Abstract
In recent years, synthetic mRNA-based applications to produce desired exogenous proteins in cells have been gaining importance. However, systemic delivery of synthetic mRNA can result in unspecific uptake into undesired cells or organs and, thereby, fail to target desired cells. Thus, local and targeted delivery of synthetic mRNA becomes increasingly important to reach the desired cell types and tissues. In this study, intradermal delivery of synthetic mRNA using a hollow microneedle injection-based method was evaluated. Furthermore, an ex vivo porcine skin model was established to analyze synthetic mRNA-mediated protein expression in the skin following intradermal delivery. Using this model, highly efficient delivery of synthetic mRNA was demonstrated, which resulted in detection of high levels of secretable humanized Gaussia luciferase (hGLuc) protein encoded by the microinjected synthetic mRNA. Interestingly, synthetic mRNA injected without transfection reagent was also able to enter the cells and resulted in protein expression. The established ex vivo porcine skin model can be used to evaluate the successful production of desired proteins after intradermal delivery of synthetic mRNAs before starting with in vivo experiments. Furthermore, the use of microneedles enables patient-friendly, painless, and efficient delivery of synthetic mRNAs into the dermis; thus, this method could be applied for local treatment of different skin diseases as well as for vaccination and immunotherapy.
Keywords: synthetic mRNA, microneedles, intradermal, delivery, protein expression
Introduction
Over the past years, interest in synthetic mRNA-based therapeutics has strongly increased in various fields, such as regenerative medicine and the prevention as well as the treatment of diseases.1 Synthetic mRNAs are applied as immunotherapeutic agents to treat cancer;2, 3 as vaccines (e.g., against HIV4 or influenza virus5 infections); for protein replacement therapies to treat asthma,6 myocardial infarction,7 or single-gene disorders;8 and for prevention of angioplasty-associated complications.9 Recently, Stadler et al.10 even succeeded in elimination of advanced tumors in mice via injection of synthetic mRNA encoding bispecific antibodies.
Several studies demonstrated that chemical modifications of synthetic mRNAs lead to reduced immunogenicity and increased stability and, thereby, improve the treatment success of synthetic mRNA-based approaches.11, 12, 13, 14 The replacement of uridine and cytidine with modified nucleosides such as pseudouridine and 5-methylcytidine resulted in highly reduced Toll-like receptor-dependent immune activation,14 and incorporation of pseudouridine enhanced the translation efficiency by diminishing RNA-dependent protein kinase (PKR) activation13. However, in recent years, further studies demonstrated that unmodified mRNAs can also be administered successfully. Thess et al.15 used sequence-engineered Epo mRNA without chemical nucleoside modifications and achieved high protein expression levels after administration in mice, cynomolgus monkeys, and pigs. Recently, Kauffman et al.16 investigated the efficacy and immunogenicity of systemically delivered lipid nanoparticles (LNPs) containing pseudouridine-modified mRNA and unmodified mRNA without codon optimization in mice. They demonstrated that pseudouridine modification of Epo mRNA did not improve the in vitro and in vivo efficacy of the mRNA-LNPs and that the unmodified mRNA had an equal immunogenicity as pseudouridine-modified mRNA. Thus, they concluded that pseudouridine modifications might be unnecessary for therapeutic applications of mRNA in the liver. These diverse outcomes in recent studies indicate that different mRNA modifications could be used for synthesis of the same therapeutic protein. In addition to mRNA modifications and purification methods, the delivery vehicle used, targeted cells and tissues, and the administration methods can result in different therapeutic efficiencies. Furthermore, the immunogenicity of generated mRNAs should be analyzed properly to select the best-suited synthetic mRNA for the desired applications.17
Because of numerous advantages compared with the classical exogenous gene delivery platforms (e.g., viral vectors or DNA plasmids), synthetic RNA-based gene delivery applications are increasingly used in the field of gene therapy and vaccination.18, 19, 20, 21 In contrast to plasmid DNA or viral vectors, synthetic mRNAs do not need to enter the cell nucleus; thus, they can be immediately translated into proteins after entering the cytosol. Consequently, no integration into the host genome occurs, which highly reduces the risk of insertional mutations. Furthermore, in non-dividing cells with an intact nuclear envelope, the transport of plasmids and viral vectors into the nucleus is strongly hindered22 and, thereby, gene delivery in post-mitotic cells is hampered, which may explain their low potency in humans.23 Viral vectors are also hindered by potential antivector immunity. Thus, in addition to transfection of mitotic cells, synthetic RNAs offer the possibility to transfect slowly or non-dividing cells.24, 25 Moreover, due to physiological degradation, exogenously delivered synthetic mRNA is transiently present in cells; thus, continuous protein overexpression-related complications are prevented.
For successful applications, a sufficient amount of synthetic mRNA needs to be delivered into target cells to efficiently express the desired protein. Cationic polymers26 or lipids27 can be used to encapsulate the negatively charged mRNA and to create an envelope protecting the synthetic mRNA against nucleases and facilitating sufficient uptake into the cells. Besides lipid- or polymer-based transport vesicles, physical methods like electroporation28 and ultrasound29 are also used to deliver nucleic acids into cells.
The in vivo application and delivery of mRNA involves further challenges, including the targeting of specific cell types and organs. Synthetic mRNA can be delivered systemically via the bloodstream;30 however, cells that cannot be reached by the systemic route need to be targeted by local administration, such as intramuscular injection or aerosol inhalation.31 In a previous study, the intramyocardial injection of modified vascular endothelial growth factor-A (VEGF-A)-encoding mRNA resulted in vascular regeneration after myocardial infarction.7 Pardi et al.32 have recently shown that intradermal injection of lipid nanoparticle-encapsulated modified mRNA leads to sufficient immunization, protecting mice and non-human primates against the Zika virus.
Intradermal application of nucleic acids is a promising strategy to treat several skin diseases or to improve wound healing by expression of desired proteins in the skin. Thereby, issues related to systemically administered nucleic acids, such as clearance from the bloodstream via the spleen, renal, and hepatic systems,33 can be eliminated. However, the stratum corneum, the outermost layer of the skin, serves as a barrier and impedes the entry of topically applied drugs.34 In the past, different strategies were developed to overcome the skin barrier to deliver nucleic acids into the skin. These include physical (e.g., microporation, microneedles, and jet injection35), active (e.g., electroporation, iontophoresis, and sonophoresis), and passive methods (e.g., nanoparticles and liposomes).36 Electroporation and sonoporation transiently permeabilize the skin using either electric pulses or low-frequency ultrasound to efficiently deliver genes into the skin.37, 38
Successful intradermal delivery of synthetic mRNAs for the production of desired therapeutic proteins has enormous potential in the field of medicine. Thus, in this study, we investigated the exogenous delivery of synthetic mRNA into the skin by using hollow microneedles. Synthetic mRNA encoding secretable humanized Gaussia luciferase (hGLuc) was synthetized by in vitro transcription (IVT). Furthermore, an ex vivo porcine skin model was established to evaluate synthetic mRNA-mediated protein expression in the skin. Using this model, the intradermal delivery of synthetic mRNA, the transfectability of skin cells, and the ability to produce hGLuc in the skin were analyzed.
