Abstract
The nuclear envelope is a physiological barrier to electrogene transfer. To understand different mechanisms of the nuclear entry for electrotransfected plasmid DNA (pDNA), the current study investigated how manipulation of the mechanisms could affect electrotransfection efficiency (eTE), transgene expression level (EL), and cell viability. In the investigation, cells were first synchronized at G2-M phase prior to electrotransfection so that the nuclear envelope breakdown (NEBD) occurred before pDNA entered the cells. The NEBD significantly increased the eTE and the EL while the cell viability was not compromised. In the second experiment, the cells were treated with a nuclear pore dilating agent (i.e., trans-1,2-cyclohexanediol). The treatment could increase the EL, but had only minor effects on eTE. Furthermore, the treatment was more cytotoxic, compared with the cell synchronization. In the third experiment, a nuclear targeting sequence (i.e., SV40) was incorporated into the pDNA prior to electrotransfection. The incorporation was more effective than the cell synchronization for enhancing the EL, but not the eTE, and the effectiveness was cell type dependent. Taken together, the data described above suggested that synchronization of the NEBD could be a practical approach to improving electrogene transfer in all dividing cells.
Keywords: electrotransfection, electroporation, nuclear import, nuclear entry
Introduction
Electrotransfection is a widely used gene delivery method because it is simple to apply, safe, and able to transfer genes to some difficult-to-transfect cells.1 The technique is also referred to as electroporation, electropermeabilization, electrogene transfer, and gene electroinjection in the literature.2 The main limitation of this technology is low efficiency, compared with viral gene delivery approaches.3 To significantly increase the efficiency, it is critical to understand how plasmid DNA (pDNA) is transferred from extracellular space to the nucleus for successful transgene expression.3 For exogenous molecules, such as pDNA, to enter the nucleus of a mammalian cell, they must overcome three physiological barriers:4 plasma membrane, cytoplasm, and nuclear envelope.2 Without help, few naked pDNA molecules can penetrate across the membrane, and less than 1/1,000 naked pDNA injected into the cytosol is effectively delivered into the nucleus.5, 6, 7 Furthermore, cytosolic injection of naked pDNA may result in no gene expression, although nuclear injection of the same pDNA can lead to transgene expression in 50%–100% of cells.8 Similar results are observed for lipid nanoparticle-mediated gene delivery.9 These observations demonstrate the importance of cytoplasmic trafficking and nuclear entry in gene delivery. At present, three routes have been proposed for pDNA entry into the nucleus: (1) direct entry when the nuclear envelope breaks down during mitosis, (2) transport through nuclear pores, and (3) direct transport across the nuclear envelope.10
The nuclear envelope consists of two layers of phospholipid membrane through which there are channels formed by nuclear pore complexes (NPCs) to allow for exchange of molecules between the nucleus and the cytosol.11 When the nuclear envelope is intact, nuclear entry is mediated by passive diffusion or facilitated transport through the channels.12 For a molecule to enter by the diffusion, it must be significantly smaller than 9 nm10, 13 or 40 kDa,14 which is the size of small proteins.15 The facilitated transport through the NPCs is limited to molecules up to 39 nm in diameter and requires signals on the imported/exported molecules. The radius of gyration of naked pDNA is ∼100 nm, which is significantly larger than the cutoff size of the channel in the NPC.16
Although mechanisms of transport across the nuclear envelope remain uncertain for non-viral gene delivery, different delivery methods have been developed based on materials with intrinsic properties for overcoming this barrier. Some peptide and polymer-based non-viral vectors, such as polylysine17 and polyethylenimine (PEI),5, 18 are capable of transfecting non-dividing cells. For example, when primary fibroblast cells in confluent monolayers are arrested at G1 by contact inhibition, polylysine-mediated gene delivery can still reach a high transfection level.17 Similar results have also been observed when pDNA is delivered via linear PEI.5 The vector has been observed inside the nucleus at early time points (as soon as 3 hr after transfection) before cell division.19 However, branched PEI complexed with pDNA is observed only in the cytoplasm.20 The average size of the non-viral vectors is 60–100 nm, which means that they cannot enter the nucleus through NPCs whose cutoff size are 20–40 nm.17, 21, 22, 23 The observations above suggest that mechanisms of the nuclear entry of the non-viral vectors are complicated and may not be unique among different cells.
Three different approaches have been used to facilitate gene transfer into the nucleus. One approach is to deliver genes of interest into cells during the M phase when the nuclear envelope breaks down. A second approach is to sufficiently dilate the channel within the NPC, using nuclear pore dilating agents.24, 25, 26 The third approach is to incorporate DNA targeting sequence (DTS) into pDNA that contains binding sites for transcription factors with nuclear localization signals (NLSs),27, 28 which can facilitate the nuclear import of pDNA through the NPC. By using these approaches individually or in combination, the current study was designed to determine how they could be used to enhance electrotransfection efficiency (eTE).
Results
Cell-Cycle Synchronization
To transfect pDNA into cells when the nuclear envelope broke down, we synchronized cells through pretreatment with 100 ng/mL nocodazole for 16 hr. Nocodazole has a higher affinity to two self-assembly sites of tubulin, which can lead to depolymerization of microtubules.29 The depolymerization does not block cells to enter the M phase but inhibits mitotic spindle formation, which arrests cells in prometaphase with the nuclear envelope breakdown (NEBD).12, 30, 31, 32 Using flow cytometry technique,33, 34, 35 we observed that the percentages of HCT116 and COS7 cells synchronized at the G2-M phase were 84.0% and 69.2%, respectively (Figure S1). To visualize the depolymerization of microtubules, we transiently transfected HCT116 cells with a plasmid encoding fusion protein of GFP-α-tubulin. In transfected cells, the depolymerization, induced by nocodazole treatment, changed the α-tubulin distribution from a fiber-like network to a diffuse pattern (Figure S2A).
Visualization of Nuclear Envelope Breakdown
The nuclear morphology was visualized by staining cells with Hoechst 33342 dye, which binds to the minor groove of double-stranded DNA, particularly in adenine- and thymine-rich locations.36, 37, 38 The pattern of the dye distribution depends on the phase in cell cycle. In non-dividing or control cells, the nuclear staining with Hoechst 33342 dye outlined the nucleus (Figures S2B and S2C). In nocodazole-treated cells that were arrested in the prometaphase, the dye was aggregated, presumably because of chromosomal condensation and segregation (Figures S2B and S2C).39, 40, 41
The NEBD in synchronized cells was visualized by using two methods to indicate the border of the nucleus: one was to label membranous structures in cells with a fluorescent dye (FM4-64FX), and the other was to express a fluorescent fusion protein, mCherry-Lamin A, in cells. In the first method, HCT116 cells were pretreated with 100 ng/mL nocodazole for 16 hr, followed by 30-min incubation with FM4-64FX in the presence of nocodazole. In non-dividing cells that were not treated with nocodazole, FM4-64FX accumulated around the nucleus to outline its border, indicating the presence of the nuclear envelope (Figure S2B). In nocodazole-treated cells that were arrested at the prometaphase, the pattern of FM4-64FX distribution was diffuse throughout the cytosol and in regions labeled with Hoechst 33342 dye (Figure S2B), indicating the NEBD in these cells. In a separate experiment, we switched the steps of synchronization and staining of cells, i.e., HCT116 cells were pre-incubated for 30 min with FM4-64FX, followed by nocodazole treatment at 100 ng/mL for 16 hr. The switch of the experimental steps had little influence on the patterns of FM4-64FX distributions (see Figures S2B and S2C), indicating that FM4-64FX transport in cells was insensitive to depolymerization of microtubules induced by nocodazole treatment. It was likely to be achieved through diffusion and fusion of vesicles in the cytoplasm.
