Abstract
Glioblastoma stem cells (GSCs) are invasive, treatment-resistant brain cancer cells that express downregulated in renal cell carcinoma (DRR), also called FAM107A, a genetic driver of GSC invasion. We developed antibody-antisense oligonucleotide (AON) conjugates to target and reduce DRR/FAM107A expression. Specifically, we used antibodies against antigens expressed on the GSCs, such as CD44 and EphA2, conjugated to chemically modified AONs against DRR/FAM107A, which were designed as chimeras of DNA and 2′-deoxy-2′-fluoro-beta-D-arabinonucleic acid (FANA) for increased nuclease stability and mRNA affinity. We demonstrate that these therapeutic conjugates successfully internalize, accumulate, and reduce DRR/FAM107A expression in patient-derived GSCs. This is the first example of an antibody-antisense strategy against cancer stem cells.
Keywords: glioblastoma stem cells, antisense oligonucleotides, targeted therapies, antibody conjugates, gene therapy, FAM107A
Introduction
Glioblastoma cancer stem cells (GSCs) are hypothesized to account, at least partially, for treatment failures in aggressive glioblastoma multiformes.1, 2, 3, 4, 5 The GSCs are highly invasive,6, 7 resistant to radiation and conventional chemotherapy,8, 9 and have the capacity to initiate new tumor growth.4, 10
Downregulated in renal cell carcinoma (DRR/FAM107A)11 is an established genetic driver of GSC invasion. It acts by regulating focal adhesion dynamics at the leading edge of migrating cells12 and by activating AKT signaling, making it an ideal anti-invasion target.12, 13, 14
Antisense oligonucleotides (AONs), which are short (15–21 nt) strands of DNA, are potent regulators of gene expression.15 There are several clinically approved AON therapeutics, including eteplirsen, a treatment for muscular dystrophy,16 and nusinersen, which is used in the treatment of spinal muscular atrophy.17
Herein, we engineered AONs for DRR/FAM107A knockdown in patient-derived GSCs. The AON is stabilized against nuclease degradation by substituting some of the DNA residues for 2′-deoxy-2′-fluoro-beta-D-arabinonucleic acid (FANA) residues, thereby increasing treatment longevity while maintaining potency.18, 19 We designed a “gapmer” AON with FANA modifications flanking the DNA core because FANA gapmers bind target mRNA with high affinity and elicit mRNA degradation while protecting the 3′ end from exonuclease degradation.20 This construct also includes a phosphorothioate (PS) backbone in lieu of the naturally occurring phosphodiester (PO) backbone for added nuclease resistance and effective RNaseH-mediated cleavage of mRNA.21, 22 Given that PS-DNA chemistry may increase immunostimulation, we incorporated a 2′-deoxy-5-methyl-cytidine at a CpG motif within the AON sequence to obviate this issue in future studies.23, 24
Antibody targeting is a powerful tool to guide therapeutic delivery to specific cell types, both as pendant groups on larger nanoparticle systems25, 26, 27, 28 and as direct antibody-drug conjugates (ADCs).29, 30, 31, 32, 33 In glioblastomas, administration of an anti-epidermal growth factor receptor (EGFR) antibody-monomethyl auristatin F ADC has resulted in a survival benefit for EGFR-amplified glioblastoma patients,34 whereas administration of an anti-EGFR antibody alone failed.35 To mediate AON delivery, we chose to investigate three monoclonal antibodies (mAbs) against three GSC surface markers: CD44, a neural stem cell marker and a marker of GSCs within the glioblastoma tumor;36, 37, 38 EphA2, a key component of cell-cell signaling that is overexpressed in many cancers and also in GSCs;39 and EGFR, which is amplified in 40%–60% of all glioblastomas.40
We developed an antibody-conjugated double-stranded AON (dsAON) therapeutic using click chemistry between an azide-modified antibody and an alkyne-modified dsAON with a unique architecture: the sense strand is modified at the 5′ and 3′ ends with functionalities for click chemistry (dibenzylcyclooctyne [DBCO]) and for imaging (cyanine 3 fluorophore [Cy3]), respectively. Whereas the sense strand is covalently conjugated to the antibody, the antisense strand is hybridized to this sense strand to facilitate its release once inside the cells. Direct conjugation of large molecules, such as antibodies to the antisense strand, can interfere with RNase H recognition of the corresponding AON:mRNA duplex.41 This approach is an improvement over the few antibody-antisense therapeutic conjugates reported, with the majority of these being non-covalent, cationic complexation42, 43 or disulfide linkages directly to the antisense strand, which are unstable and prone to degradation.44, 45 Effective delivery and gene knockdown using AONs has been demonstrated with dsDNA systems where the DNA sense strand is left unmodified and as such susceptible to nuclease-mediated digestion by endogenous cellular enzymes.46 This degradation is the necessary driving force to release the antisense strand, which is then available for hybridization with the target mRNA.46
We chose to use antibodies engineered by phage display in order to target the glioblastoma stem cells (GSCs).47 Based on immunocytochemistry data, we identified CD44 and EphA2 as the best candidates for AON delivery. We then verified that these antibodies were internalized upon binding prior to conjugating them to the dsAONs via click chemistry. We found that the CD44 mAb-dsAON conjugate significantly reduced DRR expression, which correlated with a change in cellular morphology. Thus, we can reduce expression of a key GSC target, DRR/FAM107A, using antibody-dsAON conjugates, and this provides a framework for antibody-AON conjugate testing against GSCs in vivo.
Results
AON Activity
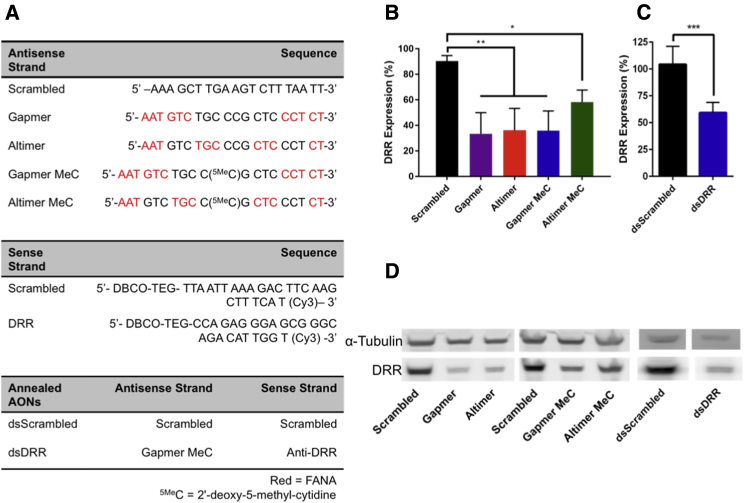
We tested four modification patterns of the DRR/FAM107A AON and compared these to a scrambled sequence control (Figure 1) in order to maximize efficacy and reduce immunogenicity: (1) gapmer motif, consisting of flanking FANA modifications with a DNA core; (2) altimer motif, where FANA substitutions were alternated every 3 nucleotides with DNA;48 (3) gapmer-MeC with a 5-methylcytidine modification at the CpG motif (5MeCG) in order to reduce immunogenicity; and (4) altimer-MeC with a similar 5MeCG modification (Figure 1A). We observed significant DRR knockdown following transfection into DRR-overexpressing (DRR+) U-251 MG glioblastoma cells with all single-stranded AONs by western blot compared to the scrambled control (Figure 1B). Although not statistically significant, we observed a trend toward greater knockdown when using gapmer-MeC compared to altimer-MeC. Therefore, all future studies were carried out using the gapmer-MeC strand.
Figure 1.
