Abstract
Sex differences in peripheral immune responses are well recognized. This is associated with sex differences in many immunological diseases. As the intestinal microbiota is known to influence the immune system, such sex differences in immune responses may be a consequence of sex-specific microbiota. Therefore, this mini-review discusses sex differences in intestinal microbiota and the possible role of microbiota in shaping sexually dimorphic immunity. Sex differences in microbiota composition are clearly found in mice studies and also in human studies. However, the lack of standardization in human studies may mask the sexual dimorphism in microbiota composition in human studies, since many factors such as age, genetic background, BMI, diet, and sex hormones appear to interfere with the sexual dimorphism in microbiota composition. Only a few mice studies found that differences in gut microbiota composition are causative for some aspects of sexually dimorphic immunity. Therefore, future studies should focus on a causal relationship between sexually dimorphic immunity and microbiota, considering the abovementioned interfering confounding factors. This would benefit the development of more sex-specific effective treatment options for immunological diseases.
Keywords: sex differences, microbiota, immunity, mice, humans, interfering factors
Introduction
Sex differences in immune responses are well recognized, but the mechanisms and reasons behind the dimorphic responses are still incompletely understood (1). A better insight in the reasons for differences in immune responses between the sexes might lead to more effective strategies to fight diseases in which sex plays a role, such as systemic lupus erythematosus and type 1 diabetes (T1D) (1).
Generally, both the innate immune response and adaptive immune response are stronger in females than in males (1). Recent research has shown that sex-specific differences in gut microbiota exist (2–17). Gut microbiota are in close contact with our intestinal immune system and play a major role in health and disease (18). As the immune system is partially shaped by gut microbiota this might be one of the reasons why sexual dimorphism develops. The interaction between intestinal microbes and the intestinal immune system appears to be reciprocal, since both are able to influence each other (19). Several gut bacteria, such as Lactobacillus plantarum and several Clostridia strains, have been shown to influence the frequency of immune cells in the intestine, such as T-regulatory cells (Tregs) (20, 21). On the other hand, the intestinal immune system is able to selectively promote the growth of specific bacteria by using mechanisms such as production of secretory immunoglobulin A (sIgA) (19). The role of sIgA in promoting specific microbiota is demonstrated by Peterson et al., who found that the production of IgA after introduction of one single bacteria into germ-free Rag−/− mice, harboring IgA-producing hybridoma cells, led to a reduced bacterial epitope expression (22). Besides influencing the intestinal immune system, gut microbiota can also shape the systemic immune system (23) (see also Figure 1).
Figure 1.
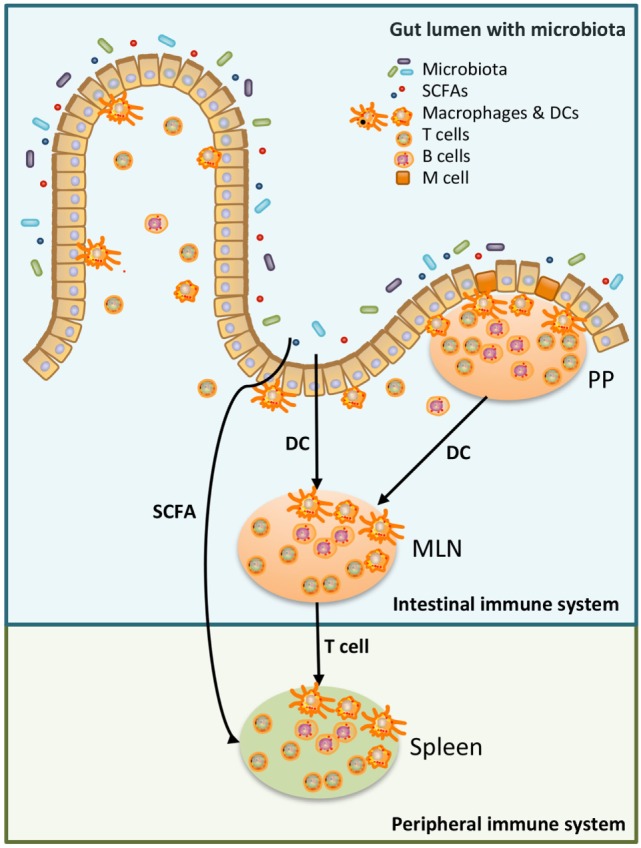
Schematic overview of the relation between microbiota, the intestinal immune system, and the peripheral immune system. Luminal microbiota and other products are continuously sampled and processed by the intestinal immune system. This could be done by specialized cells in the Peyer’s Patches (PP), microfold (M) cells, which transfer antigen to local dendritic cells (DCs). DC can present the antigen to T cells either in the PP or in the mesenteric lymph nodes (MLN). Alternatively, DC in the lamina propria sample antigen in the intestinal lumen and present antigen to T cells in the MLN (24, 25). T cells, which have recognized the antigen in the MLN, have access to the peripheral immune system and may affect the peripheral immune system (25). Moreover, metabolites produced by microbiota, such as short-chain fatty acid (SCFA), can also affect the peripheral immune system (26, 27).