In the second method, the dissolution of the nuclear envelope was visualized by using cells expressing a fluorescent fusion protein mCherry-Lamin A. It is known that Lamin A stably binds to the nuclear envelope until the early prophase when it enters the cytoplasm.30, 42 Therefore, Lamin A was served as a marker for the NEBD. It outlined the nucleus in non-dividing control cells and showed a diffuse pattern throughout dividing cells in the nocodazole-treated group (Figure S2D).
Enhancement of eTE with Cell Synchronization
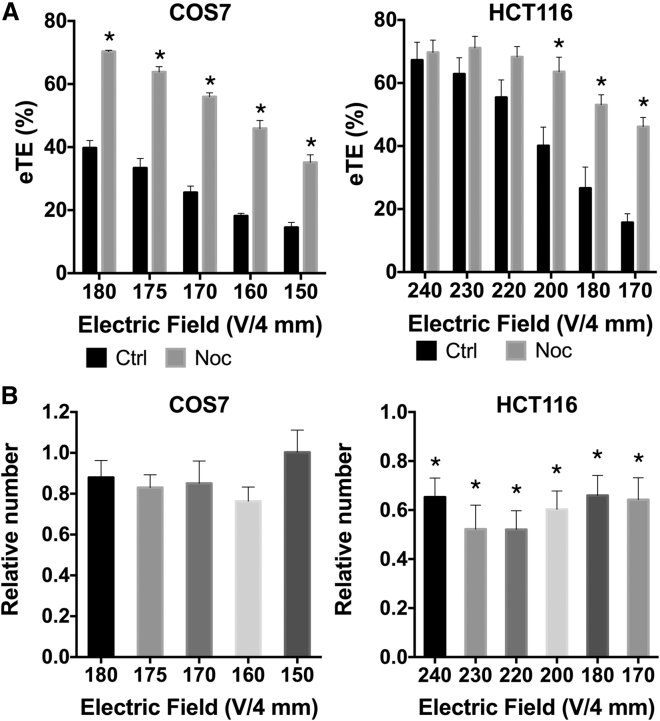
Cells were synchronized at the prometaphase through treatment with nocodazole (100 ng/mL) for 16 hr. Then the cells were collected and washed with PBS (without magnesium and calcium) to remove nocodazole and resuspended in the pulsing buffer for electrotransfection. After electrotransfection, cells were cultured in fresh medium without nocodazole for 24 hr for transgene expression. It was observed that the synchronized cells had a significantly higher eTE compared with asynchronized ones (Figure 1A). The enhancement was apparently independent of the applied field for COS7 cells but was more significant for HCT116 cells when lower electric fields (less than 200 V/4 mm) were applied. To understand whether the enhancement was due to cell size increase induced by nocodazole treatment, we compared the cell size after synchronization. The diameter of COS7 cells (mean ± SEM) was 16.1 ± 1.1 μm in the untreated control and 17.0 ± 0.6 μm in the treated group, indicating that the nocodazole treatment slightly increased the size of COS7 cells. However, the same treatment had minimal effects on the diameter of HCT116 cells (26.1 ± 0.4 versus 26.2 ± 0.3 μm). Taken together, the enhancement in eTE was not likely to be caused by cell size increase.
Figure 1.
Effects of Cell Synchronization with Nocodazole Treatment on eTE and Cell Viability
Synchronized cells were electrotransfected with eight pulses for COS7 cells and six pulses for HCT116 cells. The pulse duration and frequency were, respectively, 5 ms and 1 Hz for all cells, and the applied field strength is indicated in the plots. Both eTE and cell viability were measured at 24 hr after electrotransfection. (A) The treatment increased eTE for both HCT116 and COS7 cells. (B) Numbers of viable cells were normalized by the matched controls that were not treated with nocodazole for each applied electric field. The normalized data (i.e., the relative number) were used as a measure of cell viability. In all experiments, n = 5, *p < 0.05. All bars and error bars indicate mean and SEM, respectively.
For HCT116 cells, we also noticed that the enhancement in eTE became statistically insignificant when the field strength was higher than 200 V/4 mm (Figure 1A), indicating that cell synchronization was unnecessary when the field strength was high. It was possible that a large amount of pDNA molecules were internalized by cells exposed to the high electric field, which saturated nucleases in the cytoplasm. As a result, there was still a sufficient amount of intact pDNA molecules left in the cytoplasm that were available for nucleus entry in both asynchronized and synchronized cells when the nuclear envelope eventually broke down during mitosis. In addition to the eTE, we observed that nocodazole treatment reduced HCT116 cell viability but had statistically insignificant effects on COS7 cell viability, compared with electrotransfection alone (Figure 1B).
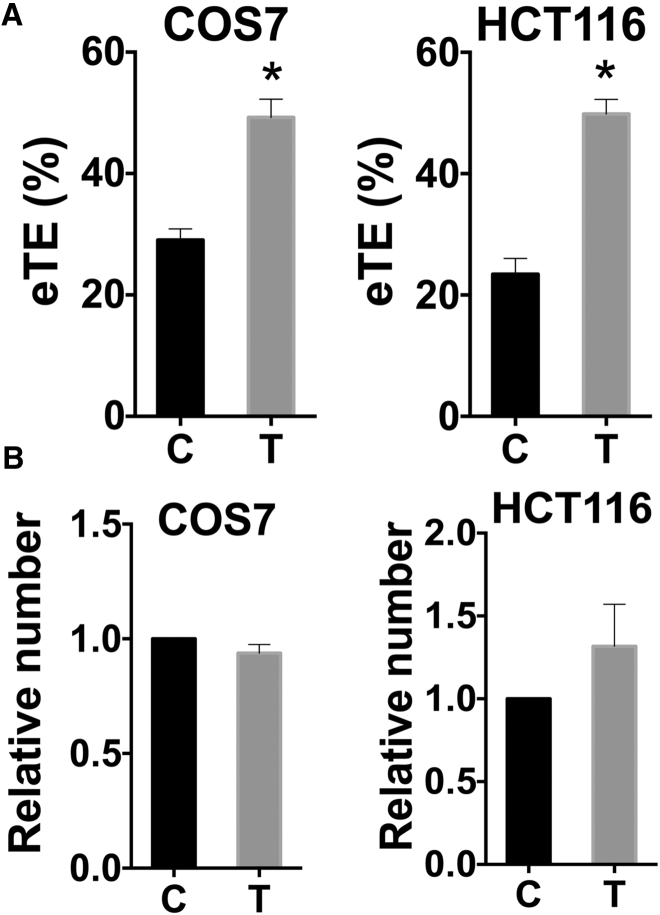
To confirm that the enhancement in eTE was due to cell synchronization instead of other side effects of nocodazole, we used an alternative method to synchronize cells to the G2-M phase, which was achieved after 8-hr release from a double-thymidine block. Again, we observed that the eTE for the synchronized cells was significantly higher than that for the control cells (Figure 2A), suggesting that the increase in eTE shown in Figure 1A was attributable to the NEBD rather than off-target effects of nocodazole. Additionally, we observed that the thymidine treatment did not affect the viability for both COS7 and HCT116 cells (Figure 2B). These data demonstrated that it was possible to significantly increase eTE through cell synchronization at prometaphase without compromising cell viability.
