Abstract
Osteoporosis (OP) is a disease characterized by bone loss, imbalance of bone metabolism and destruction of trabecular microstructure, and associated with menopause. Studies have shown that immune related lymphocytes are involved in bone metabolism. However, the molecular mechanisms hidden in the interaction of lymphocytes with OP need to be further studied. In the present study, we investigated the expression profiles and differences of lncRNAs, mRNAs, circRNAs and miRNAs in peripheral blood lymphocytes of patients with postmenopausal OP using Illumina-based complementary DNA (cDNA) deep sequencing (RNA-seq). 70 lncRNAs, 475 mRNAs, 260 circRNAs and 13 miRNAs were differentially expressed in patients with postmenopausal osteoporosis (OP group) compared with healthy controls (NC group). The functions of differentially expressed lncRNAs, circRNA, miRNA and potential targeting genes were predicted by Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Complex lncRNA-miRNA-mRNA and circRNA-miRNA-mRNA regulatory networks were constructed based on differentially expressed RNAs. Taken together, our study indicated that lncRNAs, mRNA, circRNAs and miRNA could associate with the occurrence of postmenopausal OP and may be as possible biomarkers and target genes in lymphocytes for OP.
Keywords: Osteoporosis, peripheral blood lymphocyte, lncRNAs, mRNAs, circRNAs, microRNAs
Introduction
Osteoporosis (OP) is the most common bone disease characterized by a systemic impairment of microarchitecture and bone mass that results in fragility fractures. It has been developed as the most common geriatric disease, especially in postmenopausal women [1]. Traditionally, osteoporosis has been regarded as an endocrine disease [2]; nevertheless, it is also well established that chronic inflammatory diseases are associated with OP [3]. Increasing evidences have shown a close connection between immune deficiency syndrome and OP, and although major osteoporotic risks, such as estrogen deficiency and advanced age, were observed, inflammatory background has been considered as an another pathogenetic factor [4,5]. Lymphocytes have been shown to play a role in postmenopausal OP through regulating bone metabolism [6]. Conversely, cytokines produced by osteocyte can promote lymphocyte differentiation and lead to the expansion of lymphocyte so as to further regulate the balance of bone metabolism [7]. This emphasizes an urgent demand for new biomarkers for a more reliable prediction of OP diagnosis and therapy.
Long non-coding RNAs (lncRNAs) with length longer than 200 nucleotides are defined as transcripts that are not translated into protein [8]. It is found that although lncRNA does not encode proteins, it plays an important role in regulating gene transcription through two ways: trans and cis. Trans means that lncRNA regulates the genes far away from its transcriptional point, and cis refers to the lncRNAs that lncRNA acts on the genes close to its transcriptional location [9]. Compared with microRNAs (miRNAs) those inhibit the gene expression at posttranscriptional level, lncRNAs can participate in epigenetic regulation, transcriptional and posttranscriptional regulation by different mechanisms [10]. lncRNA MEG3 inhibits osteogenic differentiation through down-regulating miR-133a-3p and its expression is increased in bone marrow stem cell of ovariectomized mice and OP patients [11]. LncRNA DANCR up-regulated in blood mononuclear cells promoted bone resorption through releasing TNF-α and IL-6 and finally resulted in OP [12]. LncRNA NTF3-5 promotes osteogenic differentiation and bone regeneration through down-regulating miR-93-3p [13].
Circular RNA (circRNA) is another non-coding RNA predominantly found in the cytoplasm in mammalian cells. Compared with linear RNAs, circRNAs are resistant to exonuclease-mediated degradation and have a stable structure due to the absence of 5’ and 3’ ends [14]. Increasing evidences have demonstrated that circRNAs can be used as a sponge of miRNA to counteract the inhibitory effect of mRNA mediated by miRNA by competing endogenous RNA (ceRNA) network [15,16]. CircRNA_007438 was up-regulated during osteoclastogenesis and targeted miRNA-6338 and miRNA-7028-3p, and circRNA_005108 was down-regulated during osteoclastogenesis and targeted miRNA-6975-3p, miRNA-6516-5p, miRNA-486b-5p and miRNA-31-3p [17]. circRNA BANP and circRNA ITCH regulated osteogenic differentiation through combining with miRNA146a and miRNA34a, which targeting PDGFRA and DUSP1, respectively [18].
So far, the expression profiles and functions of lncRNAs, mRNAs, circRNAs and miRNAs are extremely unknown in skeletal system, specifically during postmenopausal osteoporosis. To explore the underlying molecular regulation of non-coding RNAs for postmenopausal osteoporosis, differentially expressed patterns of lncRNA, mRNAs, circRNAs and miRNAs was examined in peripheral blood lymphocytes of postmenopausal OP patients and healthy controls by Illumina-based complementary DNA (cDNA) deep sequencing (RNA-seq). GO and KEGG pathway analysis were done based on the function of mRNAs that mediated by differentially expressed lncRNAs, circRNAs and miRNAs.
