SW1 and SW2 groups (two and one fish, respectively) and not in the FMC controls. CD209 antigen-like protein enhances the expression of Toll-like receptors (TLR; Geijtenbeek and Gringhuis, 2009) which in our study was only apparent for TLR8 for diet SW2.
Mapping reads against the de novo assembled S. salar reference revealed more important genes that were differentially expressed for SW1 vs. FMC and SW2 vs. FMC. Highly up-regulated was the Ig heavy chain V-III region HPC76 Fragment (M. musculus), which emphasizes the up-regulation of antigen recognition in fish fed with dietary seaweeds. Also up-regulated is T-bet (O. mykiss) which in Atlantic salmon plays an important role in Th1 T-helper cell differentiation (Kumari et al., 2015). Furthermore, among the up-regulated genes is C-C motif chemokine 19 (M. musculus) that may direct the improvement in chemotaxis and the chemokine-mediated signaling pathway which were identified through GO as important common pathways. The up-regulated nuclear receptor subfamily 1 group D member 2 (M. musculus) is a transcriptional repressor that regulates genes involved in the inflammatory response (Lam et al., 2013). The up-regulated Secretory phospholipase A2 receptor (P. abelii) is involved in cytokine production and Src-like-adapter 2 (H. sapiens) negatively regulates T-cell receptor signaling (Holland et al., 2001).
NF-kappa-B inhibitor alpha (S. salar) is both up- and down-regulated and inhibits NF-kappa-B as mediator impacting processes such as apoptosis, proliferation, differentiation, and development (Lee et al., 2014). Then, in SW1 fish NF-kappa-B activity is inhibited while in SW2 fish NF-kappa-B activity is stimulated. Besides NF-kappa-B inhibitor alpha (S. salar), also glutamate dehydrogenase 1 and serine incorporator 1 expression was in opposite direction for both experimental seaweed diets.
Among the down-regulated genes is N-acetylmuramoyl-L-alanine amidase (M. musculus) which is involved in the reduction of the defense response against Gram-positive bacterium (e.g., Zhang et al., 2016 for turbot). Also down-regulated are three transcripts associated with Heat shock cognate 71 kDa protein (G. gallus, O. latipes, and S. oedipus), which binds LPS and then mediates the LPS-induced inflammatory response including TNF secretion by monocytes, and the cytokine receptor subunit Cytokine receptor-like factor 1 (H. sapiens).
Specific Immunomodulatory Effects of 10% Laminaria digitata Inclusion
Annotation of reads to the de novo assembled reference showed that specific immunomodulatory effects of dietary Laminaria digitata include mostly down-regulated gene expression in the cytokine-mediated signaling pathway, specifically the interferon-gamma-mediated signaling pathway. The interferon-gamma-receptors are internalized after binding and through clathrin-coated pits delivered to the sorting endosome (Blouin and Lamaze, 2013). The specific modulatory effects of Laminaria digitata included the reduction of this process.
Besides MHC class I genes and a T-cell receptor, specific for the SW1 immunomodulatory effect was the expression of macrophage migration inhibitory factor (MIF), which is a widely expressed pro-inflammatory cytokine inhibiting the random migration of macrophages (Jin et al., 2007) which again emphasizes the importance of the cytokine-mediated signaling pathway.
Mapping reads against the de novo assembled S. salar reference also revealed a MHC I gene as very important and specific for SW1 vs. FMC: Major histocompatibility complex class I-related gene protein (M. musculus) which was up-regulated at fc 1,681. Second in line was sorting nexin-18 (S. salar), involved in endocytosis (Park et al., 2010), with up-regulated expression at fc 71.3. Other important up-regulated genes were C-X-C motif chemokine 13 (H. sapiens), chemotactic for B-lymphocytes, and dedicator of cytokinesis protein 2 (O. mykiss) also involved in chemotaxis by making arrangements for lymphocyte migration in response of chemokines.