Figure 2.
Effects of Cell Synchronization with Thymidine Treatment on eTE and Cell Viability
The experimental procedures were the same as those described in the legend of Figure 1, except that the cells in the treated group (T) were synchronized with double-thymidine block followed by 8-hr release. Cells in the control group (C) were asynchronized. For COS7 cells, the electrotransfection involved 8 pulses at 160 V/4 mm, 5 ms, and 1 Hz. For HCT116 cells, it involved 6 pulses at 240 V/4 mm, 5 ms, and 1 Hz. (A) The synchronization increased eTE for both HCT116 and COS7 cells. (B) The synchronization had little effect on cell viability. In all experiments, n = 4, *p < 0.05. All bars and error bars indicate mean and SEM, respectively.
Effects of Nuclear Pore Dilation on Electrotransfection
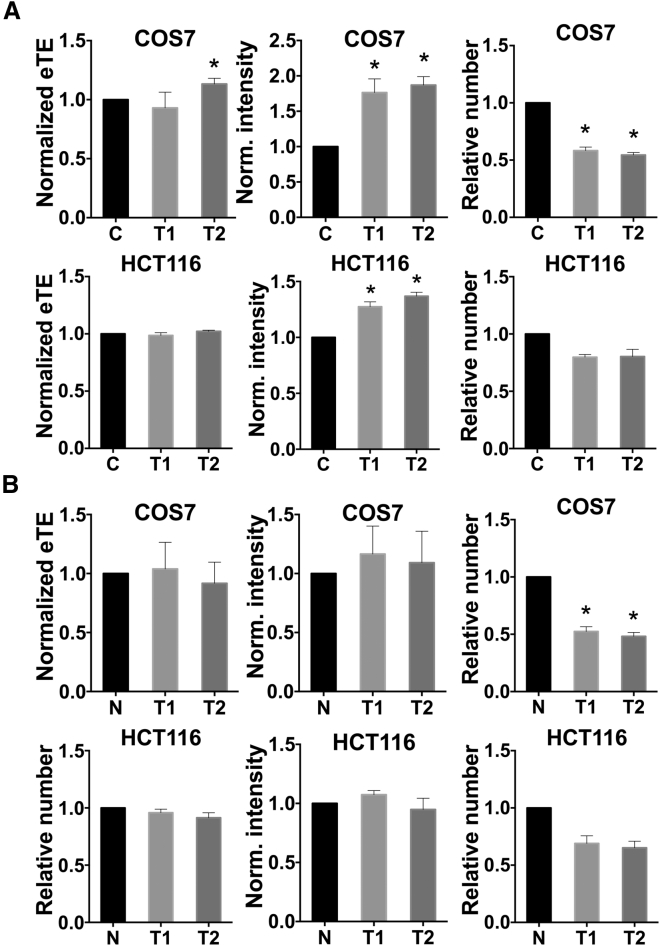
Dilation of nuclear pores may enhance the nuclear entry of pDNA. The dilating agent used in this study was trans-1,2-cyclohexanediol (TCHD),24 which is a small amphiphilic molecule that can easily cross the plasma membrane.25 The concentration and the period of TCHD treatment were optimized in a pilot study, which were 2% (w/v) and 1.5 hr, respectively, for COS7 cells, and 2% (w/v) and 2.0 hr, respectively, for HCT116 cells after electric pulsing. The treatment significantly increased the level of GFP expression (i.e., the fluorescence intensity per cell) (Figure 3A), but caused only a minor increase in the eTE for COS7 cells and had no effect on the eTE for HCT116 cells (Figure 3A).
Figure 3.
Effects of Nuclear Pore Dilation on Electrotransfection and Cell Viability
Cells were treated with a nuclear pore dilating agent, trans-1,2-cyclohexanediol (TCHD), at 2% (w/v) immediately after electrotransfection. The electrotransfection involved 8 pulses at 160 V/4 mm, 5 ms, and 1 Hz for COS7 cells, and 6 pulses at 240 V/4 mm, 5 ms, and 1 Hz for HCT116 cells. (A) Effects of TCHD treatment on eTE, level of transgene gene expression (i.e., the fluorescence intensity), and cell viability. The experiment was performed with asynchronized cells. Cells in the experimental group were treated with TCHD for two different periods (T1 and T2). T1 and T2 are 1.5 and 3 hr, respectively, for COS7 cells, and 2 and 3 hr, respectively, for HCT116 cells. Cells in the control group (C) were electrotransfected, but not post-treated with TCHD. *p < 0.01, TCHD-treated groups versus C group. (B) The experimental protocol was the same as that in (A), except that the electrotransfection was performed with cells synchronized with nocodazole treatment (N). Cells in the control group were exposed to the same treatments as those in the experimental group, except the TCHD treatment. Thus, the control group in (B) is denoted as N to indicate that it was different from the control in (A). *p < 0.01, TCHD-treated groups versus N group. In all experiments, n = 5. All bars and error bars indicate mean and SEM, respectively.
When cells were treated with nocodazole for 16 hr, not all of them were synchronized (Figure S1). Thus, we investigated whether TCHD treatment could enhance electrogene transfer in cells pretreated with nocodazole. Our data showed that the enhancement was minimal compared with that in the control group, where cells were pretreated with nocodazole only (Figure 3B), demonstrating that the nuclear pore dilation was less effective for improving electrotransfection, compared with the NEBD. The dilation of nuclear pores with TCHD also reduced the viability of cells, presumably because of disturbance of regulated exchange of molecules between the cytosol and the nucleus.
Effects of DTS on eTE
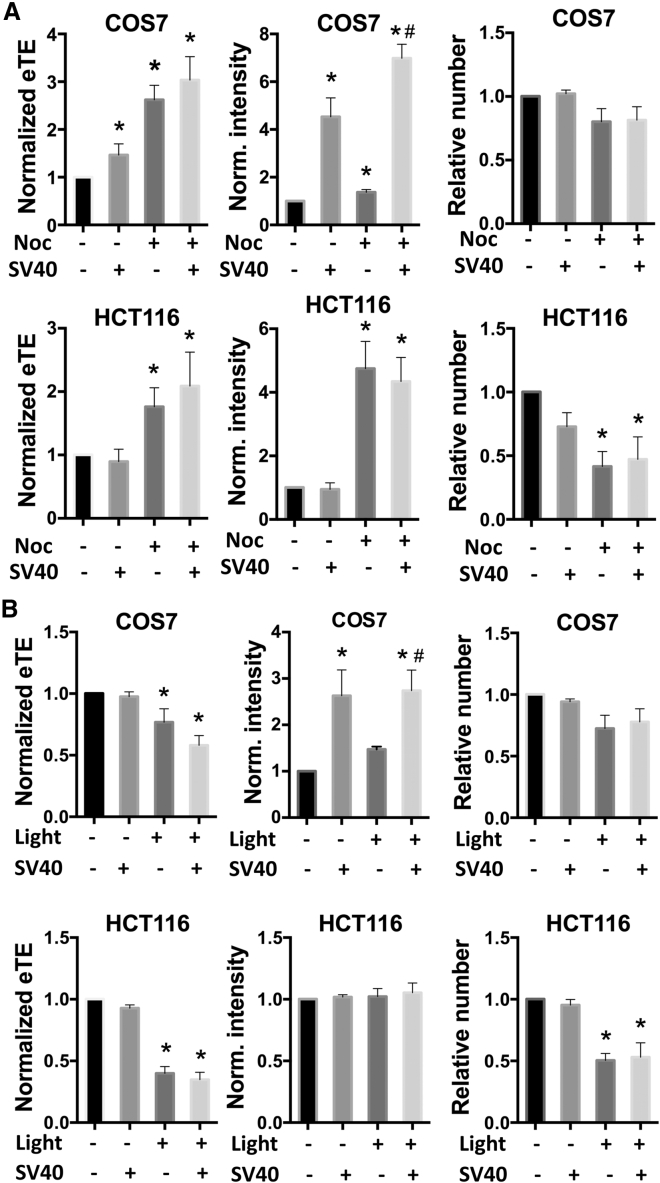
DTS SV40 in pDNA can bind to various cytosolic transcription factors with NLSs that may facilitate nuclear entry of the pDNA through NPCs.43, 44 In our experiments, we observed that the incorporation of SV40 sequence into pDNA increased the level of reporter gene expression (i.e., the fluorescence intensity per cell), compared with that with the control sequence in both asynchronized and synchronized COS7 cells (Figure 4A), but only increased the eTE for asynchronized COS7 cells. For HCT116 cells, the DTS had little effect on electrotransfection in both asynchronized and synchronized cells (Figure 4A). For both cell lines, the incorporation of DTS into pDNA did not increase the cytotoxicity of electrotransfection (see Figure 4).
