Abstract
Parkinson’s disease (PD) is one of the most prevalent forms of synucleinopathies, and it is characterized neuropathologically by the presence of intracellular inclusions composed primarily of the protein α-synuclein (α-syn) in neurons. The previous immunotherapy targeting the α-syn in PD models with monoclonal antibodies has established α-syn protein as an effective target for neuronal cell death. However, due to the essential weaknesses of antibody and the unique features of aptamers, the aptamers could represent a promising alternative to the currently used antibodies in immunotherapy for PD. In this study, the purified human α-syn was used as the target for in vitro selection of aptamers using systematic evolution by exponential enrichment. This resulted in the identification of two 58-base DNA aptamers with a high binding affinity and good specificity to the α-syn, with KD values in the nanomolar range. Both aptamers could effectively reduce α-syn aggregation in vitro and in cells and target the α-syn to intracellular degradation through the lysosomal pathway. These effects consequently rescued the mitochondrial dysfunction and cellular defects caused by α-syn overexpression. To our knowledge, this is the first study to employ aptamers to block the aberrant cellular effects of the overexpressed α-syn in cells.
Keywords: aptamer, α-synuclein, immunotherapy, Parkinson’s disease
Introduction
Parkinson’s disease (PD) is one of the most prevalent forms of synucleinopathies and neuropathologically characterized by the presence of intracellular inclusions composed primarily of the protein α-synuclein (α-syn) in neurons1. α-syn is a small, natively unfolded protein of 140 amino acids, and, in neurons, it is primarily found at the presynaptic terminal.2 In the physiological state, α-syn is an intrinsically disordered protein. The protein has the tendency to form oligomers and aggregates, which are toxic and lie at the core of its pathological function,3 although it is still in debate as to exactly which species within the aggregation process may be more toxic.4 Importantly, α-syn aggregates may propagate from cell to cell; once taken up by recipient cells, they may act as seeds for further α-syn deposition within acceptor cells.5, 6 These pathogenic α-syn species may exert their toxicity with mitochondrial damage, axonal transport dysfunction, inflammation, and other mechanisms. Thus, neutralizing the pathogenic species or facilitating their clearance will have the potential to slow down or even halt PD progression.
Immunotherapy has emerged as a promising approach to target and clear α-syn aggregate pathology in PD.7, 8 In the past, efforts to identify agents that disrupt the α-syn aggregation yielded several potential strategies, such as inhibiting the α-syn fibrillation process with small molecules,9 lowering the protein level of α-syn by RNA interference,10 or enhancing cellular degradation processes.11 In recent years, immunotherapy is more appealing, as α-syn antibodies could prevent the formation of pathogenic species, facilitate the clearance of already formed species, and even shield pathogenic forms, consequently highlighting its neuroprotective or disease-modifying capabilities.6, 12, 13, 14 However, due to the essence of antibody is protein, it has its own weaknesses. For example, the antibodies are not easily accessible to the intracellular target, significantly immunogenic, and more thermally unstable, etc.15 Therefore, there is an urgent need to overcome these weaknesses and find the antibody alternatives for application of the immunotherapy in PD.
Aptamers are short, single-stranded DNA (ssDNA) or RNA molecules that can bind to a wide range of target proteins with high affinity and specificity.16 They are considered as “chemical antibodies” and widely used as a substitute for antibodies. Compared with the conventional antibodies, aptamers have their unique features. For example, aptamers are neither immunogenic nor toxic molecules, because nucleic acids are not typically recognized by the human immune system as foreign agents; they have higher thermal stability and maintain their structures over repeated cycles of denaturation/renaturation; they can be easily generated by chemical synthesis, and can be easily labeled and adjusted.15 Moreover, the aptamers can discriminate between different conformations of the same target protein.17 Since the aptamer of Macugen was approved in 2004 by the US Food and Drug Administration (FDA),18 the number of studies on the applications of aptamers has been rapidly increased. For the neurodegenerative diseases, a DNA aptamer, called A1, was selected, which binds to β-Site amyloid precursor protein cleaving enzyme 1 (BACE1) with high affinity and good specificity, consequently inhibiting BACE1 activity and decreasing the production of beta-amyloid (Aβ).19 Furthermore, a DNA aptamer that can specifically recognize and bind to Aβ oligomer was generated in 2012 and it could be a more efficient and specific tool than antibodies in recognizing the Aβ oligomer and preventing Aβ toxicity, as well as diagnosing Alzheimer’s disease (AD).20 As for the α-syn in PD, although several DNA aptamers against α-syn have been generated,21, 22 due to the limitations of selection, these aptamers are in low affinity and specificity. As far as applying aptamers to regulate α-syn toxicity, no investigation has been reported yet.
In this study, with the optimized SELEX on recombinant α-syn, we obtained two aptamers (named as F5R1 and F5R2). The two aptamers inhibited α-syn aggregation and targeted the cellular α-syn to degradation in SK-N-SH cells and primary neurons, and rescued the mitochondrial dysfunction and cell defects induced by α-syn overexpression. To our knowledge, this is the first investigation to employ aptamers to block the aberrant cellular effects of overexpressed α-syn in cells.
Results
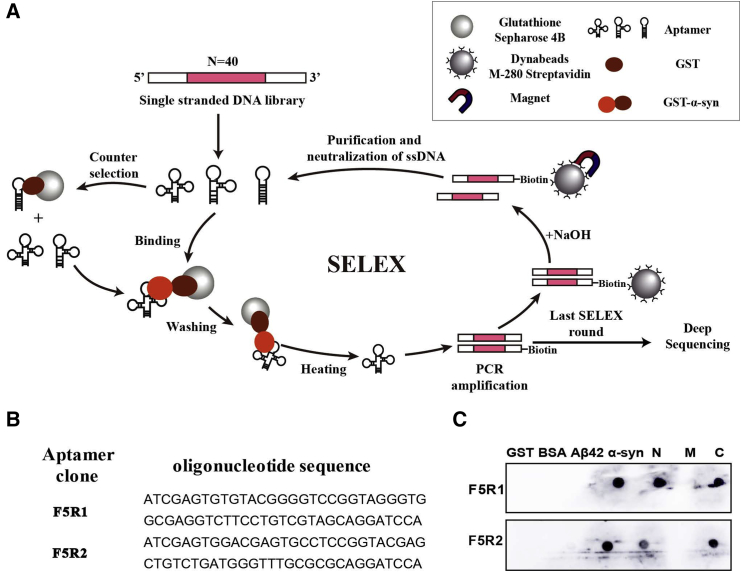
Selection of DNA Aptamers against α-Syn
In this study a glutathione-sepharose bead-based selection procedure using immobilized glutathione S-transferase (GST)-α-syn was employed, since under this condition the target protein can be kept in a near-free state, excluding the extraneous interference such as the electrophoresis from the gel-shift assay21 and conformational restriction from the aptamer blotting assay22 (Figure 1A). After the proteins were purified (Figure S1) an ssDNA library containing a 40-nt random core sequence was passed through the immobilized GST-α-syn on the beads. After 9 rounds of aptamer selection, we sequenced each clone by deep sequencing. From the total read counts, we obtained 11,019 sequences by removing the incorrect sequences from deletion and/or insertion mutations during PCR amplification and sequencing. The two sequences that most frequently appear are selected as the aptamer candidates. Then we chemically synthesized the aptamers (named as F5R1 and F5R2) with 58 bases, in which the majority of the primer binding regions were removed from the entire sequence, as these regions might be less involved in protein binding (Figure 1B). To test whether the aptamers have specificity against the α-syn protein, we used aptamer blotting with GST, Aβ1-42, and BSA as protein competitors. The aptamers of F5R1 and F5R2 all only bound to the α-syn protein (Figure 1C). Further dot blotting results showed that the aptamers could bind with N- and C termini of α-syn (Figure 1C), suggesting that the N- and C termini of α-syn are responsible for its interaction with the aptamers. Further, our competitive ELISA tests indicated that the aptamers could inhibit the binding of the antibodies to α-syn in a concentration-dependent manner (Figure S2).
Figure 1.
α-syn Aptamers Were Selected through SELEX
(A) Schematic illustration of the method used for α-syn aptamer selection. GST-tagged α-syn was immobilized on glutathione-sepharose beads. The ssDNA library was incubated with the target beads for binding. Unbound oligonucleotides were washed away, and the bound ones were released by heating at 95°C. The selected binders were amplified by PCR with biotinylated primers. ssDNAs were subsequently purified from the PCR product using streptavidin-coated magnetic beads, resulting in an enriched DNA pool, which was used in the next SELEX round. After the last round, the selected ssDNAs were sequenced by deep sequencing. (B) The aptamer candidates. After deep sequencing, the two sequences with most frequently appearing were selected as the aptamer candidates. (C) Aptamer binding specificity assay by dot blotting. Five microgram samples (α-syn, GST, Aβ42, BSA, and three domains of α-syn) were respectively immobilized onto the nitrocellulose membrane for binding of each aptamer.