DRR Expression Is Reduced following Transfection of DRR+ U-251 MG Cells with DRR Antisense Oligonucleotides
All oligonucleotide strands were delivered at a concentration of 25 nM for 24 hr, and DRR expression was measured 72 hr post-transfection. (A) Oligonucleotide sequences used were as follows: all antisense strands comprise a phosphorothioate backbone, whereas all sense strands are synthesized with a phosphodiester backbone. Red, FANA; 5MeC, 2′-deoxy-5-methyl-cytidine. (B) DRR expression following treatment with single-stranded DRR AON sequences normalized to untreated control is shown. Data were analyzed using one-way ANOVA followed by Dunnett’s post hoc test compared to Scrambled group (data are shown as mean + SD; n = 3; *p < 0.05; **p < 0.01). (C) DRR expression following treatment with double-stranded anti-DRR oligonucleotide sequences normalized to untreated control is shown. Data were analyzed using unpaired t test with Welch’s correction (data are shown as mean + SD; n ≥ 4; ***p < 0.001). (D) Representative western blot showing single-stranded and double-stranded antisense oligonucleotide DRR knockdown is shown.
We hybridized the therapeutically active gapmer-MeC antisense strand to a carrier sense strand modified with a 3′ Cy3 for imaging studies and a 5′ DBCO for click chemistry (see melting curves in Figure S1). The Cy3 and DBCO functionalities were incorporated into the sense strand during solid-phase oligonucleotide synthesis. We transfected this double-stranded AON into DRR+ GBM cells and demonstrated significant reduction of DRR expression compared to a scrambled control (Figure 1C). Representative western blots are shown (Figure 1D).
Co-expression in GSCs of DRR and Antigens for CD44, EphA2, and EGFR Antibodies
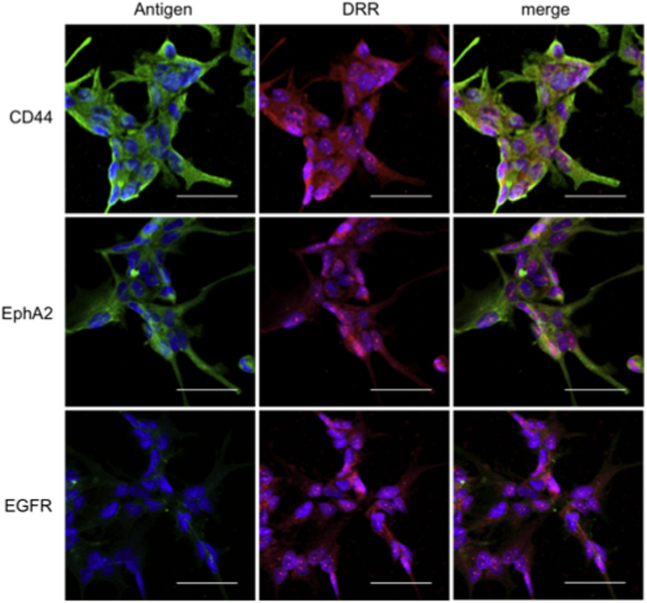
We screened the patient-derived GSCs for co-expression of DRR with potential antigens for targeted delivery of the AONs: CD44; EphA2; or EGFR. All of the GSCs used in this study strongly co-expressed DRR with CD44 and EphA2 in all fields of view yet only weakly expressed EGFR (Figure 2). Thus, CD44 mAb and EphA2 mAb were pursued for further study.
Figure 2.
Patient-Derived GSCs Strongly Co-express Antigens CD44 and EphA2 with DRR and Weakly Co-express EGFR with DRR
Representative confocal images are shown. Antigens CD44, EphA2, and EGFR (green); DRR (red); and cell nucleus (Hoechst, blue) are shown. All scale bars are 50 μM.
Internalization of CD44 and EphA2 mAbs
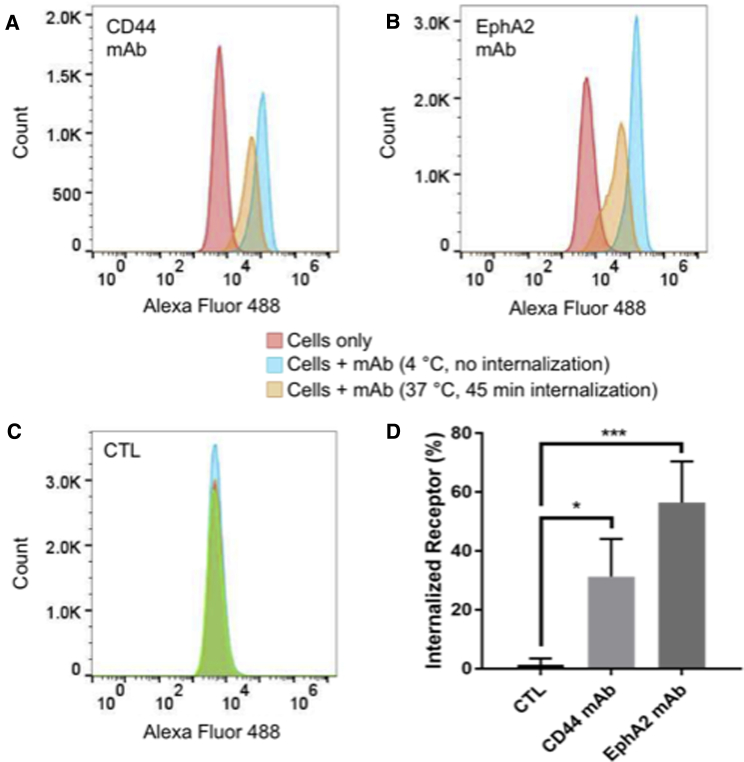
We quantified the internalization of the CD44 and EphA2 mAbs at 15, 45, and 90 min using a flow-cytometry-based assay with DRR+ U-251 MG cells. We first incubated the cells with the CD44 (Figure 3A), EphA2 (Figure 3B), or non-specific control (CTL) mAb (Figure 3C) at 4°C, incubated this sample for the given time period at 37°C in order for internalization to occur, and then incubated the cells with a fluorescently tagged secondary antibody at 4°C (see orange curves in Figures 3A–3C). We compared these to a control sample held at 4°C for the entire experiment, which would prevent internalization (see blue curves in Figures 3A–3C). Cells in the absence of antibody staining exhibited a low background fluorescence (see red curves in Figures 3A–3C). We used median fluorescent intensity (MFI) to quantify internalization. Cells allowed to internalize the antibody at 37°C (orange curves) demonstrated a reduced MFI compared to cells held at 4°C (blue curves) due to a lower amount of cell surface receptor, indicating internalization. The differences between the 15-, 45-, and 90-min time points for each mAb were not statistically significant (Figure S2). However, as we observed a trend toward greater internalization at 45 min with both mAbs, the 45-min time point is shown for comparison in Figure 3. At 45 min, we observed internalization of the EphA2 mAb and the CD44 mAb, and they were both internalized significantly more than the CTL antibody (Figure 3D).
Figure 3.
EphA2 mAbs and CD44 mAbs Are Internalized upon Binding to Cells
(A–C) Flow cytometry analysis of cell surface receptor internalization following 45-min incubation at 37°C with antibodies (A) CD44 mAb and (B) EphA2 mAb compared to (C) non-specific CTL mAb. Cells without antibodies added (cells only) and cells incubated with antibodies but held at 4°C (no internalization) curves are shown as a comparison to those cells that were allowed to internalize the mAb for 45 min at 37°C (45-min internalization). (D) Quantification of internalized receptor following 45-min incubation period for CD44 mAb and EphA2 mAb antibodies compared to CTL is shown. Data were analyzed using one-way ANOVA followed by Dunnett’s post hoc test compared to CTL group (data are shown as mean + SD; n = 3; *p < 0.05; ***p < 0.001).