Results
Synthesis of hGLuc-Encoding mRNA and Labeling with Cy3
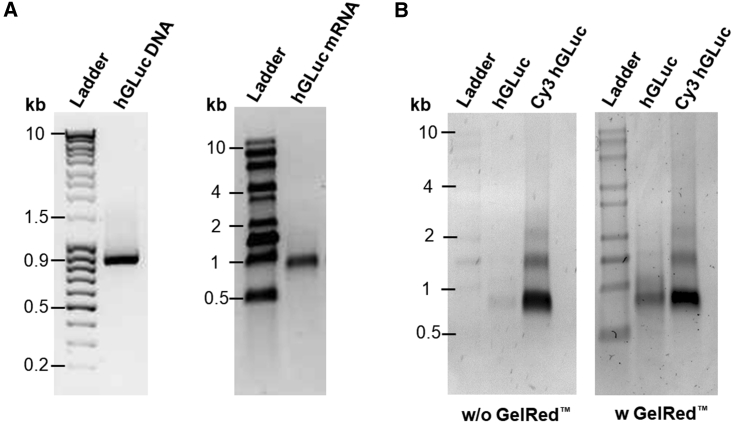
After IVT, hGLuc-encoding mRNA with a length of approximately 900 bases was obtained (Figure 1A). Additionally, to enable the detection of synthetic mRNA after intradermal delivery, hGLuc mRNA was labeled with Cy3 using Cu(I)-free azide-dibenzylcyclooctyne (DBCO) click chemistry, whereby Cy3 molecules are conjugated after IVT to the incorporated 5-azido-C3-uridine triphosphate (UTP) of synthetic mRNA. In Figure 1B, Cy3-labeled mRNA bands can be clearly seen using an UV transilluminator. The strongest fluorescence signal was detected at approximately 900 bases length. However, 2 additional weak bands at 1.5 and 2 kb can also be seen. The reason for several Cy3-labeled bands could be the amount of conjugated Cy3 molecules. Higher numbers of Cy3 molecules per mRNA can lead to a higher molecular weight. mRNA without Cy3 labeling could be clearly detected after GelRed staining, and it was at the same height as the Cy3-labeled hGLuc mRNA with the highest intensity.
Figure 1.
Analysis of the Generated PCR Product, Synthetic hGLuc mRNA, and Cy3-Labeled mRNA, by 1% Agarose Gel Electrophoresis
(A) 200 ng hGLuc DNA (left) or hGLuc mRNA (right) was loaded on 1% agarose gel. The purity and the specific length of the PCR product and synthetic mRNA were confirmed by a single band at around 900 bases. (B) First, Cy3-labeled mRNA was detected using an UV transilluminator, and then all nucleic acids were detected by GelRed staining.
Transfection of HEK293 Cells with Synthetic hGLuc mRNA Leads to High Expression Levels of hGLuc
To determine hGLuc expression after exogenous delivery of synthetic hGLuc mRNA into cells, HEK293 cells were transfected with 0.2, 0.5, or 1.5 μg of synthetic hGLuc mRNA complexed with Lipofectamine 2000. Cells transfected with either medium or medium and Lipofectamine 2000 served as controls. Luciferase activity (relative light units [RLUs]) was determined via luciferase assay in supernatants as well as in cell lysates (Figure 2).
Figure 2.
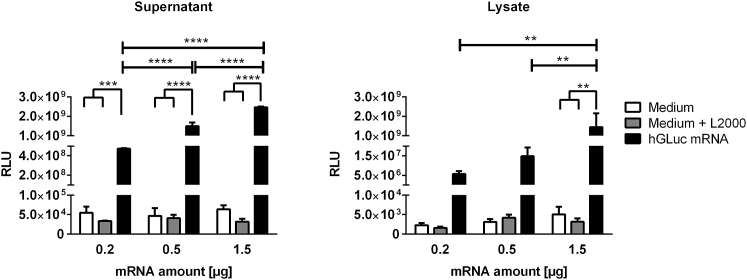
Detection of Luciferase Activity after Transfection of HEK923 Cells with Synthetic hGLuc mRNA
3 × 105 HEK293 cells were incubated for 4 hr at 37°C with 0.2, 0.5, or 1.5 μg mRNA complexed with Lipofectamine 2000. Then transfection complexes were discarded, and fresh medium was added to the cells. After 24 hr, the luciferase activity (RLUs) was detected in the supernatants and cell lysates. Cells treated with medium or medium and Lipofectamine 2000 (L2000) served as negative controls. Results are shown as mean ± SEM (n = 3). Statistical differences were determined using two-way ANOVA followed by Bonferroni’s multiple comparisons test (**p < 0.01, ***p < 0.001, ****p < 0.0001).
After transfection of HEK293 cells with 0.2, 0.5, or 1.5 μg synthetic hGLuc mRNA, significantly higher hGLuc expression was detected in supernatants compared with the supernatants of control groups (medium or medium and Lipofectamine 2000). Transfection of HEK293 cells with 0.2 μg hGLuc mRNA already resulted in significantly high expression of hGLuc, and the increase in synthetic hGLuc mRNA led to a significant increase in luciferase activity in the supernatants. In contrast, in cell lysate samples, only transfection of cells with 1.5 μg mRNA resulted in significantly higher luciferase activity compared with the control samples. Because the produced hGLuc is secretable, detection of higher hGLuc activity in the supernatant compared with the cell lysates is reasonable and demonstrates successful secretion of the protein from cells.
Establishment of a Porcine Skin Model for the Analysis of Synthetic mRNA-Mediated Protein Expression
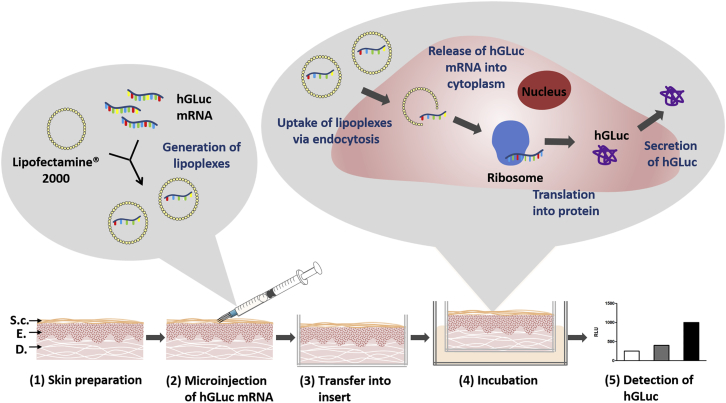
An ex vivo porcine skin model was established for the analysis of synthetic mRNA-based exogenous protein expression in the skin. Figure 3 shows the delivery strategy of synthetic mRNA into porcine skin using hollow microneedles and preparation of the skin for the analysis of protein expression. First, porcine skin was trimmed and punched into 1-mm-thick pieces with a 15-mm diameter. Microinjection of the complexed mRNA was performed using MicronJet600, a microneedle with three pyramid-shaped needles with a length of 600 μm. After the injection, a visible small bubble formation was observed, which showed that the depo remained in the dermis without any leakage on the other side of the skin samples after the injection. Skin samples were then washed with Dulbecco’s PBS (DPBS) and transferred into a ThinCert, which served as a permeable barrier between the skin and the medium underneath. The skin surface was exposed to air. Using the established porcine skin model, the skin could be maintained for up to 3 days without contamination. During this period, synthetic mRNA-mediated protein expression in the skin can be analyzed.
Figure 3.
Delivery of Synthetic hGLuc mRNA into Porcine Skin Using Hollow Microneedles and Analysis of Protein Expression
(1) Porcine skin detached from the outer side of pigs’ ears was trimmed and punched into 1-mm-thick pieces with 1.5-cm diameter and disinfected. The structure of the skin is shown schematically. S.c., stratum corneum; E, epidermis; D, dermis. (2) Lipoplexes were generated by incubation of 1.5 μg hGLuc mRNA with 1.5 μl Lipofectamine 2000 in a total volume of 35 μL OptiMEM I reduced serum-free medium for 20 min at RT. The mixture was injected into the skin using MicronJet600 microneedles. (3) After washing with DPBS, the skin was transferred into a ThinCert insert, which served as a permeable barrier between the skin and the surrounding medium. (4) The skin was incubated air-exposed in 1.5 mL human endothelial cell culture medium in one well of a 12-well plate from 24 to 72 hr at 37°C and 5% CO2. The microinjected lipoplexes can enter the cells via endocytosis. After the release of mRNA into the cytosol, mRNA is translated by ribosomes into protein. (5) After the appropriate incubation time, the produced hGLuc protein is detected by luciferase assay in the surrounding medium as well as in the skin.
