Abstract
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-related death worldwide. Prognosis is poor, and therapeutic options are limited. MicroRNAs (miRNAs) have emerged as potential therapeutic molecules against cancer. Here, we investigated the therapeutic efficacy of miR-199a-3p, an miRNA highly expressed in normal liver and downregulated in virtually all HCCs. The therapeutic value of miR-199a-3p mimic molecules was assayed in the TG221 mouse, a transgenic model highly predisposed to the development of liver cancer. Administration of miR-199a-3p mimics in the TG221 transgenic mouse showing liver cancer led to a significant reduction of number and size of tumor nodules compared to control animals. In vivo delivery confirmed protein downregulation of the miR-199a-3p direct targets, mechanistic target of rapamycin (MTOR) and p21 activated kinase 4 (PAK4), ultimately leading to the repression of FOXM1. Remarkably, the anti-tumor activity of miR-199a-3p mimics was comparable to that obtained with sorafenib. These results suggested that miR-199a-3p may be considered a promising HCC therapeutic option.
Keywords: miR-199a-3p, miRNA therapy, hepatocellular carcinoma, transgenic mouse
Introduction
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-related death worldwide.1 Despite the development of new therapeutic strategies, prognosis remains poor, and life expectancy after diagnosis of advanced cancer is approximately 6 months.2 Conventional treatments such as chemotherapy and radiation therapy have limitations, including resistance to treatments and significant toxicity.3, 4, 5 The multi-tyrosine kinase inhibitor sorafenib is the only systemic drug effective in first-line treatments in advanced-stage HCC patients.6 Very recently, new molecules (regorafenib, lenvatinib, nivolumab, and tremelimumab) have entered or are close to entering clinical use,7, 8, 9 after many failures of clinical trials in the past decade.10, 11
MicroRNAs (miRNAs) have emerged as experimental therapeutic molecules against cancer.12 miRNAs are small non-coding RNA molecules involved in the post-transcriptional modulation of gene expression, and their aberrant expression is associated with human cancer, including HCC.13, 14, 15
miR-199a-3p, the third most highly expressed miRNA in normal liver, is downregulated in virtually all HCCs, and its decrement correlates with poor prognosis.16, 17 miRNAs downregulation in liver cancer was associated to phosphorylation of exportin-5 by extracellular signal-related kinase (ERK), a mechanism that decreases export of pre-miRNAs from nucleus.18 The role of miR-199a-3p in tumorigenesis is still under investigation, but several studies have reported some of its mechanisms of action. It can inhibit cell growth through the downregulation of hypoxia inducible factor 1 alpha subunit (HIFA)19 and p21 activated kinase 4 (PAK4),17 and can promote apoptosis through Yes-associated protein 1 (YAP1) inhibition.20 Restoration of miR-199a-3p in HCC cell lines leads to reduced invasiveness and enhanced doxorubicin sensitivity by controlling the expression of the mechanistic target of rapamycin (MTOR), CD44, and MET proto-oncogene.21, 22 Recently, the ability of miR-199a-3p to suppress tumor growth, migration, invasion, and angiogenesis in HCC by targeting vascular endothelial growth factor A (VEGFA) and its receptors, hepatocyte growth factor (HGF) and matrix metallopeptidase 2 (MMP2), has been shown.23
Here, we investigated the potential therapeutic activity of miR-199a-3p in the TG221 transgenic mouse,24 a model highly predisposed to the development of liver cancer. Our study shows that miR-199a-3p can produce an anti-tumor effect similar to sorafenib or the MTOR inhibitor WYE-354, by acting on MTOR and PAK4 molecular pathways.
Results
Anti-tumor Activity of miR-199a-3p Was Similar to that of Sorafenib in the TG221 Transgenic Mouse Model
The TG221 transgenic mouse model was employed to assess the therapeutic efficacy of miR-199a-3p. In all experiments, mice were treated intraperitoneally (i.p.) with the carcinogen N-diethylnitrosamine (DEN) at 10 days of age to accelerate the appearance of liver tumors. All experiments were performed in male mice.
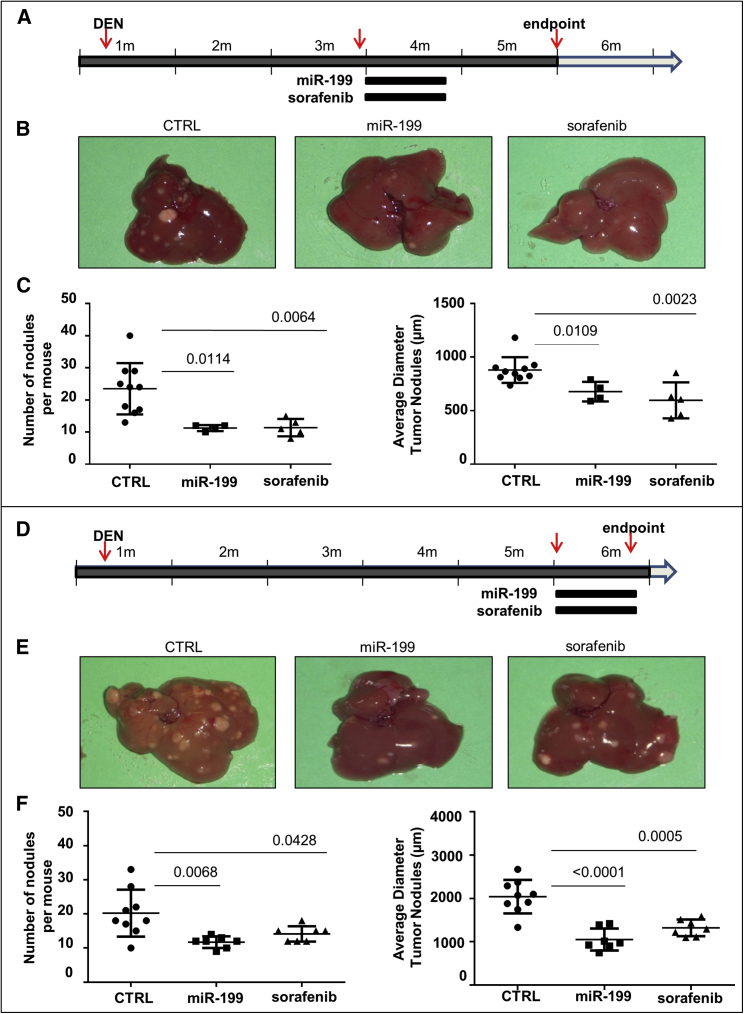
In the first experiment (Figure 1A), 4 mice (3 months old) received miR-199a-3p mimics (5 mg/Kg) by i.p. injection 3 times a week. Ten mice received scrambled oligonucleotides, and 5 mice received a daily oral administration of sorafenib (5 mg/kg). The i.p. administration was preferred to the intravenous (i.v.) route because the efficiency of miRNA delivery to liver was comparable (Figure S1) and, in case of multiple injections, more feasible and reproducible. All treatments were performed for 3 consecutive weeks. At 5 months old, 1 month after the end of treatments, mice were sacrificed, and number and size of liver lesions were assessed. Multifocal liver lesions were detected, and tumor lesions were either HCCs or dysplastic adenomas as previously described.24 Detectable tumor nodules were significantly fewer and smaller in miR-199a-3p-treated mice than in control animals (Figures 1B and 1C). Interestingly, the results obtained with miR-199a-3p were comparable to those obtained in mice treated with sorafenib.