Down-regulated genes include class I histocompatibility antigen, F10 alpha chain (G. gallus) and class I histocompatibility antigen, A9/A9 alpha chain (C. familiaris), involved in the presentation of foreign antigens to the immune system. Also down-regulated is the signal transducer and activator of transcription 1 or STAT1 (two transcripts: M. musculus and alpha/beta; H. sapiens), which stimulates the expression of genes in response to an interferon signal (Skjesol et al., 2010). STAT1 and the gamma-interferon-inducible lysosomal thiol reductase precursor (S. salar) both contribute to the down-regulated interferon-gamma-mediated signaling pathway.
Specific Immunomodulatory Effects of 10% Inclusion of a Commercial Blend of Seaweeds
Annotation of reads to the de novo assembled reference showed that specific immunomodulatory effects of including a commercial blend of seaweeds, in this case Oceanfeed®, include up-regulated gene expression in receptor-mediated endocytosis and cell adhesion, and response to lipopolysaccharide and inflammatory response: processes that are supposedly improved by the commercial blend of seaweeds. Biological processes such as NIK/NF-kappa-B signaling and defense response to virus were reduced in their functioning. Genes involved in the innate immune response were both up- and down-regulated.
By far most genes that were differentially regulated were associated with the specific modulatory effects of Oceanfeed®. Similar to Laminaria digitata, read annotation to the S. salar NCBI sequences revealed MHC class I genes being up-and downregulated, MHC class II genes were up-regulated. Also up-regulated was tumor necrosis factor receptor superfamily member 11B precursor that binds cytokine tumor necrosis factor causing cell death, which is in line with the down-regulation of anamorsin that is involved in the negative control of cell death upon cytokine withdrawal. Other up-regulated genes were Ig kappa chain V-IV region Len which is involved in antigen binding and CD209 antigen-like protein E involved in endocytosis. Proliferating cell nuclear antigen is essential for DNA replication and its expression was down-regulated.
Mapping reads against the de novo assembled S. salar reference revealed strongly up-regulated expression of important immune receptors such as T-cell receptor gamma (S. salar); cholecystokinin receptor type A (M. musculus and C. familiaris); Interleukin-13 receptor alpha-2 (S. salar); scavenger receptor cysteine-rich type 1 protein M130 (M. musculus) and protein M160 (H. sapiens); transferrin receptor protein 1 (C. familiaris); tumor necrosis factor receptor superfamily member 11B (H. sapiens); macrophage mannose receptor 1 (M. musculus); chemokine receptor-like 1 (O. mykiss); C-X-C chemokine receptor type 1 (O. cuniculus) and the nuclear receptor subfamily 5 group A member 2 (H. sapiens), which agrees with the increased responsiveness as determined by gene ontology. Other important up-regulated genes include tumor necrosis factor-inducible gene 6 protein (O. cuniculus) and the toll-like receptor 8 (S. salar) that, like other members of the TLR family, plays a fundamental role in pathogen recognition and activation of innate immunity by production of cytokines (Rebl et al., 2010; Pietretti and Wiegertjes, 2014). Important down-regulated genes include cytokine-related genes, such as the tumor necrosis factor alpha-induced protein 3 (O. mykiss) and the lipopolysaccharide-induced tumor necrosis factor-alpha (S. salar); the cytokine inducible SH2-containing protein (S. salar) and the FL cytokine receptor precursor (S. salar), and the proteasome activator complex subunit 4-like (O. niloticus). Down-regulation of the myxovirus resistance 1 (S. salar) illustrates the reduced viral immune response.