Injected Synthetic hGLuc mRNA Reaches the Dermis
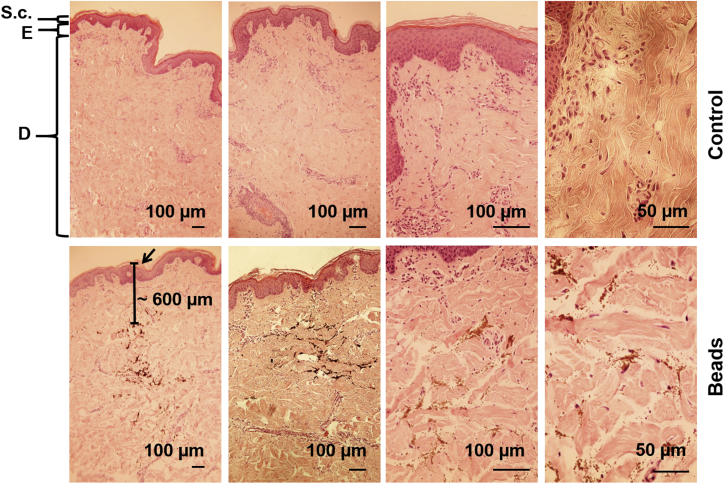
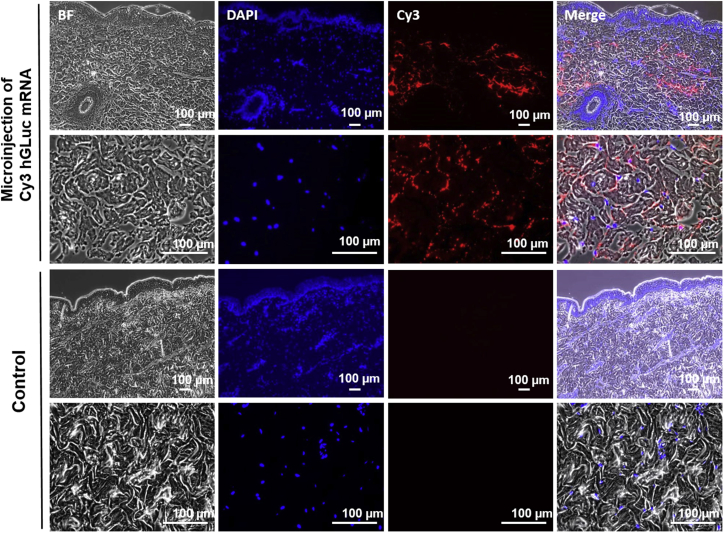
To analyze the reached injection depth by using the MicronJet600, magnetic microspheres (Figure 4) or fluorescently labeled hGLuc mRNA (Figure 5) were injected into porcine skin samples. Using magnetic microspheres, the injection depth can be determined because, in contrast to Cy3-labeled hGLuc mRNA, they do not diffuse in the tissue immediately after microinjection. The microinjection was performed with 1 μl magnetic microspheres (1-μm diameter) or lipoplexes containing 1.5 μg Cy3-labeled hGLuc mRNA in a total injection volume of 35 μl. H&E-stained sections of skin pieces showed that the microspheres reached the dermis at around 600 μm (Figure 4). The injection site is indicated by an arrow. Using fluorescence microscopy, the distribution of Cy3-labeled hGLuc mRNA was analyzed in cross-sections (Figure 5). The nuclei of the cells were stained with DAPI, whereby a homogeneous distribution of the microinjected Cy3-labeled mRNA in the dermis was detected. Furthermore, no Cy3 hGLuc mRNA could be detected in the epidermis.
Figure 4.
Analysis of Injection Depth after Delivery of Metallic Microspheres into Pig Skin
Paraffin-embedded porcine skin sections microinjected with metallic microspheres were stained with H&E. Non-injected skin pieces served as controls. The upper layer of the skin, the stratum corneum (S.c.) (20–30 μm) forms a protective barrier for the underlying epidermis (E) (50–200 μm) and the following dermis (D). The arrow indicates the injection site of the beads. The injected beads were detected approximately 600 μm under the skin surface. Nuclei, blue-purple; cytoplasm, red; collagen, light pink; erythrocytes, cherry red; microspheres, brown.
Figure 5.
Detection of Microinjected Cy3-Labeled hGLuc mRNA in Porcine Skin
Samples were fixed 4 hr after microinjection of 1.5 μg Cy3-labeled hGLuc mRNA in 4% paraformaldehyde. Paraffin-embedded tissues were sectioned at a thickness of 2.5 μm and stained with DAPI. Representative pictures of 3 experiments are presented. BF, bright field; DAPI, blue; Cy3 hGLuc mRNA, red.
Detection of Luciferase Activity after Microinjection of Synthetic hGLuc mRNA into a Porcine Ex Vivo Skin Model
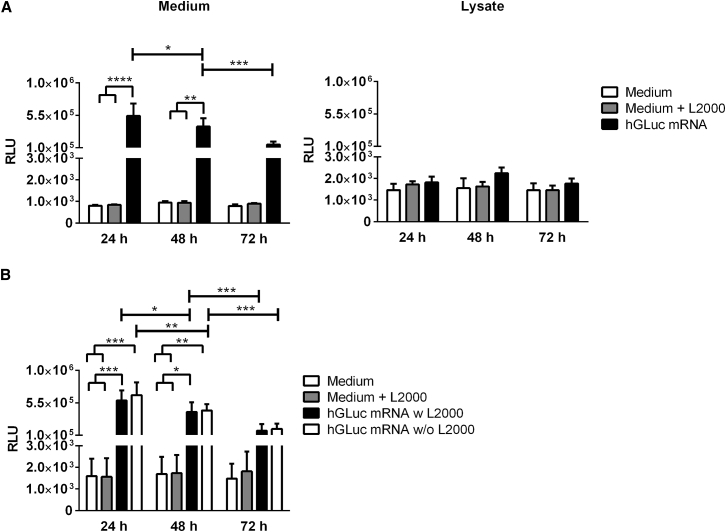
After establishing the ex vivo porcine skin model, 1.5 μg hGLuc mRNA was delivered without and with complexation with Lipofectamine 2000 by using a microinjection technique into the skin. First, the expression of hGLuc in the medium (under the ThinCert) and skin lysates was measured 24, 48, and 72 hr after microinjection of hGLuc mRNA complexed with Lipofectamine 2000 (Figure 6A). After 24 hr, the highest luciferase activity was measured in the medium because of the release of hGLuc from the basolateral side into the medium. Production of hGLuc continued over 3 days. However, a significant decrease of hGLuc was detected after each day, which can be explained by the transient presence of synthetic mRNA in the cells. In comparison, the luciferase activity in the skin lysates was not significantly different from the negative controls, which showed that the produced hGLuc was rapidly secreted into the extracellular space.
Figure 6.