Figure 1.
miR-199a-3p Mimics Controlled Liver Tumor Growth
(A) Mice were treated i.p. with the carcinogen N-diethylnitrosamine (DEN) at 10 days of age. miR-199a-3p mimics (5 mg/kg) were intraperitoneally injected three times a week for 3 weeks in 3-month-old mice. Identical schedule and amounts of scrambled oligonucleotides were used as negative controls (CTRL). Sorafenib treatment (5 mg/kg, daily oral administration) was used as a positive control for a reference drug in use against liver cancer. (B and C) At 5 months of age, mice were sacrificed, and liver lesions quantified. Number and size of nodules were significantly smaller in mice treated with miR-199a-3p and sorafenib than in the controls. (D) A similar experiment with a larger group of miR-199a-3p-treated mice and a slightly different timing of therapeutic interventions. Treatments began at 5 months of age, and mice were sacrificed immediately after the end of treatment. (E and F) In agreement with the results of the previous experiment, number and size of nodules were significantly smaller in mice treated with miR-199a-3p and sorafenib compared to the controls. In the figure, miR-199a-3p is indicated as miR-199.
In a second independent experiment (Figure 1D), the treatment schedule was slightly modified as it began when the mice were 5 months old and were sacrificed after 3 weeks, 24 hr after the end of the last treatments. Experimental groups were as follows: miR-199a-3p (seven mice), scrambled (nine mice), and sorafenib (seven mice). Measurements of anti-tumor efficacy confirmed the results of the first experiment. The number and size of nodules were significantly smaller than controls after treatment with miR-199a-3p and comparable with those obtained with sorafenib (Figures 1D–1F). A fourth group of animals (seven mice) was included to assay a possible additive or synergistic anti-tumor effect of the combination miR-199a-3p with sorafenib. The effect was not significantly higher than the single agents (Figure S2A).
Multiple Pathways Were Affected by miR-199a-3p Treatments
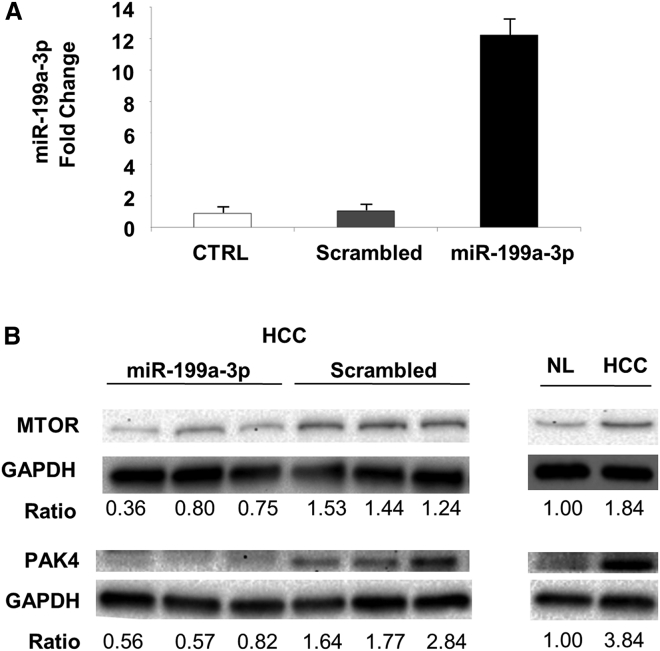
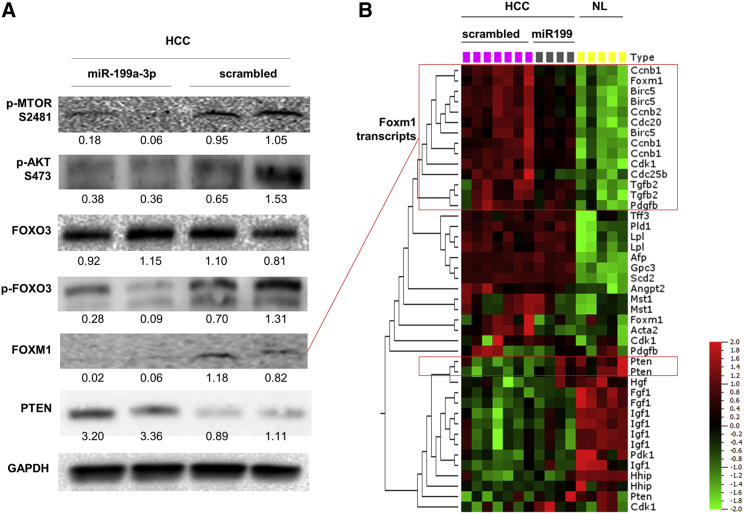
To clarify the mechanisms underlying the anti-tumor activity elicited by miR-199a-3p, we collected and analyzed livers immediately after mice sacrifice. We confirmed that liver was the organ most efficiently targeted by miR-199a-3p delivery (Figure S3). miR-199a-3p levels were considerably increased in liver tumors from mice that were injected with miRNA mimics (p = 0.006) (Figure 2A). The high level of miR-199a-3p led to a decrease of the target proteins MTOR and PAK4 (Figure 2B). Additionally, proteins of the downstream pathways, including phospho-AKT, phospho-forkhead box O3 (FOXO3), and FOXM1, were also affected (Figure 3). The reduced activity of FOXM1 was also confirmed by the analysis of differentially expressed genes in HCC and normal liver: several genes whose transcription was induced by Foxm1 were downregulated in miR-199a-3p-treated HCCs to a level that was intermediate between control HCC and normal liver (Figure 3; Table S1). Interestingly, the level of the tumor suppressor phosphatase and tensin homolog (PTEN) protein and mRNA, which was strongly downregulated in tumors (Figure S4), was restored through an indirect mechanism after treatment with miR-199a-3p (Figure 3).