Figure 4.
Effect of Incorporation of Nuclear Targeting Sequence SV40 into pDNA on Electrotransfection and Cell Viability
Two different pDNA molecules were used in the experiment, which contained SV40 (SV40+) and a control sequence (SV40−), respectively. (A) Effects of SV40 incorporation on eTE, level of transgene gene expression (i.e., the fluorescence intensity), and cell viability. The experiment was performed with either asynchronized (Noc−) or synchronized (Noc+) cells. The synchronization was achieved with nocodazole treatment. In all panels, the data were normalized by those in the baseline control group (i.e., Noc−/SV40−); n = 4, *p < 0.05, experimental group versus the baseline control group; #p < 0.05, Noc+/SV40+ group versus Noc+/SV40− group. (B) The experimental protocol was the same as that in (A) for asynchronized cells, except that endosomal escape of electrotransfected pDNA was facilitated by photochemical internalization (Light+). Cells in the Light− group were treated with the photosensitizer, but not exposed to the light. In all panels, the data were normalized by those in the baseline control group (Light−/SV40−); n = 4, *p < 0.05, experimental group versus the baseline control group; #p < 0.05, Light+/SV40+ group versus Light+/SV40− group. All bars and error bars indicate mean and SEM, respectively.
The incorporation of DTS into pDNA was less effective for improving eTE compared with cell synchronization. To understand whether it was due to entrapment of pDNA in endosomes, we pre-loaded cells with a photosensitizer and then exposed the cells to light after electrotransfection. Previous studies have shown that the photochemical reaction generates reactive oxygen species (ROS) that can cause rupture of the endosomal membrane. Therefore, the technique has been used to enhance the efficiency of gene delivery mediated by nanoparticles and some viral vectors.45, 46 However, we observed that the photochemical reaction reduced eTE and had little effect on the level of reporter gene expression in both COS7 and HCT116 cells (Figure 4B). The trends were the same for pDNA with SV40 and its matched control (Figure 4B), indicating that the endosomal escape could negatively impact electrotransfection. This observation was consistent with those in our previous study using different pDNA.47
Discussion
The study provided direct evidence showing that the nuclear envelope was a critical barrier to delivery of pDNA via electrotransfection. It also compared the effectiveness of three different approaches to improving the nuclear entry of pDNA. It was observed that synchronization of the cells at the G2-M phase prior to electrotransfection could significantly increase the eTE and the level of reporter gene expression. The increases were presumably due to the NEBD before pDNA entered the cells, which allowed passive inclusion of pDNA in the nucleus after cell mitosis.17, 48 Dilation of nuclear pores was a second approach for improving the nuclear entry, but it was less efficient than the NEBD (see Figures 1 and 3). Finally, the incorporation of DTS into pDNA could facilitate gene transfer into the nucleus in COS7 cells, but not in HCT116 cells (see Figure 4). The DTS and the cell-cycle synchronization had little toxicity to cells, whereas treatment of cells with TCHD or nocodazole could reduce cell viability. Taken together, the data indicated that pDNA transport into the nucleus could be effectively improved through either the incorporation of DTS, which was cell specific, into pDNA or the synchronization of the NEBD in dividing cells (see Figures 1 and 4).
Breakdown of Nuclear Envelope for Non-viral Gene Delivery
For most non-viral methods of gene delivery, nuclear entry of pDNA occurs mainly during mitosis when the nuclear envelope breaks down naturally. To study mechanisms of nuclear entry of electrotransfected pDNA, a previous study separated HeLa cells into different groups based on the cell size, assuming that it correlates to the phase of cell cycle.18 Although the correlation is not very precise, the authors observed that eTE was high for cells in the middle S and late S/G2 phases, compared with those in the G1 phase. In the current study, we showed that eTE and the level of reporter gene expression could be significantly increased if the electrotransfection was performed in cells after synchronization of the NEBD.49, 50 This observation was consistent with those in previous studies of non-viral gene delivery. It has been shown that the NEBD can enhance the efficiency of gene delivery by 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and dioleoyl-phosphatidylethanolamine (DOPE) (1:1 mol/mol) in HeLa cells.51 Similarly, high expression levels of reporter genes delivered by various lipofection methods are only observed in actively dividing cells.6, 17, 18, 48, 51, 52, 53, 54, 55 These studies demonstrate that non-viral gene delivery, including electrotransfection, can be enhanced through NEBD, which means that the delivery is more efficient in dividing cells than in non-dividing ones.
In addition to the NEBD, the nuclear envelope can be permeabilized by nanosecond pulsed electric field (nsPEF) because treatment of cells with nsPEF causes leak out of nuclear proteins56 and increases the efficiency of electrotransfection mediated by a microsecond pulse.57 The permeabilization is likely to be less efficient than the NEBD for the nuclear entry of pDNA and does not happen in living cells after they are treated with microsecond or longer pulses. Therefore, the NEBD is still a more effective way for improving the nuclear entry of pDNA.
Nuclear Pore Dilating Agents
Nuclear pore dilating agents, such as trans-cyclohexane-1,2-diol (TCHD), have been used to increase expression levels of transgenes delivered by non-viral vectors. TCHD is an amphipathic alcohol (116.16 molecular weight [MW]) and, upon treatment of cells, allows translocation of larger molecules from the cytosol to the nucleus in a nuclear importer-independent manner. It was suggested that TCHD could increase the permeability of the NPCs through altering interactions between phenylalanine-glycine (FG) repeats to reduce hydrophobic exclusion of NPCs.58 As a result, TCHD has been used to improve lipofection in 293T cells59 and electrotransfection in B16.F10 cells.26 In the current study, TCHD could slightly increase eTE in COS7 cells and largely increased the levels of the reporter gene expression in all treated cell lines, demonstrating that TCHD was effective for increasing the nuclear entry of pDNA via the NPCs, but not for increasing the percentage of transfected cells. It is also important to note that treatment of cells with TCHD was less effective than synchronization of the NEBD for increasing both eTE and transgene expression level. Furthermore, TCHD was toxic in vitro when used in combination with electrotransfection (see Figure 3),26 although it was apparently non-toxic in mice in vivo.59 The discrepancy could be partly due to the differences in drug concentration and treatment period. The concentration of TCHD was fixed during the treatment in in vitro studies, but decreased rapidly after administration in vivo, because of tissue clearance.