Detection of Luciferase Activity after Microinjection of Synthetic hGLuc mRNA into Porcine Skin
(A) Luciferase activity was determined in medium (n = 4) and skin lysates (n = 5) 24, 48, and 72 hr after microinjection of 1.5 μg hGLuc mRNA complexed with Lipofectamine 2000 (L200) (B) Additionally, 1.5 μg hGLuc mRNA was injected without Lipofectamine 2000 complexation, referred to as naked mRNA, into the skin (n = 3). The medium was collected, and fresh medium was added every 24 hr. Luciferase activity was determined in the collected medium. Skin pieces injected with only medium or medium plus Lipofectamine 2000 served as controls. Results are presented as mean ± SEM. Statistical differences were determined using two-way ANOVA followed by Bonferroni’s multiple comparisons test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Surprisingly, microinjection of non-complexed hGLuc mRNA, “naked mRNA,” into porcine skin showed similar luciferase activity levels as after delivery of Lipofectamine 2000-complexed hGLuc mRNA (Figure 6B). After 24 and 48 hr, microinjection of naked hGLuc mRNA resulted in significantly increased luciferase activity in the medium compared with controls without hGLuc mRNA delivery. Luciferase activity was significantly decreased after each day of incubation.
Discussion
Systemic administration of nucleic acid-based therapeutic agents leads to distribution of the drug throughout the body. This can result in non-specific uptake of nucleic acids by non-target tissues and organs. Furthermore, nucleic acids are susceptible to nucleases, and they can be rapidly cleared from the body via the spleen, renal, and hepatic systems.39 Thus, stabilization of nucleic acids against nucleases, reduction of uptake by non-specific tissues, targeting of desired cells, and reduction of excretion are required to obtain the optimal in vivo effect. To overcome these hurdles, appropriate drug delivery systems and application routes can be used. Thus, local administration of nucleic acid-based therapeutic agents at the site of the pathology is a promising approach to increase site-specific delivery, efficiency, and specificity.
In the case of synthetic mRNA-based applications, protection of synthetic mRNA from ubiquitous RNases and delivery of mRNA in a sufficient amount to the target tissue are also important to achieve therapeutically effective protein production. Therefore, various nanoparticles based on, e.g., liposomes or polymers preventing the degradation of nucleic acids have been established in recent years.40, 41 However, after systemic delivery, primary sites of accumulation of these nanoparticles are tumors, the liver, and the spleen.42 Thus, delivery of nucleic acids directly into the skin has several advantages compared with systemic delivery, such as localized treatment and prevention of rapid elimination. However, the strong barrier function of the stratum corneum prevents penetration of macromolecules, such as synthetic mRNAs, which are significantly larger than the low-molecular-weight molecules (molecular weight [MW] < 500 Da),43 which are capable of penetrating into the skin. Thus, the skin barrier must be circumvented to enable successful gene delivery. Therefore, various methods were applied to reach this aim, such as subcutaneous injection, electroporation, liquid jet injection, and gene gun delivery.43, 44
Efficient delivery of nucleic acids using electroporation was shown in several studies, mainly using plasmid DNA, whereby higher expression levels of desired proteins were detected compared with the sole direct injection of plasmid DNA into animals’ skin.37, 45, 46 However, physical methods like electroporation and ultrasound need special equipment, which impedes autonomous use by patients, and it is associated with high costs and limited usability. Furthermore, some of these systems require proprietary containers and filling systems, which might be another impediment.
Among injections, microneedles are increasingly being used for drug delivery.47 Different types of microneedles and microneedle arrays (MNAs) were established in the past, which differ in material composition and function. Because of their short length, microneedles do not reach the nerves and blood vessels embedded in the dermis, guarantee pain-free application, and highly improve patient compliance compared with the commonly used subcutaneous injection needles.33, 43, 47 MNAs can be used to generate microholes across the skin to enhance the uptake of topically applied drug formulations. Furthermore, coated or dissolvable MNAs containing the drug or hollow microneedles for infusion of liquid formulations can be applied.43 In this study, we used, for the first time, the hollow microneedle device MicronJet600 from NanoPass Technologies for delivery of synthetic mRNA into the skin and analyzed synthetic mRNA-mediated protein expression by establishing an ex vivo porcine skin model. The microneedle device MicronJet600 was used in dozens of clinical trials, among others, for improving vaccination.48, 49, 50 These studies resulted in not only an almost painless injection but, more importantly, in significant (typically x5-x10) dose sparing and even superior immunogenicity in vaccination against various infectious diseases, including seasonal51, 52 and pandemic53 influenza, herpes zoster,54 and polio.55, 56 Furthermore, application of these microneedles can warrant reliable intradermal delivery of mRNA and targeted localization of synthetic mRNA in the dermis, which are difficult to achieve using standard needles.57
A very interesting study by Li et al.58 demonstrated selective targeting of plasmid lacZ DNA to hair follicles in mice after topical application of lacZ DNA-containing liposomes. In contrast, topical application of the naked lacZ DNA plasmid did not result in gene transfer. Moreover, no cells other than follicles in the dermis and epidermis were transfected. These results indicate that the use of appropriate delivery vehicles can also enable specific targeting of hair follicles after topical application of nucleic acid-based therapeutic agents. In a later study, the same research group developed an efficient technique for ex vivo gene delivery to hair follicles.59 Here, histocultured mouse skin fragments were treated with collagenase to make hair follicles accessible to the GFP gene containing adenoviral vectors. These fragments were then grafted onto nude mice, and 75% of hair follicles expressed GFP. However, this ex vivo approach requires dissection of the skin and previous collagenase treatment. In contrast, the established microinjection method in our study is a non-invasive method that ensures quick delivery of synthetic mRNAs into the dermis without prior extensive preparation of the skin.
Histological analyses of porcine skin demonstrated that the injected synthetic mRNA reaches the dermis. At the injection site, homogeneous distribution of Cy3-labeled mRNA could be detected after injection of a small volume of 35 μl containing the synthetic mRNA. Because of the secretability of hGLuc, detection of the produced hGLuc is greatly simplified by allowing detection of hGLuc in medium. Therefore, no complex tissue lysis procedure is needed to analyze hGLuc expression in the skin. In this study, the highly sensitive enzymatic activity of hGLuc allowed direct detection of luciferase activity in the medium without the need for purification and concentration. However, the established ex vivo skin model can also be used to test the functionality of other synthetic mRNAs for intradermal applications. For detection of intracellular proteins, tissue sections can be generated and analyzed. Furthermore, the produced proteins in the skin can be determined after lysis of skin and collection of proteins.
Microinjection of 1.5 μg hGLuc mRNA into porcine skin resulted in significantly high luciferase activity in the surrounding medium after 24 and 48 hr. In contrast, skin lysates showed no significant differences in luciferase activity compared with the negative controls. Therefore, successful production of hGLuc and its secretion from cells into the extracellular matrix and, subsequently, into the surrounding medium was detected. The high luciferase activity in the medium indicates that the amount of injected hGLuc mRNA could possibly be reduced. Surprisingly, injection of naked mRNA into the skin also showed significantly higher luciferase activity in the medium compared with the controls. This showed that microinjection with MicronJet600 enables sufficient delivery of synthetic mRNA into cells without the need for a delivery vehicle. The pressure during the injection or the increased contact period of synthetic mRNA with tissue-specific cells could be reasons for the successful uptake of naked mRNA into cells.
In summary, we could successfully show efficient intradermal delivery of synthetic mRNA using MicronJet600 microneedles both with and without transfection reagent into porcine skin. Additionally, using the established ex vivo porcine skin model, different mRNA-based therapeutic agents can be easily tested regarding their functionality.
Conclusions
In recent years, synthetic mRNA emerged as a potent new drug in the field of gene therapy, which offers several advantages compared with DNA plasmids and viral vectors, including direct translation of synthetic mRNA in the cytoplasm, elimination of insertional mutagenesis, and the transient presence of exogenously expressed proteins. In this study, we demonstrated the dermal applicability of synthetic mRNAs by using hollow microneedles and a porcine skin model. Efficient and rapid intradermal delivery of synthetic mRNAs might offer new opportunities for the treatment of several skin diseases, cutaneous cancers, hyperproliferative diseases, wound healing, and vaccination.