Figure 2.
Increased miR-199a-3p Expression Correlated to MTOR Downregulation In Vivo
(A) To quantify microRNA (miRNA) molecules at 24 hr from the end of treatments, we performed a droplet digital PCR (ddPCR) on RNA isolated from tumors. A significant increase in miR-199a-3p levels was detected in liver tumors of mice treated with miRNA mimics compared to the other conditions, normal livers (NL), and liver tumors treated with scrambled oligonucleotides (p = 0.006). Data are represented as mean + SD. (B) Immunoblotting analysis of hepatocellular carcinoma (HCC) tissues treated with miR-199a-3p revealed downregulation of MTOR and p21 activated kinase 4 (PAK4) proteins compared to the controls. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal normalizer.
Figure 3.
miR-199a-3p Affected MTOR-Associated Pathway In Vivo
(A) Immunoblotting analyses of hepatocellular carcinoma (HCC) tissues treated or untreated with miR-199a-3p were performed to investigate the molecular effects of miR-199a-3p on its target genes. We found that proteins involved in the MTOR pathway (phospho-AKT, phospho-forkhead box O3 [pFOXO3], and FOXM1) were downregulated in tumors treated with miR-199a-3p. Phosphatase and tensin homolog (PTEN) protein levels were higher in treated tumors than controls. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal normalizer. (B) A hierarchical cluster analysis of differentially expressed genes in HCC and normal liver samples (NL) (experiment described in Figure 1D) was performed. Specifically, HCC of scrambled-treated mice, HCC of miR-199a-3p mice, and NL of untreated mice were compared. The analysis showed that several genes controlled by FOXM1 (Ccnb1, Cdc25b, Cdc20, cyclin-dependent kinase 1 [Cdk1], transforming growth factor-beta 2 [Tgfb2], survivin, and Foxm1 itself) were downregulated in miR-199a-3p-treated samples. The colors of the genes represent the expression values normalized using the mean expression across all samples (green, downregulated; red, upregulated). (B) miR-199a-3p is indicated as miR-199.
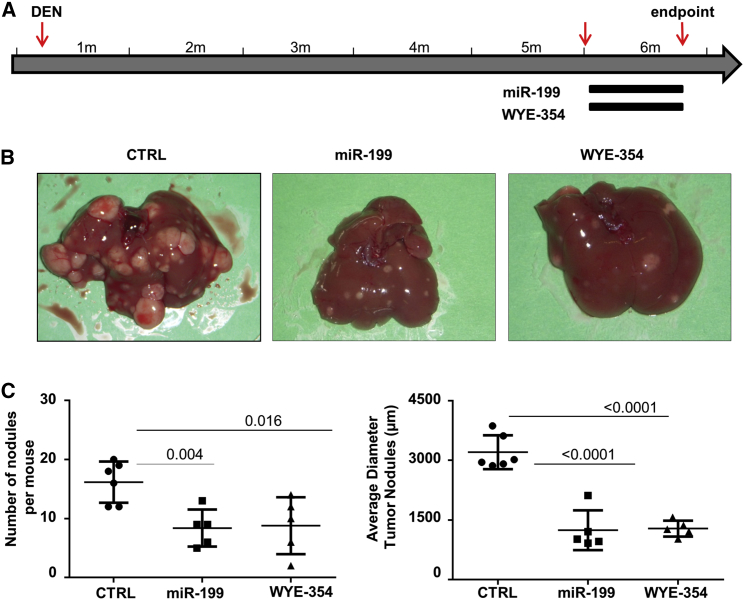
The MTOR Inhibitor WYE-354 Exhibited an Anti-tumor Activity Comparable to that of miR-199a-3p
The importance of MTOR pathway in liver tumors of the TG221 mouse was investigated by evaluating the anti-tumor efficacy of the MTOR inhibitor WYE-354, a specific MTORC1 and MTORC2 inhibitor. Five mice received a daily oral administration of WYE-354 (10 mg/kg) for 3 weeks, five mice received miR-199a-3p mimics as described above, and six mice received scrambled oligonucleotides. At the end of treatments, all mice (6 months old) were sacrificed, and number and size of liver lesions were assessed (Figure 4). Number and size of nodules were reduced in all the groups of treated mice compared to control animals, indicating a similar in vivo anti-tumor activity of miR-199a-3p or MTOR inhibitor compound. A fourth group of mice (five animals) received the combinatorial administration of miR-199a-3p and WYE-354. No significant differences were observed between single agents and their combination (Figure S2B). The drug activity was confirmed by the inhibition of phosphorylation at AKT (S-473) and MTOR (S-2481) in mice treated with WYE-354 compared to control animals (Figure S5).
Figure 4.
MTOR Inhibitor WYE-354 Had Anti-tumor Activity Comparable to that of miR-199a-3p
(A) The anti-tumor activity of the MTOR inhibitor WYE-354 was tested in TG221 mice. A group of 5-month-old mice, previously treated i.p. with the carcinogen N-diethylnitrosamine (DEN) at 10 days of age, received a daily oral administration of WYE-354 (10 mg/kg) for 3 weeks, while a group of mice received miR-199a-3p mimics and scrambled oligonucleotides (CTRL) as scheduled in previous experiments. At the end of treatments, all mice were sacrificed. (B and C) The histological evaluation of liver lesions showed a similar reduction in number and size of tumor nodules in mice treated with both miR-199a-3p and the MTOR inhibitor. In the figure, miR-199a-3p is indicated as miR-199.