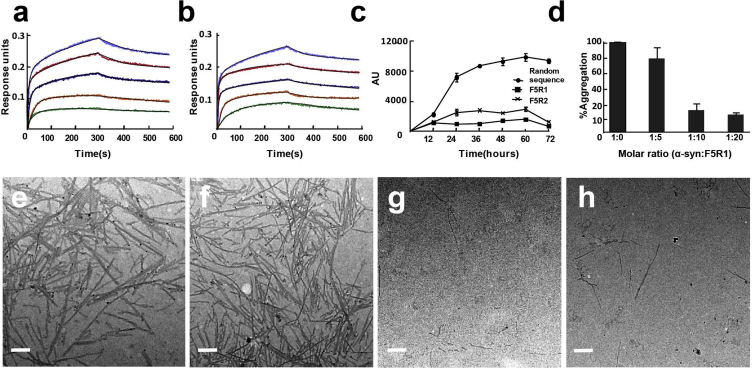
Binding Affinity of Selected Aptamers to α-Syn
To determine the binding affinities of the aptamers to α-syn, the aptamers were used as a ligand to measure its interactions with α-syn protein by biolayer interferometry (BLI). As shown in Figure 2A, binding at five different α-syn concentrations, ranging from 10 to 160 nM, readily fit a global model. The KD (equilibrium dissociation constant [KD]) value of the aptamer F5R1 was 2.40 nM, with subsidiary fast-on kinetics (Kon) of 1.42 × 105 M−1s−1 and slow-off kinetics (Koff) of 3.28 × 10−4 s−1. Similarly, for aptamer F5R2, its KD value was 3.07 nM, with fast-on kinetics (Kon) of 1.31 × 105 M−1s−1 and slow-off kinetics (Koff) of 4.02 × 10−4 s−1 (Figure 2B). These affinities are substantially higher than those of the existing DNA aptamers against α-syn (around 63 nM).22
Figure 2.
Binding Affinity of the Selected Aptamers to α-syn and Their In Vitro Inhibition of α-syn Aggregation
(A and B) BLI analysis of the aptamers F5R1 (A) and F5R2 (B) binding to α-syn, respectively. The α-syn concentrations were 10, 20, 40, 80, and 160 nM, respectively. (C) Kinetic analysis of the aggregation of α-syn in the presence of aptamers using ThT (molar ratio between the α-syn and aptamer is 1:10). (D) Dose-dependent inhibition effect of aptamer F5R1 on α-syn aggregation. The reaction mixtures were incubated at 37°C with constant agitation (1,000 rpm) for 3 days and the rate of fibrillogenesis was monitored using the thioflavin T (ThT) fluorescence assay. (E–H) TEM images of α-syn fibrils with aptamer F5R1. α-syn alone (E), α-syn with random DNA sequence (F), F5R1 (G), and F5R2 (H). Scale bar, 200 nm.
Inhibiting the α-Syn Aggregation by Aptamers In Vitro
Previous studies showed that the antibodies that recognized the N- or C terminus might inhibit the α-syn aggregation.6, 13, 23 We hypothesized that the α-syn-specific aptamers could also inhibit the self-assembly of α-syn. To test this hypothesis, thioflavin-T (ThT) binding assay was performed and demonstrated that the formation of amyloid fibrils was significantly reduced in the presence of both aptamers, and F5R1 showed better inhibition effect than F5R2 (Figure 2C). Furthermore, the dose-dependent inhibition of F5R1 was assayed, and it showed that F5R1 was efficient at an excess of 10:1 molar ratio (F5R1:α-syn) but was less efficient at 5:1 ratio (Figure 2D). Additionally, transmission electron microscopy (TEM) analysis was performed on the samples from the ThT experiment. While the fibrils formed by α-syn alone and the presence of random DNA sequence were large, broad, and had a ribbon-like shape with large quantity (Figures 2E and 2F), only a few short fibrils were detected in the presence of aptamers (Figures 2G and 2H). These results are highly correlated with the results of the ThT assay.
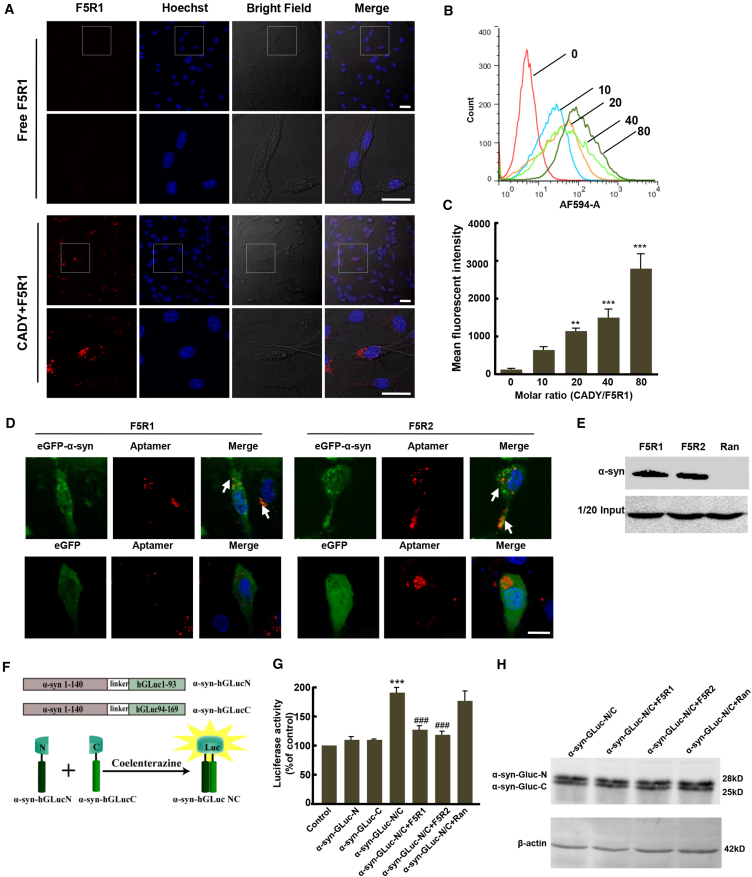
CADY-Mediated Aptamer Delivery into SK-N-SH Cells
Previous studies showed that the peptide carrier CADY could form stable complexes with nucleic acids and improved their delivery in cultured cells.24 Thus, we tried to use CADY peptide as a carrier to deliver the aptamers into the cells. First, we confirmed that the aptamers were completely associated with CADY when the molar ratio of CADY/aptamer was more than 20 and that CADY could protect the aptamer from degradation by serum nuclease (Figure S3). Next, a fixed concentration (20 nM) of the aptamers labeled with Alexa594 was complexed with CADY at a CADY/aptamer molar ratio of 40:1, and the confocal microscopy showed that the aptamers were delivered into cells with high efficiency (Figure 3A). To further confirm aptamer uptake, Alexa594-labeled aptamers with a fixed concentration of 20 nM were incubated with CADY at different CADY/aptamer molar ratios, ranging from 0:1 to 80:1 for 4 hr, then the internalization was monitored by fluorescence-activated cell sorting (FACS) (Figures 3B and 3C). The data showed that the fluorescence intensity in cells was in a dose-dependent manner, suggesting that the aptamer uptake efficiency was directly related to the molar ratio of the CADY/aptamer. For the rest of the experiments in this study, unless otherwise noted, for delivery of aptamers into SK-N-SH cells, the molar ratio of CADY/aptamer at 40:1 was used. Additionally, we confirmed that aptamer uptake could not cause cytotoxicity in the SK-N-SH cells (Figure S4).
Figure 3.
Aptamers Were Delivered by CADY into Cultured SK-N-SH Cells and Inhibited α-syn Aggregation
(A) The CADY/Alexa594-labeled F5R1 complexes were formed at molar ratio of 20/1 and overlaid onto SK-N-SH cells for 4 hr at 37°C. Cells were then washed and the fluorescence was detected by confocal laser microscopy. Scale bars, 25 μm. (B and C) The CADY/F5R1 complexes were formed at different molar ratios and overlaid onto SK-N-SH cells for 4 hr at 37°C. Cells were then washed and trypsinized, and the fluorescent cells were counted by flow cytometry. Results are reported for 100,000 cells (n = 5). (D) SK-N-SH cells were transfected with EGFP-α-syn constructs or EGFP vector as control. After 6 hr, the aptamers of F5R1 and F5R2 labeled with Alexa594 complexed with CADY were used to treat these cells, respectively. The EGFP-α-syn co-localization with F5R1 or F5R2 was detected by confocal laser microscopy. Arrows showed the co-localization spot. Scale bar, 10 μm. (E) SK-N-SH cells were pre-treated with CADY/biotinylated-aptamer complexes (aptamer at 100 nM and the CADY/aptamer molar ratio at 40; random DNA sequence served as the control), and then transfected with pcDNA3.1-α-syn for α-syn overexpression. Streptavidin magnetic beads were incubated with protein lysate overnight at 4°C. Samples were resolved by using SDS-PAGE and α-syn was detected by western blotting. (F) Principle of bioluminescent-protein complementation assay based on G. princeps luciferase. (G) SK-N-SH cells with/without CADY/aptamer complex pre-treatment were co-transfected with constructs of α-syn-hGLucN and α-syn-hGLucC. After transfection for 24 hr, the luciferase activity from protein complementation was measured in an automated plate reader at 480 nm with substrate coelenterazine (20 μM). Data are presented as the mean ± SD (one-way ANOVA) ***p < 0.001 compared with control group (n = 6); ###p < 0.001 compared with α-syn-hGLuN/C group. (H) SK-N-SH cells with/without CADY/aptamer complex pre-treatment were transfected with constructs of α-syn-hGLucN and α-syn-hGLucC to show the expression of each protein. Immunoblots were probed with antibody against α-syn.
Aptamers Inhibited α-Syn Aggregation in SK-N-SH Cells
To further investigate whether aptamers also can recognize intracellular α-syn and block its aggregation in living SK-N-SH cells, first, the aptamers labeled with Alexa Flour-594 were delivered into EGFP-α-syn overexpressing cells, and the confocal laser scanning data showed that both F5R1 and F5R2 were co-localized with α-syn in cytoplasm (Figure 3D). We next confirmed that aptamers directly bound to α-syn in cells with a pull-down assay using biotinylated aptamers as affinity capture agents (Figure 3E), whereas no binding was observed in the random DNA sequence group.