Materials and methods
Patients and samples
The total number of peripheral blood lymphocyte samples we used for this study is six, including three patients with postmenopausal osteoporosis (OP group; age range 52-58 years) and three healthy controls (Control group; age range 49-50 years). Osteoporosis was defined by spine or hip bone mineral density (BMD) T-score ≤ -2.5 SD and healthy control was defined by spine or hip bone mineral density (BMD) T-score ≥ -1.0 SD. The detailed characteristics of the study subjects are summarized in Table 1. All patient samples were obtained at the time of diagnosis or relapse and with informed consent at the Second Affiliated Hospital of Harbin Medical University.
Table 1.
Characteristics of the study subjects
| Traits | OP group (n=3) | NC group (n=3) |
|---|---|---|
| Age (years) | 55.6±3.2 | 49.7±0.58 |
| Height (cm) | 161.3±7.77 | 163.3±1.53 |
| Weight (kg) | 55±2.60 | 65±4.3 |
| Menarche age (years) | 16.7±1.53 | 15.3±1.15 |
| Menopause age (years) | 49.7±1.53 | 46.7±2.08 |
| Forearm T-score | -1.03±1.39 | 0.63±1.33 |
| Hip T-score | -2.47±0.46 | 0.97±1.47 |
| Femoral neck T-score | -2.63±0.72 | 0.33±0.87 |
| Femur T-score | -1.9±0.89 | 0.67±0.63 |
Isolation of peripheral blood lymphocytes
Thirty milliliters of fresh heparinized elbow venous blood samples from patients’ was suspended with PBS and added into the lymphocyte separation solution with the same volume. After centrifugation at 950×g for 20 min, the cells at the interface were moved into a new tube containing 10 ml PBS and then centrifugation at 250×g for 10 min. Cells were then suspended with 5 ml PBS and centrifugation at 250×g for 10 min. The prepared peripheral blood lymphocytes were resuspended with 1 ml Trizol, subsequently moved into liquid nitrogen for 10 min and stored at -80°C until used.
RNA isolation and RNA-seq analysis
Total RNA was isolated from the peripheral blood lymphocytes using Trizol reagent (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s protocol. 5 μg of RNA was used as input material for the RNA sample preparations. Sequencing libraries were generated using RNase R digested and rRNA-depleted RNAs or NEBNext Multiplex Small RNA Library Prep Set for Illumina (NEB, USA) by NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations. After cluster generation on a cBot Cluster Generation System using HiSeq PE Cluster Kit v4 cBot or TruSeq SR Cluster Kit v3-cBot-HS (Illumia), the library preparations were sequenced on an Illumina Hiseq 2500 platform.
Differential expression analysis
Differential miRNA and circRNA expression analysis of two samples was performed using the DEGseq [19]. Cuffdiff provides statistical routines for determining differential expression in digital transcript or gene expression data using a model based on the negative binomial distribution [20]. P-value < 0.05 were assigned as differentially expressed.
Target gene prediction and competing endogenous RNA (ceRNA) network analysis
Predicting the target genes and binding sites of miRNA were performed by miRanda (Enright et al, 2003) [21]. Cis role of target gene prediction was performed by searching coding genes 100 k/10k downstream and upstream of lncRNA and trans role of target gene prediction was identified by calculating the expressed correlation between lncRNAs and coding genes with custom scripts. The lncRNA/circRNA-miRNA-mRNA ceRNA network was constructed according to the common target miRNAs of the lncRNA/circRNAs and mRNAs.
GO and KEGG enrichment analysis
Gene Ontology (GO) enrichment analysis was used on the target gene candidates of differentially expressed miRNAs and the source genes of differential circRNAs. We used KEGG Orthology Based Annotation System [22] software to test the statistical enrichment of the target gene candidates in KEGG pathways. GO terms and KEGG pathways with a P-value < 0.05 were considered significantly enriched by differential expressed genes.