This mini-review describes the current knowledge on sexually dimorphism in systemic and intestinal immunity and the interplay with gut microbiota. Recent exchange studies of microbiota between male and female mice have shed light on the complex relationship between microbiota, immunity, and genetics of the host. Ultimately this knowledge might lead to sex-specific strategies to manage disease.
Sex Differences in Peripheral Immunity
Both the innate immune arm and adaptive immune arm have sex-specific differences as comprehensively reviewed by Klein et al. (1). Females were found to have a higher number of cells of the adaptive immune system such as T-helper (Th) cells and B cells (1), and a higher percentage of IL-2-producing lymphocytes (28). Males, on the other hand, were shown to have less phagocytic capacity of both macrophages and neutrophils, but a higher frequency of natural killer (NK) cells (1). Moreover, the percentage of tumor necrosis factor alpha (TNF-α), interleukin 1 beta (IL-1β), and IL-12-producing monocytes was found to be higher in males (28). These differences are held responsible for the sex bias in many immunological diseases, as the higher immune reactivity in females may contribute to their higher risk to develop autoimmune diseases, such as rheumatoid arthritis and systemic lupus erythematosus and their higher resistance to various infections as compared with males (1, 29).
Sex hormones (progesterone, estrogen, and testosterone) may play a role in the induction of sex difference in immune responses. Most immune cells in both males and females express receptors for these hormones (30), while circulating hormone levels are very different between the sexes. The impact of sex hormones on immunity is, for instance, illustrated by differences in the immune status between the follicular and the luteal phase of the ovarian cycle in women (31–33). For example, the number of peripheral Tregs is higher during the follicular phase as compared with the luteal phase (33), while the number of Th2 cells is higher in the luteal phase as compared with the follicular phase (31, 32). This is associated with low levels of progesterone and estrogen in the follicular phase and high levels of these hormones in the luteal phase (33). These temporary shifts in sex hormone levels are thought to underlie the fast and profound changes in immune status during the ovarian cycle. Also changes in the immune system after menopause in women suggest a role for sex hormones in regulating immune responses. Menopause is associated with decreased estrogen and progesterone levels (34) and has a substantial impact on the peripheral immune system, with an increase in pro-inflammatory serum markers (IL-1, IL-6, and TNF-α) and a decrease in Th cells and B cells (35).
Sexual Dimorphism in Intestinal Immunity
Many pathogens enter the human body via the gastrointestinal tract. Not surprisingly the human body has developed very efficacious but specific strategies to prevent invasion of harmful invaders into the periphery (36), while at the same time the defense system should tolerate the trillions of commensal bacteria that are required for digestion of food, vitamin production (37), and production of immunological active molecules such as short-chain fatty acids (SCFAs) (38). The first line of defense is accomplished by forming a physical barrier of epithelial cells, covered by a mucus layer (36). Below and in between this barrier the body’s largest population of immune cells is located; approximately 80% of all immune cells are located here (39). The ileum contains special immune sampling sites that are not covered by mucus, which are called the Peyer’s patches (PP) (40). On top of the PP specialized epithelial cells, called microfold (M) cells, sample luminal content, which is transported to the dome of the PP for processing and recognition by the intestinal immune system (41). However, also in between the epithelial cells in the small intestine immune sampling occurs. This is done by specialized antigen-presenting cells (APCs) such as dendritic cells (DCs) and macrophages. These DCs and macrophages are present just below the surface of the epithelium and constantly sample the gut lumen. They can engulf antigens and present them to other cells, and depending on the type of antigen they either activate or attenuate an immune response (24). After encountering an antigen, DCs are known to migrate to the PP or mesenteric lymph nodes (MLN) to present their antigens