Synthesis of mAb-dsDRR and mAb-dsScrambled Conjugates
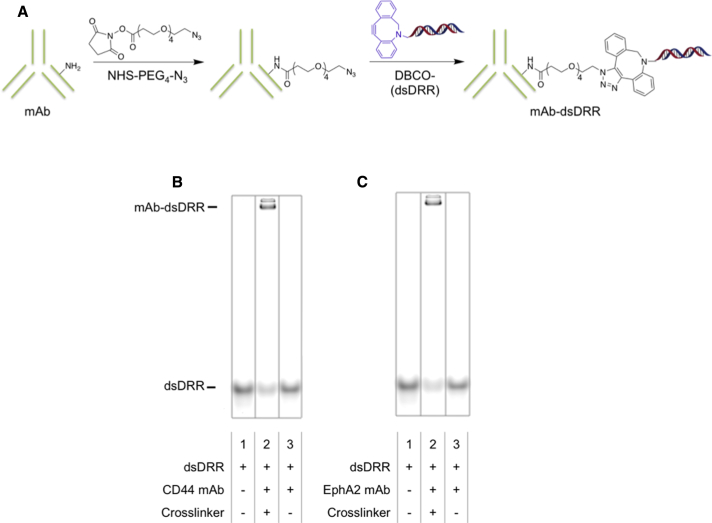
For antibody-mediated delivery of dsAONs, we first modified each of CD44 and EphA2 antibodies with an azide functional group by covalently bonding N-hydroxysuccinimide (NHS)-PEG4-N3, a crosslinker with an NHS-activated ester at one end and an azide at the other, to one of the lysines therein (Figure 4A). The azide modification enabled click conjugation of the DBCO-modified sense oligonucleotide to the antibody following hybridization with either the gapmer-MeC (dsDRR) or a scrambled sequence (dsScrambled; Figure 4A). We determined conjugation efficiency by gel electrophoresis followed by ImageJ quantification to be 60% for dsDRR conjugation to both CD44 mAb (Figure 4B) and EphA2 mAb (Figure 4C). We observed the appearance of two bands for both the CD44 mAb-dsDRR and the EphA2 mAb-dsDRR conjugates (lane 2 of Figures 4B and 4C), which we expect is due to the conjugation of either one or two AON strands per antibody. To confirm the absence of non-specific aggregation or adsorption, the dsDRR was mixed together with either the CD44 or EphA2 mAb (without prior azide modification of the antibodies) and no evidence of conjugation was detected (lane 3 of Figures 4B and 4C).
Figure 4.
CD44 mAb and EphA2 mAb Can Be Efficiently Conjugated to dsDRR Using Click Chemistry
(A) Scheme of antibody modification with dsDRR. (B) 10% PAGE analysis of CD44 mAb-dsDRR conjugation is shown: lane 1: dsDRR only; lane 2: CD44 mAb conjugated to dsDRR via NHS-PEG4-N3 crosslinker; are lane 3: dsDRR and CD44 mAb combined without crosslinker present. (C) 10% PAGE analysis of EphA2 mAb-dsDRR conjugation is shown: lane 1: dsDRR only; lane 2: EphA2 mAb conjugated to dsDRR via NHS-PEG4-N3 crosslinker; and lane 3: dsDRR and EphA2 mAb combined without crosslinker present.
To further verify the conjugation of the AONs to the antibodies, we reacted azide-modified mAb with the DRR sense strand, and this conjugate was analyzed by MALDI-TOF mass spectrometry (Figure S3). Experimental results closely matched the expected conjugate mass Mobt = 160.0 kDa versus Mth = 160.2 kDa.
DRR Knockdown and Cellular Uptake of mAb-dsDRR Conjugates
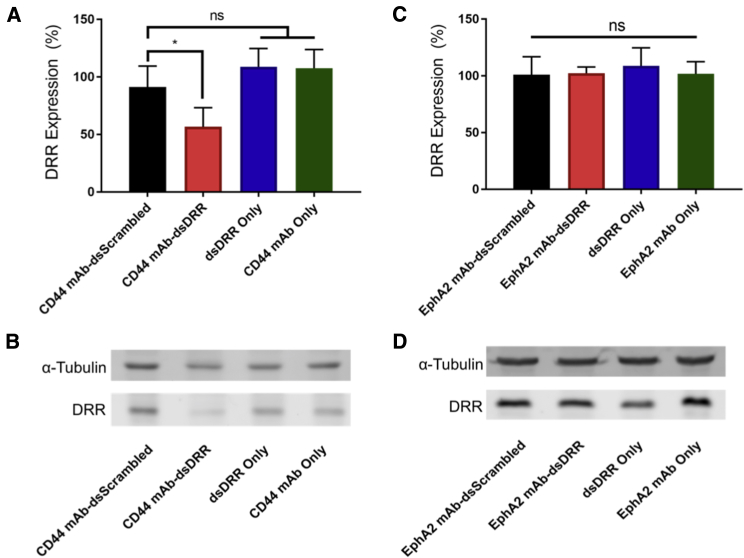
To determine the knockdown efficiency of the EphA2 mAb-dsDRR and CD44 mAb-dsDRR conjugates, we incubated patient-derived GSCs with each formulation for 72 hr at 150 nM and compared to mAb-dsScrambled, dsDRR alone, and antibody alone controls. We quantified DRR protein expression using western blot analysis normalized first to α-tubulin and then to a no treatment control. The CD44 mAb-dsDRR conjugate significantly reduced DRR expression in patient-derived GSCs compared to the CD44 mAb-dsScrambled control (Figures 5A and 5B). We observed no knockdown with the negative controls (dsDRR or CD44 mAb alone). Unexpectedly, treatment with EphA2 mAb-dsDRR conjugate did not reduce DRR expression relative to any of the EphA2 mAb-dsScrambled, dsDRR, or EphA2 mAb controls (Figures 5C and 5D).
Figure 5.
DRR Expression of Patient-Derived GSCs after Treatment with CD44 mAb-dsDRR (150 nM) or EphA2 mAb-dsRR (150 nM) Normalized to Untreated Control
Cells were treated for 24 hr, and DRR expression was measured 72 hr post-treatment. (A) Quantification of DRR expression following CD44 mAb-dsDRR treatment is shown. Data were analyzed using one-way ANOVA followed by Dunnett’s post hoc test compared to CD44 mAb-dsScrambled (data are shown as mean + SD; n ≥ 4; *p < 0.05). (B) Representative western blot shows DRR knockdown following treatment with CD44 mAb-dsDRR. (C) Quantification of DRR expression following EphA2 mAb-dsDRR treatment is shown. Data were analyzed using one-way ANOVA followed by Dunnett’s post hoc test compared to EphA2 mAb-dsScrambled (data are shown as mean + SD; n ≥ 3). (D) Representative western blot showing DRR expression following treatment with EphA2 mAb-dsDRR is shown.