Materials and Methods
Ethics Statement
The study was performed in accordance with the Federation of European Laboratory Animal Science Associations (FELASA) and American Association for Laboratory Animal Science (AALAS) recommendations for the care and use of laboratory animals. The Animal Care and Welfare Commissioner of the University of Tübingen approved the protocols and procedures.
Animals
German Landrace pigs (70 ± 10 kg) were obtained from a local, specific pathogen-free breeding facility (Benz, Germany). The animals were mainly female and 4–6 months old. Pigs were anesthetized with an intramuscular (i.m.) injection of atropine (0.05 mg kg−1, Dr. Franz Koehler Chemie, Bensheim, Germany) and azaperone (2 mg kg−1, Janssen-Cilag, Neuss, Germany), followed by i.m. injection of midazolam (0.05 mg kg−1, Ratiopharm, Ulm, Germany) and xylazine (30–50 mg kg−1, Eurovet Animal Health, Bladel, the Netherlands). Euthanasia was performed by intravenous injection of a lethal dose of potassium chloride (1 mmol kg−1, B. Braun Melsungen, Melsungen, Germany), and the ears were explanted.
Synthesis of mRNA
The plasmid pEX-A2 containing the secretable hGLuc coding sequence was produced by Eurofins Genomics (Ebersberg, Germany). Synthesis of hGLuc-encoding mRNA was performed by IVT according to our previously established protocol.60 Briefly, the plasmid insert was amplified by PCR using the HotStar HiFidelity polymerase kit (QIAGEN, Hilden, Germany) and 0.7 μM of each forward (5′-TTGGACCCTCGTACAGAAGCTAATACG-3′) and reverse primer (5′-T120-CTTCCTACTCAGGCTTTATTCAAAGACCA-3′). Primers were purchased from ELLA Biotech (Martinsried, Germany) in high-performance liquid chromatography (HPLC) grade. PCR was run using the following cycling protocol: initial activation step at 94°C for 3 min, followed by 25 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 1 min, extension at 72°C for 1 min, and final extension at 72°C for 5 min. During the PCR, a poly T-tail of 120 thymidines (T) was added to the insert. The PCR product was purified using the MinElute PCR purification kit (QIAGEN) according manufacturer’s instructions.
The DNA product was then in vitro-transcribed into mRNA using the MEGAscript T7 kit (Thermo Fisher Scientific, Waltham, USA). The IVT reaction was performed at 37°C for 4 hr, and the mixture contained 7.5 mM ATP, 1.875 mM guanosine triphosphate (GTP) (both from the MEGAscript T7 kit), 7.5 mM 5-Methyl-CTP, 7.5 mM pseudo-UTP (both from TriLink BioTechnologies, San Diego, USA), 2.5 mM 3′-O-Me-m7G(5′)ppp(5′)G RNA cap structure analog (New England Biolabs, Frankfurt am Main, Germany), 40 U RiboLock RNase inhibitor (Thermo Fisher Scientific), and 1.5 μg PCR product. Afterward, the template DNA was removed by adding 1 μL TURBO DNase (from the MEGAscript T7 kit) and incubation at 37°C for 15 min. Then the reaction mixture was purified using the RNeasy MinElute cleanup kit (QIAGEN) according to the manufacturer’s instructions. Afterward, the mRNA was treated for 30 min at 37°C with 5 U Antarctic phosphatase and 1× Antarctic phosphatase reaction buffer in a total volume of 45.5 μl (New England Biolabs, Frankfurt am Main, Germany). Subsequently, mRNA was purified using the RNeasy Mini Kit according to the manufacturer’s instructions. The concentration of mRNA was determined using a photometer (BioPhotometer, Eppendorf, Hamburg, Germany). The quality and purity of the PCR product and the synthetic mRNA were assessed by 1% agarose gel electrophoresis and staining with 1× GelRed (Biotium, Fermont, USA). The peqGOLD range mix DNA ladder (0.08–10 kb, VWR, Darmstadt, Germany) and the RNA ladder (0.5–10 kb, Thermo Fisher Scientific) were used as size markers.
Cy3 Labeling of Synthetic hGLuc mRNA
Cy3 labeling of synthetic mRNA was performed via Cu(I)-free (DBCO) click chemistry. First, 5-azido-C3-UTP was incorporated during the IVT into the synthetic mRNA, and, subsequently, Cy3 was conjugated to 5-azido-C3-UTP by incubation with DBCO-Sulfo-Cy3. Therefore, during the IVT, 1.9 mM 5-azido-C3-UTP (Jena Bioscience, Jena, Germany) and 5.6 mM pseudo-UTP were used instead of 7.5 mM pseudo-UTP. The IVT reaction mixture was purified using the RNeasy MinElute cleanup kit (QIAGEN) according to the manufacturer’s instructions. Afterward, 5-azido-C3-UTP-modified mRNA was incubated with a 5-fold molar excess of DBCO-Sulfo-Cy3 (Jena Bioscience) in a total amount of 40 μl, adjusted with nuclease-free water, at 37°C for 1 hr. The molar amount of DBCO-sulfo-Cy3 in relation to the amount of azide-labeled mRNA was calculated using the manufacturer’s data sheet. The reaction mixture was purified again using the RNeasy MinElute cleanup kit (QIAGEN), and the purity of the mRNA was verified by 1% agarose gel electrophoresis and staining with 1× GelRed in Tris-borate-EDTA (TBE) for 30 min at room temperature (RT). The concentration was determined using a photometer, and the mRNA was stored at −80°C.
Cultivation of Cells
HEK293 cells were cultivated in DMEM with high glucose and L-glutamine with 10% heat-inactivated FBS at 37°C and 5% CO2. The medium was changed every 3–4 days. Cells were passaged when they reached 80% confluency. Cells were washed once with DPBS (without Ca2+/Mg2+) and detached with 0.05% trypsin-EDTA. All cell culture reagents were obtained from Thermo Fisher Scientific.
Transfection of HEK293 Cells with Synthetic hGLuc mRNA
To perform transfection experiments, 3 × 105 HEK293 cells were seeded per well of a 6-well plate and incubated overnight at 37°C with 5% CO2. The next day, 0.2, 0.5, or 1.5 μg hGLuc mRNA and 1 μl (for 0.2 and 0.5 μg mRNA) or 1.5 μl (for 1.5 μg mRNA) Lipofectamine 2000 (Thermo Fisher Scientific) were added, respectively, to 1 mL OptiMEM I reduced serum-free medium (Thermo Fisher Scientific) and incubated for 20 min at RT to generate transfection complexes. HEK293 cells were washed once with DPBS (w/o Ca2+/Mg2+), and transfection complexes were added to the cells and incubated at 37°C and 5% CO2 for 4 hr. Subsequently, the transfection complexes were aspirated, 2 mL cell culture medium was added to each well, and cells were incubated for 24 hr at 37°C and 5% CO2. Luciferase activity was determined in both the supernatant and the cell lysate using a luciferase assay.
Collection of Supernatant and Generation of Cell Lysates for Detection of Luciferase Activity
After 24 hr of transfection, 2 mL supernatant was collected to determine the secreted luciferase in the supernatant. To determine the luciferase activity in HEK293 cells, cells were washed once with DPBS (w/o Ca2+/Mg2+) and lysed by adding 350 μL of 1× luciferase cell culture lysis reagent (Promega, Mannheim, Germany) per well of a 6-well plate. After 15 min of incubation at RT, cells were scraped, transferred to protein low-binding reaction tubes, and stored on ice before vortexing the samples for 10–15 s. Subsequently, lysates were centrifuged at 12,000 × g for 2 min at 4°C. The supernatants were stored at −80°C until performance of luciferase activity measurements. For the luciferase assay, 40 μL of each supernatant or lysate was used.