Facilitated Transport through NPCs
One approach to overcoming the nuclear envelope barrier is to enhance facilitated transport through NPCs. It can be achieved with NLS molecules.60, 61 The NLS molecules can be exogenous or endogenous peptides or proteins that initiate active transport of attached cargos into the nucleus through direct interactions with the nuclear import system.62 In the current study, we observed that incorporation of a nuclear targeting sequence, SV40, into pDNA could improve electrotransfection in COS7, but not HCT116 cells (see Figure 4A), although SV40 has been considered as a universal DTS for mammalian cells.27, 28 It is possible that HCT116 cells do not contain NLS molecules that can bind to SV40, or that the binding affinities and nuclear import system are less optimal in this cell line. If the affinities are low, pDNA will dissociate from the nuclear import system before it crosses the NPCs.63 It is also unknown how efficiently pDNA with the DTS can dissociate from the NLS molecules upon reaching the intra-nuclear space, and how the NLS molecules will affect biological activities of pDNA, which can potentially limit gene delivery and expression efficiencies.64 Data from synchronized COS7 cells shown in Figure 4 demonstrated that SV40 was more effective for increasing gene expression level in COS7 cells than enhancing the eTE. Quantitatively, the observation could not be explained solely by the difference in intra-nuclear concentrations between pDNA with SV40 and the control pDNA without the sequence. Potentially, it might suggest that the pDNA with SV40 sequence had a higher transcription efficiency than its matched control. It can be laborious to design and synthesize nuclear target vectors with specific NLS or those that can bind to endogenous NLS molecules for each type of cell.65 Thus, the use of the NLS for improving the transport of DNA into the nucleus has achieved only limited success in applications.66
Conclusions
This study elucidated the nuclear envelope as a main physical barrier to pDNA transport in cells that has critically limited the efficiency of electrotransfection. The barrier could be circumvented, without significantly compromising cell viability, by using NLS-mediated delivery or synchronization of the NEBD. It is also worth mentioning that although the study was performed with only two pulse sequences, its conclusions were likely to be valid for other microsecond and millisecond pulse sequences, because changes in the pulses should have little influence on mechanisms of the nuclear entry of naked pDNA. Results from the study could be useful for gene therapy applications that involve in vitro or ex vivo transfection, such as modification of T cells for immunotherapy. Electrotransfection can become a favorable choice for immunotherapy applications because it has been reported that efficiencies of many viral and non-viral methods for gene delivery are low in immune cells.67 Additionally, electrotransfection has been successfully used in transfecting cells that have been considered to be difficult to transfect.1 In future studies, experimental conditions will be optimized to further improve eTE so that the technology can be more widely implemented for clinical applications.
Materials and Methods
Cell Culture
COS7 (African green monkey fibroblast-like kidney) and HCT116 (human colorectal carcinoma) cell lines were obtained from ATCC (Manassas, VA, USA). COS7 cells were cultured in high-glucose DMEM (GIBCO, Grand Island, NY, USA), supplemented with 10% (v/v) fetal bovine serum and 1% penicillin-streptomycin (15140-122; GIBCO). HCT116 cells were cultured in McCoy medium with 10% FBS and 1% penicillin-streptomycin. Cells were passaged every 2–3 days and were incubated at 37°C in 5% CO2 and 95% air.
Cell Synchronization
We first prepared stock solution of nocodazole (M1404; Sigma-Aldrich, St. Louis, MO) with DMSO (5 mg/mL) and then added it to cell culture medium to make the final solution (100 ng/mL). The control solution was prepared by adding the equivalent volume of DMSO to cell culture medium without nocodazole. The solution of thymidine (T1895; Sigma-Aldrich) was prepared with cell culture medium (2 mM). The control solution for thymidine was fresh cell culture medium Cell synchronization was achieved with two methods. In the first method, cells were incubated with nocodazole at a concentration of 100 ng/mL for 16 hr. Thereafter, the synchronized cells were collected via trypsinization followed by neutralization with medium and then washed with PBS (without calcium or magnesium) to remove nocodazole. In the second method,68, 69 cells were first incubated with 2 mM thymidine for 16 hr and then briefly washed three times with fresh medium, followed by incubation at 37°C in fresh medium containing no thymidine for 8 hr. Thereafter, the cells were treated again with thymidine for an additional 16 hr and released for 8 hr in fresh cell culture medium at 37°C. In the no treatment control group, the cells were treated with the control solutions, and all other experimental steps were the same as those in the treatment group.
Electrotransfection
Each transfection was performed with 106 cells. The cells were resuspended in 100 μL of pulsing buffer (Hepes buffered saline [HeBS]) with 6 μg of pDNA on ice. In most experiments, we used pEGFP-N1 (Clontech, Palo Alto, CA, USA), unless indicated specifically. In some experiments, we used pDNA with the SV40 sequence (pDD805) or its matched control that was generously provided by Dr. David Dean at University of Rochester. The cell suspension was transferred to electroporation cuvettes with two parallel plate electrodes spaced 4 mm apart. Cells were electrotransfected with the BTX ECM 830 Square Wave Electroporation System (Harvard Apparatus, Holliston, MA, USA). Unless indicated specifically, COS7 cells were treated with 8 electric pulses at 160 V/4 mm, 5-ms duration, and 1-Hz frequency; HCT116 cells were treated with 6 electric pulses at 240 V/4 mm, 5-ms duration, and 1-Hz frequency. The cuvettes were kept at room temperature for 10 min following the pulse application to allow the cells to recover before pipetting them to a six-well plate with full cell culture medium. The eTE and cell viability were measured at 24 hr after electrotransfection with flow cytometry.
Visualization of Microtubule Depolymerization
HCT116 cells were seeded at a density of 0.5 × 106 cells/well in a six-well plate. On the next day, cells were transfected with a plasmid encoding the fusion protein GFP-α-tubulin (pBABE-GFP plasmid, kindly provided by Dr. Terry Lechler at Duke University). The cells were transfected via Lipofectamine 2000 (11668019; Invitrogen) at a ratio of 1:3 (DNA:Lipofectamine) for 4 hr. Then the cells were transferred to full medium and cultured for 24 hr. Hoechst 33342 dye (H1399; Molecular Probes) was prepared as a stock solution of 10 mg/mL in ddH2O and used at a final concentration of 1 μg/mL to stain the nucleus prior to imaging.
Visualization of Nuclear Envelope Dissolution
Two methods were used for the visualization of the nuclear envelope in cells. The first one was based on the FM4-64FX dye (F34653; Invitrogen, Carlsbad, CA, USA). It was added to the cell culture medium at a concentration of 5 μg/mL. The cells were incubated with the dye for 30 min either at the end of the nocodazole treatment or prior to the treatment. The dye initially bound to the plasma membrane and eventually reached the nuclear envelope via endocytosis and intracellular transport. Meanwhile, the nucleus was stained with Hoechst 33342 for 30 min prior to imaging. The second method was based on expression of a Lamin A fusion protein, in which the cells were transfected with a plasmid encoding mCherry-Lamin A (Plasmid 55068; Addgene, Cambridge, MA, USA). The procedures were the same as those described above for pBABE-GFP transfection. For imaging the dissolution of the nuclear envelope, fluorescence images were acquired with an Andor XD revolution spinning disk confocal microscope (Andor Technology) equipped with 40× and 100× oil-immersion objectives in the Light Microscopy Core Facility (LMCF) at Duke University.