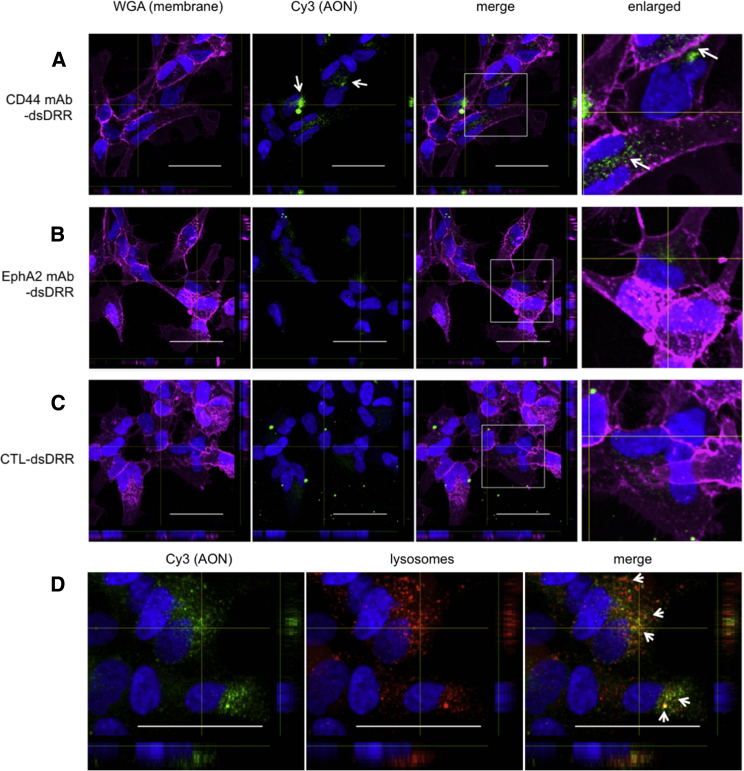
To better understand this discrepancy between the knockdown observed for CD44 mAb-dsDRR and EphA2 mAb-dsDRR, we examined cellular uptake with patient-derived GSCs. We analyzed uptake of the conjugates by confocal microscopy of Cy3-labeled dsDRR after a 3-hr incubation with the mAb-dsDRR conjugates at 75 nM and 150 nM (Figure S4). At 150 nM, we observed considerable uptake of CD44 mAb-dsDRR (Figure 6A), whereas we observed minimal uptake of both EphA2 mAb-dsDRR (Figure 6B) and control, immunoglobulin G (IgG)-dsDRR (Figure 6C). This suggests that the lack of knockdown observed for EphA2 mAb-dsDRR is correlated to a reduced cell uptake of the EphA2 mAb-dsDRR conjugate.
Figure 6.
Antisense Oligonucleotides Conjugated to CD44 mAb Are Taken up by GSCs and Trafficked into the Endolysosomal Pathway
All treatments were at a concentration of 150 nM, and cells were fixed and imaged 3 hr post-treatment. (A) Uptake of CD44 mAb-dsDRR (white arrows) compared to (B) EphA2 mAb-dsDRR and (C) CTL-dsDRR after 3 hr incubation at 37°C is shown. CTL is a non-specific human IgG. (D) Colocalization (white arrows) of CD44 mAb-dsDRR with the lysosomal compartments following a 2-hr pulse and 1-hr chase. Cell membrane (wheat germ agglutinin [WGA], magenta), cell nucleus (Hoechst, blue), AON (Cy3, green), and lysosome (Dextran647, red) are shown. Representative z stack images are shown. All scale bars are 50 μM.
To gain greater insight into the fate of the CD44 mAb-dsDRR conjugate, we incubated the patient-derived GSCs with the CD44 mAb-dsDRR conjugate and a lysosomal marker for 2 hr and then incubated the GSCs cells with fresh media for an additional 1 hr to allow for complete internalization. The dsDRR Cy3 signal almost completely colocalized with the lysosomal marker (Figure 6D), suggesting that the majority of the conjugate was trafficked into the endolysosomal pathway, likely with a small percentage trafficked to the cytoplasm to account for the gene knockdown observed.
Cellular Morphology following DRR Knockdown
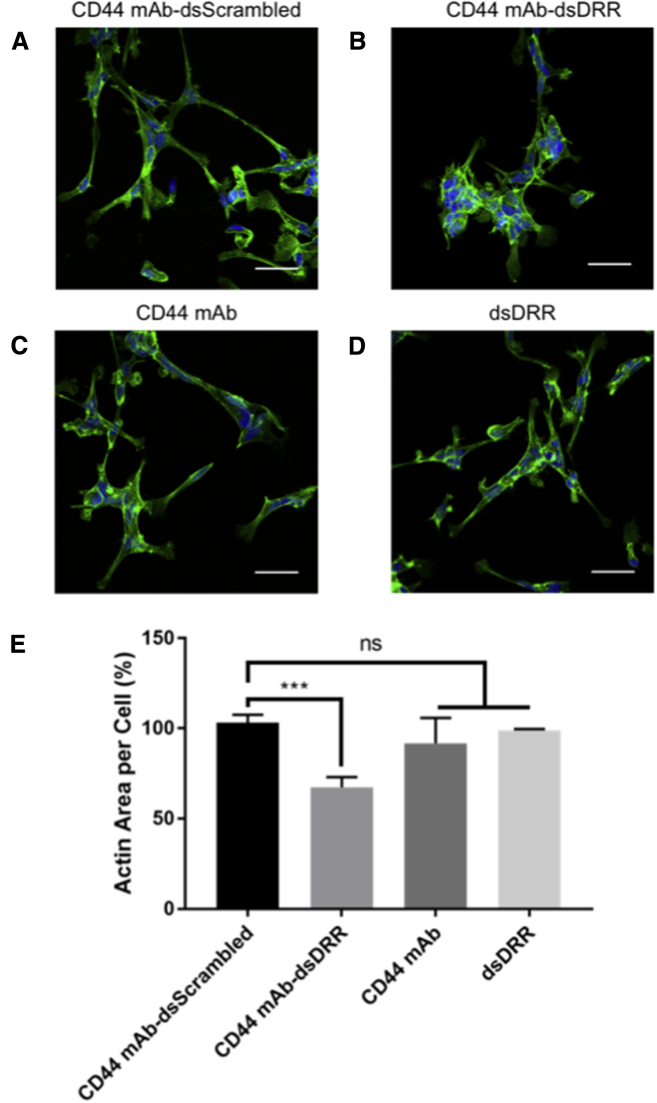
DRR knockdown often results in a change in cell phenotype from spindle- to rounder-shaped cells, reflecting a less invasive morphology.12 Whereas untreated GSCs have a spindle-shaped morphology, we found that GSCs treated with CD44 mAb-dsDRR exhibit a rounder cell morphology with fewer, shorter projections and centralized focal adhesions, suggesting reduced invasive capacity (Figure 7A). We quantified this effect by measuring cell area per nucleus, with actin staining delineating the cell membrane and normalized to an untreated control. Although the cell area was variable due to the complex cell bundles, we observed a significant decrease in cell area per nucleus with the CD44 mAb-dsDRR treatment relative to controls, including the CD44 mAb alone, dsDRR alone, and CD44 mAb-dsScrambled (Figure 7B).
Figure 7.
GSCs Treated with CD44 mAb-dsDRR (150 nM) Have a Rounder Shape, Fewer Projections, and Centralized Focal Adhesions Relative to the Spindle-Shaped Cells of the Control Treatments
Cells were treated for 24 hr and imaged 72 hr post-treatment. (A) Cells treated with CD44 mAb-dsScrambled are shown. (B) Cells treated with CD44 mAb-dsDRR are shown. (C) Cells treated with CD44 mAb alone are shown. (D) Cells treated with dsDRR alone are shown. Representative z stack images are shown. All scale bars are 50 μM. Cell nucleus (Hoechst, blue) and actin (Phalloidin Alexa 488, green) are shown. (E) Change in cellular morphology is quantified as actin area per cell normalized to a no treatment control. Data were analyzed using one-way ANOVA compared to CD44 mAb-dsScrambled with Dunnett’s post hoc correction (data are shown as mean + SD; n ≥ 3; ***p < 0.001).
Discussion
Here, we report, for the first time, an AON-antibody conjugate constructed via covalent click conjugation through a non-therapeutic, sense carrier strand and show how each component of the system is optimized for maximum knockdown of DRR.