Porcine Skin Preparation
Porcine skin from the outer side of the animal’s ear was used for the intradermal injection. The skin was detached directly post mortem or after storage of the ear for 24 hr at 4°C, washed with 0.9% NaCl solution (Fresenius Kabi, Bad Homburg, Germany), and trimmed to 1-mm thickness using a dermatome (Acculan 3Ti dermatome, B. Braun, Melsungen, Germany). Afterward, round pieces with a diameter of 1.5 cm were punched out and used for microinjection. The surface of the punched skin pieces was disinfected using an iodine solution (0.5 M I2, Fluca Analytical, Seelze, Germany) and rinsed once or twice with DPBS (w/o Ca2+/Mg2+). Additionally, the skin was incubated for 30 min in an antibiotic solution composed of 250 μg/mL gentamicin (Sigma-Aldrich, St. Louis, USA) and 1.25 μg/mL amphotericin B (PromoCell, Heidelberg, Germany) in DMEM (Thermo Fisher Scientific) and then washed once or twice with DPBS (w/o Ca2+/Mg2+). The residual DPBS was removed from the surface with a sterile swab prior to intradermal injection.
Microinjection of Synthetic hGLuc mRNA into Porcine Skin
For the intradermal injection of synthetic mRNA, the hollow microneedle device MicronJet600 from NanoPass Technologies (Nes Ziona, Israel) was used. The hGLuc mRNA transfection complexes were drawn, using a cannula (B. Braun, Melsungen, Germany), into a 1-mL syringe (Luer-Lok Tip, BD Biosciences, Heidelberg, Germany). All air bubbles were removed, and the microneedle device was attached to the syringe. After injection of synthetic mRNA into the skin, each skin piece was washed once with 1 mL DPBS (w/o Ca2+/Mg2+).
To perform the microinjections, 1.5 μg of synthetic hGLuc mRNA or Cy3-labeled hGLuc mRNA was complexed with 1.5 μL Lipofectamine 2000 in OptiMEM I reduced serum-free medium in a total volume of 35 μL. The appropriate volumes of OptiMEM I reduced serum-free medium or OptiMEM I reduced serum-free medium with Lipofectamine 2000 served as controls.
Skin pieces were transferred into ThinCert cell culture inserts with a pore size of 1 μm (Greiner Bio-One, Frickenhausen, Germany) and then placed in one well of a 12-well plate containing 1.5 mL human endothelial cell culture medium (VascuLife EnGS-Mv microvascular endothelial kit without hydrocortisone hemisuccinate, CellSystems, Troisdorf, Germany). The medium under the inserts was collected every 24 hr, and fresh medium was added every 24 hr until a total incubation time of 72 hr. Furthermore, in additional experiments, skin pieces were collected after 24, 48, or 72 hr of incubation. The luciferase activity in the medium and skin lysates was analyzed using a luciferase assay. For this purpose, 40 μL of each sample was used.
Preparation of Skin Lysates for Detection of Luciferase Activity
After the respective incubation times, skin samples were cut into small pieces and frozen in liquid nitrogen. The tissue was homogenized using the Minilys homogenizer (Peqlab, Erlangen, Germany). For this purpose, 0.8 g zirconium oxide beads (Precellys, Bertin Instruments, Montigny-le-Bretonneux, France) with a diameter of 1.4 mm were used. After addition of 400 μL 1× luciferase cell culture lysis reagent (Promega, Mannheim, Germany) to each skin sample, homogenization was performed at 5,000 rpm six times for 20 s. Afterward, the homogenates were centrifuged for 15 min at 13,000 rpm and 4°C. The supernatants were transferred into protein low-binding reaction tubes. The homogenization and centrifugation procedures were repeated one additional time with 400 μL 1× luciferase cell culture lysis reagent. The lysates were pooled and stored at −80°C. The luciferase assay was performed with 40 μL of each lysate.
Luciferase Assay
After the delivery of synthetic hGLuc mRNA, the expression of hGLuc in supernatants, cells, and skin samples was determined using a luciferase assay. For this purpose, the luciferase substrate coelenterazine was used. In the presence of luciferase, the substrate is oxidized under emission of light. Therefore, 40 μL of each sample was transferred in triplicates in a 96-well plate (Nunc Maxisorp, Thermo Fisher Scientific). Luciferase activity was measured using a Mithras LB 940 multimode microplate reader (Berthold Technologies, Bad Wildbad, Germany). To measure luminescence, 100 μL of 20 μg/mL coelenterazine (Carl Roth, Karlsruhe, Germany) in DPBS (w/o Ca2+/Mg2+) was injected automatically per sample, and luminescence was detected in RLUs.
Detection of Delivered Synthetic hGLuc mRNA in the Skin
To determine the depth of the microinjection in the skin, either magnetic beads or Cy3-labeled hGLuc mRNA were injected. Using histological sections, the specific skin layer reached by the microneedles was identified. Therefore, 1 μL magnetic beads (10 μg) (Dynabeads MyOne Streptavidin T1, 1 μm, Thermo Fisher Scientific) in 35 μL nuclease-free water were injected into the skin using microneedles. Additionally, 1.5 μg of Cy3-labeled hGLuc mRNA and 1.5 μL of Lipofectamine 2000 were complexed in 35 μL of OptiMEM I reduced serum-free medium for 20 min at RT and injected into the skin using hollow microneedles. Non-injected skin pieces served as negative controls. After injection, the tissue was washed twice with DPBS (w/o Ca2+/Mg2+) and fixed in 4% paraformaldehyde (PFA) (Merck, Darmstadt, Germany) in DPBS (w/o Ca2+/Mg2+) overnight at 4°C.
Histological Analysis
Preparation of Paraffin-Embedded Sections
Skin samples were placed into an embedding cassette and transferred in a 4% PFA solution. After dehydration, samples were incubated in clearing agent, followed by infiltration and embedding in paraffin. The infiltrated tissues were then embedded into wax blocks. The steps were performed using a tissue-embedding machine and an automatic tissue processor. Paraffin specimens were cut into 2.5-μm-thick sections and mounted on SuperFrost microscope slides (R. Langenbrinck, Emmendingen, Germany), dried overnight at RT, and stored at 4°C until staining. Prior to staining, sections were deparaffinized in xylene (100% xylene, mixture of isomers, AnalaR NORMAPUR, VWR, Darmstadt, Germany) four times for 5 min, rehydrated using a graded ethanol (AnalaR NORMAPUR, VWR, Darmstadt, Germany) series (99%, 96%, and 70%) for 2 min each, washed twice in water, and boiled for 2 min in 1× Tris-EDTA buffer (pH 9). The slides were then cooled down under running tap water, washed three times with DPBS (w/o Mg2+ and Ca2+) for 2 min, and dried. Stained and mounted samples were stored at 4°C in the dark.
Histochemical Staining
Tissue sections were stained either with the fluorescent dye DAPI or with H&E. DAPI staining was performed using Vectashield mounting medium containing DAPI (Vector Laboratories, Burlingame, CA, USA). Stained and mounted samples were stored at 4°C in the dark.
To detect Cy3-labeled mRNA, fluorescence images were taken using the Axiovert135 microscope (Carl Zeiss, Oberkochen, Germany) and analyzed with AxioVision Rel 4.8 software. Light microscope images were taken with a camera (Canon EOS 550D) and analyzed using the software EOS Digital Solution (Canon).
Statistical Analysis
Data are shown as means ± SEM. Statistical analysis of the data was performed using GraphPad Prism version 6. Two-way ANOVA for repeated measurements and Bonferroni’s multiple comparison test were applied. p < 0.05 was considered statistically significant.