Next, we employed a protein-fragment complementation assay (PCA)25, 26, 27 to investigate whether the aptamers could inhibit the formation of α-syn aggregates in cells (Figure 3F). Twenty-four hr after co-transfection of the α-syn-hGLucN and α-syn-hGLucC constructs into the SK-N-SH cells, the reconstituted luciferase activity was almost 2-fold as high as that in control cells. However, the pre-treatment with the aptamers of F5R1 or F5R2 prevent this increase in luciferase activity, respectively. In contrast, the random DNA sequence pre-treatment did not show such an effect (Figure 3G). Additionally, aptamers at various concentrations (from 1 to 20 nmol/L) complexed with CADY in the pre-treatment caused the decrease in luciferase activity in an aptamer concentration-dependent manner (Figure S5). To exclude the possibility that the decreased luciferase activity was due to the aptamers delivered into the cells downregulating the α-syn level, we further confirmed that the intracellular protein level of α-syn-hGLuN and α-syn-hGLuC did not show any change between the indicated groups (Figure 3H). Collectively, these data suggested that the aptamers inhibited the α-syn oligomerization in cells, and, for the rest of the experiments regarding aptamer pre-treatment to the SK-N-SH cells, the aptamer concentration of 20 nmol/L was used.
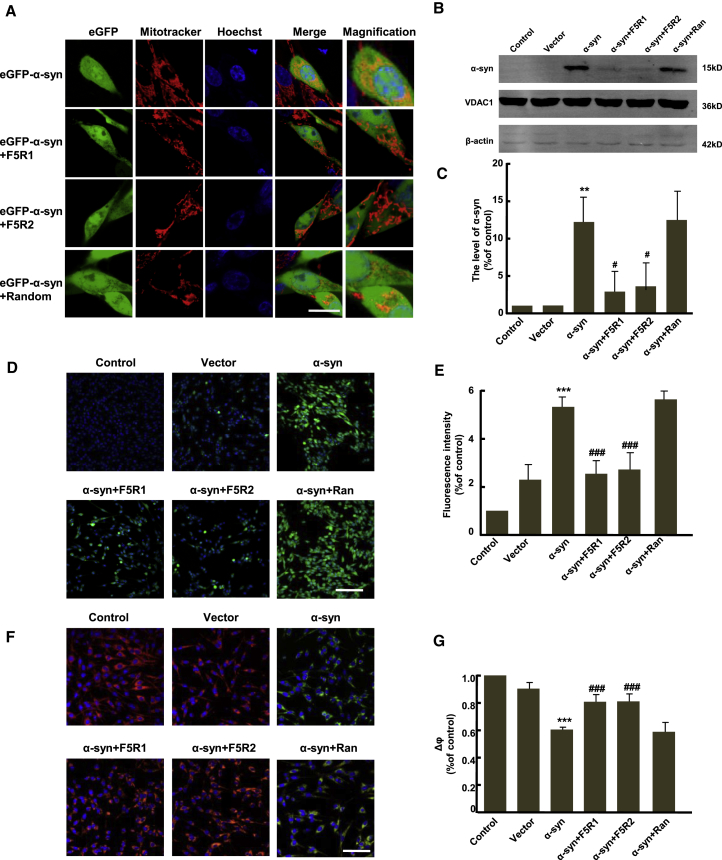
Aptamers Protected against α-Syn-Induced Mitochondria Dysfunction
Previous studies showed that aggregated α-syn was more strongly associated with mitochondria,28 and these aggregates augmented oxidative stress and suppressed mitochondrial and cellular functions.29 So we further tested whether these aptamers could block the association of α-syn with mitochondria and consequently suppress the oxidative stress. Figure 4A shows, in non-treated or random DNA sequence-treated groups, intense co-localization of α-syn (green) with the mitotracker (red) was detected. However, in the aptamer treatment groups, less mitochondrial localization of α-syn was observed. Further evidence for mitochondrial association of α-syn was achieved by immunoblotting (Figures 4B and 4C). These results suggested that the aptamers of F5R1 and F5R2 could block the association of α-syn with mitochondria since the aptamers could inhibit the α-syn aggregation in cells.
Figure 4.
The Aptamers of F5R1 and F5R2 Prevented α-syn Binding to Mitochondria and Rescued Mitochondrial Dysfunction
(A) After aptamer pretreatment, SK-N-SH cells were transfected with EGFP-α-syn cDNA. 24 hr after transfection, cells were stained by mitotracker (red) for marking mitochondria. Merge shows overlaid images of mitochondria and EGFP-α-syn. Nuclei were counter-stained with Hoechst (blue). Scale bar, 20 μm. (B) After aptamer pretreatment, SK-N-SH cells were transfected with pCDNA3.1-α-syn for overexpression of α-syn. Twenty-four hr after transfection, the mitochondria in the cells were isolated, and the α-syn associated with mitochondria was measured by immunoblotting. The random DNA sequence group was used as a negative control. After 24 hr, mitochondria were purified, and its purity was tested by immunoblotting the mitochondria extracts against a mitochondrial marker, VDAC1, and a cytoplasmic marker, β-actin. VDAC1 also serves as mitochondrial loading control. (C) Quantitative analysis of the protein level of α-syn within mitochondria. Data are presented as the mean ± SD (one-way ANOVA) **p < 0.01 compared with vector group; #p < 0.5 compared with α-syn overexpressing group (n = 3). (D–G) Cells were treated as in (B), and the mitochondrial function was assayed 24 hr post-transfection. (D) The Mitochondrial reactive oxygen species (ROS) were visualized by DCF-DA (scale bar, 50 μm). (E) Quantitative analysis of DCF-DA fluorescence intensity by high-content screening in SK-N-SH cells. Data are presented as the mean ± SD (one-way ANOVA) ***p < 0.001 compared with vector group; ###p < 0.001 compared with α-syn overexpression group (n = 6). (F) Δφm detected with JC-1 in transfected SK-N-SH cells. (G) Quantitative analysis of red fluorescence intensity by high-content screening in SK-N-SH cells. Data are presented as the mean ± SD (one-way ANOVA) ***p < 0.001 compared with vector group; ###p < 0.001 compared with α-syn overexpression group (n = 6).
Next, we evaluated intracellular reactive oxygen species (ROS) levels and Δφm in SK-N-SH cells by DCF-DA and JC-1 staining. The ROS level increased about 2-fold in the α-syn overexpression group compared with the blank vector group. However, the F5R1 or F5R2 pre-treatment almost completely eliminated the increase of ROS level by α-syn. The aptamer effects on ROS were not observed in the random DNA sequence pre-treatment group (Figures 4D and 4E). Similarly, aptamers of the F5R1 and F5R2 also could rescue the membrane potential decline induced by α-syn (Figures 4F and 4G). Collectively, F5R1 and F5R2 could prevent the α-syn-induced increase in ROS and decrease in mitochondrial membrane potential and, therefore, potentially rescue the mitochondrial dysfunction caused by α-syn overexpression.
Aptamers Induced Lysosomal Degradation of α-Syn
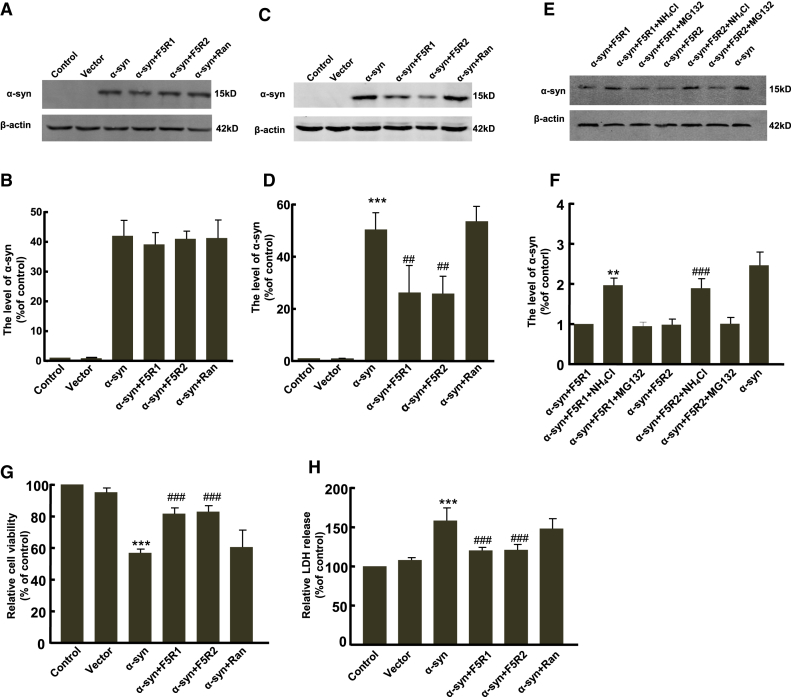
Since previous studies indicated that aptamer could effectively target the protein to intracellular degradation,30 we hypothesized that F5R1 and F5R2 delivered into the cells could induce the α-syn degradation. To address our hypothesis, pcDNA3.1-α-syn constructs were transfected into the SK-N-SH cells with or without aptamer pre-treatment, and we found that, after the transfection for 24 hr, regardless of the aptamer pre-treatment, the α-syn protein level did not show any significant change (Figures 5A and 5B); interestingly, 48 hr after the transfection, both F5R1 and F5R2 pre-treatments resulted in a significant decrease in α-syn level when compared to the untreated groups, whereas the random DNA sequence pre-treatment did not have such an effect (Figures 5C and 5D). These results suggested that the α-syn aptamers of F5R1 and F5R2 effectively targeted α-syn to intracellular degradation.
Figure 5.