The antisense therapeutic strand was optimized for effective knockdown first through phosphorothioate modification of the backbone and FANA modification of specific nucleotides and then by 5-methylation of the CpG containing oligonucleotide. FANA modification has been shown to stabilize the especially sensitive flanking regions of the AON whereas an unmodified gap consisting of 9 DNA nt allows for efficient endonucleolytic RNase H activity.49 We found a trend toward more potent DRR knockdown with the gapmer motif compared to the alternating oligonucleotide FANA-modified altimer. CpG motifs trigger an innate immune response;23, 50 yet methylation of the cytosine residue in strands containing this motif can decrease this immune response, thereby decreasing off-target effects.24, 51 Whereas the immune response was not examined herein, with an ultimate view of testing this strategy in vivo, we confirmed the activity of the methylated CpG therapeutic strand and used it in all of the in vitro studies.
We chose a targeted antibody-mediated approach for AON delivery because, like many polyanions, AONs do not readily cross the cell membrane, especially at low concentrations.52 To determine the optimal antibody for targeted delivery, first we screened the GSCs for antigen expression and then for antibody internalization. Whereas CD44, EphA2, and EGFR are all expressed in many patient-derived GSCs,36, 39, 40, 53 only CD44 and EphA2 were strongly co-expressed with DRR. Notably, EGFR amplification is often lost in vitro, which correlates with our observations.54 By flow cytometry, we saw that both CD44 and EphA2 mAbs were internalized into GBM cells. CD44 mAb-dsDRR more effectively knocked down DRR expression than EphA2 mAb-dsDRR, which was corroborated by greater accumulation of dsDRR in the GSCs when delivered with CD44 mAb. The difference in uptake and efficacy observed might be due to slight differences in internalization pathway or route of endosomal escape, which are difficult to observe using established techniques. Although the inhibitory activity of the engineered antibodies used is not reported, it is important to note that both CD44 and EphA2 antibodies have been reported to play a role in maintaining stem-like characteristics of CSCs.39, 55 In our experiments, we did not observe off-target effects in terms of DRR/FAM107A expression or cell morphology following treatment with CD44 or EphA2 antibodies alone.
To gain insight into the trafficking of the CD44 mAb-dsDRR, we used a pulse-chase experiment and found that the internalized CD44 mAb-dsDRR colocalized with the lysosomes. Whereas it is recognized that antibody-oligonucleotide conjugates are typically trafficked into the endolysosomal pathway,45, 56, 57, 58, 59 the DRR knockdown observed demonstrates that some dsDRR is trafficked into the cytoplasm. Whereas the Cy3 signal associated with the duplex sense strand is colocalized with the lysosomes, some of the antisense strand may have dissociated from the sense strand and diffused into the cytoplasm. To further increase the potency of knockdown, incorporation of moieties to prevent lysosomal accumulation and facilitate early endosomal escape in order to optimize knockdown is being actively pursued.58
We show the first example of an antibody-antisense therapeutic conjugate used to modulate the genetic expression of cancer stem cells. With sufficient dsDRR uptake, we observe both gene knockdown and a change in cell morphology, consistent with a less invasive phenotype: we observe a clear distinction from spindle-shaped cells without treatment to a more rounded morphology with CD44 mAb-dsDRR treatment. This lays the foundation for future studies, where the goal will be to knockdown the DRR gene in GSCs in vivo and thereby reduce tissue invasion and cell metastasis.
Materials and Methods
Cell Lines
GBM DRR+ cells (U-251 MG glioblastoma cells with stable transfection of DsredDRR fusion protein) were cultured as previously described.12 Patient-derived GSCs were provided by the Petrecca lab at McGill University following consent from the patients and approval by the hospital ethics committee. GSCs were expanded as neurosphere in complete neurocult-proliferation media (Neurocult NS-A Proliferation kit [Stem Cell 05751], 20 ng/mL recombinant EGF, 20 ng/mL recombinant basic fibroblast growth factor (bFGF), and 2 μg/mL heparin).
Antibodies
CD44 and EphA2 mAbs were provided by the Toronto Recombinant Antibody Centre (TRAC).47 EGFR mAb (cetuximab) is a clinically available formulation (Erbitux 2 mg/mL; Eli Lilly). CTL antibody (IgG from human serum) was purchased from Sigma and used as received (cat. no. I4506).
AON Synthesis
Nucleoside 3′-phosphoramidites and gene-machine-compatible reagents were purchased from ChemGenes and used as received, and all DBCO-TEG (cat. no. 10-1941) and cyanine 3 CPGs (cat. no. 20-5913-41) were purchased from GlenResearch. All AONs were synthesized on an Applied Biosystems (ABI) 3400 DNA synthesizer at 1 μmol scale using Uny-linker CPG as solid support, except for the sense delivery oligonucleotide, which utilized cyanine 3 CPG as the solid support. The synthesis cycle conditions were as previously described,14 with the exception that 0.02 M I2 in 1:2:10 pyridine/water/tetrahydrofuran (THF) was used for the Cy3/DBCO-modified sense delivery oligonucleotides. When a phosphorothioate backbone was needed, a 0.10 M solution of ([dimethylamino-methylidene]amino)-3H-1,2,4-dithiozaoline-3-thione (DDTT) in Py:MeCN (9:1) was used for the oxidation step instead of the aqueous I2 solution. Deprotection and cleavage from the solid support was achieved in concentrated aqueous NH4OH for 48 hr at room temperature. Purifications were performed by high-performance liquid chromatography (HPLC), using a Protein Pak DEAE 5PW analytical anion-exchange column. A stationary phase of Milli-Q water and a mobile phase of 1 M LiClO4 in water was used for analysis and purification using a gradient of 0%−50% over 46 min. Following purification, excess LiClO4 salts were removed using NAP-25 Sephadex size-exclusion columns. The oligonucleotides were analyzed by liquid chromatography-mass spectrometry (LC-MS) using a Dionex Ultimate 3000 coupled to a Bruker Maxis Impact QTOF in negative ESI mode. Samples were run through an Acclaim RSLC 120 C18 column (2.2 μM 120A 2.1 × 50 mm) using a gradient of 98% mobile phase A (100 mM hexafluoroisopropanol [HFIP] and 5 mM tetraethylammonium [TEA] in H2O) and 2% mobile phase B (MeOH) to 40% mobile phase A and 60% mobile phase B in 8 min. The data were processed and deconvoluted using the Bruker DataAnalysis software version 4.1 (Table S1).
AON Duplex Formation
Sense and AON strands were annealed in annealing buffer (10 mM Tris [pH 7.5–8.0], 50 mM NaCl, and 1 mM EDTA) by heating at 95°C for 2 min followed by slow cooling to room temperature over 1 hr. The annealed oligonucleotides were stored at 4°C until use. Duplex formation was assessed by native PAGE in Tris/Borate/EDTA buffer followed by imaging on a Typhoon FLA 9500 biomolecular imager.
Preparation of mAb-dsDRR and mAb-dsScrambled Conjugates
mAbs were modified with NHS-PEG4-N3 (Thermo 26130) according to the provided protocol from Thermo Fisher Scientific. Briefly, a 100-mM stock solution of NHS-PEG4-N3 was prepared in DMSO (Sigma 472301). 10 equivalents (eq.) of this solution was added to the mAb in PBS (Sigma D8537), and this reaction was shaken for 1 hr at room temperature (RT). After purification by dialysis for 24 hr against PBS, 2 eq. of dsDRR or dsScrambled was added to the N3-modified mAb, and this reaction was allowed to proceed for 3 hr at 37°C. The resulting product was stored at −80°C until use or at 4°C for up to 24 hr. Conjugation efficiency was analyzed by 10% PAGE.
DRR Knockdown Assays: Lipofectamine 2000 Transfection Protocol
This protocol was adapted from Anzahaee et al.14 Briefly, DRR+ GBM cells were seeded at 120,000 cells/well in a 6-well plate and allowed to adhere overnight. AONs were complexed to Lipofectamine 2000 (Thermo 11668027; at 3 μL/well) for 20 min in Opti-MEM media (Thermo 31985062) according to the Lipofectamine 2000 reagent protocol and added to the cells in DMEM media (11995605) for a final DMEM:Opti-MEM ratio of 1.5:0.5 and AON concentration of 25 nM. At 24 hr, an additional 1 mL DMEM media was added. After a total of 72 hr of incubation at 37°C, cells were collected and lysed using 0.1% NP-40 (Fluka 74385). Protein expression was assessed via western blot analysis of DRR with beta-tubulin as a loading control.