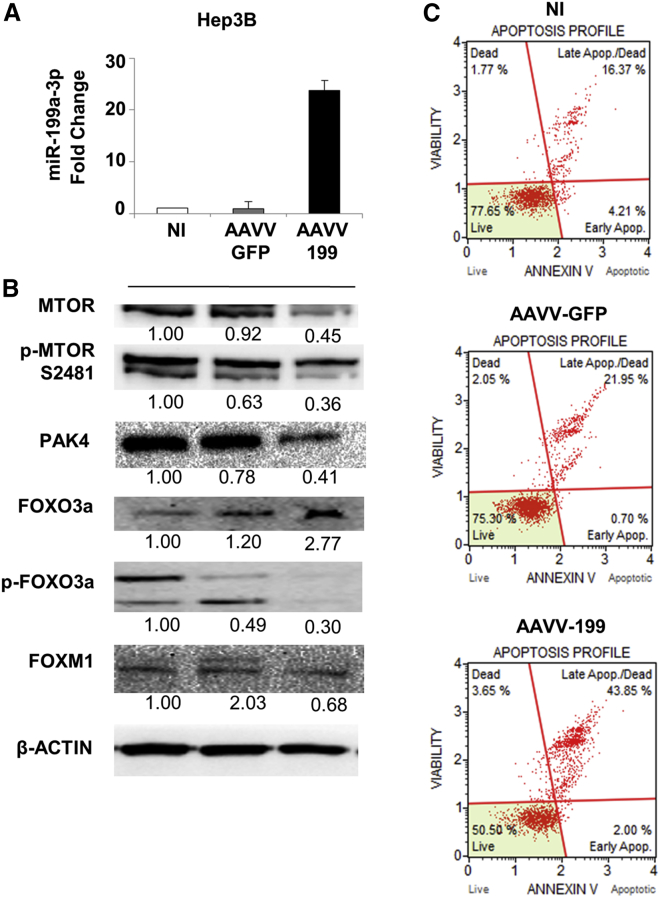
Enforced Expression of miR-199a-3p Induced Apoptosis and Deregulation of the MTOR and PAK4 Signaling Pathways in Human HCC Cells In Vitro
To confirm the biological and molecular effects of miR-199a-3p in human cells, we infected the HCC Hep3B and HepG2 cells with an adeno-associated viral vector (AAVV) expressing miR-199a-3p (AAVV-199). The level of miR-199a-3p and its biological effects were evaluated 120 hr post-infection, time needed to allow gene expression from an AAVV. The upregulation of miR-199a-3p caused an increase in cell apoptosis. Weak effects were observed for the control AAVV-GFP (Figure 5). Molecular analyses confirmed the downregulation of MTOR and its phosphorylated form, PAK4, as well as the downregulation of phospho-FOXO3A and FOXM1 proteins (Figure 5). The association between FOXO3A and FOXM1 was confirmed by the enforced overexpression of FOXO3A in Hep3B cells, which induced a decrease in FOXM1 levels compared to the control (Figure S6). The same apoptotic effect was observed in HepG2 cells after miR-199a-3p upregulation (Figure S7).
Figure 5.
Overexpression of miR-199a-3p Increased Apoptosis Levels in Hep3B Cells by Affecting MTOR and PAK4 Pathways
miR-199a-3p expression increased in Hep3B cells by an adeno-associated viral vector (AAVV) expressing miR-199a-3p (AAVV-199). As experimental controls, the cells were infected with a control virus AAVV- GFP or uninfected. Cells were collected and analyzed 120 hr after infection. (A) The expression levels of miR-199a-3p in the infected cells were determined by qPCR, and proteins were analyzed using immunoblot. miR-199a-3p levels were significantly higher in cells infected with AAVV-199 compared to the controls (p < 0.0001). All the data reported are an average of the experiment performed in triplicate (mean + SD). (B) The increased expression of miR-199a-3p-related genes involved in the MTOR and PAK4 pathways. In samples infected with AAVV-199, immunoblotting analyses revealed a downregulation of MTOR and its phosphorylated form p-2481, PAK4, the phosphorylated form of forkhead box O3 (FOXO3a), and FOXM1 compared to the controls (AAVV-GFP, NI). (C) Cells treated with miR-199a-3p showed a considerable increase in the percentage of total apoptosis compared to control cells. Notably, the control virus AAVV-GFP did not induce appreciable changes in cell viability (live cells in the plot) compared to the uninfected control, indicating the non-toxicity of the virus itself and the specificity of the effect mediated by AAVV-199.
Discussion
miRNA-based therapies have become of great interest. Restoration of tumor suppressor miRNAs or inhibition of oncogenic miRNAs are approaches that have been used in several pre-clinical models, including liver cancer.25 The anti-tumor activity of miR-199a-3p has been previously tested in subcutaneous and orthotopic HCC mouse models,17, 19, 23, 26 confirming the tumor-suppressing activity of miR-199a-3p. In the present work, we expanded the evaluation of the anti-tumor activity of miR-199a-3p using the TG221 transgenic mouse.24 Liver of these mice are characterized by the presence of steatohepatitis, which resembles the human condition named non-alcoholic steatohepatitis (NASH) and develops spontaneous cancers at 9–12 months of age. In this mouse, the treatment with the carcinogen DEN accelerates the appearance of tumors that become visible at 3–4 months of age in all male mice.
We performed various independent experiments to assess the in vivo anti-cancer activity of miR-199a-3p in liver tumors of the TG221 mice. Irrespectively of their design, all experiments indicated that miR-199a-3p induced a significant reduction of number and size of tumor nodules, proving the potential usefulness of this miRNA mimic as a therapeutic agent.
As mechanism of action, we have shown that both MTOR and PAK4 oncoproteins are downregulated following miR-199a-3p-enforced expression by in vivo mimics delivery into mouse tumors or in vitro AAVV infection of human HCC cell lines. Because the importance of these proteins in cancer is well established,27, 28 these data indicate that their downregulation is important for the induction of miR-199a-3p anti-tumor activity.
MTOR is an essential factor of the PI3K-AKT-PTEN molecular pathway, important for cell survival, proliferation, and switch to a glycolytic metabolism. Albeit MTOR gene is mutated at lower frequency than other elements of the pathway in human cancer, it is a target of small molecules that are employed as anti-tumor agents in certain cancers of the kidney and breast.