Nuclear Pore Dilation
TCHD (141712; Sigma-Aldrich) was first dissolved in full culture medium at the concentration of 5% (w/v) or 430 mM and then diluted to 2% with the same medium before being used. For cell treatment, the TCHD solution (2%, 500 μL) was added to the electroporation cuvettes containing pulsed cells at room temperature immediately after pulsing. After 10 min, the cell suspension was pipetted to six-well plates containing the TCHD solution (2%, 1 mL per well) and incubated at 37°C for the indicated periods. The treatment was blocked by dilution of the TCHD solution in each well with 5 mL of fresh medium without TCHD.
Photochemical Internalization
The photochemical internalization (PCI) protocol was followed according to a procedure described in our previous study.47 Briefly, PCI was performed with an amphiphilic, sulfonated photosensitizer, TPPS2a (T40637; Frontier Scientific, Logan, UT, USA). TPPS2a was added to the culture medium at a concentration of 0.4 μg/mL from a stock dissolved in DMSO at 2 mg/mL. After 18 hr, cells were washed three times with cell culture medium and incubated in photosensitizer-free cell culture medium for 4 hr to remove the photosensitizer from the plasma membrane. Then the cells were electrotransfected with different types of pDNA indicated in the Results. At 10 min after application of electric field, the cells were exposed to blue light (375–550 nm with a peak at 435 nm) at an irradiance of 1.5 mW/cm2 for 2 min. The light was delivered by two Osram L 18/67 bulbs.
Cell Counting and Size Measurement
Single-cell suspension was collected, mixed with trypan blue solution, and pipetted to a hemocytometer. The hemocytometer was inserted into the Countess II FL automated cell counter (Thermo Fisher Scientific, Waltham, MA, USA) to determine the concentration and the size distribution of viable cells. The histogram was used to determine mean and SEM of the cell size.
Flow Cytometry
The cell culture medium in the six-well plates was aspirated, and the adherent cells were washed with PBS without Ca2+ and Mg2+, trypsinized, and resuspended in a medium containing propidium iodide (PI) (5 μg/mL). The flow cytometer (BD FACSCanto II, Becton Dickinson, Franklin Lakes, NJ, USA) was set to collect 10,000 events for each sample. Control cells were used to correct for autofluorescence in flow cytometry analysis and were prepared under the same conditions as those for experimental cells, except that no plasmid was added to the pulsing buffer. Forward and side light scatter gating were used to exclude debris and isolate the cell populations of interest. The apparent eTE was defined as the number of viable cells expressing GFP (PI-negative, GFP-positive) out of the total number of viable cells (PI-negative). The level of GFP expression was quantified by using the geometric mean of the fluorescence intensity per cell that was PI-negative and GFP-positive. Viability of cells for each sample was determined by collecting events for 20 s under the “medium” flow rate setting in the flow cytometer, and applying forward light scatter gating and fluorescence detection to isolate the live cell population only (i.e., PI-negative). The viability was defined as the number of live cells in the treated group normalized by the non-treated control in each experiment.70 Cell cycle was determined by flow cytometry measurement after cells were fixed in 70% ethanol and stained with PI.34, 71 The flow cytometer settings were kept constant for all experiments. FlowJo software was used in all data analysis.
Statistical Analysis
The mean and SEM are reported for all data. The Mann-Whitney U test was used to compare data between two groups. The difference was considered to be statistically significant if the p value was less than 0.05.
Author Contributions
L.D.C., C.-C.C., L.W., and M.M. conducted the experiments; L.D.C. and F.Y. designed the experiments and wrote the paper.
Conflicts of Interest
There is no conflict of interest.
Acknowledgments
This research was supported partly by funding from the NIH (GM098520), National Science Foundation (BES-0828630), and Duke University Pharmacological Sciences Training Program (PSTP) (T32 GM 007105).
Footnotes
Supplemental Information includes two figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.02.009.
Supplemental Information
References
- 1.Heller R., Heller L.C. Gene electrotransfer clinical trials. Adv. Genet. 2015;89:235–262. doi: 10.1016/bs.adgen.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Henshaw J.W., Yuan F. Field distribution and DNA transport in solid tumors during electric field-mediated gene delivery. J. Pharm. Sci. 2008;97:691–711. doi: 10.1002/jps.21000. [DOI] [PubMed] [Google Scholar]
- 3.Mellott A.J., Forrest M.L., Detamore M.S. Physical non-viral gene delivery methods for tissue engineering. Ann. Biomed. Eng. 2013;41:446–468. doi: 10.1007/s10439-012-0678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adler A.F., Leong K.W. Emerging links between surface nanotechnology and endocytosis: impact on nonviral gene delivery. Nano Today. 2010;5:553–569. doi: 10.1016/j.nantod.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollard H., Remy J.S., Loussouarn G., Demolombe S., Behr J.P., Escande D. Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. J. Biol. Chem. 1998;273:7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- 6.Zabner J., Fasbender A.J., Moninger T., Poellinger K.A., Welsh M.J. Cellular and molecular barriers to gene transfer by a cationic lipid. J. Biol. Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 7.Labat-Moleur F., Steffan A.M., Brisson C., Perron H., Feugeas O., Furstenberger P., Oberling F., Brambilla E., Behr J.P. An electron microscopy study into the mechanism of gene transfer with lipopolyamines. Gene Ther. 1996;3:1010–1017. [PubMed] [Google Scholar]
- 8.Capecchi M.R. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980;22:479–488. doi: 10.1016/0092-8674(80)90358-x. [DOI] [PubMed] [Google Scholar]
- 9.Cornelis S., Vandenbranden M., Ruysschaert J.-M., Elouahabi A. Role of intracellular cationic liposome–DNA complex dissociation in transfection mediated by cationic lipids. DNA Cell Biol. 2002;21:91–97. doi: 10.1089/104454902753604961. [DOI] [PubMed] [Google Scholar]
- 10.Pérez-Martínez F.C., Guerra J., Posadas I., Ceña V. Barriers to non-viral vector-mediated gene delivery in the nervous system. Pharm. Res. 2011;28:1843–1858. doi: 10.1007/s11095-010-0364-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karp G. Cellular reproduction. In: Witt K., Stat M., McFadden P., editors. Cell and Molecular Biology: Concepts and Experiments. Sixth Edition. John Wiley & Sons; 2009. pp. 475–532. [Google Scholar]