Results
Differential expression analysis of lncRNAs, mRNAs, circRNA and miRNAs
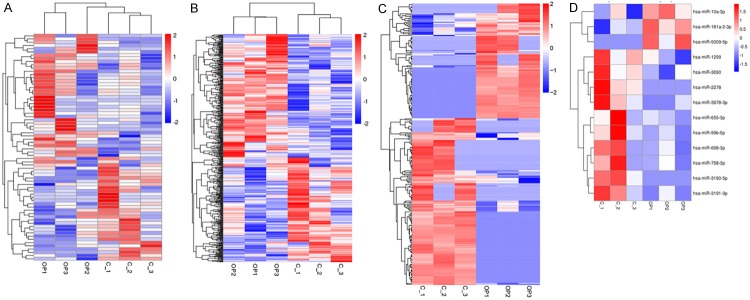
Hierarchical clustering revealed lncRNAs, mRNAs, circRNA and miRNAs expression in OP and normal peripheral blood lymphocyte samples. A total of 70 significantly expressed lncRNAs were obtained, including 39 up-regulated lncRNAs and 31 down-regulated lncRNAs (Figure 1A). Meanwhile, top 5 up-regulated lncRNAs, including lncRNA_000180 (2.57-fold), lncRNA_000386 (2.50-fold), lncRNA RP11-885N19.6 (2.17-fold), lncRNA AP006222.2 (2.16-fold) and lncRNA_000285 (2.01-fold), and down-regulated lncRNAs, including lncRNA_000283 (-3.61-fold), lncRNA_000416 (-3.43-fold), lncRNA RP11-525A16.4 (-2.17-fold), lncRNA_000483 (-1.99-fold) and lncRNA_000647 (-1.96-fold). Moreover, a total of 475 mRNAs were detected to be differentially expressed (Figure 1B). Among them, 264 and 211 were up-regulated and down-regulated in OP groups compared with control groups, respectively. Meanwhile, top 5 up-regulated mRNAs, including HLA-DQA1 (5.25-fold), FOLR3 (4.61-fold), COX14 (4.60-fold), NXF3 (4.39-fold) and HLA-DRB5 (4.36-fold), and down-regulated mRNAs, including COQ10A (-12.5-fold), ZBTB37 (-12.0-fold), UQCC1 (-11.2-fold), PPBP (-4.91-fold) and RETN (-3.20-fold).
Figure 1.
Expression profiles of mRNAs, lncRNAs, circRNAs and miRNAs. Hierarchical clustering of all differentially expressed mRNAs (A), lncRNAs (B), circRNAs (C) and miRNAs (D) in peripheral blood lymphocytes of postmenopausal OP groups and control groups.
In the same way, a cluster was generated and analyzed with hierarchical clustering for the often differentially regulated 260 circRNAs (Figure 1C). 106 of these circRNAs were up-regulated and 154 were down-regulated in OP groups. Meanwhile, top 5 up-regulated circRNAs, including circRNA_0010452 (4.64-fold), circRNA_0022348 (4.21-fold), circRNA_0015566 (4.13-fold), circRNA_0003323 (4.12-fold) and circRNA_0013121 (4.08-fold), and down-regulated circRNAs, including circRNA_0021739 (-8.51-fold), circRNA_0011269 (-4.41-fold), circRNA_0019693 (-4.29-fold), circRNA_0005245 (-4.12-fold) and circRNA_0010349 (-3.98-fold). A total of 13 miRNAs were specifically dysregulated, including 3 up-regulated miRNAs and 10 down-regulated miRNAs, respectively, in OP patients compared with controls (Figure 1D). Meanwhile, top up-regulated miRNAs, including miR-181a-2-3p (1.15-fold), miR-10a-3p (0.81-fold) and miR-5009-5p (0.79fold), and down-regulated miRNAs, including miR-3190-5p (-1.03-fold), miR-3690 (-0.98fold), miR-1299 (-0.97-fold), miR-668-3p (-0.90fold) and miR-655-3p (-0.87-fold).
Functional enrichment analysis
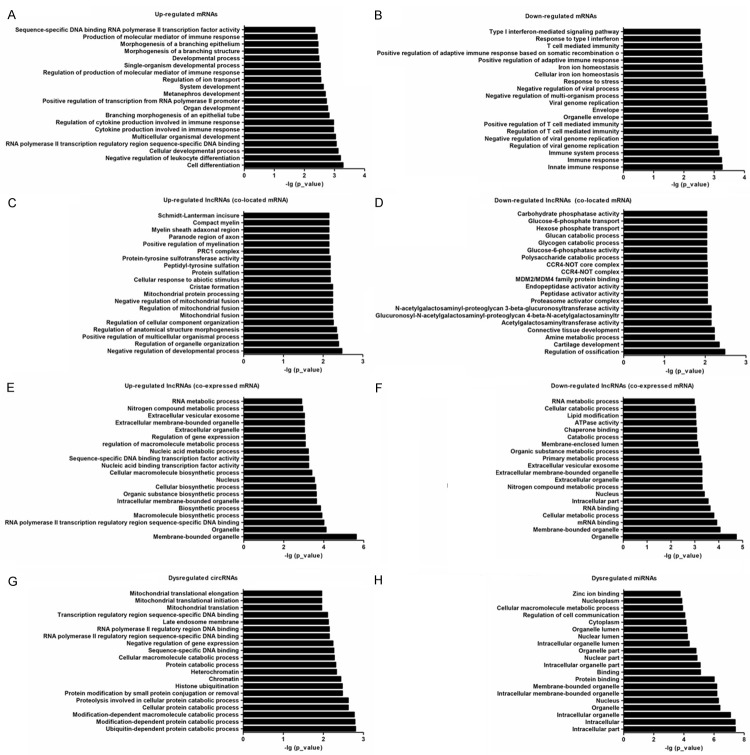
Gene ontology (GO) enrichment analysis of significantly differentially expressed mRNAs showed that the up-regulated mRNAs were associated with 371 biological processes (BPs) such as cell differentiation, 29 molecular functions (MFs) such as sequence-specific DNA binding RNA polymerase II transcription factor activity, and 14 cellular components (CCs) such as Set1C/COMPASS complex (Figure 2A). Moreover, down-regulated mRNAs were associated with 155 BC such as innate immune response, 22 MF such as ceramidase activity, and 15 CC such as organelle envelope (Figure 2B).