to lymphocytes (36, 40). Macrophages, on the other hand, are resident and stay in the lamina propria where they have a role as intestinal innate effector cells and also communicate with other local immune cells (42). The cytokine milieu induced by luminal antigens will regulate intestinal immune cell plasticity. In case of food proteins or commensal bacteria, intestinal epithelial cells are stimulated and produce factors such as thymic stromal lymphopoietin and transforming growth factor beta (TGB-β) and retinoic acid (RA), which can condition specific CD103+ DCs to develop a tolerogenic phenotype (24). Besides, specific anti-inflammatory CX3CR1+ macrophages are able to condition CD103+ DCs to become tolerogenic by transferring soluble antigens to these DCs (43). Subsequently, these CD103+ DCs can promote the differentiation of naïve Th cells into Tregs by producing RA and TGB-β. Tregs can produce IL-10 and are important in controlling other Th responses, preventing inflammation and promoting tolerance (24, 40). However, in case of pathogens, intestinal epithelial cells and APCs will produce different factors, such as IL-1β, IL-6, and IL-23, which create a pro-inflammatory microenvironment (40, 44). This will polarize both DCs and macrophages to a pro-inflammatory phenotype (42). Subsequently, DCs will migrate to the induction sites and differentiate naïve Th cells into, e.g., Th1 or Th17 cells which will lead to inflammation and the elimination of the bacteria (44).
Despite the close interaction between the intestinal immune system and the peripheral immune system, sexual dimorphism in the intestinal immune responses is addressed in not more than a few studies (45–47). For that reason, our group studied the effect of sex on several intestinal immune cells (T cells, DCs, macrophages, and NK cells) in the PP of mice. We demonstrated that sexual dimorphism indeed also exists in the intestinal immune system and can be visualized in the PP. Males had a lower percentage of T cells, but a higher percentage of Th1 cell in the PP. Furthermore, males had a higher percentage of CD80+ DCs and NK cells in the PP as compared with females (45). Thus overall, male mice showed an enhanced intestinal innate immune arm and a reduced adaptive immune arm as compared with females. In another study in rats, a lower percentage of T cells was found in the MLN of males as compared with females, while also a lower percentage of macrophages was found in the male MLN as compared with the female MLN (46). In addition, many sexually dimorphically expressed genes in the small intestine and colon of mice related to immunological functions were found (47, 48).
Sexual Dimorphism in Intestinal Microbiome
The intestinal immune system is in close contact with trillions of microbes, mainly anaerobic bacteria, together called the microbiome (49). This microbiome supports the host on various levels. Bacteria ferment dietary components, such as complex carbohydrates, that cannot be metabolized by human enzymes (37, 50). Moreover, commensal microbiota compete for luminal substrates with pathogens, preventing the growth of harmful pathogenic bacteria and thereby protect the host from pathogenic infections (37, 50). Other bacteria produce vitamins for the host, such as biotin and vitamin K, or modulate the sensitivity to hormones involved in the host energy storage, such as insulin and leptin (37, 50–52). Moreover, microbiota or microbial fermentation products, such as the SCFA butyrate, have been shown to influence immune cells (20, 21, 53). For example, some members of the microbiota, such as Lactobacillus plantarum and several Clostridia strains, were found to induce regulatory responses in T cells (20, 21), while others were able to stimulate specific Th17-cell responses (54). Furthermore, the SCFA butyrate has been shown to induce the differentiation of Tregs in the colon (53). The numbers and diversity of bacterial species vary along the gastrointestinal tract with a gradual increase in numbers from stomach toward the distal part of the colon (55, 56). During recent years, it has been found that a disbalance in intestinal microbiota communities may be involved in the pathogenesis of some immunological Western diseases, such as inflammatory bowel disease (IBD) and metabolic syndrome (57). Both IBD and metabolic syndrome may have a sex bias in prevalence (58, 59), which might be explained by differences in both intestinal immunity and by microbiota differences between the sexes.