PAK4, which is an effector for small GTPases activated by the HGF receptor MET29, 30 such as RAC or cell division control protein 42 (CDC42), is at the node of various oncogenic pathways.28, 31 PAK4 can activate ERK, AKT, and WNT pathways involved in cell survival and proliferation.32 PAK4 can also activate signal transducer and activator of transcription 3 (STAT3) and maintain the stem cell phenotype.33 Finally, PAK4 interacts and phosphorylates several proteins that mediate migration, invasion, angiogenesis, and vascular permeability.28, 29, 31 PAK4 mutations and amplification have been reported in human malignancies.28 Various PAK4 inhibitors have been developed, but none have yet been validated for clinical use.28
In the absence of useful PAK4-specific small-molecule inhibitors, we tested the anti-tumor effect of MTOR inhibition by using the drug WYE-354, a second generation inhibitor able to block both MTORC1 and MTORC234 in comparison with miR-199a-3p. It has been shown that WYE-354 has anti-tumor activity in vitro and in vivo in mouse xenografts of human colon, gallbladder, and prostate cancer,35, 36 and is being tested in early-phase trials.37 In our model, miR-199a-3p and the MTOR inhibitor compound exhibited a comparable anti-tumor activity. These results confirm first the importance of this pathway for tumor development and second the importance of its control for the anti-tumor activity mediated by miR-199a-3p. Interestingly, a strong anti-tumor activity of the WYE-354 inhibitor was previously observed in PTEN null prostatic and glioma cell xenograft models.35 It is notable that liver tumors of the TG221 model presented a significant PTEN downregulation.
By reducing MTOR and PAK4 functions, miR-199a-3p led to a reduced phosphorylation and degradation of the tumor suppressor protein FOXO3,38 ultimately leading to the inhibition of the oncogenic FOXM1 transcription factor. An association between FOXO3 and FOXM1 has been previously reported39, 40 and was also confirmed in the present study. The indirect modulation of FOXM1 by miR-199a-3p was confirmed by the analysis of genes whose transcription is controlled by FOXM1: several genes involved in cell-cycle progression, such as Ccnb1, Cdc25b, Cdc20, and cyclin-dependent kinase 1 (Cdk1),41, 42 as well as transforming growth factor-beta 2 (Tgfb2) and survivin (Birc5), were all downregulated in miR-199a-3p-treated samples. The essential role of FOXM1 in hepatic tumor progression has been previously demonstrated in vivo by showing its upregulation in DEN-induced liver tumors.43 Additionally, it has been shown that FOXM1 depletion led to a reduction in the number of tumors in HRas transgenic mice.44 In humans, FOXM1 is overexpressed in several solid tumors, including liver,45 and correlates with poor prognosis of liver cancer patients.46 Our results indicate that FOXM1 may represent a critical downstream effector of liver tumorigenesis discovered in the TG221 mouse model, and miR-199a-3p exhibits an anti-tumor action through its inhibition via indirect mechanisms that result from the MTOR and PAK4 controlled pathways.
This study also revealed that miR-199a-3p mimics had an antitumor effect comparable to sorafenib, the only systemic drug currently approved as first line for advanced HCC. Although the therapeutic use of miR-199-3p in place of sorafenib cannot be suggested, it should be noted that miRNA mimics do not show the serious toxicity frequently associated with sorafenib,47, 48 possibly suggesting a testing of miR-199a-3p mimics for patients intolerant to sorafenib. Furthermore, miR-199a-3p was shown to increase susceptibility to doxorubicin-induced apoptosis,21 suggesting a possible use in combination with other chemotherapeutic agents.
In conclusion, having shown that miR-199a-3p administration led to a reduction of in vivo tumor growth comparable to that obtained with sorafenib, and induced growth inhibition and pro-apoptotic activity in human HCC cells, the present study suggests that miR-199a-3p mimics can represent therapeutic molecules potentially useful for the treatment of HCC. In addition, the study highlighted the role of miR-199a-3p in the regulation of MTOR and PAK4 pathways, ultimately leading to inhibition of FOXM1, indicating these oncoproteins as critical effectors of liver cancer and potential direct targets of anti-HCC therapies.
Materials and Methods
In Vivo Mouse Studies
The TG221 transgenic mouse was used in all experiments.24 Mice were maintained in vented cabinets at 25°C with 12-hr light-dark cycle, with food and water ad libitum. To facilitate tumor development, 10-day newborn male mice received one i.p. injection of DEN (Sigma-Aldrich, St. Louis, MO, USA) (7.5 mg/kg body weight). The study was performed according with the Guidelines for the Care and Use of Laboratory Animals of the Italian Ministry of Health. To comply with the 2010/63/EU directive of the European Parliament and Council, enforced by the Italian law requiring a minimized number of experimental animals, G*Power (http://www.gpower.hhu.de/) was used to define the sample size for long-term experiments. For short-term experiments, the smallest number of mice sufficient to perform statistical analyses was used. All animals were randomly assigned to different treatment groups at the start of the studies. The protocol for animal experimentation was approved by the Italian Ministry of Health (approval no. 55/2015-PR released on January 29, 2015).
RNA Oligonucleotides for In Vivo Therapies
miR-199a-3p and scrambled unmodified single-stranded RNA oligonucleotides were obtained from Axolabs (Kulmbach, Germany). In vivo delivery was facilitated by the use of lipid nanoparticles as vehicle. The injections were performed i.p. three times a week for a period of 3 weeks, and oligonucleotides were administered to a final concentration of 5 mg/kg. Mice were randomly enrolled for treatments.
Lipid Nanoparticles
The lipid components of the nanoparticles were 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dimyristoyl-sn-glycerol, methoxypolyethylene glycol (DMG-PEG; Mw 2,000; Avanti Polar Lipids, Alabaster, AL, USA), and linoleic acid (Sigma-Aldrich). The molar ratio of DOPE:linoleic acid:DMG-PEG was 50:48:2. The preparation of empty nanoparticles was performed as previously described.49
Anti-tumor Drugs
Sorafenib tosylate (Cat No. S1040; Selleckchem, Houston, TX, USA) was dissolved in a 50:50 Cremophor EL and ethanol solution for in vivo experiments. WYE-354 (Cat No. S1266; Selleckchem) was dissolved in dimethyl sulfoxide (DMSO) for in vitro experiments and in 4% DMSO, 30% PEG 300, and 5% Tween 80 for in vivo experiments as indicated by manufacturer’s instructions. The mice received a daily oral administration of sorafenib or WYE-354 at the final concentration of 5 and 10 mg/kg, respectively.