The Aptamers of F5R1 and F5R2 Enhanced Lysosomal Degradation of α-syn and Rescued the Cell Defects
(A) SK-N-SH cells pre-treated with F5R1, F5R2 or random DNA sequence were transfected the α-syn or vector control vectors and incubated for 24 hr. The extracts were separated by SDS-PAGE and blotted onto PVDF membrane. The membrane was blocked and probed with the α-syn specific polyclonal antibody. β-actin served as the loading control. (B) Quantitative analysis of the total protein level of α-syn from (A). (C) SK-N-SH cells were similarly treated as in (A) except for incubation time (48 hr). The cell extracts were immunoblotted with the α-syn polyclonal antibody. β-actin served as the loading control. (D) Quantitative analysis of the total protein level of α-syn from (C). Data are presented as the mean ± SD (one-way ANOVA) ***p < 0.001 compared with vector group; ##p < 0.01 compared with α-syn overexpression group (n = 3). (e) Cells were treated similarly as in (C), except for treated with NH4Cl or MG132 after 6 hr of transfection. The cell extracts were immunoblotted with the α-syn polyclonal antibody. (F). Quantitative analysis of the total protein level of α-syn from (E). Data are presented as the mean ± SD (one-way ANOVA) **p < 0.01 compared with α-syn+F5R1 group; ###p < 0.001 compared with α-syn+F5R2 group (n = 5). (G and H) Cells were treated similarly as in (E), Forty-eight hr after transfection, the cell viability was assessed by MTT assay (G) and LDH assay (H). Data are presented as the mean ± SD (one-way ANOVA) ***p < 0.001 compared with vector group; ###p < 0.001 compared with α-syn overexpression group (n = 6).
Next, because a previous report showed that α-syn underwent degradation in lysosomes once perturbed by antibodies,14 we hypothesized that F5R1 and F5R2 delivered into the cells could induce the α-syn degradation by lysosomes. To test this hypothesis, we examined the effect of a specific inhibitor of lysosomes (NH4Cl) and a proteasome inhibitor (MG132) on α-syn degradation, respectively. As shown in Figures 5E and 5F, the drug of NH4Cl significantly inhibited the effect of the aptamers of F5R1 and F5R2 on α-syn degradation. However, the proteasome inhibitor MG132 exerted no significant effect on α-syn degradation. Taken together, we provided evidence to support that the α-syn aptamers of F5R1 and F5R2 can effectively target α-syn to intracellular degradation, likely through the lysosomal pathway.
Aptamers Reduced the Cytotoxicity Caused by α-Syn Overexpression
Because F5R1 and F5R2 could reduce the α-syn oligomerization, preserve mitochondrial function, and facilitate the cellular α-syn degradation, we then tested whether F5R1 and F5R2 could reduce the cytotoxicity and rescue the cell defects with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and lactate dehydrogenase (LDH) release assays. It showed that overexpression of α-syn resulted in a 30% decrease in cell viability compared with the blank vector group. However, the F5R1 or F5R2 pre-treatment could significantly increase the cell viability compared with the α-syn-alone group. Further, the minimal effective concentration to rescue the cell defects was estimated at 15 nM (Figure S6). In contrast, such an effect was not observed in the random DNA sequence-treatment group (Figure 5G). Similarly, these two aptamers also decreased LDH release in SK-N-SH cells as compared to the α-syn-only group, and the random DNA sequence treatment had no such effect (Figure 5H). These data collectively demonstrated that the α-syn aptamers of F5R1 and F5R2 were able to rescue cell dysfunction.
Aptamers Delivered into Primary Neurons Induced α-Syn Degradation and Rescued the Mitochondria Dysfunction and Cell Defects in Primary Neurons
To further investigate the effect of aptamers on reducing α-syn toxicity in primary neurons, primary neurons with aptamer pre-treatment were infected with lentiviral particle-containing LV-α-syn, then the cellular α-syn protein level, mitochondria function, and primary neuronal viability were measured.
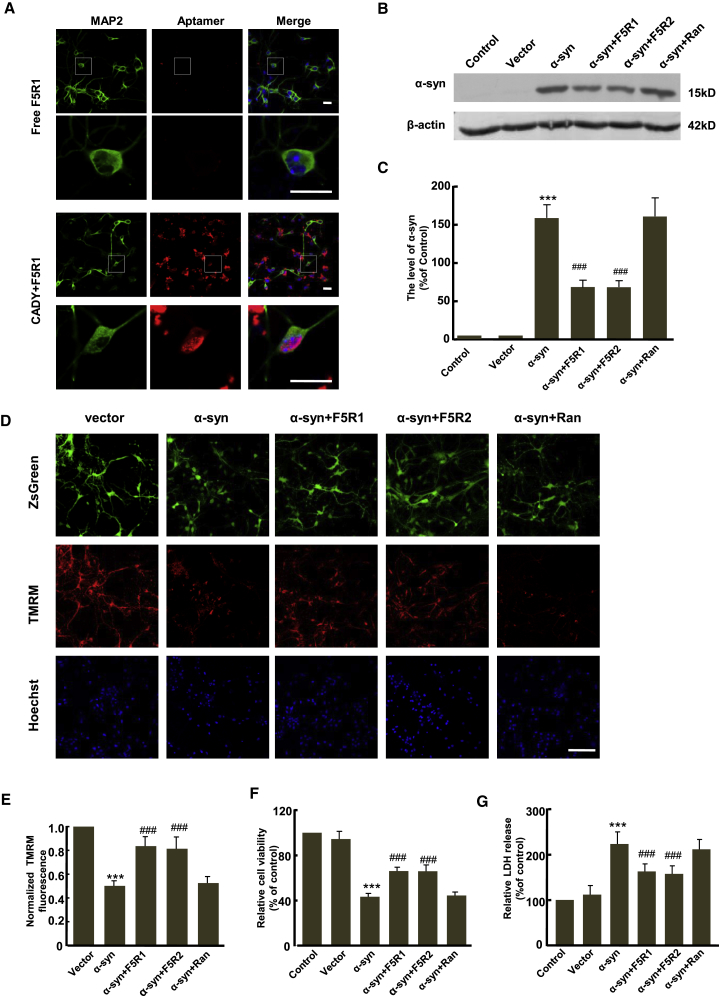
First, we confirmed that that aptamers could be delivered into the primary neurons (Figure 6A) by Alexa594-labeled aptamer F5R1 uptake assay. Next, to further test whether aptamers also can induce α-syn degradation, the primary neurons at 3 DIV (days in vitro) were treated with the CADY/aptamer complex for 2 hr and then infected with the LV-α-syn virus. At 8 DIV, total neuronal proteins were extracted, and the immunoblotting showed that, as we expected, both F5R1 and F5R2 could reduce the α-syn protein level more than 50%, whereas the random DNA sequence had no such effect (Figures 6B and 6C).
Figure 6.
Aptamers were Delivered into Cultured Primary Neurons and Rescued the Cell Toxicity Caused by α-syn Overexpression
(A) Alexa594-labeled aptamer F5R1 (10 nM) was complexed with CADY peptide (the molar ratio of CADY/F5R1 at 20), and then incubated with the 3 DIV (day in vitro) primary neurons for 2 hr. The fluorescence was detected by confocal laser microscopy (scale bar, 25 μm). The neurons are stained with MAP2 in green. (B) The 3 DIV primary neurons were treated with CADY/aptamer complex for 2 hr as in (A), then were infected with LV-α-syn cDNA. At 8 DIV, total neuronal proteins were extracted, and the human α-syn was detected by immunoblotting. β-actin was examined as a loading control. (C) Quantitative analysis of α-syn level from (B). Data are presented as the mean ± SD (one-way ANOVA) ***p < 0.001 compared with vector group; ###p < 0.001 compared with α-syn overexpression group (n = 6). (D) The primary neurons were treated as described in (B). At 8 DIV, the mitochondrial transmembrane potential (Δφm) was detected by Δφm-sensitive probe TMRM under confocal laser microscopy. ZsGreen was used to confirm the virus vector working well in neurons (scale bar, 100 μm). (E). Reactive fluorescence intensity of TMRM in mitochondria from 3 coverslips was quantitative analyzed. Data are presented as the mean ± SD (one-way ANOVA) ***p < 0.001 compared with vector; ###p < 0.001 compared with α-syn overexpression group (n = 3). (F and G) Primary neurons were treated as described in (B). At 8 DIV, cell viability and cytotoxicity were detected in primary neurons with the MTT (F) and LDH (G) assays. Data are presented as the mean ± SD (one-way ANOVA) ***p < 0.001 compared with vector; ###p < 0.001 compared with α-syn overexpression group (n = 6).
To examine the protective effects of F5R1 and F5R2 on the mitochondrial function, the primary neurons were treated similarly as in Figure 6B, and the mitochondrial transmembrane potential (Δφm) was detected by Δφ-sensitive probe TMRM. As shown in Figures 6D and 6E, a decrease in TMRM intensity in the α-syn overexpression group was observed compared with the vector control group. Notably, both F5R1 and F5R2 treatments could suppress the decline in TMRM fluorescence, and the random DNA sequence had no such effect. These results suggested that the decrease in membrane potential in α-syn overexpressing primary neurons could be prevented by the aptamers. Next, the viability and apoptosis of primary neurons with the aptamer treatment were measured as a marker of cellular functions. Figures 6F and 6G showed that α-syn overexpression in the primary neurons reduced cell viability and increased cell apoptosis, and the effects were reversed by F5R1 or F5R2, but not random DNA sequence.