It is only recently that studies demonstrated pertinent sex differences in microbiota composition (2–17) that possibly might explain differences in not only intestinal immunity but also peripheral immunity between the sexes. Most studies have been performed in rodents. Female mice were found to have an increased microbiota diversity as compared with male mice (4, 9, 17). In addition, when focusing on bacterial composition, many bacterial species were found to have a higher abundance in one of the two sexes, but in the studies performed, different bacterial species were found to be enriched in either of the sexes. For example, Sheng et al. found a higher relative abundance of S24-7, but a lower relative abundance of Bacteroidaceae, Rikenellaceae, Lactobacillaceae, and Verrucomicrobiaceae in male as compared with female C57BL/6 WT mice (12). Kozik et al. found that in WT B6.129S mice, males had a higher relative abundance of Ruminococcaceae, Ruminococcus, and Anaerostipes, whereas Peptostreptococcaceae was higher in females (9). These studies may indicate that sex differences in the microbiome may depend on strain, i.e., genetic background. A similar conclusion was drawn by Org et al., who investigated sex differences in gut microbiota composition in 89 different inbred strains of mice and when analyzing the strains separately, strain-specific sex differences in microbiota composition were found (5). However, this group also found that in the total cohort the phylum Actinobacteria and Tenericutes were more abundant in male than female mice. In addition, at genus level they identified Allobaculum, Anaeroplasma, and Erwinia to be more abundant in males than females, whereas SMB53, Dorea, Coprococcus, and Ruminococcus were more abundant in female mice (5).
Not only strain but also diet was found to interfere with sex effects regarding microbiota composition in mice (2, 12, 13). For instance, administration of a high fat diet to C57BL/6 mice for 81 days induced a change in microbiota composition, but the magnitude of this effect was significantly different between males and females. The high fat diet increased the abundance of several Ruminococcaceae and Lachnospiraceae, but the affected members within these bacterial families differed in males and females (13). In addition, Sheng et al. found that in C57BL/6 mice the effect of a western diet (WD), defined as a high fat and carbohydrate diet for 4 months, significantly reduced the relative abundance of Erysipelotrichaceae in males only, whereas this WD significantly reduced the relative abundance of Lachnospiraceae in females only (12).
Not only mice studies but also human studies investigated the effect of sex on the gut microbiome. Similar to the mice studies, some human studies also found sex differences in the microbiome (2, 6–8, 16), while others did not (60–62). Mueller et al. found a higher level of the Bacteroides–Prevotella phylogenetic group in men as compared with women (6), while Li et al. found a higher abundance of some Clostridia, Bacteroidetes, and Proteobacteria species in men as compared with women (7). Haro et al. found no significant differences in microbiota diversity, however, at the genera level they found that men had a lower relative abundance of the Bilophila genus and a higher relative abundance of Veillonella and Methanobrevibacter genera compared with women (16). Finally, Dominianni et al. found that men had a higher relative abundance of Bacteroidetes as compared with women (8).
The challenge in identifying pertinent human sex differences is standardization of the studies, as it is known that factors such as age, genetic background, BMI, diet, and sex hormones can influence sexual dimorphism in microbiota (2, 4, 5, 16, 63) (Figure 2). The interaction between diet and sex was clearly shown in humans by Bolnick et al. who found that in humans the effect of diet was significantly different between men and women (2). For example, the relative abundance of Parabacteroides had a significant positive correlation with the amount of saturated fatty acid intake in females, while this effect was absent in males (2). In addition, the reproductive condition of females (e.g., menstrual cycle, the use of oral contraceptives and menopause), which is often not taken into account, may affect the microbiome, as effects of sex hormones on the gut microbiota have been shown by Org et al. in mice (5). Moreover, Yurkovetskiy et al. also found effects of sex hormones on the microbiota. They found that the microbiota of non-obese diabetic (NOD) mice was not different between males and females before puberty; however, after puberty male mice had a significantly less diverse microbiota, while castration of the male mice reversed these sex differences (4). Similarly, recent studies have shown that the microbiota composition changes with age (63, 64). We found that aging had a different effect in male and female mice; aged male mice had a higher abundance of Lachnospira pectinoschiza et rel. as compared with young male mice, while old females had a higher abundance of Olsenella et rel. and Prevotella ruminicola et rel. as compared with young females (63). Finally, Haro et al. found that men had a lower relative abundance of the Bacteroides genus than women, but only when their BMI was above 33 (16). The interaction between sex, immune responses, microbiota, genetic background, and environmental factors, such as diet, seemed to be in line with the results of Zelinkova and der Woude, who found that the possible sex bias in susceptibility to IBD might be related to geographical factors (e.g., genetic background and/or environmental factors) (59).
Figure 2.