DRR Knockdown Assays: Treatment with mAb Conjugate
GSCs were seeded at 250,000 cells/well in poly(l-ornithine)-, poly-L-ornithine (PLO)- (Sigma P4957), and laminin (Thermo CB-40232)-coated 6-well plates and allowed to adhere overnight in Neurocult NS-A proliferation media (Stem Cell 05751). mAb conjugates (or AONs or mAb alone) were added to 500 μL Opti-MEM media and added to the cells for a final Opti-MEM:Neurocult ratio of 1.5:0.5 v/v and mAb-AON concentration of 150 nM. 24 hr later, an additional 1 mL Neurocult media was added. After a total of 72 hr of incubation at 37°C, cells were collected and lysed using 0.1% NP-40 (Sigma 74385). Protein expression was assessed via western blot analysis of DRR with α-tubulin as a loading control.
mAb Internalization Assay
This protocol was adapted from Mumper et al.60 Briefly, the internalization of the CD44 and EphA2 mAbs were determined after measuring surface levels of mAb after various incubation periods at 37°C. CD44 mAb at 4 μg/mL in 2% FBS (Sigma F1051) in PBS (FACS buffer) was added to an equivalent volume of 1 × 106 cells in FACS buffer for one hour on ice at 4°C. The cells were then washed 3× in ice cold FACS buffer and suspended in 200 μL at 37°C for the indicated time. A control sample was held at 4°C throughout the experiment. At the indicated time point, cells were removed from 37°C, quenched with 1 mL ice cold FACS buffer, and then spun to a pellet and resuspended in 2 μg/mL goat anti-human IgG, Alexa 488 (Thermo A-11013) in FACS buffer. Cells were washed 2× with ice cold FACS buffer, 1× with ice cold PBS, and then fixed with 2% paraformaldehyde (PFA) (Bioshop PAR070) in PBS and stored at 4°C until analysis on a BD Accuri C6 flow cytometer. Internalized receptor was quantified using the median fluorescence intensity calculated from FlowJo 10 software.
Immunocytochemistry (CD44, EphA2, and DRR)
GSCs were seeded at 20,000 cells/well in a PLO/laminin-coated 8-well coverglass plate and allowed to adhere overnight at 37°C. The cells were then fixed using 4% PFA. Cells were permeabilized using 1% Triton X-100 (Sigma T-9284) and then stained with either Epha2 mAb or CD44 mAb together with a DRR antibody provided by the Petrecca lab at McGill University. Secondary antibodies were anti-human IgG-Alexa 488 and goat anti-rabbit IgG-Alexa Fluor 647 (Thermo A-21245). The cells were then counterstained with Hoechst 33342 and images were captured on an Olympus FV1000 confocal microscope.
Cellular Uptake
GSCs were seeded at 20,000 cells/well in a PLO/laminin-coated 8-well coverglass plate and allowed to adhere overnight at 37°C. The cells were then treated with the indicated formulation for 3 hr at 37°C, washed with PBS, and fixed using 4% PFA. The cells were stained with wheat germ agglutinin (WGA)-Alexa 488 or 647 (Thermo W11261 or W32466), counterstained with Hoechst (Invitrogen H1399), and imaged on an Olympus FV1000 confocal microscope.
Lysosomal Accumulation
GSCs were seeded at 20,000 cells/well in a PLO/laminin-coated 8-well coverglass plate and allowed to adhere overnight at 37°C. The cells were then treated with the CD44 mAb-dsDRR at 150 nM and the lysosomal marker Dextran Alexa Fluor 647 (Thermo D22914) at 25 μg/mL for 2 hr at 37°C (pulse). The media was then replaced, and the cells were incubated for an additional 1 hr at 37°C (chase) and then washed with PBS and fixed using 4% PFA. The cells were then stained with WGA-Alexa 488 and counterstained with Hoechst, and images were captured on an Olympus FV1000 confocal microscope.
Cellular Morphology following DRR Knockdown
GSCs were seeded at 15,000 cells/well in a PLO/laminin-coated 8-well coverglass plate and allowed to adhere overnight at 37°C. The cells were then treated with the indicated formulation for 72 hr at 37°C, washed with PBS, and fixed using 4% PFA. The cells were then stained with mouse anti-vinculin antibody (Sigma V9131) and anti-mouse Alexa Fluor 647 (Thermo A-21236) antibodies, followed by Alexa 488 phalloidin (Thermo A-12379) to label actin filaments. The cells were counterstained with Hoechst, and images were captured on an Olympus FV1000 confocal microscope.
Author Contributions
A.E.A. conceived and performed experiments and wrote the first and subsequent drafts of the manuscript. E.M.-A. synthesized and characterized all oligonucleotide materials and provided major revisions of the first draft. P.U.L. provided cell lines, advice, and feedback. A.M. performed flow cytometry experiments. S.M.-M. conceptualized fundamental work on the project and provided feedback on the manuscript. M.J.D., K.P., and M.S.S. secured funding, supervised and guided the research, and participated in writing the manuscript.
Acknowledgments
We are grateful to the Canadian Institute for Health Research (CHRP to M.S.S., M.J.D., and K.P. [CPG-140191]) and the Natural Sciences & Engineering Council of Canada (CHRP to M.S.S., M.J.D., and K.P. [CHRPJ 478471-15] and a CGSD to A.E.A.). We thank Professors Dev Sidhu and Jason Moffat and Dr. Jarrett Adams of the TRAC facility for thoughtful discussions and providing us with CD44 mAbs and EphA2 mAbs. We thank Dr. Alex Wahba (McGill University) for MS characterization of AONs. We thank members of the Shoichet, Damha, and Petrecca labs for thoughtful review of this manuscript.
Footnotes
Supplemental Information includes Supplemental Materials and Methods, four figures, and one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.04.004.
Contributor Information
Masad J. Damha, Email: masad.damha@mcgill.ca.
Molly S. Shoichet, Email: molly.shoichet@utoronto.ca.