Aptamers Rescued the Neurite Morphology Change Caused by α-Syn
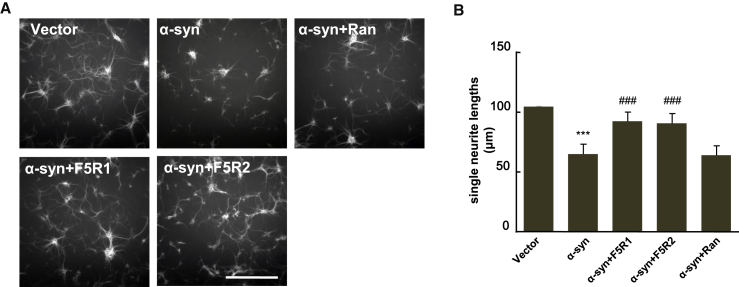
It was reported that overexpression of α-syn resulted in reduced neurite extension in neuronal cells, which is a detrimental factor for neurite integrity.31, 32 Here, we investigated whether aptamers could rescue the impairment of neurite outgrowth caused by α-syn. The primary neurons at 3 DIV were first treated by CADY/aptamer complex and then they were infected with the LV-α-syn virus. After a 5-day culture, the single neurite length was measured. The mean single neurite length (60 μm) was significantly reduced in the neurons overexpressing α-syn as compared with the blank vector group (100 μm). On the other hand, the neurite length in the F5R1- (93 μm) and F5R2- (89 μm) treated groups were obviously longer than that in the α-syn overexpression group, whereas there was no statistically significant difference between the random DNA sequence group and the α-syn overexpression group (Figure 7). These results implied that α-syn aptamers rescued the impairment of neurite outgrowth caused by α-syn and stabilized neurite morphology.
Figure 7.
Aptamers Rescued the Neurite Morphology Change of the Primary Neuron Infected with LV-α-syn
(A) The aptamers of F5R1 and F5R2 (10 nM each) was complexed with CADY peptide (the molar ratio of CADY/F5R1 or CADY/F5R2 at 20) and incubated with primary neurons at 3 DIV for 2 hr, then were infected with LV-α-syn vectors. At 8 DIV, an immunocytochemistry of the primary neurons was performed. Neurons were stained against MAP2, and the random DNA sequence group served as the negative control (scale bar, 200 μm). (B) Quantification of single neurite lengths of primary neurons. Data are presented as the mean ± SD (one-way ANOVA) ***p < 0.001 compared with vector; ###p < 0.001 compared with α-syn overexpression group (n = 6).
Discussion
This study demonstrated that α-syn aptamers were able to inhibit α-syn aggregation and promote clearance of α-syn in human α-syn overexpressing cell lines and primary neurons. Aptamers of F5R1 and F5R2, two 58-base-long nucleotides, were selected under the optimized SELEX process and demonstrated a high affinity and specificity to bind to α-syn against its N- and C termini. These aptamers were efficient in reducing α-syn aggregation in vitro and in cells and further targeted the α-syn to intracellular degradation through the lysosomal pathway, consequently rescuing the mitochondrial dysfunction and cellular defects caused by α-syn overexpression (Figure 8). To our knowledge, it is the first to employ the aptamers to block the aberrant cellular effects of overexpressed α-syn in cells.
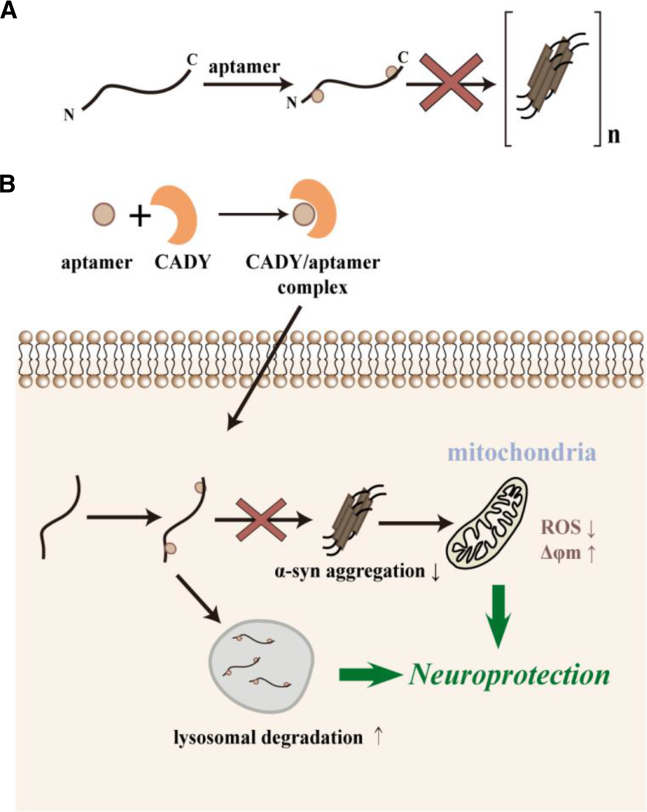
Figure 8.
The Aptamers Exert Neuroprotective Effects against α-syn-Induced Cell Dysfuction by Inhibiting α-syn Aggregation and Promoting α-syn Degradation through Lysosomes
The selected two aptamers possessed the high affinity and specificity to bind to the N- and C termini of α-syn and inhibited the α-syn aggregation process in vitro (A). After these aptamers were delivered into the cells, they were efficient in reducing α-syn aggregation in vivo, and further targeted the α-syn to intracellular degradation through lysosomal pathway, consequently rescuing the mitochondrial dysfunction and cellular defects caused by α-syn overexpression (B).
SELEX is a common process involving the progressive isolation of highly selective ssDNA/RNA from a combinatorial single-stranded oligonucleotide library. Previously, although several aptamers to α-syn have been selected,21, 22 their affinities are relatively low (the KD value was around 63 nM), probably because of the extraneous inferences during the selection process, such as the α-syn conformational restriction from the membrane blotting22 and the electrophoresis from the gel-shift assay.21 In this study, the selection process was optimized: the glutathione-sepharose bead-based selection procedure using immobilized GST-α-syn was employed since, under this condition, the target protein could be kept in a near-free state (Figure 1A). Consequently, the selected aptamers of F5R1 and F5R2 showed higher affinity (their KD value were 2.40 nM and 3.07 nM, respectively).
Although in past decades the mechanisms of α-syn aggregation have been extensively studied, a consensus view of the aggregation mechanism is still elusive. Interestingly, the NMR residual dipolar coupling (RDC) data suggested that there exists long-range interaction between the N- and C termini in the α-syn.33 Under conditions that increase the long-range interactions, the α-syn aggregation is enhanced; however, conditions that weaken such interactions are associated with reduced aggregation.34, 35 Thus, it seems that long-range interactions may facilitate the α-syn aggregation process.36 In such scenarios, since the aptamers may bind with the N- and C termini of α-syn and potentially shield these two fragments, the long-range interaction within α-syn probably would be weakened or blocked, so its aggregation is inhibited. Certainly, more structure-function data are still needed to support this hypothesis.
The major limitation of the therapeutic application of nucleic acids is the poor permeability of the cell membrane and their restricted biodistribution.24, 37, 38 As a secondary amphipathic peptide derived from PPTG1 peptide, CADY was used to rapidly deliver the siRNA into the challenging cell lines (including primary neurons) and, consequently, a long-term silencing response was achieved, with 60% knockdown of the target gene after 5 days.24 In this study, it was shown that the CADY was thoroughly associated with the aptamers with a molar ratio >20, as already reported for other noncovalent peptide carriers.39 Furthermore, the CADY peptide could protect aptamers from serum degradation and efficiently deliver the aptamers into the SK-N-SH cells and primary neurons with no cytotoxicity.
Currently, although it is not clear which form of the α-syn protein is more cytotoxic, massive neuropathological, biochemical, and genetic evidence have shown that α-syn oligomers and aggregates are critical for the death of dopaminergic neurons and the progression of PD.3, 4, 40 Particularly, previous studies showed that the aggregated α-syn bound to the mitochondria of α-syn overexpressing SHSY cells, resulting in mitochondrial impairments via increasing ROS and decreasing Δφm,29 which plays a role in death of the dopaminergic neurons in PD.41, 42 In this study, the data demonstrated that both F5R1 and F5R2 aptamers could recognize cellular α-syn, block its aggregation in cells, and inhibit binding of α-syn to mitochondria. Functionally, the mitochondrial dysfunction was rescued by the aptamer treatment. These data strongly support the previous findings in co-localization of α-syn within the mitochondria of α-syn overexpressing cells and functional significance for α-syn association with mitochondria.
In previous studies, the DNA aptamer against ErbB-2 effectively targeted the protein to intracellular degradation in lysosomes in cells and animals.30 More passive administration of α-syn antibodies to cell and animal models have demonstrated that the antibody directed α-syn protein clearance via the lysosomal pathway.8, 12, 14 In agreement with these findings, our data showed that the levels of α-syn in cell lysate were decreased after aptamer treatment, which was possibly attributed to the aptamer targeting α-syn to intracellular degradation through the lysosomal pathway.
In summary, aptamers against the protein of α-syn were selected, and we demonstrated that these aptamers inhibited α-syn aggregation and further targeted the cellular α-syn to degradation through the lysosomal pathway, consequently rescuing the mitochondrial dysfunction and cell defects in the α-syn overexpressing SK-N-SH cells and primary neurons. With future experiments in animals, these aptamers could be developed as a unique and potential strategy to target α-syn in the context of PD therapy.
Materials and Methods
Preparation of α-Synuclein
The α-syn gene was subcloned into the pGEX-4T-1 vector, and the fusion protein GST-α-syn was expressed in Escherichia coli BL21 (DE3), which was induced by the addition of 0.2 mM isopropyl-1-thiob-D-galactopyranoside (IPTG) at 28°C. The cells were disrupted by sonification, and the supernatant was collected by centrifugation (18,000 g, 10 min, 4°C). Then the fusion protein GST-α-syn was purified on glutathione-sepharose 4B according to the manufacturer’s instructions (GE Healthcare, Boston, MA). The purified GST-fusion proteins were desalted on Vivaspin 6 column from GE Healthcare, followed by dialysis into binding buffer (PBS, 1 mM MgCI2, pH 7.4) to remove the free glutathione. To cleave the GST tag from the fusion protein and recover the proteins of α-syn and GST separately, thrombin was used for cleavage with on-column settings according to the manufacturer’s instructions (GE Healthcare, Boston, MA).