Figure 2.
GO analysis of the biological function of mRNAs, lncRNAs, circRNAs and miRNAs. Go terms of up- (A) and down-regulated mRNAs (B), up- (C) and down-regulated lncRNA co-located mRNAs (D), up- (E) and down-regulated lncRNA co-expressed mRNAs (F), up- and down-regulated mRNAs producing differentially expressed circRNAs (G) and targeting by miRNAs (H) with top 20.
Functional enrichment analysis was also applied on the differentially expressed lncRNA-targeted mRNAs, including co-located and co-expressed mRNAs. The up-regulated lncRNAtargeted mRNAs (co-located) were associated with 130 BPs such as negative regulation of developmental process, 13 MFs such as protein-tyrosine sulfotransferase activity, and 10 CCs such as PRC1 complex (Figure 2C). Moreover, down-regulated lncRNA-targeted mRNAs (co-located) were associated with 132 BPs such as regulation of ossification, 29 MFs such as acetylgalactosaminyltransferase activity, and 14 CCs such as proteasome activator complex (Figure 2D). The up-regulated lncRNA-targeted mRNAs (co-expressed) were associated with 142 BPs such as macromolecule biosynthetic process, 27 MFs such as nucleic acid binding transcription factor activity, and 31 CCs such as membrane-bounded organelle (Figure 2E). Moreover, down-regulated lncRNA-targeted mRNAs (co-expressed) were associated with 138 BPs such as cellular metabolic process, 33 MFs such as mRNA binding, and 43 CCs such as organelle (Figure 2F).
Functional enrichment analysis applied on the differentially expressed circRNAs showed that the up-regulated and down-regulated mRNAs producing differentially expressed circRNAs were associated with 52 BPs such as ubiquitin-dependent protein catabolic process, 16 MFs such as sequence-specific DNA binding, and 19 CCs such as chromatin (Figure 2G). Functional enrichment analysis applied on the differentially expressed miRNAs showed that the up-regulated and down-regulated mRNAs targeted by miRNAs were associated with 243 BPs such as regulation of cell communication, 43 MFs such as protein binding, and 55 CCs such as intracellular part (Figure 2H).
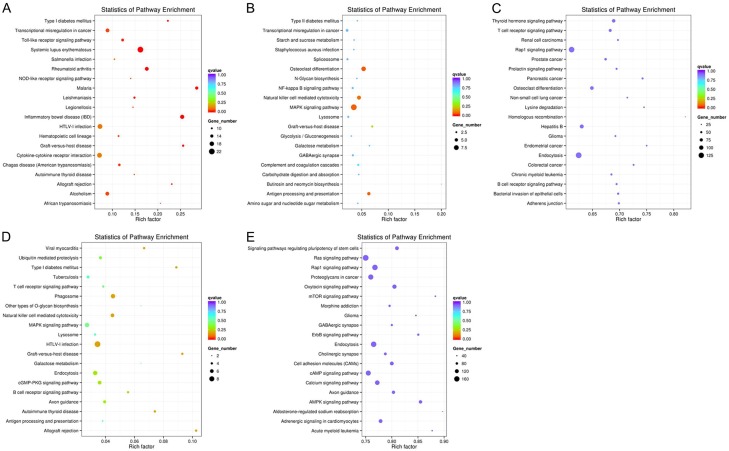
KEGG pathway enrichment analysis for significantly differentially expressed mRNAs showed that up-regulated mRNAs were associated with 23 pathways such as Systemic lupus erythematosus, Inflammatory bowel disease (IBD) and Graft-versus-host disease, and down-regulated mRNAs were associated with 14 pathways such as Rheumatoid arthritis, Malaria and Legionellosis (Figure 3A). With regard of differentially expressed lncRNAtargeted mRNAs, including co-located and co-expressed mRNAs, up- and down-regulated lncRNA-targeted mRNAs (co-located) were associated with pathways such as Osteoclast differentiation and MAPK signaling pathway (Figure 3B). Moreover, up- and down-regulated lncRNAtargeted mRNAs (co-expressed) were associated with pathways such as Thyroid hormone signaling pathway and T cell receptor signaling pathway (Figure 3C). With regard of differentially expressed circRNAs and miRNAs, up- and down-regulated mRNAs producing differentially expressed circRNAs were associated with pathways such as Allograft rejection and Graft-versus-host disease, respectively (Figure 3D). The up- and down-regulated mRNAs targeted by miRNAs were associated with pathways such as AMPK signaling pathway and Oxytocin signaling pathway, respectively (Figure 3E).