Recombinant AAVV-199
To obtain an AAVV expressing miR-199a-3p, all the plasmids required for AAVV generation (pAAV-IRES-GFP, pAAV-DJ, and pHelper) were purchased from Cell Biolabs (San Diego, CA, USA). The cassette expressing the miR-199a-3p sequence was obtained from the pIRES-miR-199a plasmid50 and cloned upstream of the IRES-GFP sequence into pAAV-IRES-GFP using XbaI sites. The infectious recombinant AAVV-199 was generated by a 1:1:1 (molar ratio) triple transfection of pAAV-199 with helper plasmids (pAAV-DJ and pHelper) into 293FT cells. Viral production and titration were performed as previously reported.51
Cell Culture
The HCC cell lines Hep3B (HB-8064) and HepG2 (HB-8065) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The human embryonic kidney cells 293FT were obtained from Invitrogen (Carlsbad, CA, USA). Cell lines were propagated and maintained in Dulbecco’s modified Iscove’s medium (IMDM) supplemented with 10% fetal bovine serum (FBS), 0.1% gentamycin, and 1% L-glutamine (Sigma-Aldrich). In vitro cell transfections were performed using Lipofectamine 2000 (Invitrogen). A FOXO3 (untagged)-human plasmid (Cat No. SC119227; OriGene Technologies, Rockville, MD, USA) was used for transfection in Hep3B cells.
Apoptosis Assay
The Muse Annexin V and Dead Cell Assay kit (Cat. No. MCH100105; Merck Millipore, Burlington, MA, USA) was used to measure viable, apoptotic, and dead cells in a Muse Cell Analyzer instrument (Cat No. 0500-3115; Merck Millipore). All assays were performed in triplicate.
Western Blot Analyses
Cell cultures were washed with PBS and collected at the defined time point. Tissue samples were collected, immediately frozen in liquid nitrogen, and stored at −80°C until protein extraction. Samples were dissolved by repeated syringing in radioimmune precipitation (RIPA) buffer (R0278; Sigma-Aldrich) containing phosphatase and protease inhibitors (P2850 and P8340; Sigma-Aldrich). Lysates were centrifuged at 8,000 × g for 10 min at 4°C to pellet the debris, and supernatants were collected and analyzed by western blot. Rabbit polyclonal antibodies against PTEN (#9552; Cell Signaling, Danvers, MA, USA), AKT (C6E7, #4691; Cell Signaling), phospho-AKT (Ser473, D9E, #4060; Cell Signaling), MTOR (7C10, #2983; Cell Signaling), phospho-MTOR (Ser2481, #2974; Cell Signaling), FOXO3A (D19A7, #12829; Cell Signaling), phospho-FOXO3A (#2599; Cell Signaling), FOXM1 (TA327372; OriGene Technologies), and PAK4 (#3242; Cell Signaling) were diluted following the manufacturer’s instructions and incubated at 4°C for 16 hr. Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal antibody (Clone 2D9, TA802519; OriGene Technologies) or anti-β-actin monoclonal antibody (13E5, #4979; Cell Signaling) were used as housekeeping genes. A horseradish-peroxidase-conjugated secondary antibody (#7074; Cell Signaling) was used for chemiluminescent detection. For signal detection, Clarity Western ECL Substrate (Cat No. 170-5060; Bio-Rad, Laboratories, Hercules, CA, USA) was used according to the manufacturer’s instructions. Digital images were acquired with Chemidoc (Bio-Rad Laboratories). Signals were quantified by ImageJ software (https://imagej.nih.gov/ij/), and protein expression levels were normalized according to housekeeping protein expression.
Reverse Transcriptase Droplet Digital PCR
Total RNA was extracted from cells and from frozen liver tissues using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. ddPCR was used to measure the expression level of miRNAs. For qPCR analysis, 5 ng of purified RNA was retro-transcribed using TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). cDNA was used for amplification in a 20-μL reaction volume containing ddPCR Supermix for Probes (Bio-Rad Laboratories) and the TaqMan miRNA PCR probe set specific for miR-199a-3p (assay ID002304; Applied Biosystems). Droplets generation, cycling conditions for TaqMan assays, and count of positive droplets were performed as previously described.52 The relative abundance of miRNAs was normalized using the TaqMan Assays for RNAs U6 (assay ID001973; Applied Biosystems).
Gene Expression
RNAs were hybridized on Agilent Whole Mouse Gene Expression Microarray (G4852A, 8x60K; Agilent Technologies, Palo Alto, CA, USA), and one-color gene expression was performed according to the manufacturer’s protocol. Labeled cRNA was synthesized from 100 ng of total RNA using the Low RNA Input Linear Amplification kit (Agilent Technologies) in the presence of cyanine 3-cytosine triphosphate (CTP) (Perkin-Elmer Life Sciences, Boston, MA, USA). Images generated by Agilent scanner and Feature Extraction 10.5 software (Agilent Technologies) were used to obtain the microarray raw data. Qlucore Omics Explorer software (QOE) (https://www.qlucore.com/) (Qlucore AB, Lund, Sweden) was used to analyze microarray data.
Histological Procedures
Tissue samples from at least two representative fragments of each lobe of the liver were taken at autopsy and fixed in 10% phosphate-buffered formalin for 12–24 h and embedded in paraffin. Serial sections were stained with H&E for the histological determination of nodules number and dimension. ImageJ software was used for assessment of nodule sizes.
Statistical Analysis
Statistical significance of group similarity was resolved using a 2-tailed Student’s t test. A p value threshold <0.05 was considered significant. When appropriate, group value was expressed in terms of mean ± SD. GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA) was used for data analysis. No samples or animals were excluded from the analyses. None of the investigators were blinded to group allocations.
Author Contributions
E.C., S.S., and M.N. conceived and designed research; E.C., L.D.A., P.G., C.S., B.K.E., M.R., A.C., F.M., and S.U. performed the experiments; E.C., L.D.A., S.S., and M.N. analyzed the data; E.C., S.S., and M.N. wrote the manuscript; C.B. and B.Z. performed bioinformatics and statistical analyses; L.G., S.B., and G.A. performed histological analyses; A.F., L.G., and L.M.N. contributed reagents and materials; all authors read and approved the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by funds from the Italian Association for Cancer Research (AIRC IG grants 15615 and 20055) and the University of Ferrara to M.N. We wish to thank Fernanda Mora and Augusto Bevilacqua for technical and administrative support.
Footnotes
Supplemental Information includes seven figures and one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.04.002.
Contributor Information
Elisa Callegari, Email: elisa.callegari@unife.it.
Massimo Negrini, Email: ngm@unife.it.