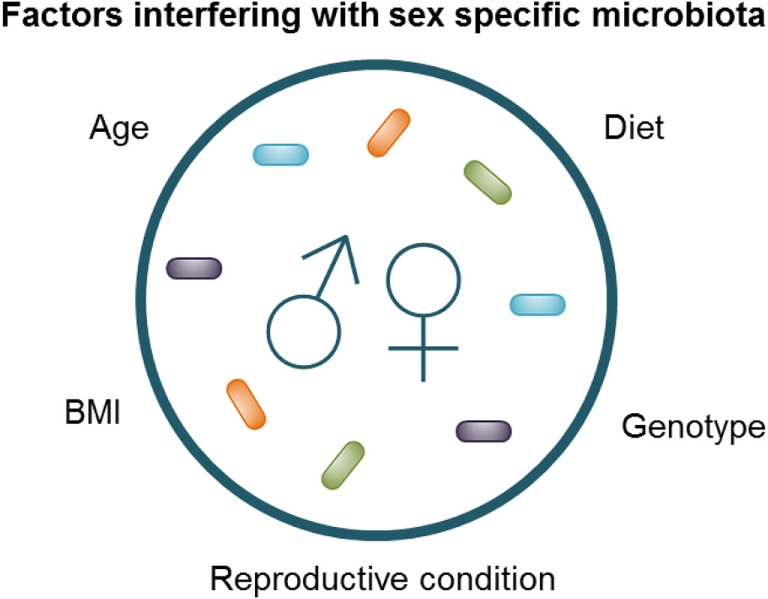
Factors interfering with sex-specific microbiota composition.
Interaction Between Microbiome and Immune System
In view of the well-known interplay between microbiota and immunity, it seems likely that the sex differences in microbiota may be (partly) responsible for the sex differences in immune responses. To determine causal relations, Fransen et al. performed a microbiota transfer study in germ-free mice by transferring male microbiota into female germ-free mice and female microbiota into germ-free male mice (47). Fransen observed striking effects and sex-dependent differences within 4 weeks after microbiota transfer. For instance, germ-free male recipients of male microbiota had higher percentages of RORγt+Foxp3+ cells in the PPs and MLN as compared with germ-free male recipients of female microbiota. On the other hand, there were also differences that were independent of the microbiome. It was shown that males in general had higher percentages of conventional Tregs (47), irrespective of whether they received the microbiota from male or female mice. These sex differences may be caused by sex hormones or the presence of the Y or X chromosome.
Further evidence for casual relations between sexual dimorphism in microbiota and effects on immunity follows from studies in NOD mice, in which female mice have a higher chance to spontaneously develop autoimmune T1D than males. This sex bias disappears when the mice are raised under germ-free conditions, demonstrating the influence of microbiota on sex differences in development of autoimmunity and thus on sex differences in immune responses (3, 4). This was confirmed in a microbiota transfer study by Markle et al. (3). They showed that transplantation of microbiota from conventional NOD males to germ-free NOD females resulted in protection of the female mice against T1D. Since they also found this transplantation increased testosterone levels in the female mice (3), these findings suggest that both microbiota and sex hormones may be involved in the sex bias in this autoimmune disease and therefore in immune responses.
Concluding Remarks and Future Perspectives
Present knowledge shows that sex has an effect on the microbiota composition in both mice and humans. Mice studies demonstrate that this may partly explain sexually dimorphic immunity. However, there are also effects of sex hormones and genetics on the immune system. In humans, pertinent sex differences in microbiota and effects on immunity are more difficult to proof. The absence of sex differences in several human microbiota analyses may be caused by the fact that interfering factors like reproductive condition (e.g., menstrual cycle, the use of oral contraceptives and menopause), genetic background, and diet are not taken into account but may influence the microbiota composition and immune system. A simple experiment in mice, which investigated sex differences in microbiota composition in multiple genetically distinct mice strains, revealed that sex-dependent differences were pertinent in most strains, but the specific species that differed between male and female mice were dependent on the genetic background of the strains (5). This illustrates the complexity of the interplay between microbiota, immunity, and genetics of the host and the need for highly controlled human studies in which confounding factors are as much as possible excluded. Further insight into the causal relationship between sex, microbiome, and immunity is required. This could include studies in germ-free mice, in which the effect of a single sex-specific bacterial species on the immune system could be studied. It should also include studies investigating the function of sex-specific microbiota composition and immune system, for example, in challenge models like colitis (IBD) or gastrointestinal infections, such as Salmonella. In these studies, the abovementioned interfering factors need to be taken into account. Such studies may result in the development of more tailored sex-specific treatment strategies. Ultimately, this knowledge might lead to sex-specific strategies to manage diseases.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol (2016) 16:626–38. 10.1038/nri.2016.90 [DOI] [PubMed] [Google Scholar]