Author Contributions
S.G. and M.A.-A. conceived and designed the experiments. S.G. and M.P. performed the experiments with support from H.S., M.A.-A., and H.P.W. and analyzed the data. E.K., Y.L., D.L., H.P.W., and C.S. contributed reagents/materials/analysis tools. S.G. and M.A.-A. wrote the paper. M.A.-A. supervised the project.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
The authors would like to thank the German Heart Foundation/German Foundation of Heart Research for Grant F/38/13; the European Social Funds in Baden-Württemberg, Germany; and the Ministry of Science, Research, and the Arts of the State of Baden-Württemberg (MWK-BW). We acknowledge support from the Deutsche Forschungsgemeinschaft and the Open Access Publishing Fund of the University of Tübingen. Furthermore, we thank Prof. Falko Fend from the Institute of Pathology and Neuropathology (Department of General and Molecular Pathology, University Hospital Tübingen) for their support during preparation of paraffin-embedded skin samples, histological sections, and H&E staining.
References
- 1.Steinle H., Behring A., Schlensak C., Wendel H.P., Avci-Adali M. Concise Review: Application of In Vitro Transcribed Messenger RNA for Cellular Engineering and Reprogramming: Progress and Challenges. Stem Cells. 2017;35:68–79. doi: 10.1002/stem.2402. [DOI] [PubMed] [Google Scholar]
- 2.Weide B., Carralot J.P., Reese A., Scheel B., Eigentler T.K., Hoerr I., Rammensee H.G., Garbe C., Pascolo S. Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J. Immunother. 2008;31:180–188. doi: 10.1097/CJI.0b013e31815ce501. [DOI] [PubMed] [Google Scholar]
- 3.Weide B., Garbe C., Rammensee H.G., Pascolo S. Plasmid DNA- and messenger RNA-based anti-cancer vaccination. Immunol. Lett. 2008;115:33–42. doi: 10.1016/j.imlet.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Pardi N., Secreto A.J., Shan X., Debonera F., Glover J., Yi Y., Muramatsu H., Ni H., Mui B.L., Tam Y.K. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 2017;8:14630. doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petsch B., Schnee M., Vogel A.B., Lange E., Hoffmann B., Voss D., Schlake T., Thess A., Kallen K.J., Stitz L., Kramps T. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012;30:1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 6.Mays L.E., Ammon-Treiber S., Mothes B., Alkhaled M., Rottenberger J., Müller-Hermelink E.S., Grimm M., Mezger M., Beer-Hammer S., von Stebut E. Modified Foxp3 mRNA protects against asthma through an IL-10-dependent mechanism. J. Clin. Invest. 2013;123:1216–1228. doi: 10.1172/JCI65351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zangi L., Lui K.O., von Gise A., Ma Q., Ebina W., Ptaszek L.M., Später D., Xu H., Tabebordbar M., Gorbatov R. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michel T., Kankura A., Salinas Medina M.L., Kurz J., Behring A., Avci-Adali M., Nolte A., Schlensak C., Wendel H.P., Krajewski S. In Vitro Evaluation of a Novel mRNA-Based Therapeutic Strategy for the Treatment of Patients Suffering from Alpha-1-Antitrypsin Deficiency. Nucleic Acid Ther. 2015;25:235–244. doi: 10.1089/nat.2015.0537. [DOI] [PubMed] [Google Scholar]
- 9.Abraham M.K., Nolte A., Reus R., Behring A., Zengerle D., Avci-Adali M., Hohmann J.D., Peter K., Schlensak C., Wendel H.P., Krajewski S. In vitro Study of a Novel Stent Coating Using Modified CD39 Messenger RNA to Potentially Reduce Stent Angioplasty-Associated Complications. PLoS ONE. 2015;10:e0138375. doi: 10.1371/journal.pone.0138375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stadler C.R., Bahr-Mahmud H., Celik L., Hebich B., Roth A.S., Roth R.P. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat. Med. 2017;23:815–817. doi: 10.1038/nm.4356. [DOI] [PubMed] [Google Scholar]
- 11.Gallie D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 12.Stepinski J., Waddell C., Stolarski R., Darzynkiewicz E., Rhoads R.E. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl (3′-deoxy)GpppG. RNA. 2001;7:1486–1495. [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kauffman K.J., Mir F.F., Jhunjhunwala S., Kaczmarek J.C., Hurtado J.E., Yang J.H., Webber M.J., Kowalski P.S., Heartlein M.W., DeRosa F., Anderson D.G. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials. 2016;109:78–87. doi: 10.1016/j.biomaterials.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weissman D., Karikó K. mRNA: Fulfilling the Promise of Gene Therapy. Mol. Ther. 2015;23:1416–1417. doi: 10.1038/mt.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 20.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson Ö., Thompson J. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alberer M., Gnad-Vogt U., Hong H.S., Mehr K.T., Backert L., Finak G., Gottardo R., Bica M.A., Garofano A., Koch S.D. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet. 2017;390:1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- 22.Zou S., Scarfo K., Nantz M.H., Hecker J.G. Lipid-mediated delivery of RNA is more efficient than delivery of DNA in non-dividing cells. Int. J. Pharm. 2010;389:232–243. doi: 10.1016/j.ijpharm.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean D.A., Strong D.D., Zimmer W.E. Nuclear entry of nonviral vectors. Gene Ther. 2005;12:881–890. doi: 10.1038/sj.gt.3302534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Tendeloo V.F., Ponsaerts P., Lardon F., Nijs G., Lenjou M., Van Broeckhoven C., Van Bockstaele D.R., Berneman Z.N. Highly efficient gene delivery by mRNA electroporation in human hematopoietic cells: superiority to lipofection and passive pulsing of mRNA and to electroporation of plasmid cDNA for tumor antigen loading of dendritic cells. Blood. 2001;98:49–56. doi: 10.1182/blood.v98.1.49. [DOI] [PubMed] [Google Scholar]
- 25.Kofler R.M., Aberle J.H., Aberle S.W., Allison S.L., Heinz F.X., Mandl C.W. Mimicking live flavivirus immunization with a noninfectious RNA vaccine. Proc. Natl. Acad. Sci. USA. 2004;101:1951–1956. doi: 10.1073/pnas.0307145101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fotin-Mleczek M., Duchardt K.M., Lorenz C., Pfeiffer R., Ojkić-Zrna S., Probst J., Kallen K.J. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J. Immunother. 2011;34:1–15. doi: 10.1097/CJI.0b013e3181f7dbe8. [DOI] [PubMed] [Google Scholar]
- 27.Martinon F., Krishnan S., Lenzen G., Magné R., Gomard E., Guillet J.G., Lévy J.P., Meulien P. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 1993;23:1719–1722. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto M., Takemoto T. Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. Sci. Rep. 2015;5:11315. doi: 10.1038/srep11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delalande A., Kotopoulis S., Postema M., Midoux P., Pichon C. Sonoporation: mechanistic insights and ongoing challenges for gene transfer. Gene. 2013;525:191–199. doi: 10.1016/j.gene.2013.03.095. [DOI] [PubMed] [Google Scholar]
- 30.Youn H., Chung J.K. Modified mRNA as an alternative to plasmid DNA (pDNA) for transcript replacement and vaccination therapy. Expert Opin. Biol. Ther. 2015;15:1337–1348. doi: 10.1517/14712598.2015.1057563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kormann M.S., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 32.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCaffrey J., Donnelly R.F., McCarthy H.O. Microneedles: an innovative platform for gene delivery. Drug Deliv. Transl. Res. 2015;5:424–437. doi: 10.1007/s13346-015-0243-1. [DOI] [PubMed] [Google Scholar]