Supplemental Information
References
- 1.Lacroix M., Abi-Said D., Fourney D.R., Gokaslan Z.L., Shi W., DeMonte F., Lang F.F., McCutcheon I.E., Hassenbusch S.J., Holland E. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J. Neurosurg. 2001;95:190–198. doi: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
- 2.Nieder C., Grosu A.L., Astner S., Molls M. Treatment of unresectable glioblastoma multiforme. Anticancer Res. 2005;25:4605–4610. [PubMed] [Google Scholar]
- 3.Chen J., McKay R.M., Parada L.F. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 5.Gilbertson R.J., Rich J.N. Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nat. Rev. Cancer. 2007;7:733–736. doi: 10.1038/nrc2246. [DOI] [PubMed] [Google Scholar]
- 6.Molina J.R., Hayashi Y., Stephens C., Georgescu M.-M. Invasive glioblastoma cells acquire stemness and increased Akt activation. Neoplasia. 2010;12:453–463. doi: 10.1593/neo.10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paw I., Carpenter R.C., Watabe K., Debinski W., Lo H.-W. Mechanisms regulating glioma invasion. Cancer Lett. 2015;362:1–7. doi: 10.1016/j.canlet.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suvà M.L., Rheinbay E., Gillespie S.M., Patel A.P., Wakimoto H., Rabkin S.D., Riggi N., Chi A.S., Cahill D.P., Nahed B.V. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lathia J.D., Hitomi M., Gallagher J., Gadani S.P., Adkins J., Vasanji A., Liu L., Eyler C.E., Heddleston J.M., Wu Q. Distribution of CD133 reveals glioma stem cells self-renew through symmetric and asymmetric cell divisions. Cell Death Dis. 2011;2:e200. doi: 10.1038/cddis.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradshaw A., Wickremsekera A., Tan S.T., Peng L., Davis P.F., Itinteang T. Cancer stem cell hierarchy in glioblastoma multiforme. Front. Surg. 2016;3:21. doi: 10.3389/fsurg.2016.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L., Darling J., Zhang J.-S., Liu W., Qian J., Bostwick D., Hartmann L., Jenkins R., Bardenhauer W., Schutte J. Loss of expression of the DRR 1 gene at chromosomal segment 3p21.1 in renal cell carcinoma. Genes Chromosomes Cancer. 2000;27:1–10. doi: 10.1002/(sici)1098-2264(200001)27:1<1::aid-gcc1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 12.Le P.U., Angers-Loustau A., de Oliveira R.M.W., Ajlan A., Brassard C.L., Dudley A., Brent H., Siu V., Trinh G., Mölenkamp G. DRR drives brain cancer invasion by regulating cytoskeletal-focal adhesion dynamics. Oncogene. 2010;29:4636–4647. doi: 10.1038/onc.2010.216. [DOI] [PubMed] [Google Scholar]
- 13.Dudley A., Sater M., Le P.U., Trinh G., Sadr M.S., Bergeron J., Deleavey G.F., Bedell B., Damha M.J., Petrecca K. DRR regulates AKT activation to drive brain cancer invasion. Oncogene. 2014;33:4952–4960. doi: 10.1038/onc.2013.436. [DOI] [PubMed] [Google Scholar]
- 14.Anzahaee M.Y., Deleavey G.F., Le P.U., Fakhoury J., Petrecca K., Damha M.J. Arabinonucleic acids: 2′-stereoisomeric modulators of siRNA activity. Nucleic Acid Ther. 2014;24:336–343. doi: 10.1089/nat.2014.0496. [DOI] [PubMed] [Google Scholar]
- 15.Dias N., Stein C.A. Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer Ther. 2002;1:347–355. [PubMed] [Google Scholar]
- 16.Aartsma-Rus A., Krieg A.M. FDA approves eteplirsen for Duchenne muscular dystrophy: the next chapter in the eteplirsen saga. Nucleic Acid Ther. 2017;27:1–3. doi: 10.1089/nat.2016.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein C.A., Castanotto D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 2017;25:1069–1075. doi: 10.1016/j.ymthe.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deleavey G.F., Damha M.J. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012;19:937–954. doi: 10.1016/j.chembiol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Ferrari N., Bergeron D., Tedeschi A.-L., Mangos M.M., Paquet L., Renzi P.M., Damha M.J. Characterization of antisense oligonucleotides comprising 2′-deoxy-2′-fluoro-β-D-arabinonucleic acid (FANA): specificity, potency, and duration of activity. Ann. N Y Acad. Sci. 2006;1082:91–102. doi: 10.1196/annals.1348.032. [DOI] [PubMed] [Google Scholar]
- 20.Lok C.-N., Viazovkina E., Min K.-L., Nagy E., Wilds C.J., Damha M.J., Parniak M.A. Potent gene-specific inhibitory properties of mixed-backbone antisense oligonucleotides comprised of 2′-deoxy-2′-fluoro-D-arabinose and 2′-deoxyribose nucleotides. Biochemistry. 2002;41:3457–3467. doi: 10.1021/bi0115075. [DOI] [PubMed] [Google Scholar]
- 21.Agrawal S., Kandimalla E.R. Antisense therapeutics: is it as simple as complementary base recognition? Mol. Med. Today. 2000;6:72–81. doi: 10.1016/s1357-4310(99)01638-x. [DOI] [PubMed] [Google Scholar]
- 22.Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014;24:374–387. doi: 10.1089/nat.2014.0506. [DOI] [PubMed] [Google Scholar]
- 23.Krieg A.M., Yi A.-K., Matson S., Waldschmidt T.J., Bishop G.A., Teasdale R., Koretzky G.A., Klinman D.M. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Q., Temsamani J., Iadarola P.L., Jiang Z., Agrawal S. Effect of different chemically modified oligodeoxynucleotides on immune stimulation. Biochem. Pharmacol. 1996;51:173–182. doi: 10.1016/0006-2952(95)02177-9. [DOI] [PubMed] [Google Scholar]
- 25.Owen S.C., Patel N., Logie J., Pan G., Persson H., Moffat J., Sidhu S.S., Shoichet M.S. Targeting HER2+ breast cancer cells: lysosomal accumulation of anti-HER2 antibodies is influenced by antibody binding site and conjugation to polymeric nanoparticles. J. Control. Release. 2013;172:395–404. doi: 10.1016/j.jconrel.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 26.Chen F., Hong H., Zhang Y., Valdovinos H.F., Shi S., Kwon G.S., Theuer C.P., Barnhart T.E., Cai W. In vivo tumor targeting and image-guided drug delivery with antibody-conjugated, radiolabeled mesoporous silica nanoparticles. ACS Nano. 2013;7:9027–9039. doi: 10.1021/nn403617j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley R.S., Day E.S. Frizzled7 antibody-functionalized nanoshells enable multivalent binding for Wnt signaling inhibition in triple negative breast cancer cells. Small. 2017;13:1700544. doi: 10.1002/smll.201700544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sivaram A.J., Wardiana A., Howard C.B., Mahler S.M., Thurecht K.J. Recent advances in the generation of antibody-nanomaterial conjugates. Adv. Healthc. Mater. 2018;7:1700607. doi: 10.1002/adhm.201700607. [DOI] [PubMed] [Google Scholar]
- 29.Senter P.D., Sievers E.L. The discovery and development of brentuximab vedotin for use in relapsed Hodgkin lymphoma and systemic anaplastic large cell lymphoma. Nat. Biotechnol. 2012;30:631–637. doi: 10.1038/nbt.2289. [DOI] [PubMed] [Google Scholar]
- 30.Lambert J.M., Chari R.V.J. Ado-trastuzumab Emtansine (T-DM1): an antibody-drug conjugate (ADC) for HER2-positive breast cancer. J. Med. Chem. 2014;57:6949–6964. doi: 10.1021/jm500766w. [DOI] [PubMed] [Google Scholar]
- 31.Lyon R.P., Setter J.R., Bovee T.D., Doronina S.O., Hunter J.H., Anderson M.E., Balasubramanian C.L., Duniho S.M., Leiske C.I., Li F., Senter P.D. Self-hydrolyzing maleimides improve the stability and pharmacological properties of antibody-drug conjugates. Nat. Biotechnol. 2014;32:1059–1062. doi: 10.1038/nbt.2968. [DOI] [PubMed] [Google Scholar]
- 32.Beck A., Goetsch L., Dumontet C., Corvaïa N. Strategies and challenges for the next generation of antibody-drug conjugates. Nat. Rev. Drug Discov. 2017;16:315–337. doi: 10.1038/nrd.2016.268. [DOI] [PubMed] [Google Scholar]