- 2.Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Org E, Parks B, et al. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat Commun (2014) 5:4500. 10.1038/ncomms5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markle JGM, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kamkczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science (2013) 339:1084–8. 10.1126/science.1233521 [DOI] [PubMed] [Google Scholar]
- 4.Yurkovetskiy L, Burrows M, Khan A, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity (2013) 39:400–12. 10.1016/j.immuni.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Org E, Mehrabian M, Parks BW, Shipkova P, Liu X, Drake TA, et al. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes (2016) 7:313–22. 10.1080/19490976.2016.1203502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol (2006) 72:1027–33. 10.1128/AEM.72.2.1027-1033.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A (2008) 105:2117–22. 10.1073/pnas.0712038105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominianni C, Sinha R, Goedert JJ, Pei Z, Yang L, Hayes RB, et al. Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS One (2015) 10:1–14. 10.1371/journal.pone.0124599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozik AJ, Nakatsu CH, Chun H, Jones-Hall YL. Age, sex, and TNF associated differences in the gut microbiota of mice and their impact on acute TNBS colitis. Exp Mol Pathol (2017) 103:311–9. 10.1016/j.yexmp.2017.11.014 [DOI] [PubMed] [Google Scholar]
- 10.Gomez A, Luckey D, Yeoman CJ, Marietta EV, Berg Miller ME, Murray JA, et al. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PLoS One (2012) 7:e36095. 10.1371/journal.pone.0036095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fushuku S, Fukuda K. Gender difference in the composition of fecal flora in laboratory mice, as detected by denaturing gradient gel electrophoresis (DGGE). Exp Anim (2008) 57:489–93. 10.1538/expanim.57.489 [DOI] [PubMed] [Google Scholar]
- 12.Sheng L, Jena PK, Liu HX, Kalanetra KM, Gonzalez FJ, French SW, et al. Gender differences in bile acids and microbiota in relationship with gender dissimilarity in steatosis induced by diet and FXR inactivation. Sci Rep (2017) 7:1748. 10.1038/s41598-017-01576-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bridgewater LC, Zhang C, Wu Y, Hu W, Zhang Q, Wang J, et al. Gender-based differences in host behavior and gut microbiota composition in response to high fat diet and stress in a mouse model. Sci Rep (2017) 7:10776. 10.1038/s41598-017-11069-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie G, Wang X, Zhao A, Yan J, Chen W, Jiang R, et al. Sex-dependent effects on gut microbiota regulate hepatic carcinogenic outcomes. Sci Rep (2017) 7:45232. 10.1038/srep45232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernbom N, Norrung B, Saadbye P, Molbak L, Vogensen FK, Licht TR. Comparison of methods and animal models commonly used for investigation of fecal microbiota: effects of time, host and gender. J Microbiol Methods (2006) 66:87–95. 10.1016/j.mimet.2005.10.014 [DOI] [PubMed] [Google Scholar]
- 16.Haro C, Rangel-Zuniga OA, Alcala-Diaz JF, Gomez-Delgado F, Perez-Martinez P, Delgado-Lista J, et al. Intestinal microbiota is influenced by gender and body mass index. PLoS One (2016) 11:e0154090. 10.1371/journal.pone.0154090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao L, Feng Q, Liang S, Sonne SB, Xia Z, Qiu X, et al. A catalog of the mouse gut metagenome. Nat Biotechnol (2015) 33:1103–8. 10.1038/nbt.3353 [DOI] [PubMed] [Google Scholar]
- 18.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science (2012) 336:1268–73. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature (2012) 489:231–41. 10.1038/nature11551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smelt MJ, de Haan BJ, Bron PA, van Swam I, Meijerink M, Wells JM, et al. L. plantarum, L. salivarius, and L. lactis attenuate Th2 responses and increase Treg frequencies in healthy mice in a strain dependent manner. PLoS One (2012) 7:e47244. 10.1371/journal.pone.0047244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature (2013) 500:232–6. 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- 22.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe (2007) 2:328–39. 10.1016/j.chom.2007.09.013 [DOI] [PubMed] [Google Scholar]