Figure 3.
KEGG pathway enrichment analysis of mRNAs, lncRNAs, circRNAs and miRNAs. KEGG pathway enrichment analysis of up- and down-regulated mRNAs (A), up- and down-regulated lncRNA co-located mRNAs (B) and co-expressed mRNAs (C), up- and down-regulated mRNAs producing differentially expressed circRNAs (D) and tar10.10.0ge00t01ing by miRNAs (E) with top 20.
Construction of ceRNA network
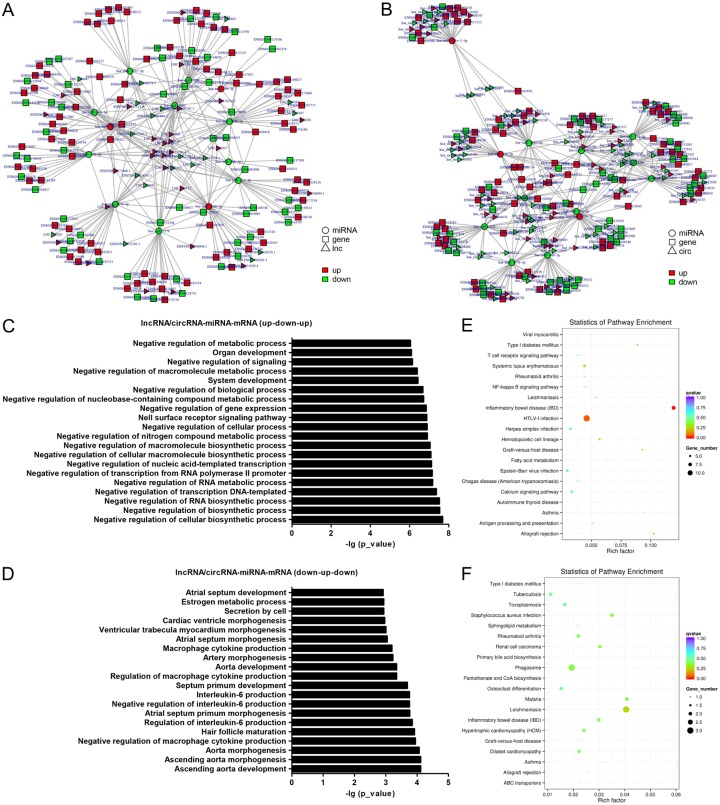
Through merging the common targeted miRNAs, we constructed a network of lncRNAs-miRNAs-mRNAs with a total of 70 lncRNAs, 13 miRNAs and 459 mRNAs (Figure 4A), and circRNAs-miRNAs-mRNAs with a total of 260 circRNAs, 13 miRNAs and 459 mRNAs (Figure 4B). Moreover, lncRNAs and circRNAs were found to share a common binding site of miRNA response elements. For example, lncRNA_000406, lncRNA ENST00000613780.4, circRNA_0016760 and circRNA_0018034 are ceRNA of miR-10a-3p targeting IL2RB, SOX4, PLCXD1, EPG5, IKZF2, PTAFR, TGFBR3, LMTK2 and PVRL1. Whereas, lncRNA_000037, lncRNA_000655, lncRNA_000187, lncRNA_000361, circRNA_0022625, circRNA_0025901 and circRNA_0004790 are ceRNA of miR-1299 targeting ADAMTS10, MSR1, ADTRP, PVRL1, STEAP4, TOX, PLCXD1, C5AR2 and RBP7.
Figure 4.
lncRNAs/circRNAs-miRNAs-mRNAs regulation network. lncRNAs-miRNAs-mRNAs with a total of 49 lncRNAs, 13 miRNAs and 315 mRNAs (A) and circRNAs-miRNAs-mRNAs with a total of 228 circRNAs, 13 miRNAs and 315 mRNAs (B). Go annotation of lncRNAs/circRNA-miRNAs-mRNAs with a mode of up-down-up (C) or down-up-down (D) with top 20. KEGG pathway enrichment analysis of lncRNAs/circRNA-miRNAs-mRNAs with a mode of up-down-up (E) or down-up-down (F) with top 20.