Supplemental Information
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A., Center M.M., DeSantis C., Ward E.M. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol. Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 3.Vardy J., Rourke S., Tannock I.F. Evaluation of cognitive function associated with chemotherapy: a review of published studies and recommendations for future research. J. Clin. Oncol. 2007;25:2455–2463. doi: 10.1200/JCO.2006.08.1604. [DOI] [PubMed] [Google Scholar]
- 4.Vardy J., Tannock I. Cognitive function after chemotherapy in adults with solid tumours. Crit. Rev. Oncol. Hematol. 2007;63:183–202. doi: 10.1016/j.critrevonc.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Kim Y., Joo K.M., Jin J., Nam D.H. Cancer stem cells and their mechanism of chemo-radiation resistance. Int. J. Stem Cells. 2009;2:109–114. doi: 10.15283/ijsc.2009.2.2.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F., de Oliveira A.C., Santoro A., Raoul J.L., Forner A., SHARP Investigators Study Group Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 7.Bruix J., Qin S., Merle P., Granito A., Huang Y.H., Bodoky G., Pracht M., Yokosuka O., Rosmorduc O., Breder V., RESORCE Investigators Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- 8.Duffy A.G., Ulahannan S.V., Makorova-Rusher O., Rahma O., Wedemeyer H., Pratt D., Davis J.L., Hughes M.S., Heller T., ElGindi M. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma. J. Hepatol. 2017;66:545–551. doi: 10.1016/j.jhep.2016.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sprinzl M.F., Galle P.R. Current progress in immunotherapy of hepatocellular carcinoma. J. Hepatol. 2017;66:482–484. doi: 10.1016/j.jhep.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Llovet J.M., Hernandez-Gea V. Hepatocellular carcinoma: reasons for phase III failure and novel perspectives on trial design. Clin. Cancer Res. 2014;20:2072–2079. doi: 10.1158/1078-0432.CCR-13-0547. [DOI] [PubMed] [Google Scholar]
- 11.Llovet J.M., Zucman-Rossi J., Pikarsky E., Sangro B., Schwartz M., Sherman M., Gores G. Hepatocellular carcinoma. Nat. Rev. Dis. Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 12.Catela Ivkovic T., Voss G., Cornella H., Ceder Y. microRNAs as cancer therapeutics: a step closer to clinical application. Cancer Lett. 2017;407:113–122. doi: 10.1016/j.canlet.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 14.Negrini M., Ferracin M., Sabbioni S., Croce C.M. MicroRNAs in human cancer: from research to therapy. J. Cell Sci. 2007;120:1833–1840. doi: 10.1242/jcs.03450. [DOI] [PubMed] [Google Scholar]
- 15.Peng Y., Croce C.M. The role of microRNAs in human cancer. Signal Transduct. Target. Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murakami Y., Yasuda T., Saigo K., Urashima T., Toyoda H., Okanoue T., Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 17.Hou J., Lin L., Zhou W., Wang Z., Ding G., Dong Q., Qin L., Wu X., Zheng Y., Yang Y. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Sun H.L., Cui R., Zhou J., Teng K.Y., Hsiao Y.H., Nakanishi K., Fassan M., Luo Z., Shi G., Tili E. ERK activation globally downregulates miRNAs through phosphorylating exportin-5. Cancer Cell. 2016;30:723–736. doi: 10.1016/j.ccell.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia X.Q., Cheng H.Q., Qian X., Bian C.X., Shi Z.M., Zhang J.P., Jiang B.H., Feng Z.Q. Lentivirus-mediated overexpression of microRNA-199a inhibits cell proliferation of human hepatocellular carcinoma. Cell Biochem. Biophys. 2012;62:237–244. doi: 10.1007/s12013-011-9263-8. [DOI] [PubMed] [Google Scholar]
- 20.Ren K., Li T., Zhang W., Ren J., Li Z., Wu G. miR-199a-3p inhibits cell proliferation and induces apoptosis by targeting YAP1, suppressing Jagged1-Notch signaling in human hepatocellular carcinoma. J. Biomed. Sci. 2016;23:79. doi: 10.1186/s12929-016-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fornari F., Milazzo M., Chieco P., Negrini M., Calin G.A., Grazi G.L., Pollutri D., Croce C.M., Bolondi L., Gramantieri L. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184–5193. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- 22.Henry J.C., Park J.K., Jiang J., Kim J.H., Nagorney D.M., Roberts L.R., Banerjee S., Schmittgen T.D. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem. Biophys. Res. Commun. 2010;403:120–125. doi: 10.1016/j.bbrc.2010.10.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh A., Dasgupta D., Ghosh A., Roychoudhury S., Kumar D., Gorain M., Butti R., Datta S., Agarwal S., Gupta S. MiRNA199a-3p suppresses tumor growth, migration, invasion and angiogenesis in hepatocellular carcinoma by targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 2017;8:e2706. doi: 10.1038/cddis.2017.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callegari E., Elamin B.K., Giannone F., Milazzo M., Altavilla G., Fornari F., Giacomelli L., D’Abundo L., Ferracin M., Bassi C. Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology. 2012;56:1025–1033. doi: 10.1002/hep.25747. [DOI] [PubMed] [Google Scholar]
- 25.Callegari E., Gramantieri L., Domenicali M., D’Abundo L., Sabbioni S., Negrini M. MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. 2015;22:46–57. doi: 10.1038/cdd.2014.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan J., Liu Z., Xiao M., Hao F., Wang C., Chen Y., Lu Y., Liang J. MicroRNA-199a-3p inhibits tumorigenesis of hepatocellular carcinoma cells by targeting ZHX1/PUMA signal. Am. J. Transl. Res. 2017;9:2457–2465. [PMC free article] [PubMed] [Google Scholar]