- 23.Molloy MJ, Bouladoux N, Belkaid Y. Intestinal microbiota: shaping local and systemic immune responses. Semin Immunol (2012) 24:58–66. 10.1016/j.smim.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pabst O, Mowat AM. Oral tolerance to food protein. Mucosal Immunol (2012) 5:232–9. 10.1038/mi.2012.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macpherson AJ, Smith K. Mesenteric lymph nodes at the center of immune anatomy. J Exp Med (2006) 203:497–500. 10.1084/jem.20060227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Correa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MA. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol (2016) 5:e73. 10.1038/cti.2016.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuno M, Noto D, Kaga N, Chiba A, Miyake S. The dual role of short fatty acid chains in the pathogenesis of autoimmune disease models. PLoS One (2017) 12:e0173032. 10.1371/journal.pone.0173032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouman A, Schipper M, Heineman M, Faas M. Gender difference in the non-specific and specific immune response in humans. Am J Reprod Immunol (2004) 52:19–26. 10.1111/j.1600-0897.2004.00177.x [DOI] [PubMed] [Google Scholar]
- 29.Ngo ST, Steyn FJ, McCombe PA. Gender differences in autoimmune disease. Front Neuroendocrinol (2014) 35:347–69. 10.1016/j.yfrne.2014.04.004 [DOI] [PubMed] [Google Scholar]
- 30.Faas MM, Vos P, Melgert BN. Sex hormones and immune regulation. Brain Immune (2011). [Google Scholar]
- 31.Bouman A, Moes H, Heineman MJ, de Leij LFMH, Faas MM. The immune response during the luteal phase of the ovarian cycle: increasing sensitivity of human monocytes to endotoxin. Fertil Steril (2001) 76:555–9. 10.1016/S0015-0282(01)01971-9 [DOI] [PubMed] [Google Scholar]
- 32.Faas M, Bouman A, Moesa H, Heineman MJ, de Leij L, Schuiling G. The immune response during the luteal phase of the ovarian cycle: a Th2-type response? Fertil Steril (2000) 74:1008–13. 10.1016/S0015-0282(00)01553-3 [DOI] [PubMed] [Google Scholar]
- 33.Oertelt-Prigione S. Immunology and the menstrual cycle. Autoimmun Rev (2012) 11:A486–92. 10.1016/j.autrev.2011.11.023 [DOI] [PubMed] [Google Scholar]
- 34.Koebele SV, Bimonte-Nelson HA. Modeling menopause: the utility of rodents in translational behavioral endocrinology research. Maturitas (2016) 87:5–17. 10.1016/j.maturitas.2016.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gameiro CM, Romão F, Castelo-Branco C. Menopause and aging: changes in the immune system – a review. Maturitas (2010) 67:316–20. 10.1016/j.maturitas.2010.08.003 [DOI] [PubMed] [Google Scholar]
- 36.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol (2010) 10:159–69. 10.1038/nri2710 [DOI] [PubMed] [Google Scholar]
- 37.Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol (2010) 10:735–44. 10.1038/nri2850 [DOI] [PubMed] [Google Scholar]
- 38.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol (2013) 14:676–84. 10.1038/ni.2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furness JB, Kunze WA, Clerc N. Nutrient tasting and signaling mechanisms in the gut. II. The intestine as a sensory organ: neural, endocrine, and immune responses. Am J Physiol (1999) 277:G922–8. 10.1152/ajpgi.1999.277.5.G922 [DOI] [PubMed] [Google Scholar]
- 40.Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol (2003) 3:331–41. 10.1038/nri1057 [DOI] [PubMed] [Google Scholar]
- 41.Brayden DJ, Jepson MA, Baird AW. Keynote review: intestinal Peyer’s patch M cells and oral vaccine targeting. Drug Discov Today (2005) 10:1145–57. 10.1016/S1359-6446(05)03536-1 [DOI] [PubMed] [Google Scholar]
- 42.Cerovic V, Bain CC, Mowat AM, Milling SW. Intestinal macrophages and dendritic cells: what’s the difference? Trends Immunol (2014) 35:270–7. 10.1016/j.it.2014.04.003 [DOI] [PubMed] [Google Scholar]
- 43.Mazzini E, Massimiliano L, Penna G, Rescigno M. Oral tolerance can be established via gap junction transfer of fed antigens from CX3CR1(+) macrophages to CD103(+) dendritic cells. Immunity (2014) 40:248–61. 10.1016/j.immuni.2013.12.012 [DOI] [PubMed] [Google Scholar]