- 12.Güttinger S., Laurell E., Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y., Huang L. Barriers to systemic gene delivery. In: Yeo Y., editor. Nanoparticulate Drug Delivery Systems: Strategies, Technologies, and Applications. First Edition. John Wiley & Sons; 2013. pp. 52–80. [Google Scholar]
- 14.Cohen S., Au S., Panté N. How viruses access the nucleus. Biochim. Biophys. Acta. 2011;1813:1634–1645. doi: 10.1016/j.bbamcr.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Hausman G.M., Cooper R.E. The nuclear envelope and traffic between the nucleus and cytoplasm. In: Cooper G.M., editor. The Cell: A Molecular Approach. Sixth Edition. Sinauer Associates; 2013. pp. 345–358. [Google Scholar]
- 16.Latulippe D.R., Zydney A.L. Radius of gyration of plasmid DNA isoforms from static light scattering. Biotechnol. Bioeng. 2010;107:134–142. doi: 10.1002/bit.22787. [DOI] [PubMed] [Google Scholar]
- 17.Zauner W., Brunner S., Buschle M., Ogris M., Wagner E. Differential behaviour of lipid based and polycation based gene transfer systems in transfecting primary human fibroblasts: a potential role of polylysine in nuclear transport. Biochim. Biophys. Acta. 1999;1428:57–67. doi: 10.1016/s0304-4165(99)00022-7. [DOI] [PubMed] [Google Scholar]
- 18.Brunner S., Fürtbauer E., Sauer T., Kursa M., Wagner E. Overcoming the nuclear barrier: cell cycle independent nonviral gene transfer with linear polyethylenimine or electroporation. Mol. Ther. 2002;5:80–86. doi: 10.1006/mthe.2001.0509. [DOI] [PubMed] [Google Scholar]
- 19.Matsumoto Y., Itaka K., Yamasoba T., Kataoka K. Intranuclear fluorescence resonance energy transfer analysis of plasmid DNA decondensation from nonviral gene carriers. J. Gene Med. 2009;11:615–623. doi: 10.1002/jgm.1338. [DOI] [PubMed] [Google Scholar]
- 20.Wightman L., Kircheis R., Rössler V., Carotta S., Ruzicka R., Kursa M., Wagner E. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. J. Gene Med. 2001;3:362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 21.Feldherr C.M., Akin D. Signal-mediated nuclear transport in proliferating and growth-arrested BALB/c 3T3 cells. J. Cell Biol. 1991;115:933–939. doi: 10.1083/jcb.115.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Görlich D., Mattaj I.W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 23.Whittaker G., Bui M., Helenius A. The role of nuclear import and export in influenza virus infection. Trends Cell Biol. 1996;6:67–71. doi: 10.1016/0962-8924(96)81017-8. [DOI] [PubMed] [Google Scholar]
- 24.Vandenbroucke R.E., Lucas B., Demeester J., De Smedt S.C., Sanders N.N. Nuclear accumulation of plasmid DNA can be enhanced by non-selective gating of the nuclear pore. Nucleic Acids Res. 2007;35:e86. doi: 10.1093/nar/gkm440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lentacker I., Vandenbroucke R.E., Lucas B., Demeester J., De Smedt S.C., Sanders N.N. New strategies for nucleic acid delivery to conquer cellular and nuclear membranes. J. Control. Release. 2008;132:279–288. doi: 10.1016/j.jconrel.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 26.Pasquet L., Bellard E., Rols M.P., Golzio M., Teissie J. Post-pulse addition of trans-cyclohexane-1,2-diol improves electrotransfer mediated gene expression in mammalian cells. Biochem. Biophys. Rep. 2016;7:287–294. doi: 10.1016/j.bbrep.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dean D.A. Cell-specific targeting strategies for electroporation-mediated gene delivery in cells and animals. J. Membr. Biol. 2013;246:737–744. doi: 10.1007/s00232-013-9534-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dean D.A. Import of plasmid DNA into the nucleus is sequence specific. Exp. Cell Res. 1997;230:293–302. doi: 10.1006/excr.1996.3427. [DOI] [PubMed] [Google Scholar]
- 29.Xu K., Schwarz P.M., Ludueña R.F. Interaction of nocodazole with tubulin isotypes. Drug Dev. Res. 2002;55:91–96. [Google Scholar]
- 30.Georgatos S.D., Pyrpasopoulou A., Theodoropoulos P.A. Nuclear envelope breakdown in mammalian cells involves stepwise lamina disassembly and microtubule-drive deformation of the nuclear membrane. J. Cell Sci. 1997;110:2129–2140. doi: 10.1242/jcs.110.17.2129. [DOI] [PubMed] [Google Scholar]
- 31.Gönczy P. Nuclear envelope: torn apart at mitosis. Curr. Biol. 2002;12:R242–R244. doi: 10.1016/s0960-9822(02)00781-9. [DOI] [PubMed] [Google Scholar]
- 32.Salina D., Bodoor K., Eckley D.M., Schroer T.A., Rattner J.B., Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- 33.Jackman J., O’Connor P.M. Methods for synchronizing cells at specific stages of the cell cycle. Curr. Protoc. Cell Biol. 2001;Chapter 8 doi: 10.1002/0471143030.cb0803s00. Unit8.3. [DOI] [PubMed] [Google Scholar]
- 34.Harper J.V. Synchronization of cell populations in G1/S and G2/M phases of the cell cycle. Methods Mol. Biol. 2005;296:157–166. doi: 10.1385/1-59259-857-9:157. [DOI] [PubMed] [Google Scholar]
- 35.Zieve G.W., Turnbull D., Mullins J.M., Mcintosh J.R. Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor nocodazole. Nocodazole accumulated mitotic cells. Exp. Cell Res. 1980;126:397–405. doi: 10.1016/0014-4827(80)90279-7. [DOI] [PubMed] [Google Scholar]
- 36.Mocharla R., Mocharla H., Hodes M.E. A novel, sensitive fluorometric staining technique for the detection of DNA in RNA preparations. Nucleic Acids Res. 1987;15:10589. doi: 10.1093/nar/15.24.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latt S.A., Stetten G. Spectral studies on 33258 Hoechst and related bisbenzimidazole dyes useful for fluorescent detection of deoxyribonucleic acid synthesis. J. Histochem. Cytochem. 1976;24:24–33. doi: 10.1177/24.1.943439. [DOI] [PubMed] [Google Scholar]
- 38.Singh R., Kalra R.S., Hasan K., Kaul Z., Cheung C.T., Huschtscha L., Reddel R.R., Kaul S.C., Wadhwa R. Molecular characterization of collaborator of ARF (CARF) as a DNA damage response and cell cycle checkpoint regulatory protein. Exp. Cell Res. 2014;322:324–334. doi: 10.1016/j.yexcr.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 39.Jordan M.A., Thrower D., Wilson L. Effects of vinblastine, podophyllotoxin and nocodazole on mitotic spindles. Implications for the role of microtubule dynamics in mitosis. J. Cell Sci. 1992;102:401–416. doi: 10.1242/jcs.102.3.401. [DOI] [PubMed] [Google Scholar]
- 40.Reece J.B., Urry L.A., Cain M.L., Wasserman S.A., Minorsky P.V., Jackson R.B. The cell cycle. In: Cummings B., editor. Campbell Biology. Pearson Education; 2011. pp. 206–223. [Google Scholar]
- 41.Choi H.J., Fukui M., Zhu B.T. Role of cyclin B1/Cdc2 up-regulation in the development of mitotic prometaphase arrest in human breast cancer cells treated with nocodazole. PLoS ONE. 2011;6:e24312. doi: 10.1371/journal.pone.0024312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beaudouin J., Gerlich D., Daigle N., Eils R., Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. doi: 10.1016/s0092-8674(01)00627-4. [DOI] [PubMed] [Google Scholar]