RNAseq Validation and Analyses on the Physiological Process Level
RNAseq data in this study were not validated by RT-qPCR. Unlike with microarray data, the verification of RNAseq data by RT-qPCR is often done but not necessarily required. RNAseq and RT-qPCR are two different methods with a different dynamic range of sensitivities. RNAseq has a much broader linear range than microarrays and the statistical analyses by DESeq are very stringent. RT-qPCR itself is often not even well validated according to MIQE guidelines. Validation using RT-qPCR on the same RNA samples as assayed in the RNA-seq analysis only validates the technology, it does not validate the conclusion about the treatments or conditions (Fang and Cui, 2011). Moreover, RNAseq represents a high-throughput transcriptomic approach, specifically useful when analysing on the level of physiological processes (e.g., Palstra et al., 2015; Rurangwa et al., 2015) which was also the main aim of this study. This paper is aimed to be a first step toward a project on the immune modulating effects of seaweeds in which this study can be followed up by dedicated RT-qPCR studies to validate the results obtained on a new and larger set of samples.
Conclusion
In conclusion, Atlantic salmon fed with dietary seaweeds showed more homogenous growth but was not different in FCR. In general no major differences in performance were found in comparison with fishmeal fed fish. However, the dietary seaweeds provoke a more efficient immune response, which involves more efficient identification of the infection site and processing and presentation of antigens. Specific Laminaria digitata effects included reduction of interferon-gamma-mediated signaling. The commercial blend of seaweeds improved immune processes such as receptor-mediated endocytosis and cell adhesion, and increased the expression of genes involved in response to lipopolysaccharide and inflammatory response. It can be concluded that the inclusion of seaweeds such as Laminaria digitata, can have important modulatory effects on the immune capacity of Atlantic salmon resulting in a more efficient immune response.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
AP and JK: conception and design of the study. AP, JK, ABG, and RD: acquisition of data. AP, JK, and RD: analysis and interpretation of data. AP, JK, RD, and MP: drafted the manuscript and reviewed. All authors read and approved the final manuscript.
Conflict of Interest Statement
RD was employed by company ZF-screens B.V. The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank S. Jong-Raadsen and H. Jansen (ZF-screens-B.V.) for technical support related to RNA sequencing. The authors also thank A. van Gool, Y. van Es, and A. Hofman (Wageningen Marine Research) for animal care. They and R. Benito, D. Mes, K. de Hoon, N. de Visser, and S. Neitzel are acknowledged for assistance with sampling. Part of this study was presented as poster at the Aquaculture Europe congress in Rotterdam (2015).
Funding. This study was funded as DLO project within the TripleP@Sea sub-theme KB MUPs Feed Component 2 (KB-24-001-019).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.00625/full#supplementary-material
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2014). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni M., Romano N., Finoia M. G., Abellic L., Scapigliati G., Tiscar P. G. (2005). Short- and long-term effects of a dietary yeast b-glucan (Macrogard) and alginic acid (Ergosan) preparation on immune response in sea bass (Dicentrarchus labrax). Fish Shellfish Immunol. 18 311–325. 10.1016/j.fsi.2004.08.003 [DOI] [PubMed] [Google Scholar]
- Bansemir A., Blume M., Schröder S., Lindequist U. (2006). Screening of cultivated seaweeds for antibacterial activity against fish pathogenic bacteria. Aquaculture 252 79–84. 10.1016/j.aquaculture.2005.11.051 [DOI] [Google Scholar]