Functional enrichment analysis showed that lncRNAs/circRNAs-miRNAs-mRNAs with a mode of up-down-up were associated with BPs such as negative regulation of cellular biosynthetic process, MFs such as binding, and CCs such as nuclear chromatin (Figure 4C). lncRNAs/circRNAs-miRNAs-mRNAs with a mode of down-up-down were associated with BPs such as ascending aorta development, MFs such as 17-beta-hydroxysteroid dehydrogenase (NAD+) activity, and CCs such as rhabdomere (Figure 4D).
KEGG pathway enrichment analysis for lncRNAs/circRNAs-miRNAs-mRNAs with a mode of up-down-up showed association with pathways such as Inflammatory bowel disease (IBD), HTLV-I infection and Allograft rejection (Figure 4E). Whereas, lncRNAs/circRNAs-miRNAs-mRNAs with a mode of down-up-down were associated with pathways such as Leishmaniasis, Malaria and Staphylococcus aureus infection (Figure 4F).
Discussion
Bone formation and absorption maintain a dynamic balance to maintain bone health. The disorder of bone metabolism causes bone related diseases, especially osteoporosis, which can lead to a decrease in bone density, the deterioration of bone microstructures and increase the risk of fracture [23,24]. Recently, the functions of non-coding RNAs such as miRNAs, lncRNAs and circRNAs have been described in many diseases including cancers [25,26]. In our study, the expression profiles of lncRNA, mRNA, circRNA and miRNA were detected using RNA-seq analysis, which acts as a relatively novel platform for gene expression data. The RNA-seq is to determine the sequence of all the transcripts expressed by a particular organization at a specific time and to analyze the changes in the structure of the transcriptional sequence, at the same time, the expression of mRNA and gene polymorphism at the genomic level are also analyzed [27].
A total of 70 lncRNAs, 475 mRNAs, 260 circRNAs and 13 miRNAs were significantly expressed in postmenopausal OP groups compared with control groups. Among differentially expressed lncRNAs, lncRNA MALAT1 had been found overexpressed in osteosarcoma patients and associated with prognosis, tumor size, clinic stages, distant metastasis and clinicopathology [28], and knockdown of lncRNA MIAT enhanced osteogenic differentiation of human adipose-derived stem cells [29]. circRNA_0010763 was up-regulated in our OP groups compared with control, whereas it was up-regulated in osteoclasts compared with monocytes and down-regulated in osteoclastogenesis [17]. miR-99b-5p and miR-758-3p were down-regulated in our OP groups compared with control, whereas they were previously showed up-regulated during osteoclast differentiation [30] and in the rapidly proliferating osteosarcoma cell lines [31]. miR-3190-5p was down-regulated in our OP groups compared with control and in MC3T3-E1 cells exposed to fluoride, one of the most potent agent for OP [32]. These results suggest an important role of the special RNAs in bone metabolism. In addition, we reported that TMEM176B up-regulated in our OP groups compared with control was related to osteogenic differentiation of myoblasts [33]. CENPM, CENPA, RRM2, SHCBP1, KIFC1, DTL, TOP2A, and SLC2A1 were up-regulated, whereas CSTA was down-regulated, in our OP groups compared with control. However, microarray analysis used in the previous study showed a contrary expression in osteoporotic bone tissue compared with us [34]. These confused findings may be exampled by the differences in measurement techniques and sample sources.
GO enrichment analysis showed that significantly differentially expressed mRNAs were associated with 526 BPs, 51 MFs and 29 CCs, including immune system process, G-protein alpha-subunit binding and cell-cell adherens junction, lncRNA targeted co-located mRNAs were associated with 262 BPs, 42 MFs and 24 CCs, including sulfation, acetylgalactosaminyltransferase activity and PRC1 complex, and lncRNA targeted co-expressed mRNAs were associated with 280 BPs, 60 MFs and 74 CCs, including organic substance metabolic process, protein domain specific binding and membrane-bounded organelle. The differentially expressed mRNAs producing differentially expressed circRNAs were associated with 52 BPs, 16 MFs and 19 CCs, including ubiquitin-dependent protein catabolic process, sequence-specific DNA binding and chromatin. The differentially expressed mRNAs targeted by miRNAs were associated with 243 BPs, 43 MFs and 55 CCs, including regulation of cell communication, protein binding and intracellular part. Immune response-related biological processes were covered by many differentially expressed RNAs in postmenopausal OP, which were agreement with our description that a relationship is existed between osteoporosis and immune system, and other studies were also stated [7].