- 43.Vernon M.M., Dean D.A., Dobson J. DNA targeting sequence improves magnetic nanoparticle-based plasmid DNA transfection efficiency in model neurons. Int. J. Mol. Sci. 2015;16:19369–19386. doi: 10.3390/ijms160819369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young J.L., Benoit J.N., Dean D.A. Effect of a DNA nuclear targeting sequence on gene transfer and expression of plasmids in the intact vasculature. Gene Ther. 2003;10:1465–1470. doi: 10.1038/sj.gt.3302021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hellum M., Høgset A., Engesaeter B.Ø., Prasmickaite L., Stokke T., Wheeler C., Berg K. Photochemically enhanced gene delivery with cationic lipid formulations. Photochem. Photobiol. Sci. 2003;2:407–411. doi: 10.1039/b211880g. [DOI] [PubMed] [Google Scholar]
- 46.Prasmickaite L., Høgset A., Selbo P.K., Engesaeter B.O., Hellum M., Berg K. Photochemical disruption of endocytic vesicles before delivery of drugs: a new strategy for cancer therapy. Br. J. Cancer. 2002;86:652–657. doi: 10.1038/sj.bjc.6600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cervia L.D., Chang C.-C., Wang L., Yuan F. Distinct effects of endosomal escape and inhibition of endosomal trafficking on gene delivery via electrotransfection. PLoS One. 2017;12:e0171699. doi: 10.1371/journal.pone.0171699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fasbender A., Zabner J., Zeiher B.G., Welsh M.J. A low rate of cell proliferation and reduced DNA uptake limit cationic lipid-mediated gene transfer to primary cultures of ciliated human airway epithelia. Gene Ther. 1997;4:1173–1180. doi: 10.1038/sj.gt.3300524. [DOI] [PubMed] [Google Scholar]
- 49.Golzio M., Teissié J., Rols M.-P. Cell synchronization effect on mammalian cell permeabilization and gene delivery by electric field. Biochim. Biophys. Acta. 2002;1563:23–28. doi: 10.1016/s0005-2736(02)00369-3. [DOI] [PubMed] [Google Scholar]
- 50.Yorifuji T., Tsuruta S., Mikawa H. The effect of cell synchronization on the efficiency of stable gene transfer by electroporation. FEBS Lett. 1989;245:201–203. doi: 10.1016/0014-5793(89)80221-2. [DOI] [PubMed] [Google Scholar]
- 51.Tseng W.C., Haselton F.R., Giorgio T.D. Mitosis enhances transgene expression of plasmid delivered by cationic liposomes. Biochim. Biophys. Acta. 1999;1445:53–64. doi: 10.1016/s0167-4781(99)00039-1. [DOI] [PubMed] [Google Scholar]
- 52.Mortimer I., Tam P., MacLachlan I., Graham R.W., Saravolac E.G., Joshi P.B. Cationic lipid-mediated transfection of cells in culture requires mitotic activity. Gene Ther. 1999;6:403–411. doi: 10.1038/sj.gt.3300837. [DOI] [PubMed] [Google Scholar]
- 53.Brunner S., Sauer T., Carotta S., Cotten M., Saltik M., Wagner E. Cell cycle dependence of gene transfer by lipoplex, polyplex and recombinant adenovirus. Gene Ther. 2000;7:401–407. doi: 10.1038/sj.gt.3301102. [DOI] [PubMed] [Google Scholar]
- 54.Wilke M., Fortunati E., van den Broek M., Hoogeveen A.T., Scholte B.J. Efficacy of a peptide-based gene delivery system depends on mitotic activity. Gene Ther. 1996;3:1133–1142. [PubMed] [Google Scholar]
- 55.Brisson M., Huang L. Liposomes: conquering the nuclear barrier. Curr. Opin. Mol. Ther. 1999;1:140–146. [PubMed] [Google Scholar]
- 56.Thompson G.L., Roth C.C., Kuipers M.A., Tolstykh G.P., Beier H.T., Ibey B.L. Permeabilization of the nuclear envelope following nanosecond pulsed electric field exposure. Biochem. Biophys. Res. Commun. 2016;470:35–40. doi: 10.1016/j.bbrc.2015.12.092. [DOI] [PubMed] [Google Scholar]
- 57.Guo S., Jackson D.L., Burcus N.I., Chen Y.-J., Xiao S., Heller R. Gene electrotransfer enhanced by nanosecond pulsed electric fields. Mol. Ther. Methods Clin. Dev. 2014;1:14043. doi: 10.1038/mtm.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ribbeck K., Görlich D. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 2002;21:2664–2671. doi: 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Griesenbach U., Wilson K.M., Farley R., Meng C., Munkonge F.M., Cheng S.H., Scheule R.K., Alton E.W. Assessment of the nuclear pore dilating agent trans-cyclohexane-1,2-diol in differentiated airway epithelium. J. Gene Med. 2012;14:491–500. doi: 10.1002/jgm.2643. [DOI] [PubMed] [Google Scholar]
- 60.Adam S.A., Marr R.S., Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katayama T., Niidome Y. Use of Synthetic Peptides for Non-Viral Gene Delivery. In: Taira K., Kataoka K., Niidome T., editors. Non-viral Gene Therapy: Gene Design and Delivery. Springer Science & Business Media; 2005. pp. 87–102. [Google Scholar]
- 62.Kobiler O., Drayman N., Butin-Israeli V., Oppenheim A. Virus strategies for passing the nuclear envelope barrier. Nucleus. 2012;3:526–539. doi: 10.4161/nucl.21979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Evans E.A., Calderwood D.A. Forces and bond dynamics in cell adhesion. Science. 2007;316:1148–1153. doi: 10.1126/science.1137592. [DOI] [PubMed] [Google Scholar]
- 64.Zanta M.A., Belguise-Valladier P., Behr J.P. Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc. Natl. Acad. Sci. USA. 1999;96:91–96. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cartier R., Reszka R. Utilization of synthetic peptides containing nuclear localization signals for nonviral gene transfer systems. Gene Ther. 2002;9:157–167. doi: 10.1038/sj.gt.3301635. [DOI] [PubMed] [Google Scholar]
- 66.Hébert E. Improvement of exogenous DNA nuclear importation by nuclear localization signal-bearing vectors: a promising way for non-viral gene therapy? Biol. Cell. 2003;95:59–68. doi: 10.1016/s0248-4900(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 67.Carlsten M., Childs R.W. Genetic manipulation of NK cells for cancer immunotherapy: techniques and clinical implications. Front. Immunol. 2015;6:266. doi: 10.3389/fimmu.2015.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boulay K., Ghram M., Viranaicken W., Trépanier V., Mollet S., Fréchina C., DesGroseillers L. Cell cycle-dependent regulation of the RNA-binding protein Staufen1. Nucleic Acids Res. 2014;42:7867–7883. doi: 10.1093/nar/gku506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sibani S., Price G.B., Zannis-Hadjopoulos M. Decreased origin usage and initiation of DNA replication in haploinsufficient HCT116 Ku80+/- cells. J. Cell Sci. 2005;118:3247–3261. doi: 10.1242/jcs.02427. [DOI] [PubMed] [Google Scholar]
- 70.Ross D.D., Joneckis C.C., Ordóñez J.V., Sisk A.M., Wu R.K., Hamburger AW Nora R.E., Nora R.E. Estimation of cell survival by flow cytometric quantification of fluorescein diacetate/propidium iodide viable cell number. Cancer Res. 1989;49:3776–3782. [PubMed] [Google Scholar]
- 71.Pozarowski P., Darzynkiewicz Z. Checkpoint Controls and Cancer. Humana Press; 2004. Analysis of cell cycle by flow cytometry; pp. 301–311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.