- Blouin C. M., Lamaze C. (2013). Interferon gamma receptor: the beginning of the journey. Front. Immunol. 4:267 10.3389/fimmu.2013.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero-Solares A., Hall J. R., Xue X., Eslamloo K., Taylor R. G., Parrish C. C., et al. (2017). The dietary replacement of marine ingredients by terrestrial animal and plant alternatives modulates the antiviral immune response of Atlantic salmon (Salmo salar). Fish Shellfish Immunol. 64 24–38. 10.1016/j.fsi.2017.02.040 [DOI] [PubMed] [Google Scholar]
- Dettleff P., Moen T., Santi N., Martinez V. (2017). Transcriptomic analysis of spleen infected with infectious salmon anemia virus reveals distinct pattern of viral replication on resistant and susceptible Atlantic salmon (Salmo salar). Fish Shellfish Immunol. 61 187–193. 10.1016/j.fsi.2017.01.005 [DOI] [PubMed] [Google Scholar]
- Eslamloo K., Xue X., Hall J. R., Smith N. C., Caballero-Solares A., Parrish C. C., et al. (2017). Transcriptome profiling of antiviral immune and dietary fatty acid dependent responses of Atlantic salmon macrophage-like cells. BMC Genomics 18:706 10.1186/s12864-017-4099-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Cui X. (2011). Design and validation issues in RNA-seq experiments. Brief. Bioinform. 12 280–287. 10.1093/bib/bbr004 [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T. B., Gringhuis S. I. (2009). Signalling through C-type lectin receptors: shaping immune responses. Nat. Rev. Immunol. 9 465–479. 10.1038/nri2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland S. J., Liao X. C., Mendenhall M. K., Zhou X., Pardo J., Chu P., et al. (2001). Functional cloning of Src-like adapter protein-2 (SLAP-2), a novel inhibitor of antigen receptor signaling. J. Exp. Med. 194 1263–1276. 10.1084/jem.194.9.1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev D. B., Roach J. C., Mackenzie S., Planas J. V., Goetz F. W. (2005). Endotoxin recognition: in fish or not in fish? FEBS Lett. 579 6519–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H. J., Xiang L.-X., Shao J. Z. (2007). Molecular cloning and identification of macrophage migration inhibitory factor (MIF) in teleost fish. Dev. Comp. Immunol. 31 1131–1144. 10.1016/j.dci.2007.02.004 [DOI] [PubMed] [Google Scholar]
- Kaattari S. L., Irwin M. J. (1985). Salmonid spleen and anterior kidney harbor populations of lymphocytes with different B cell repertoires. Dev. Comp. Immunol. 9 433–444. 10.1016/0145-305X(85)90006-0 [DOI] [PubMed] [Google Scholar]
- Kumari J., Zhang Z., Swain T., Chi H., Niu C., Bøgwald J. (2015). Transcription factor T-Bet in Atlantic salmon: characterization and gene expression in mucosal tissues during Aeromonas salmonicida infection. Front. Immunol. 6:345 10.3389/fimmu.2015.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M. T., Cho H., Lesch H. P., Gosselin D., Heinz S., Tanaka-Oishi Y., et al. (2013). Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498 511–515. 10.1038/nature12209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Umasuthan N., Whang I., Saranya Revathy K., Lee S., De Zoysa M., et al. (2014). Two NF-kB inhibitor-alpha (IkBa) genes from rock bream (Oplegnathus fasciatus): molecular characterization, genomic organization and mRNA expression analysis after immune stimulation. Fish Shellfish Immunol. 41 633–642. 10.1016/j.fsi.2014.10.015 [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. (2009). 1000 Genome project data processing subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 25 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie S., Balasch J. C., Novoa B., Ribas L., Roher N., Krasnov A. (2008). Comparative analysis of the acute response of the trout, O. mykiss, head kidney to in vivo challenge with virulent and attenuated infectious hematopoietic necrosis virus and LPS-induced inflammation. BMC Genomics 9:141 10.1186/1471-2164-9-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie S., Liarte C., Iliev D., Planas J. V., Tort L., Goetz F. W. (2004). Characterization of a highly inducible novel CC chemokine from differentiated rainbow trout (Oncorhynchus mykiss) macrophages. Immunogenetics 56 611–615. 10.1007/s00251-004-0698-6 [DOI] [PubMed] [Google Scholar]