Design of a DNA Library
The starting point of the selection process was a 40-nt completely random ssDNA library (5′-GATGTGGTAGGTGATCGAGTG-N40-GCAGGATCCATCCACCTCTA-3′) that was chemically synthesized by Invitrogen (Carlsbad, CA) and high-performance liquid chromatography (HPLC) purified. A 20-mer primer-binding region was flanking the randomized region at both ends, respectively. To fold the structure, the synthesized oligonucleotides were heated at 95°C for 3 min and then gradually cooled to 25°C at a rate of 5°C per min in PBS.
Selection of Aptamers
Iterative rounds of aptamer selection and amplification during the systematic evolution of ligands by exponential enrichment (SELEX) process were modified from previous protocols.43 Primers that anneal to the 5′-and 3′-sequences flanking the random core region used during the selection were: Apt_5′, GATGTGGTAGGTGATCGAGTG, and Apt_3′, TAGAGGTGGATGGATCCTGC; in non-biotinylated and 5′-biotinylated forms respectively (HPLC-purified). A 1 nmol aliquot of DNA library was incubated with GST-α-syn immobilized on glutathione-sepharose beads in PBS for 45 min at 4°C. The unbound DNA was separated and removed by centrifugation at 500 g for 5 min. The bound sequences were released by heating to 95°C for 2 min and PCR-amplified using biotinylated primers. A 100 μL aliquot of the PCR product containing enriched biotinylated double-stranded DNA was equilibrated with 100 μL of 2 × binding and washing buffer (10 mM Tris-HCI, 1 mM EDTA, 2 M NaCI, pH7.5) and 1 mg of M-280 streptavidin magnetic beads (Invitrogen, Carlsbad, CA) for 10 min. By magnetic separation, non-biotinylated ssDNA was recovered and dissociated from the immobilized complementary biotinylated strand using 100 μL of 150 mM NaOH for 5 min. Then, nine cycles were performed with counter selection using immobilized GST magnetic beads at rounds two and five. During the last round of SELEX, the recovered DNA molecules were PCR amplified using non-biotinylated primers and subjected to be sequenced by deep sequencing (WuXi AppTec, Shanghai, China).
Dot Blotting
5 μg proteins of BSA, Aβ1-42, GST, α-syn, and its three truncates, N terminus (residues 1–60), NAC domain (residues 61–95), and C terminus (residues 96–140) (synthesized by Thermo Fisher Scientific, Waltham, MA), were respectively immobilized onto nitrocellulose filter membrane. After blocking with 0.5% fat-free milk in Tris-buffered saline Tween-20 (TBST) for 1 hr, the membrane was incubated with 100 nM aptamer labeled with FITC overnight at room temperature. The membrane was washed three times with TBST and then incubated with FITC antibody that was labeled with HRP (Takara, Japan) for 4 hr. Dots were visualized using enhanced chemiluminescence.
Evaluation of the Aptamer Binding Affinity by BLI
The BLI experiments were performed on a FortéBio (Menlo Park, CA) Octet QK biosensor using streptavidin (SA) sensors to determine binding affinity of aptamers against α-syn. Assays were performed in 96-well microplates at 25°C. All volumes were 200 μL. After loading biotinylated aptamers onto SA sensors, a baseline was established in the binding buffer (50 mMTris/100 mMNaCl, pH 7.0) prior to monitoring the association at various α-syn concentrations. Dissociation subsequently was measured in buffer only. The KD of aptamer was calculated with Octet software (version 8.0) from the association and dissociation curve of the aptamer binding with α-syn.
ThT Fluorescence Assay of Fibril Formation In Vitro
α-syn was dissolved to a concentration of 360 μM (around 5 mg/mL) in the assembly buffer (50 mMTris/100 mMNaCl, pH 7.0). The α-syn protein was mixed with or without aptamer at a 1:10 ratio, the reaction mixtures were incubated at 37°C with constant agitation (1,000 rpm) as described by Luk et al.,44 and the rate of fibrillogenesis was monitored using the thioflavin T (ThT) fluorescence assay (excitation at 450 nm, 2.5 nm slit, and emission at 480 nm, 5 nm slit). ThT was added to a 500-fold diluted sample, and fluorescence was measured using a fluorometer from PerkinElmer EnVision (Waltham, MA).
Transmission Electron Microscopy Assay
Samples (10 μL) from the α-syn ThT fluorescence assay were placed on 400-mesh copper grids covered by carbon-stabilized Formvar film (SP I Supplies, West Chester, PA). After 2 min, excess fluid was removed and the grids were negatively stained with 10 μL of 2% uranyl acetate solution for 2 min. Finally, excess fluid was removed and the samples were viewed by a JEM electron microscope operating (Tokyo, Japan) at 80 kV.
Lentiviral Vector Production
For lentiviral expression of α-syn in primary neurons, the human α-syn gene was cloned into the lentiviral vector pLVX-IRES-ZsGreen1 to generate pLVX-IRES-ZsGreen1/α-syn. Lentiviral particles were generated by PEI-mediated co-transfection of HEK293T cells with pLVX-IRES-Green1/α-syn (or empty pLVX-IRES-Green1, as control), psPAX2 (Packaging plasmid), and pMD2G (Envelope plasmid). Virus (LV-α-syn) was collected after 72 hr, and titers up to 3–4 × 106 infectious units/mL were obtained.
Cell Culture, CADY-Mediated Aptamer Treatment, and Protein Overexpression
Human neuroblastoma SK-N-SH cells were cultured in 6-well plates with DMEM with 10% FBS, 50 U/mL penicillin, and 50 mg/mL streptomycin. All media were from GIBCO (VA). CADY peptide (Ac-GLWRALWRLLRSLWRLLWRA-cysteamide, 1 mg) was synthesized by Thermo Fisher Scientific (Waltham, MA) and dissolved into 10 μL dimethylsulfoxide (DMSO, Sigma, St. Louis, MO) rapidly, and then DNase-free water was added to get the CADY solution at 2 mg/mL (0.3 mM) with sonication for 10 min. To obtain CADY/aptamer complexes, naked aptamer (5 μmol/l stock solution) was mixed with CADY peptide (100 μM stock solution) and incubated for 37°C for 30 min. For aptamer delivery into SK-N-SH cells, the cells were overlaid with 200 μL preformed complexes (20 nM aptamer; the CADY/aptamer molar ratio at 40) for 5 min, and then 400 μL of DMEM was added. After incubation for 2 hr, 1 mL of medium containing 16% FBS was added to obtain a final FBS concentration of 10%. For α-syn overexpression, 3 μg pcDNA3.1 blank vector, pcDNA3.1-α-syn, pEGFP-N1 blank vector, or pEGFP-N1-α-syn were diluted in 1 mL of DMEM separately. Then, 9 μg polyethyleneimine (PEI; Polysciences, Warrington, PA) was added to the DNA solution. The PEI/DNA solution was incubated at 37°C for 30 min to allow the PEI and DNA to form a nanoparticle complex. The pre-prepared PEI/DNA solution was then added to the plate well, and SK-N-SH cells were grown in the transfection mixture for another 24 hr or 48 hr at 37°C and in 5% CO2.
Primary neuronal cultures were prepared from brains of C57BL/6 mouse embryos (days 14.5–15.5). All experiments were authorized by the Institutional Animal Care and Use Committee of the Capital Medical University of Science and Technology (approval No. AEEI-2016-057) and performed according to the NIH Guide for the Care and Use of Laboratory Animals. Briefly, dissociated neurons were plated onto poly-L-lysine (Sigma, St. Louis, MO) -coated coverslips or dishes at 20,000-40,000 cells/cm2 or 70,000–100,000 cells/cm2, respectively. The cells were cultured in Neurobasal medium (GIBCO) supplemented with l-glutamine (0.5 mM) and 50× B27 supplement (for a final concentration of 1× ; GIBCO). For aptamer delivery into primary neurons, after 3–4 days of cell culture, half of the medium was discarded and the CADY/aptamer complexes (aptamer at 10 nM and the CADY/aptamer molar ratio at 20) were added to the dishes. 2 hr later, the primary neurons were infected with lentivirus overexpressing α-syn (LV-α-syn) or control lentivirus.
Serum Nuclease Protection Assay
Naked aptamer (100 fmol) or the CADY/aptamer complexes formed as described earlier were incubated in DMEM containing 50% FBS at 37°C for 24 hr. Proteinase K (200 μg/mL) was added into the mixture and incubated for 1 hr at 37°C to stop the reaction and digestion of CADY peptide. The aptamer was extracted with phenol:chloroform and analyzed on a 1.5% agarose gel.
Biotinylated Aptamer Pull-Down Assay
SK-N-SH cells were pre-treated with CADY/biotinylated-aptamer complexes (aptamer at 100 nM and the CADY/aptamer molar ratio at 40; random DNA sequence, ATCGAGTG-N40-GCAGGATCCA, served as control) and then transfected with pcDNA3.1-α-syn for α-syn overexpression. After transfection for 18 hr, cells were washed three times with PBS and lysed (10 mmol/L Tris-HCl pH 7.5; 200 mmol/L NaCl; 5 mmol/L EDTA acid; 0.1% Triton X-100; mammalian protease inhibitors). Streptavidin magnetic beads (10 μL; Invitrogen, Carlsbad,CA) were incubated with 500 μg of protein lysate overnight at 4°C. The streptavidin beads were washed three times in BW buffer (5 mM Tris-HCl, 0.5 mM EDTA, 1M NaCl). Samples were resolved by using SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA). The membrane was blocked with 3% (w/v) milk, and an α-syn-specific mouse polyclonal antibody (BD Biosciences, Franklin Lakes, NJ) was used to detect α-syn. After three washing steps, an HRP-labeled secondary antibody (Takara, Japan) was used for colorimetric staining.