- 33.McCombs J.R., Owen S.C. Antibody drug conjugates: design and selection of linker, payload and conjugation chemistry. AAPS J. 2015;17:339–351. doi: 10.1208/s12248-014-9710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Bent M., Gan H.K., Lassman A.B., Kumthekar P., Merrell R., Butowski N., Lwin Z., Mikkelsen T., Nabors L.B., Papadopoulos K.P. Efficacy of depatuxizumab mafodotin (ABT-414) monotherapy in patients with EGFR-amplified, recurrent glioblastoma: results from a multi-center, international study. Cancer Chemother. Pharmacol. 2017;80:1209–1217. doi: 10.1007/s00280-017-3451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neyns B., Sadones J., Joosens E., Bouttens F., Verbeke L., Baurain J.F., D’Hondt L., Strauven T., Chaskis C., In’t Veld P. Stratified phase II trial of cetuximab in patients with recurrent high-grade glioma. Ann. Oncol. 2009;20:1596–1603. doi: 10.1093/annonc/mdp032. [DOI] [PubMed] [Google Scholar]
- 36.Brown D.V., Filiz G., Daniel P.M., Hollande F., Dworkin S., Amiridis S., Kountouri N., Ng W., Morokoff A.P., Mantamadiotis T. Expression of CD133 and CD44 in glioblastoma stem cells correlates with cell proliferation, phenotype stability and intra-tumor heterogeneity. PLoS ONE. 2017;12:e0172791. doi: 10.1371/journal.pone.0172791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown D.V., Daniel P.M., D’Abaco G.M., Gogos A., Ng W., Morokoff A.P., Mantamadiotis T. Coexpression analysis of CD133 and CD44 identifies proneural and mesenchymal subtypes of glioblastoma multiforme. Oncotarget. 2015;6:6267–6280. doi: 10.18632/oncotarget.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anido J., Sáez-Borderías A., Gonzàlez-Juncà A., Rodón L., Folch G., Carmona M.A., Prieto-Sánchez R.M., Barba I., Martínez-Sáez E., Prudkin L. TGF-β receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 39.Binda E., Visioli A., Giani F., Lamorte G., Copetti M., Pitter K.L., Huse J.T., Cajola L., Zanetti N., DiMeco F. The EphA2 receptor drives self-renewal and tumorigenicity in stem-like tumor-propagating cells from human glioblastomas. Cancer Cell. 2012;22:765–780. doi: 10.1016/j.ccr.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liffers K., Lamszus K., Schulte A. EGFR amplification and glioblastoma stem-like cells. Stem Cells Int. 2015;2015:427518. doi: 10.1155/2015/427518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lima W.F., Nichols J.G., Wu H., Prakash T.P., Migawa M.T., Wyrzykiewicz T.K., Bhat B., Crooke S.T. Structural requirements at the catalytic site of the heteroduplex substrate for human RNase H1 catalysis. J. Biol. Chem. 2004;279:36317–36326. doi: 10.1074/jbc.M405035200. [DOI] [PubMed] [Google Scholar]
- 42.Bäumer S., Bäumer N., Appel N., Terheyden L., Fremerey J., Schelhaas S., Wardelmann E., Buchholz F., Berdel W.E., Müller-Tidow C. Antibody-mediated delivery of anti-KRAS-siRNA in vivo overcomes therapy resistance in colon cancer. Clin. Cancer Res. 2015;21:1383–1394. doi: 10.1158/1078-0432.CCR-13-2017. [DOI] [PubMed] [Google Scholar]
- 43.Bäumer N., Appel N., Terheyden L., Buchholz F., Rossig C., Müller-Tidow C., Berdel W.E., Bäumer S. Antibody-coupled siRNA as an efficient method for in vivo mRNA knockdown. Nat. Protoc. 2016;11:22–36. doi: 10.1038/nprot.2015.137. [DOI] [PubMed] [Google Scholar]
- 44.Uckun F.M., Qazi S., Dibirdik I., Myers D.E. Rational design of an immunoconjugate for selective knock-down of leukemia-specific E2A-PBX1 fusion gene expression in human Pre-B leukemia. Integr. Biol. 2013;5:122–132. doi: 10.1039/c2ib20114c. [DOI] [PubMed] [Google Scholar]
- 45.Sugo T., Terada M., Oikawa T., Miyata K., Nishimura S., Kenjo E., Ogasawara-Shimizu M., Makita Y., Imaichi S., Murata S. Development of antibody-siRNA conjugate targeted to cardiac and skeletal muscles. J. Control. Release. 2016;237:1–13. doi: 10.1016/j.jconrel.2016.06.036. [DOI] [PubMed] [Google Scholar]
- 46.Astriab-Fisher A., Fisher M.H., Juliano R., Herdewijn P. Increased uptake of antisense oligonucleotides by delivery as double stranded complexes. Biochem. Pharmacol. 2004;68:403–407. doi: 10.1016/j.bcp.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 47.Hornsby M., Paduch M., Miersch S., Sääf A., Matsuguchi T., Lee B., Wypisniak K., Doak A., King D., Usatyuk S. A high through-put platform for recombinant antibodies to folded proteins. Mol. Cell. Proteomics. 2015;14:2833–2847. doi: 10.1074/mcp.O115.052209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min K.-L., Viazovkina E., Galarneau A., Parniak M.A., Damha M.J. Oligonucleotides comprised of alternating 2′-deoxy-2′-fluoro-β-D-arabinonucleosides and D-2′-deoxyribonucleosides (2‘F-ANA/DNA ’altimers’) induce efficient RNA cleavage mediated by RNase H. Bioorg. Med. Chem. Lett. 2002;12:2651–2654. doi: 10.1016/s0960-894x(02)00439-0. [DOI] [PubMed] [Google Scholar]
- 49.Champoux J.J., Schultz S.J. Ribonuclease H: properties, substrate specificity and roles in retroviral reverse transcription. FEBS J. 2009;276:1506–1516. doi: 10.1111/j.1742-4658.2009.06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klinman D.M., Yamshchikov G., Ishigatsubo Y. Contribution of CpG motifs to the immunogenicity of DNA vaccines. J. Immunol. 1997;158:3635–3639. [PubMed] [Google Scholar]
- 51.Henry S., Stecker K., Brooks D., Monteith D., Conklin B., Bennett C.F. Chemically modified oligonucleotides exhibit decreased immune stimulation in mice. J. Pharmacol. Exp. Ther. 2000;292:468–479. [PubMed] [Google Scholar]
- 52.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 53.Persano L., Rampazzo E., Basso G., Viola G. Glioblastoma cancer stem cells: role of the microenvironment and therapeutic targeting. Biochem. Pharmacol. 2013;85:612–622. doi: 10.1016/j.bcp.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 54.Pandita A., Aldape K.D., Zadeh G., Guha A., James C.D. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer. 2004;39:29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 55.Chanmee T., Ontong P., Kimata K., Itano N. Key roles of hyaluronan and its CD44 receptor in the stemness and survival of cancer stem cells. Front. Oncol. 2015;5:180. doi: 10.3389/fonc.2015.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cuellar T.L., Barnes D., Nelson C., Tanguay J., Yu S.-F., Wen X., Scales S.J., Gesch J., Davis D., van Brabant Smith A. Systematic evaluation of antibody-mediated siRNA delivery using an industrial platform of THIOMAB-siRNA conjugates. Nucleic Acids Res. 2015;43:1189–1203. doi: 10.1093/nar/gku1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu B. Exploring cell type-specific internalizing antibodies for targeted delivery of siRNA. Brief. Funct. Genomics Proteomics. 2007;6:112–119. doi: 10.1093/bfgp/elm015. [DOI] [PubMed] [Google Scholar]
- 58.Varkouhi A.K., Scholte M., Storm G., Haisma H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 59.Bäumer N., Berdel W.E., Bäumer S. Immunoprotein-mediated siRNA delivery. Mol. Pharm. 2017;14:1339–1351. doi: 10.1021/acs.molpharmaceut.6b01039. [DOI] [PubMed] [Google Scholar]
- 60.Glatt D.M., Beckford Vera D.R., Parrott M.C., Luft J.C., Benhabbour S.R., Mumper R.J. The interplay of antigen affinity, internalization, and pharmacokinetics on CD44-positive tumor targeting of monoclonal antibodies. Mol. Pharm. 2016;13:1894–1903. doi: 10.1021/acs.molpharmaceut.6b00063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.