- Martin S. A. M., Król E. (2017). Nutrigenomics and immune function in fish: new insights from omics technologies. Dev. Comp. Immunol. 75 86–98. 10.1016/j.dci.2017.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rubio L., Evensen Ø., Krasnov A., Jørgensen S. M., Wadsworth S., Ruohonen K., et al. (2014). Effects of functional feeds on the lipid composition, salmon transcriptomic responses and pathology in heart of Atlantic (Salmo salar L.) before and after experimental challenge with Piscine Myocarditis Virus (PMCV). BMC Genomics 15:462 10.1186/1471-2164-15-462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rubio L., Morais S., Evensen Ø., Wadsworth S., Ruohonen K., Vecino J. L. G.et al. (2012). Functional feeds inflammation reduce heart salmon pathology in Atlantic (Salmo salar L.) following experimental challenge with Atlantic salmon reovirus (ASRV). PLoS One 7:e40266 10.1371/journal.pone.0040266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez-Acuña G., Gonçalves A. T., Valenzuela-Muñoz V., Pino-Marambio J., Wadsworth S., Gallardo-Escárate C. (2015). Transcriptome immunomodulation of in-feed additives in Atlantic salmon Salmo salar infested with sea lice Caligus rogercresseyi. Fish Shellfish Immunol. 47 450–460. 10.1016/j.fsi.2015.09.009 [DOI] [PubMed] [Google Scholar]
- Palstra A. P., Beltran S., Burgerhout E., Brittijn S. A., Magnoni L. J., Henkel C. V. (2013). Deep RNA sequencing of the skeletal muscle transcriptome in swimming fish. PLoS One 8:e53171 10.1371/journal.pone.0053171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra A. P., Fukaya K., Chiba H., Dirks R. P., Planas J. V., Ueda H. (2015). The olfactory transcriptome and progression of sexual maturation in homing chum salmon Oncorhynchus keta. PLoS One 10:e0137404 10.1371/journal.pone.0137404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Kim Y., Lee S., Park J. J., Park Z. Y., Sun W., et al. (2010). SNX18 shares a redundant role with SNX9 and modulates endocytic trafficking at the plasma membrane. J. Cell Sci. 123 1742–1750. 10.1242/jcs.064170 [DOI] [PubMed] [Google Scholar]
- Peixoto M. J., Salas-Leitón E., Pereira L. F., Queiroz A., Magalhães F., Pereira R. (2016). Role of dietary seaweed supplementation on growth performance, digestive capacity and immune and stress responsiveness in European seabass (Dicentrarchus labrax). Aquacult. Rep. 3 189–197. 10.1016/j.aqrep.2016.03.005 [DOI] [Google Scholar]
- Pietretti D., Wiegertjes G. F. (2014). Ligand specificities of Toll-like receptors in fish: indications from infection studies. Dev. Comp. Immunol. 43 205–222. 10.1016/j.dci.2013.08.010 [DOI] [PubMed] [Google Scholar]
- Rajapakse N., Kim S.-K. (2011). Nutritional and digestive health benefits of seaweed. Adv. Food Nutr. Res. 64 17–28. 10.1016/B978-0-12-387669-0.00002-8 [DOI] [PubMed] [Google Scholar]
- Rebl A., Goldammer T., Seyfert H.-M. (2010). Toll-like receptor signaling in bony fish. Vet. Immunol. Immunopathol. 134 139–150. 10.1016/j.vetimm.2009.09.021 [DOI] [PubMed] [Google Scholar]
- Reverter M., Bontemps N., Lecchini D., Banaigs B., Sasal P. (2014). Use of plant extracts in fish aquaculture as an alternative to chemotherapy: current status and future perspectives. Aquaculture 433 50–61. 10.1016/j.aquaculture.2014.05.048 [DOI] [Google Scholar]