- 44.Omenetti S, Pizarro TT. The Treg/Th17 axis: a dynamic balance regulated by the gut microbiome. Front Immunol (2015) 6:639. 10.3389/fimmu.2015.00639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elderman M, van Beek A, Brandsma E, de Haan B, Savelkoul H, de Vos P, et al. Sex impacts Th1, Treg, and DCs in both the intestinal and systemic immunity in a mouse strain and location dependent manner. Biol Sex Differ (2016) 7:21. 10.1186/s13293-016-0075-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shastri P, McCarville J, Kalmokoff M, Brooks SP, Green-Johnson JM. Sex differences in gut fermentation and immune parameters in rats fed an oligofructose-supplemented diet. Biol Sex Differ (2015) 6:13. 10.1186/s13293-015-0031-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fransen F, van Beek AA, Borghuis T, Meijer B, Hugenholtz F, van der Gaast-de Jongh C, et al. The impact of gut microbiota on gender-specific differences in immunity. Front Immunol (2017) 8:754. 10.3389/fimmu.2017.00754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steegenga WT, Mischke M, Lute C, Boekschoten MV, Pruis MG, Lendvai A, et al. Sexually dimorphic characteristics of the small intestine and colon of prepubescent C57BL/6 mice. Biol Sex Differ (2014) 5:11. 10.1186/s13293-014-0011-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science (2006) 312:1355–9. 10.1126/science.1124234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prakash S, Rodes L, Coussa-Charley M, Tomaro-Duchesneau C. Gut microbiota: next frontier in understanding human health and development of biotherapeutics. Biologics (2011) 5:71–86. 10.2147/BTT.S19099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kreznar JH, Keller MP, Traeger LL, Rabaglia ME, Schueler KL, Stapleton DS, et al. Host genotype and gut microbiome modulate insulin secretion and diet-induced metabolic phenotypes. Cell Rep (2017) 18:1739–50. 10.1016/j.celrep.2017.01.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cani PD, Knauf C. How gut microbes talk to organs: the role of endocrine and nervous routes. Mol Metab (2016) 5:743–52. 10.1016/j.molmet.2016.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature (2013) 504:446–50. 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- 54.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe (2008) 4:337–49. 10.1016/j.chom.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol (2016) 14:20–32. 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johansson ME. Mucus layers in inflammatory bowel disease. Inflamm Bowel Dis (2014) 20:2124–31. 10.1097/MIB.0000000000000117 [DOI] [PubMed] [Google Scholar]
- 57.Parekh PJ, Balart LA, Johnson DA. The influence of the gut microbiome on obesity, metabolic syndrome and gastrointestinal disease. Clin Transl Gastroenterol (2015) 6:e91. 10.1038/ctg.2015.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pradhan AD. Sex differences in the metabolic syndrome: implications for cardiovascular health in women. Clin Chem (2014) 60:44–52. 10.1373/clinchem.2013.202549 [DOI] [PubMed] [Google Scholar]
- 59.Zelinkova Z, der Woude CJ. Gender and inflammatory bowel disease. J Clin Cell Immunol (2014) 5:245–50. 10.4172/2155-9899.1000245 [DOI] [Google Scholar]
- 60.Dicksved J, Floistrup H, Bergstrom A, Rosenquist M, Pershagen G, Scheynius A, et al. Molecular fingerprinting of the fecal microbiota of children raised according to different lifestyles. Appl Environ Microbiol (2007) 73:2284–9. 10.1128/AEM.02223-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lay C, Rigottier-Gois L, Holmstrom K, Rajilic M, Vaughan EE, de Vos WM, et al. Colonic microbiota signatures across five northern European countries. Appl Environ Microbiol (2005) 71:4153–5. 10.1128/AEM.71.7.4153-4155.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature (2012) 486:207–14. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elderman M, Sovran B, Hugenholtz F, Graversen K, Huijskes M, Houtsma E, et al. The effect of age on the intestinal mucus thickness, microbiota composition and immunity in relation to sex in mice. PLoS One (2017) 12:e0184274. 10.1371/journal.pone.0184274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garcia-Pena C, Alvarez-Cisneros T, Quiroz-Baez R, Friedland RP. Microbiota and aging. A review and commentary. Arch Med Res (2017) 48(8):681–9. 10.1016/j.arcmed.2017.11.005 [DOI] [PubMed] [Google Scholar]