- 34.Trommer H., Neubert R.H. Overcoming the stratum corneum: the modulation of skin penetration. A review. Skin Pharmacol. Physiol. 2006;19:106–121. doi: 10.1159/000091978. [DOI] [PubMed] [Google Scholar]
- 35.Villemejane J., Mir L.M. Physical methods of nucleic acid transfer: general concepts and applications. Br. J. Pharmacol. 2009;157:207–219. doi: 10.1111/j.1476-5381.2009.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zakrewsky M., Kumar S., Mitragotri S. Nucleic acid delivery into skin for the treatment of skin disease: Proofs-of-concept, potential impact, and remaining challenges. J. Control. Release. 2015;219:445–456. doi: 10.1016/j.jconrel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L., Nolan E., Kreitschitz S., Rabussay D.P. Enhanced delivery of naked DNA to the skin by non-invasive in vivo electroporation. Biochim. Biophys. Acta. 2002;1572:1–9. doi: 10.1016/s0304-4165(02)00270-2. [DOI] [PubMed] [Google Scholar]
- 38.Tezel A., Dokka S., Kelly S., Hardee G.E., Mitragotri S. Topical delivery of anti-sense oligonucleotides using low-frequency sonophoresis. Pharm. Res. 2004;21:2219–2225. doi: 10.1007/s11095-004-7674-6. [DOI] [PubMed] [Google Scholar]
- 39.Mastrobattista E., Hennink W.E., Schiffelers R.M. Delivery of nucleic acids. Pharm. Res. 2007;24:1561–1563. doi: 10.1007/s11095-007-9349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guan S., Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017;24:133–143. doi: 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- 41.Wang T., Upponi J.R., Torchilin V.P. Design of multifunctional non-viral gene vectors to overcome physiological barriers: dilemmas and strategies. Int. J. Pharm. 2012;427:3–20. doi: 10.1016/j.ijpharm.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 42.Bozzuto G., Molinari A. Liposomes as nanomedical devices. Int. J. Nanomedicine. 2015;10:975–999. doi: 10.2147/IJN.S68861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alkilani A.Z., McCrudden M.T., Donnelly R.F. Transdermal Drug Delivery: Innovative Pharmaceutical Developments Based on Disruption of the Barrier Properties of the stratum corneum. Pharmaceutics. 2015;7:438–470. doi: 10.3390/pharmaceutics7040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brambilla D., Luciani P., Leroux J.C. Breakthrough discoveries in drug delivery technologies: the next 30 years. J. Control. Release. 2014;190:9–14. doi: 10.1016/j.jconrel.2014.03.056. [DOI] [PubMed] [Google Scholar]
- 45.Heller R., Cruz Y., Heller L.C., Gilbert R.A., Jaroszeski M.J. Electrically mediated delivery of plasmid DNA to the skin, using a multielectrode array. Hum. Gene Ther. 2010;21:357–362. doi: 10.1089/hum.2009.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heller R., Schultz J., Lucas M.L., Jaroszeski M.J., Heller L.C., Gilbert R.A., Moelling K., Nicolau C. Intradermal delivery of interleukin-12 plasmid DNA by in vivo electroporation. DNA Cell Biol. 2001;20:21–26. doi: 10.1089/10445490150504666. [DOI] [PubMed] [Google Scholar]
- 47.Kim Y.C., Park J.H., Prausnitz M.R. Microneedles for drug and vaccine delivery. Adv. Drug Deliv. Rev. 2012;64:1547–1568. doi: 10.1016/j.addr.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levin Y., Kochba E., Hung I., Kenney R. Intradermal vaccination using the novel microneedle device MicronJet600: Past, present, and future. Hum. Vaccin. Immunother. 2015;11:991–997. doi: 10.1080/21645515.2015.1010871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hung I.F., Levin Y., To K.K., Chan K.H., Zhang A.J., Li P., Li C., Xu T., Wong T.Y., Yuen K.Y. Dose sparing intradermal trivalent influenza (2010/2011) vaccination overcomes reduced immunogenicity of the 2009 H1N1 strain. Vaccine. 2012;30:6427–6435. doi: 10.1016/j.vaccine.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 50.Lee H.J., Choi H.J., Kim D.R., Lee H., Jin J.E., Kim Y.R., Lee M.S., Cho S.N., Kang Y.A. Safety and efficacy of tuberculin skin testing with microneedle MicronJet600™ in healthy adults. Int. J. Tuberc. Lung Dis. 2016;20:500–504. doi: 10.5588/ijtld.15.0678. [DOI] [PubMed] [Google Scholar]
- 51.Levin Y., Kochba E., Shukarev G., Rusch S., Herrera-Taracena G., van Damme P. A phase 1, open-label, randomized study to compare the immunogenicity and safety of different administration routes and doses of virosomal influenza vaccine in elderly. Vaccine. 2016;34:5262–5272. doi: 10.1016/j.vaccine.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Levin Y., Kochba E., Kenney R. Clinical evaluation of a novel microneedle device for intradermal delivery of an influenza vaccine: are all delivery methods the same? Vaccine. 2014;32:4249–4252. doi: 10.1016/j.vaccine.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 53.Hung I.F., Levin Y., To K.K. Quantitative and qualitative analysis of antibody response after dose sparing intradermal 2009 H1N1 vaccination. Vaccine. 2012;30:2707–2708. doi: 10.1016/j.vaccine.2011.12.069. [DOI] [PubMed] [Google Scholar]
- 54.Beals C.R., Railkar R.A., Schaeffer A.K., Levin Y., Kochba E., Meyer B.K., Evans R.K., Sheldon E.A., Lasseter K., Lang N. Immune response and reactogenicity of intradermal administration versus subcutaneous administration of varicella-zoster virus vaccine: an exploratory, randomised, partly blinded trial. Lancet Infect. Dis. 2016;16:915–922. doi: 10.1016/S1473-3099(16)00133-X. [DOI] [PubMed] [Google Scholar]
- 55.Troy S.B., Kouiavskaia D., Siik J., Kochba E., Beydoun H., Mirochnitchenko O., Levin Y., Khardori N., Chumakov K., Maldonado Y. Comparison of the Immunogenicity of Various Booster Doses of Inactivated Polio Vaccine Delivered Intradermally Versus Intramuscularly to HIV-Infected Adults. J. Infect. Dis. 2015;211:1969–1976. doi: 10.1093/infdis/jiu841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anand A., Zaman K., Estívariz C.F., Yunus M., Gary H.E., Weldon W.C., Bari T.I., Steven Oberste M., Wassilak S.G., Luby S.P. Early priming with inactivated poliovirus vaccine (IPV) and intradermal fractional dose IPV administered by a microneedle device: A randomized controlled trial. Vaccine. 2015;33:6816–6822. doi: 10.1016/j.vaccine.2015.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flynn P.M., Shenep J.L., Mao L., Crawford R., Williams B.F., Williams B.G. Influence of needle gauge in Mantoux skin testing. Chest. 1994;106:1463–1465. doi: 10.1378/chest.106.5.1463. [DOI] [PubMed] [Google Scholar]
- 58.Li L., Lishko V., Hoffman R.M. Liposome targeting of high molecular weight DNA to the hair follicles of histocultured skin: a model for gene therapy of the hair growth processes. In Vitro Cell. Dev. Biol. Anim. 1993;29A:258–260. doi: 10.1007/BF02633949. [DOI] [PubMed] [Google Scholar]
- 59.Saito N., Zhao M., Li L., Baranov E., Yang M., Ohta Y., Katsuoka K., Penman S., Hoffman R.M. High efficiency genetic modification of hair follicles and growing hair shafts. Proc. Natl. Acad. Sci. USA. 2002;99:13120–13124. doi: 10.1073/pnas.192453799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Avci-Adali M., Behring A., Steinle H., Keller T., Krajeweski S., Schlensak C., Wendel H.P. In vitro synthesis of modified mRNA for induction of protein expression in human cells. J. Vis. Exp. 2014;93:e51943. doi: 10.3791/51943. [DOI] [PMC free article] [PubMed] [Google Scholar]