Isolation of Mitochondria
Mitochondria were isolated from SK-N-SH cells using the Mitochondria/Cytosol Fractionation Kit (Applygen Technologies, Beijing, China) as reported previously.45 Briefly, cells were digested with 0.25% trypsin and suspended the cells in Mito-Cyto extraction buffer and grinded 30–40 times on ice. The cell lysate was centrifuged at 800 g for 5 min at 4°C to pellet the nucleus and cell debris. The supernatant was collected and centrifuged at 800 g for 5 min at 4°C again, and the supernatant was collected and centrifuged at 12,000 g for 10 min at 4°C to pellet mitochondria.
Measurement of ROS and Δφm
Cells were incubated with 10 μM dichlorofluorescein diacetate (DCF-HA, Sigma, St. Louis, MO) in the dark at 37°C for 30 min then washed three times with PBS. Green fluorescence intensity was determined by high content analysis (Cellomics Array Scan Infinity; Thermo Scientific, Waltham, MA) at 488/530 nm. The Δφm was measured using JC-1 (Sigma, St. Louis, MO), a dual-emission membrane potential-sensitive probe that exists as a green fluorescent monomer at a low Δφm and shows red/orange fluorescence in the aggregate form at a higher Δφm. JC-1 (1.3 μg/mL) was added to cultured cells in plate wells after washing twice with PBS for 30 min at 37°C. The change of fluorescence at 488/530 nm was monitored by high-content screening, and the ratio of red/green fluorescence intensity was determined. The Δφm of primary neurons was measured by loading the tetramethyl rhodamine methyl ester (TMRM; 10 nM; Invitrogen, Carlsbad, CA) in the medium.
Measurement of Cell Viability
The viability of SK-N-H cells was measured by the MTT assay, which involved counting the relative number of live cells based on the conversion of MTT (Sigma, St. Louis, MO) to formazan crystals. The cytotoxicity was measured by lactate dehydrogenase (LDH) assay, which was performed using a cytotoxicity detection kit (Roche, Mannheim, Germany). A 100 μL aliquot of culture supernatant was used to determine LDH release, with 100 μL of preservation solution used as a blank to correct the optical density reading at 490 nm.
Western Blot
Cell or mitochondria pellet was resuspended in RIPA with protease inhibitor (PI) for 30 min on ice. After four freeze-thaw cycles, the samples were centrifuged at 12,000 g for 15 min at 4°C. Protein concentrations were determined using a bicinchoninic acid protein assay kit (Pierce Biotechnology, Rockford, IL). Proteins were resolved by 10% SDS-PAGE and transferred to a PVDF membrane, which was blocked and then incubated with anti-α-syn (BD Biosciences, Franklin Lakes, NJ), anti-β-actin (Beijing Guanxingyu, China), and anti-VDAC1 (Abcam, Cambridge, UK) for 2 hr at room temperature. After washing three times with TTBS (20 mM Tris-HCl, 500 mM NaCl, 0.05% Tween20, pH7.5), membranes were incubated with IRDy800 mouse secondary antibody (LI-COR, Lincoln, NE) at room temperature for 1 hr. Protein bands were detected using an Odyssey Infrared Imaging system (LI-COR, Lincoln, NE) and analyzed with ImageJ software (National Institutes of Health, Bethesda, MD).
Immunofluorescence and Confocal Microscopy
The coverslips containing probed cells were washed three times with DMEM, then images were acquired using a Leica TCS-SP5 confocal microscope (Leica Microsystems, Tokyo, Japan) at room temperature. Fluorescence Alexa594 and TMRM images were acquired using 590 nm excitation and 617 nm emission, and mitotracker red images were acquired using 543 nm excitation and 579 nm emission. The EGFP images were acquired using 488 nm excitation and 516 nm emission.
Gaussia Luciferase PCA
SK-N-SH cells were cultured in 96-well plates with opti-MEM (Invitrogen, Carlsbad, CA) with 5% FBS at 37°C. The cells with/without CADY/aptamer complex pretreatment were co-transfected with constructs of α-syn-hGLucN and α-syn-hGLucC. After transfection for 24 hr, cells were washed with PBS and replaced with opti-MEM. Luciferase activity from protein complementation was measured for live cells in an automated plate reader at 480 nm following the injection of the cell permeable substrate, coelenterazine (20 μM; Sigma, St. Louis, MO), with a signal integration time of 2 s.
Neurite Length Measurement
The Cellomics ArrayScan VTI HCS reader high-content imaging system (Thermo Fisher Scientific, Waltham, MA) was used for automated image acquisition and morphometric analyses as previously described.46 Image analysis was performed using the vHCS Scan software package with a manually optimized version of the Cellomics Neural Profiling Bioapplication for neurite outgrowth analysis. MAP2 protein-positive expression was analyzed in Target Activation Bioapplication. Output from high content image analysis included measurement of neurite outgrowth (neurites per neuron, neurite length per neuron). The single neurite length was determined by dividing the neurite length of the entire neuron by the number of neurites.
Author Contributions
J.Z. designed the experiments and wrote the paper; Yuan Zheng, J.Q., F.X., and Yan Zheng conducted experiments and analyzed the data; B.Y., Y.C., and H.Y. made constructive suggestions for this experiment and interpreted the data.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31571202, 31271136, and 81371398), the Importation and Development of High-Caliber Talents Project of Beijing Municipal Institutions (CIT&TCD201504087), the Clinic-Basic Fund of Capital Medical University (17JL34), and the Project of Construction of Innovative Teams and Teacher Career Development for Universities and Colleges Under Beijing Municipality (IDHT20140514).
Footnotes
Supplemental Information includes six figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.02.011.
Supplemental Information
References
- 1.Jellinger K.A. Neuropathological spectrum of synucleinopathies. Mov. Disord. 2003;18(Suppl 6):S2–S12. doi: 10.1002/mds.10557. [DOI] [PubMed] [Google Scholar]
- 2.Iwai A., Masliah E., Yoshimoto M., Ge N., Flanagan L., de Silva H.A., Kittel A., Saitoh T. The precursor protein of non-A beta component of Alzheimer’s disease amyloid is a presynaptic protein of the central nervous system. Neuron. 1995;14:467–475. doi: 10.1016/0896-6273(95)90302-x. [DOI] [PubMed] [Google Scholar]
- 3.Wood S.J., Wypych J., Steavenson S., Louis J.C., Citron M., Biere A.L. alpha-synuclein fibrillogenesis is nucleation-dependent. Implications for the pathogenesis of Parkinson’s disease. J. Biol. Chem. 1999;274:19509–19512. doi: 10.1074/jbc.274.28.19509. [DOI] [PubMed] [Google Scholar]
- 4.Bousset L., Pieri L., Ruiz-Arlandis G., Gath J., Jensen P.H., Habenstein B., Madiona K., Olieric V., Böckmann A., Meier B.H., Melki R. Structural and functional characterization of two alpha-synuclein strains. Nat. Commun. 2013;4:2575. doi: 10.1038/ncomms3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J.Y., Englund E., Holton J.L., Soulet D., Hagell P., Lees A.J., Lashley T., Quinn N.P., Rehncrona S., Björklund A. Lewy bodies in grafted neurons in subjects with Parkinson’s disease suggest host-to-graft disease propagation. Nat. Med. 2008;14:501–503. doi: 10.1038/nm1746. [DOI] [PubMed] [Google Scholar]
- 6.Tran H.T., Chung C.H., Iba M., Zhang B., Trojanowski J.Q., Luk K.C., Lee V.M. A-synuclein immunotherapy blocks uptake and templated propagation of misfolded α-synuclein and neurodegeneration. Cell Rep. 2014;7:2054–2065. doi: 10.1016/j.celrep.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergström A.L., Kallunki P., Fog K. Development of Passive Immunotherapies for Synucleinopathies. Mov. Disord. 2016;31:203–213. doi: 10.1002/mds.26481. [DOI] [PubMed] [Google Scholar]
- 8.Valera E., Spencer B., Masliah E. Immunotherapeutic Approaches Targeting Amyloid-β, α-Synuclein, and Tau for the Treatment of Neurodegenerative Disorders. Neurotherapeutics. 2016;13:179–189. doi: 10.1007/s13311-015-0397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frydman-Marom A., Shaltiel-Karyo R., Moshe S., Gazit E. The generic amyloid formation inhibition effect of a designed small aromatic β-breaking peptide. Amyloid. 2011;18:119–127. doi: 10.3109/13506129.2011.582902. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi M., Suzuki M., Fukuoka M., Fujikake N., Watanabe S., Murata M., Wada K., Nagai Y., Hohjoh H. Normalization of Overexpressed α-Synuclein Causing Parkinson’s Disease By a Moderate Gene Silencing With RNA Interference. Mol. Ther. Nucleic Acids. 2015;4:e241. doi: 10.1038/mtna.2015.14. [DOI] [PubMed] [Google Scholar]
- 11.He Q., Koprich J.B., Wang Y., Yu W.B., Xiao B.G., Brotchie J.M., Wang J. Treatment with Trehalose Prevents Behavioral and Neurochemical Deficits Produced in an AAV α-Synuclein Rat Model of Parkinson’s Disease. Mol. Neurobiol. 2016;53:2258–2268. doi: 10.1007/s12035-015-9173-7. [DOI] [PubMed] [Google Scholar]