KEGG pathway enrichment analysis showed that differentially expressed RNAs were associated with pathways such as Type II diabetes mellitus, Osteoclast differentiation, Rheumatoid arthritis, NF-kB, MAPK, AMPK, Toll-like receptor, T and B cell receptor signaling pathways, while PPAR and Insulin signaling pathways were not significant difference between OP and control, which was partly consistent with the previous study in the mandible of ovariectomized mice [35]. In addition, also reported that the differentially expressed genes were associated with MAPK, PPAR, Toll-like receptor and Type I diabetes mellitus signaling pathways [34]. OP is highly prevalent in patients with rheumatoid arthritis who are also accompanied with activation of Toll-like receptors [36]. MAPK/NF-kB signaling pathway has been found to be associated with trabecular structure and osteoclastogenic differentiation in diabetes-related osteoporosis [37].
ceRNAs are of great importance in regulating gene expression [38]. Here, this is our first time to construct lncRNA/circRNA-miRNA-mRNA ceRNA network of OP based on our RNA-seq data and we found 70 lncRNAs (e.g. MIAT, C15orf54, RP11-96H19.1 and RP11-13A1.1), 260 circRNAs (circRNA_0004790, circRNA_0025901, circRNA_0008189 and circRNA_0022625), 13 miRNAs (e.g. miR-3678-3p, miR-31905p, miR-1299 and miR-181a-2-3p) and 459 mRNAs (e.g. RUNX3, COL5A3, TMEM8A and TLR4). Moreover, GO and KEGG pathway analysis showed that lncRNA/circRNA-miRNA-mRNA ceRNA network was associated with negative regulation of biosynthetic and metabolic process and Inflammatory bowel disease (IBD) signaling pathway, etc. Our results indicate an important role of lncRNAs and circRNAs in the occurrence of OP through competing miRNA response elements.
In summary, our findings show significant differences in the expression of lncRNAs, mRNAs, circRNAs, and miRNAs between postmenopausal OP patients and healthy controls, and the functions of the RNAs were also identified based on our RNA-seq analysis. These differentially expressed RNAs may be as possible biomarkers and target genes in lymphocytes for OP.
Acknowledgements
This work was supported by the Financial Scheme for Application Technology Research and Development Project of Harbin Science and Technology Bureau (2017AB9BS037).
Disclosure of conflict of interest
None.
References
- 1.Wade SW, Strader C, Fitzpatrick LA, Anthony MS, O’Malley CD. Estimating prevalence of osteoporosis: examples from industrialized countries. Arch Osteoporos. 2014;9:182. doi: 10.1007/s11657-014-0182-3. [DOI] [PubMed] [Google Scholar]
- 2.Mirza F, Canalis E. Management of endocrine disease: Secondary osteoporosis: pathophysiology and management. Eur J Endocrinol. 2015;173:R131–151. doi: 10.1530/EJE-15-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montalcini T, Romeo S, Ferro Y, Migliaccio V, Gazzaruso C, Pujia A. Osteoporosis in chronic inflammatory disease: the role of malnutrition. Endocrine. 2013;43:59–64. doi: 10.1007/s12020-012-9813-x. [DOI] [PubMed] [Google Scholar]
- 4.Zhao R. Immune regulation of osteoclast function in postmenopausal osteoporosis: a critical interdisciplinary perspective. Int J Med Sci. 2012;9:825–832. doi: 10.7150/ijms.5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tella SH, Gallagher JC. Prevention and treatment of postmenopausal osteoporosis. J Steroid Biochem Mol Biol. 2014;142:155–170. doi: 10.1016/j.jsbmb.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyce BF, Xing L. Bruton and Tec: new links in osteoimmunology. Cell Metab. 2008;7:283–285. doi: 10.1016/j.cmet.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Faienza MF, Ventura A, Marzano F, Cavallo L. Postmenopausal osteoporosis: the role of immune system cells. Clin Dev Immunol. 2013;2013:575936. doi: 10.1155/2013/575936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online. 2014;16:11. doi: 10.1186/1480-9222-16-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Li Y, Zhang Y, Ma L, Lin L, Meng J, Jiang L, Wang L, Zhou P, Zhang Y. LncRNA MEG3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR133a-3p. Biomed Pharmacother. 2017;89:1178–1186. doi: 10.1016/j.biopha.2017.02.090. [DOI] [PubMed] [Google Scholar]
- 12.Tong X, Gu PC, Xu SZ, Lin XJ. Long non-coding RNA-DANCR in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. Biosci Biotechnol Biochem. 2015;79:732–737. doi: 10.1080/09168451.2014.998617. [DOI] [PubMed] [Google Scholar]