- 27.Bhat M., Sonenberg N., Gores G.J. The mTOR pathway in hepatic malignancies. Hepatology. 2013;58:810–818. doi: 10.1002/hep.26323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radu M., Semenova G., Kosoff R., Chernoff J. PAK signalling during the development and progression of cancer. Nat. Rev. Cancer. 2014;14:13–25. doi: 10.1038/nrc3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paliouras G.N., Naujokas M.A., Park M. Pak4, a novel Gab1 binding partner, modulates cell migration and invasion by the Met receptor. Mol. Cell. Biol. 2009;29:3018–3032. doi: 10.1128/MCB.01286-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aspenström P., Fransson A., Saras J. Rho GTPases have diverse effects on the organization of the actin filament system. Biochem. J. 2004;377:327–337. doi: 10.1042/BJ20031041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dart A.E., Wells C.M. P21-activated kinase 4—not just one of the PAK. Eur. J. Cell Biol. 2013;92:129–138. doi: 10.1016/j.ejcb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Tyagi N., Bhardwaj A., Singh A.P., McClellan S., Carter J.E., Singh S. p-21 activated kinase 4 promotes proliferation and survival of pancreatic cancer cells through AKT- and ERK-dependent activation of NF-κB pathway. Oncotarget. 2014;5:8778–8789. doi: 10.18632/oncotarget.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyagi N., Marimuthu S., Bhardwaj A., Deshmukh S.K., Srivastava S.K., Singh A.P., McClellan S., Carter J.E., Singh S. p-21 activated kinase 4 (PAK4) maintains stem cell-like phenotypes in pancreatic cancer cells through activation of STAT3 signaling. Cancer Lett. 2016;370:260–267. doi: 10.1016/j.canlet.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vilar E., Perez-Garcia J., Tabernero J. Pushing the envelope in the mTOR pathway: the second generation of inhibitors. Mol. Cancer Ther. 2011;10:395–403. doi: 10.1158/1535-7163.MCT-10-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L., Zhu Y.R., Wang S., Zhao S. Autophagy inhibition sensitizes WYE-354-induced anti-colon cancer activity in vitro and in vivo. Tumour Biol. 2016;37:11743–11752. doi: 10.1007/s13277-016-5018-x. [DOI] [PubMed] [Google Scholar]
- 36.Weber H., Leal P., Stein S., Kunkel H., García P., Bizama C., Espinoza J.A., Riquelme I., Nervi B., Araya J.C. Rapamycin and WYE-354 suppress human gallbladder cancer xenografts in mice. Oncotarget. 2015;6:31877–31888. doi: 10.18632/oncotarget.5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dienstmann R., Rodon J., Serra V., Tabernero J. Picking the point of inhibition: a comparative review of PI3K/AKT/mTOR pathway inhibitors. Mol. Cancer Ther. 2014;13:1021–1031. doi: 10.1158/1535-7163.MCT-13-0639. [DOI] [PubMed] [Google Scholar]
- 38.Sarbassov D.D., Guertin D.A., Ali S.M., Sabatini D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 39.Nestal de Moraes G., Bella L., Zona S., Burton M.J., Lam E.W. Insights into a critical role of the FOXO3a-FOXM1 axis in DNA damage response and genotoxic drug resistance. Curr. Drug Targets. 2016;17:164–177. doi: 10.2174/1389450115666141122211549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao F., Lam E.W. Role of the forkhead transcription factor FOXO-FOXM1 axis in cancer and drug resistance. Front. Med. 2012;6:376–380. doi: 10.1007/s11684-012-0228-0. [DOI] [PubMed] [Google Scholar]
- 41.Laoukili J., Kooistra M.R., Brás A., Kauw J., Kerkhoven R.M., Morrison A., Clevers H., Medema R.H. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 42.Wang X., Kiyokawa H., Dennewitz M.B., Costa R.H. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc. Natl. Acad. Sci. USA. 2002;99:16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gusarova G.A., Wang I.C., Major M.L., Kalinichenko V.V., Ackerson T., Petrovic V., Costa R.H. A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J. Clin. Invest. 2007;117:99–111. doi: 10.1172/JCI27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kopanja D., Pandey A., Kiefer M., Wang Z., Chandan N., Carr J.R., Franks R., Yu D.Y., Guzman G., Maker A., Raychaudhuri P. Essential roles of FoxM1 in Ras-induced liver cancer progression and in cancer cells with stem cell features. J. Hepatol. 2015;63:429–436. doi: 10.1016/j.jhep.2015.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu M., Tang Z., Meng F., Tai M., Zhang J., Wang R., Liu C., Wu Q. Elevated expression of FoxM1 promotes the tumor cell proliferation in hepatocellular carcinoma. Tumour Biol. 2016;37:1289–1297. doi: 10.1007/s13277-015-3436-9. [DOI] [PubMed] [Google Scholar]
- 46.Sun H., Teng M., Liu J., Jin D., Wu J., Yan D., Fan J., Qin X., Tang H., Peng Z. FOXM1 expression predicts the prognosis in hepatocellular carcinoma patients after orthotopic liver transplantation combined with the Milan criteria. Cancer Lett. 2011;306:214–222. doi: 10.1016/j.canlet.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 47.Blanchet B., Billemont B., Barete S., Garrigue H., Cabanes L., Coriat R., Francès C., Knebelmann B., Goldwasser F. Toxicity of sorafenib: clinical and molecular aspects. Expert Opin. Drug Saf. 2010;9:275–287. doi: 10.1517/14740330903510608. [DOI] [PubMed] [Google Scholar]
- 48.Lamarca A., Feliu J., Barriuso J. Severe toxicity caused by sorafenib in hepatocellular carcinoma match the data from renal cell carcinoma. Br. J. Cancer. 2012;106:1246. doi: 10.1038/bjc.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang X., Schwind S., Yu B., Santhanam R., Wang H., Hoellerbauer P., Mims A., Klisovic R., Walker A.R., Chan K.K. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: a novel therapeutic strategy in acute myeloid leukemia. Clin. Cancer Res. 2013;19:2355–2367. doi: 10.1158/1078-0432.CCR-12-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Callegari E., Elamin B.K., D’Abundo L., Falzoni S., Donvito G., Moshiri F., Milazzo M., Altavilla G., Giacomelli L., Fornari F. Anti-tumor activity of a miR-199-dependent oncolytic adenovirus. PLoS ONE. 2013;8:e73964. doi: 10.1371/journal.pone.0073964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moshiri F., Callegari E., D’Abundo L., Corrà F., Lupini L., Sabbioni S., Negrini M. Inhibiting the oncogenic mir-221 by microRNA sponge: toward microRNA-based therapeutics for hepatocellular carcinoma. Gastroenterol. Hepatol. Bed Bench. 2014;7:43–54. [PMC free article] [PubMed] [Google Scholar]
- 52.Miotto E., Saccenti E., Lupini L., Callegari E., Negrini M., Ferracin M. Quantification of circulating miRNAs by droplet digital PCR: comparison of EvaGreen- and TaqMan-based chemistries. Cancer Epidemiol. Biomarkers Prev. 2014;23:2638–2642. doi: 10.1158/1055-9965.EPI-14-0503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.