- 12.Bae E.J., Lee H.J., Rockenstein E., Ho D.H., Park E.B., Yang N.Y., Desplats P., Masliah E., Lee S.J. Antibody-aided clearance of extracellular α-synuclein prevents cell-to-cell aggregate transmission. J. Neurosci. 2012;32:13454–13469. doi: 10.1523/JNEUROSCI.1292-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Games D., Valera E., Spencer B., Rockenstein E., Mante M., Adame A., Patrick C., Ubhi K., Nuber S., Sacayon P. Reducing C-terminal-truncated alpha-synuclein by immunotherapy attenuates neurodegeneration and propagation in Parkinson’s disease-like models. J. Neurosci. 2014;34:9441–9454. doi: 10.1523/JNEUROSCI.5314-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masliah E., Rockenstein E., Mante M., Crews L., Spencer B., Adame A., Patrick C., Trejo M., Ubhi K., Rohn T.T. Passive immunization reduces behavioral and neuropathological deficits in an alpha-synuclein transgenic model of Lewy body disease. PLoS ONE. 2011;6:e19338. doi: 10.1371/journal.pone.0019338. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Qu J., Yu S., Zheng Y., Zheng Y., Yang H., Zhang J. Aptamer and its applications in neurodegenerative diseases. Cell. Mol. Life Sci. 2017;74:683–695. doi: 10.1007/s00018-016-2345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 17.Rhie A., Kirby L., Sayer N., Wellesley R., Disterer P., Sylvester I., Gill A., Hope J., James W., Tahiri-Alaoui A. Characterization of 2′-fluoro-RNA aptamers that bind preferentially to disease-associated conformations of prion protein and inhibit conversion. J. Biol. Chem. 2003;278:39697–39705. doi: 10.1074/jbc.M305297200. [DOI] [PubMed] [Google Scholar]
- 18.Bunka D.H., Stockley P.G. Aptamers come of age - at last. Nat. Rev. Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 19.Liang H., Shi Y., Kou Z., Peng Y., Chen W., Li X., Li S., Wang Y., Wang F., Zhang X. Inhibition of BACE1 Activity by a DNA Aptamer in an Alzheimer’s Disease Cell Model. PLoS ONE. 2015;10:e0140733. doi: 10.1371/journal.pone.0140733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahimi F., Bitan G. Selection of aptamers for amyloid beta-protein, the causative agent of Alzheimer’s disease. J. Vis. Exp. 2010;(39):1955. doi: 10.3791/1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsukakoshi K., Harada R., Sode K., Ikebukuro K. Screening of DNA aptamer which binds to alpha-synuclein. Biotechnol. Lett. 2010;32:643–648. doi: 10.1007/s10529-010-0200-5. [DOI] [PubMed] [Google Scholar]
- 22.Tsukakoshi K., Abe K., Sode K., Ikebukuro K. Selection of DNA aptamers that recognize α-synuclein oligomers using a competitive screening method. Anal. Chem. 2012;84:5542–5547. doi: 10.1021/ac300330g. [DOI] [PubMed] [Google Scholar]
- 23.Näsström T., Gonçalves S., Sahlin C., Nordström E., Screpanti Sundquist V., Lannfelt L., Bergström J., Outeiro T.F., Ingelsson M. Antibodies against alpha-synuclein reduce oligomerization in living cells. PLoS ONE. 2011;6:e27230. doi: 10.1371/journal.pone.0027230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crombez L., Aldrian-Herrada G., Konate K., Nguyen Q.N., McMaster G.K., Brasseur R., Heitz F., Divita G. A new potent secondary amphipathic cell-penetrating peptide for siRNA delivery into mammalian cells. Mol Ther. 2009;17:95–103. doi: 10.1038/mt.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Outeiro T.F., Putcha P., Tetzlaff J.E., Spoelgen R., Koker M., Carvalho F., Hyman B.T., McLean P.J. Formation of toxic oligomeric alpha-synuclein species in living cells. PLoS ONE. 2008;3:e1867. doi: 10.1371/journal.pone.0001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putcha P., Danzer K.M., Kranich L.R., Scott A., Silinski M., Mabbett S., Hicks C.D., Veal J.M., Steed P.M., Hyman B.T., McLean P.J. Brain-permeable small-molecule inhibitors of Hsp90 prevent alpha-synuclein oligomer formation and rescue alpha-synuclein-induced toxicity. J. Pharmacol. Exp. Ther. 2010;332:849–857. doi: 10.1124/jpet.109.158436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danzer K.M., Ruf W.P., Putcha P., Joyner D., Hashimoto T., Glabe C., Hyman B.T., McLean P.J. Heat-shock protein 70 modulates toxic extracellular α-synuclein oligomers and rescues trans-synaptic toxicity. FASEB J. 2011;25:326–336. doi: 10.1096/fj.10-164624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parihar M.S., Parihar A., Fujita M., Hashimoto M., Ghafourifar P. Mitochondrial association of alpha-synuclein causes oxidative stress. Cell. Mol. Life Sci. 2008;65:1272–1284. doi: 10.1007/s00018-008-7589-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parihar M.S., Parihar A., Fujita M., Hashimoto M., Ghafourifar P. Alpha-synuclein overexpression and aggregation exacerbates impairment of mitochondrial functions by augmenting oxidative stress in human neuroblastoma cells. Int. J. Biochem. Cell Biol. 2009;41:2015–2024. doi: 10.1016/j.biocel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Mahlknecht G., Maron R., Mancini M., Schechter B., Sela M., Yarden Y. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. Proc. Natl. Acad. Sci. USA. 2013;110:8170–8175. doi: 10.1073/pnas.1302594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takenouchi T., Hashimoto M., Hsu L.J., Mackowski B., Rockenstein E., Mallory M., Masliah E. Reduced neuritic outgrowth and cell adhesion in neuronal cells transfected with human alpha-synuclein. Mol. Cell. Neurosci. 2001;17:141–150. doi: 10.1006/mcne.2000.0923. [DOI] [PubMed] [Google Scholar]
- 32.Koch J.C., Bitow F., Haack J., d’Hedouville Z., Zhang J.N., Tönges L., Michel U., Oliveira L.M., Jovin T.M., Liman J. Alpha-Synuclein affects neurite morphology, autophagy, vesicle transport and axonal degeneration in CNS neurons. Cell Death Dis. 2015;6:e1811. doi: 10.1038/cddis.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClendon S., Rospigliosi C.C., Eliezer D. Charge neutralization and collapse of the C-terminal tail of alpha-synuclein at low pH. Protein Sci. 2009;18:1531–1540. doi: 10.1002/pro.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung Y.H., Eliezer D. Residual structure, backbone dynamics, and interactions within the synuclein family. J. Mol. Biol. 2007;372:689–707. doi: 10.1016/j.jmb.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paleologou K.E., Schmid A.W., Rospigliosi C.C., Kim H.Y., Lamberto G.R., Fredenburg R.A., Lansbury P.T., Jr., Fernandez C.O., Eliezer D., Zweckstetter M., Lashuel H.A. Phosphorylation at Ser-129 but not the phosphomimics S129E/D inhibits the fibrillation of alpha-synuclein. J. Biol. Chem. 2008;283:16895–16905. doi: 10.1074/jbc.M800747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rospigliosi C.C., McClendon S., Schmid A.W., Ramlall T.F., Barré P., Lashuel H.A., Eliezer D. E46K Parkinson’s-linked mutation enhances C-terminal-to-N-terminal contacts in alpha-synuclein. J. Mol. Biol. 2009;388:1022–1032. doi: 10.1016/j.jmb.2009.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Fougerolles A., Vornlocher H.P., Maraganore J., Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akhtar S., Benter I.F. Nonviral delivery of synthetic siRNAs in vivo. J. Clin. Invest. 2007;117:3623–3632. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris M.C., Gros E., Aldrian-Herrada G., Choob M., Archdeacon J., Heitz F., Divita G. A non-covalent peptide-based carrier for in vivo delivery of DNA mimics. Nucleic Acids Res. 2007;35:e49. doi: 10.1093/nar/gkm053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu K.P., Weinstock D.S., Narayanan C., Levy R.M., Baum J. Structural reorganization of alpha-synuclein at low pH observed by NMR and REMD simulations. J. Mol. Biol. 2009;391:784–796. doi: 10.1016/j.jmb.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norris K.L., Hao R., Chen L.F., Lai C.H., Kapur M., Shaughnessy P.J., Chou D., Yan J., Taylor J.P., Engelender S. Convergence of Parkin, PINK1, and α-Synuclein on Stress-induced Mitochondrial Morphological Remodeling. J. Biol. Chem. 2015;290:13862–13874. doi: 10.1074/jbc.M114.634063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durcan T.M., Fon E.A. The three 'P’s of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev. 2015;29:989–999. doi: 10.1101/gad.262758.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shum K.T., Chan C., Leung C.M., Tanner J.A. Identification of a DNA aptamer that inhibits sclerostin’s antagonistic effect on Wnt signalling. Biochem. J. 2011;434:493–501. doi: 10.1042/BJ20101096. [DOI] [PubMed] [Google Scholar]
- 44.Luk K.C., Song C., O’Brien P., Stieber A., Branch J.R., Brunden K.R., Trojanowski J.Q., Lee V.M. Exogenous alpha-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. Proc. Natl. Acad. Sci. USA. 2009;106:20051–20056. doi: 10.1073/pnas.0908005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X.D., Sun H., Liu G.T. 5-Bromotetrandrine enhances the sensitivity of doxorubicin-induced apoptosis in intrinsic resistant human hepatic cancer Bel7402 cells. Cancer Lett. 2010;292:24–31. doi: 10.1016/j.canlet.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Harrill J.A., Freudenrich T.M., Machacek D.W., Stice S.L., Mundy W.R. Quantitative assessment of neurite outgrowth in human embryonic stem cell-derived hN2 cells using automated high-content image analysis. Neurotoxicology. 2010;31:277–290. doi: 10.1016/j.neuro.2010.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.