- 13.Peng W, Zhu SX, Wang J, Chen LL, Weng JQ, Chen SL. Lnc-NTF3-5 promotes osteogenic differentiation of maxillary sinus membrane stem cells via sponging miR-93-3p. Clin Implant Dent Relat Res. 2018;20:110–121. doi: 10.1111/cid.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 15.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 16.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 17.Dou C, Cao Z, Yang B, Ding N, Hou T, Luo F, Kang F, Li J, Yang X, Jiang H, Xiang J, Quan H, Xu J, Dong S. Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci Rep. 2016;6:21499. doi: 10.1038/srep21499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu X, Li M, Jin Y, Liu D, Wei F. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet. 2017;18:100. doi: 10.1186/s12863-017-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 20.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 23.Bliuc D, Alarkawi D, Nguyen TV, Eisman JA, Center JR. Risk of subsequent fractures and mortality in elderly women and men with fragility fractures with and without osteoporotic bone density: the dubbo osteoporosis epidemiology Study. J Bone Miner Res. 2015;30:637–646. doi: 10.1002/jbmr.2393. [DOI] [PubMed] [Google Scholar]
- 24.Bliuc D, Nguyen ND, Nguyen TV, Eisman JA, Center JR. Compound risk of high mortality following osteoporotic fracture and refracture in elderly women and men. J Bone Miner Res. 2013;28:2317–2324. doi: 10.1002/jbmr.1968. [DOI] [PubMed] [Google Scholar]
- 25.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 26.Yang C, Wu D, Gao L, Liu X, Jin Y, Wang D, Wang T, Li X. Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget. 2016;7:13479–13490. doi: 10.18632/oncotarget.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ansorge WJ. Next-generation DNA sequencing techniques. N Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 28.Gao KT, Lian D. Long non-coding RNA MALAT1 is an independent prognostic factor of osteosarcoma. Eur Rev Med Pharmacol Sci. 2016;20:3561–3565. [PubMed] [Google Scholar]
- 29.Jin C, Zheng Y, Huang Y, Liu Y, Jia L, Zhou Y. Long non-coding RNA MIAT knockdown promotes osteogenic differentiation of human adipose-derived stem cells. Cell Biol Int. 2017;41:33–41. doi: 10.1002/cbin.10697. [DOI] [PubMed] [Google Scholar]
- 30.Franceschetti T, Dole NS, Kessler CB, Lee SK, Delany AM. Pathway analysis of microRNA expression profile during murine osteoclastogenesis. PLoS One. 2014;9:e107262. doi: 10.1371/journal.pone.0107262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauvrak SU, Munthe E, Kresse SH, Stratford EW, Namlos HM, Meza-Zepeda LA, Myklebost O. Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. Br J Cancer. 2013;109:2228–2236. doi: 10.1038/bjc.2013.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Zhang X, Zhao Z, Xu H. Preliminary analysis of MicroRNAs expression profiling in MC3T3-E1 cells exposed to fluoride. Biol Trace Elem Res. 2017;176:367–373. doi: 10.1007/s12011-016-0833-x. [DOI] [PubMed] [Google Scholar]
- 33.Yano M, Kawao N, Tamura Y, Okada K, Kaji H. A novel factor, Tmem176b, induced by activinlike kinase 2 signal promotes the differentiation of myoblasts into osteoblasts. Exp Clin Endocrinol Diabetes. 2014;122:7–14. doi: 10.1055/s-0033-1357129. [DOI] [PubMed] [Google Scholar]
- 34.Trost Z, Trebse R, Prezelj J, Komadina R, Logar DB, Marc J. A microarray based identification of osteoporosis-related genes in primary culture of human osteoblasts. Bone. 2010;46:72–80. doi: 10.1016/j.bone.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 35.Hao L, Fu J, Tian Y, Wu J. Systematic analysis of lncRNAs, miRNAs and mRNAs for the identification of biomarkers for osteoporosis in the mandible of ovariectomized mice. Int J Mol Med. 2017;40:689–702. doi: 10.3892/ijmm.2017.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goh FG, Midwood KS. Intrinsic danger: activation of Toll-like receptors in rheumatoid arthritis. Rheumatology (Oxford) 2012;51:7–23. doi: 10.1093/rheumatology/ker257. [DOI] [PubMed] [Google Scholar]
- 37.Li XJ, Zhu Z, Han SL, Zhang ZL. Bergapten exerts inhibitory effects on diabetes-related osteoporosis via the regulation of the PI3K/AKT, JNK/MAPK and NF-kappaB signaling pathways in osteoprotegerin knockout mice. Int J Mol Med. 2016;38:1661–1672. doi: 10.3892/ijmm.2016.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang M, Zhong Z, Lv M, Shu J, Tian Q, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7:47186–47200. doi: 10.18632/oncotarget.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]