- Rozas-Serri M., Peña A., Maldonado L. (2018). Transcriptomic profiles of post-smolt Atlantic salmon challenged with Piscirickettsia salmonis reveal a strategy to evade the adaptive immune response and modify cell-autonomous immunity. Dev. Comp. Immunol. 81 348–362. 10.1016/j.dci.2017.12.023 [DOI] [PubMed] [Google Scholar]
- Rurangwa E., Sipkema D., Kals J., Ter Veld M., Forlenza M., Bacanu G. M. (2015). Impact of a novel protein meal on the gastrointestinal microbiota and host transcriptome of larval zebrafish Danio rerio. Front. Physiol. 6:133 10.3389/fphys.2015.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem M., Rexroad C. E., III, Wang J., Thorgaard G. H., Yao J. (2010). Characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches. BMC Genomics 11:564 10.1186/1471-2164-11-564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas I., Lockhart K., Bowden T. J., Collet B., Secombes C. J., Ellis A. E. (2004). An assessment of immunostimulants as Mx inducers in Atlantic salmon (Salmo salar L.) parr and the effect of temperature on the kinetics of Mx responses. Fish Shellfish Immunol. 17 159–170. 10.1016/j.fsi.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Sheikhzadeh N., Pashaki A. K., Nofouzi K., Heidarieh M., Tayefi-Nasrabadi H. (2012). Effects of dietary Ergosan on cutaneous mucosal immune response in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 32 407–410. 10.1016/j.fsi.2011.11.028 [DOI] [PubMed] [Google Scholar]
- Skjesol A., Hansen T., Shi C.-Y., Thim H. L., Jørgensen J. B. (2010). Structural and functional studies of STAT1 from Atlantic salmon (Salmo salar). BMC Immunol. 11:17 10.1186/1471-2172-11-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler-Vila A., Coughlan S., Guiry M. D., Kraan S. (2009). The red alga Porphyra dioica as a fish-feed ingredient for rainbow trout (Oncorhynchus mykiss): effects on growth, feed efficiency, and carcass composition. J. Appl. Phycol. 21 617–624. 10.1007/s10811-009-9423-z [DOI] [Google Scholar]
- Swain P., Nayak S. K., Nanda P. K., Dash S. (2008). Biological effects of bacterial lipopolysaccharide (endotoxin) in fish: a review. Fish Shellfish Immunol. 25 191–201. 10.1016/j.fsi.2008.04.009 [DOI] [PubMed] [Google Scholar]
- Tacchi L., Bickerdike R., Douglas A., Secombes C. J., Martin S. A. M. (2011). Transcriptomic responses to functional feeds in Atlantic salmon (Salmo salar). Fish Shellfish Immunol. 31 704–715. 10.1016/j.fsi.2011.02.023 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L. M. P., Araújo M., Batista S., Peixoto M. J., Sousa-Pinto I., Brotas V. (2016). Carotenoid deposition, flesh quality and immunological response of Nile tilapia fed increasing levels of IMTA-cultivated Ulva spp. J. Appl. Phycol. 28 691–701. 10.1007/s10811-015-0590-9 [DOI] [Google Scholar]
- Valente L. M. P., Gouveia A., Rema P., Matos J., Gomes E. F., Pinto I. S. (2006). Evaluation of three seaweeds Gracilaria bursa-pastoris, Ulva rigida and Gracilaria cornea as dietary ingredients in European sea bass (Dicentrarchus labrax) juveniles. Aquaculture 252 85–91. 10.1016/j.aquaculture.2005.11.052 [DOI] [Google Scholar]
- van Kampen E. J., Zijlstra W. G. (1961). Standardization of hemoglobinometry II. The hemiglobincyanide method. Clin. Chim. Acta 6 538–544. 10.1016/0009-8981(61)90145-0 [DOI] [PubMed] [Google Scholar]
- Wilke T., Faulkner S., Murphy L., Kealy L., Kraan S., Brouns F. (2015). Seaweed enrichment of feed supplied to farm-raised Atlantic salmon (Salmo salar) is associated with higher total fatty acid and LC n-3 PUFA concentrations in fish flesh. Eur. J. Lipid Sci. Technol. 117 767–772. 10.1002/ejlt.201400166 [DOI] [Google Scholar]
- Zhang L., Gao C., Liu F., Song L., Su B., Li C. (2016). Characterization and expression analysis of a peptidoglycan recognition protein gene, SmPGRP2 in mucosal tissues of turbot (Scophthalmus maximus L.) following bacterial challenge. Fish Shellfish Immunol. 56 367–373. 10.1016/j.fsi.2016.07.029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.


