Summary
Background
Germline pathogenic variants in the E-cadherin gene (CDH1) are strongly associated with the development of hereditary diffuse gastric cancer. There is a paucity of data to guide risk assessment and management of families with hereditary diffuse gastric cancer that do not carry a CDH1 pathogenic variant, making it difficult to make informed decisions about surveillance and risk-reducing surgery. We aimed to identify new candidate genes associated with predisposition to hereditary diffuse gastric cancer in affected families without pathogenic CDH1 variants.
Methods
We did whole-exome sequencing on DNA extracted from the blood of 39 individuals (28 individuals diagnosed with hereditary diffuse gastric cancer and 11 unaffected first-degree relatives) in 22 families without pathogenic CDH1 variants. Genes with loss-of-function variants were prioritised using gene-interaction analysis to identify clusters of genes that could be involved in predisposition to hereditary diffuse gastric cancer.
Findings
Protein-affecting germline variants were identified in probands from six families with hereditary diffuse gastric cancer; variants were found in genes known to predispose to cancer and in lesser-studied DNA repair genes. A frameshift deletion in PALB2 was found in one member of a family with a history of gastric and breast cancer. Two different MSH2 variants were identified in two unrelated affected individuals, including one frameshift insertion and one previously described start-codon loss. One family had a unique combination of variants in the DNA repair genes ATR and NBN. Two variants in the DNA repair gene RECQL5 were identified in two unrelated families: one missense variant and a splice-acceptor variant.
Interpretation
The results of this study suggest a role for the known cancer predisposition gene PALB2 in families with hereditary diffuse gastric cancer and no detected pathogenic CDH1 variants. We also identified new candidate genes associated with disease risk in these families.
Funding
UK Medical Research Council (Sackler programme), European Research Council under the European Union's Seventh Framework Programme (2007–13), National Institute for Health Research Cambridge Biomedical Research Centre, Experimental Cancer Medicine Centres, and Cancer Research UK.
Introduction
Gastric cancer is the fourth most common cancer globally. The best characterised inherited gastric cancer is the diffuse type, which has the hallmark of multiple foci of signet-ring cells.1 The term hereditary diffuse gastric cancer is used to describe families with a history of diffuse gastric cancer that meet the criteria of at least two cases of gastric cancer in first-degree or second-degree relatives regardless of age of onset (with one confirmed case of diffuse gastric cancer); one case of diffuse gastric cancer diagnosed before age 40 years; or a personal or family history of diffuse gastric cancer and lobular breast cancer, including one case diagnosed before age 50 years.2, 3
Germline mutations in the E-cadherin gene (CDH1) explain 25–30% of hereditary diffuse gastric cancer cases, with more than 100 pathogenic germline variants currently described within this gene.4 For families with hereditary diffuse gastric cancer and known pathogenic CDH1 mutations, guidelines exist for risk assessment, disease management, surveillance (including regular endoscopies), and risk-reducing therapies (including prophylactic gastrectomy).5, 6 However, for families with no pathogenic variant in CDH1, the risk assessment is uncertain and, therefore, making decisions about and assessing the efficacy of risk-reducing strategies is challenging.
Research in context.
Evidence before this study
Knowledge of factors causing predisposition to hereditary diffuse gastric cancer in families with no pathogenic variants in CDH1 is limited by the rarity of the disease, which makes doing large-scale association studies difficult. In 2015, Hansford and colleagues described variants in DNA repair-related genes in 144 families with hereditary diffuse gastric cancer without CDH1 pathogenic variants. These genes included PALB2, BRCA2, and ATM, which are associated with breast cancer risk. Further investigation of these genes, other known cancer-predisposing genes, and genes associated with DNA repair will aid in the disease management of families with hereditary diffuse gastric cancer without CDH1 pathogenic variants, whose risk of disease development is currently unknown.
Added value of this study
This study is one of the largest germline, whole-exome sequencing analysis of families with hereditary diffuse gastric cancer without CDH1 mutation to date. Both affected and unaffected individuals were recruited from families with hereditary diffuse gastric cancer, providing the opportunity to look for protein-affecting variants that segregate with phenotype. We used a unique approach to pathway analysis that involved clustering of physically interacting genes that were enriched for variants in these families and annotating them with a Gene Ontology term. Additionally, combining findings from this study with data from previously published studies allowed a more complete analysis of the role of the cancer predisposition genes PALB2 and BRCA2.
Implications of all the available evidence
We identified a cluster of interacting genes involved in DNA repair that could be associated with predisposition to hereditary diffuse gastric cancer, in particular, PALB2. These findings should help guide future studies seeking to elucidate the clinical implications of genes that have not been previously associated with hereditary diffuse gastric cancer. Identification of these genes could provide families with hereditary diffuse gastric cancer without CDH1 pathogenic variants with improved information about the risks associated with their disease and allow them to make informed decisions about risk reduction and disease management.
Other familial cancer syndromes that have been linked to gastric cancer predisposition include Lynch syndrome, which is characterised by mutations in DNA mismatch repair genes; Peutz-Jeghers syndrome caused by mutations in STK11; and Li-Fraumeni syndrome, which is associated with germline TP53 mutations.2, 7, 8, 9 Diffuse gastric cancer does not appear to be over-represented in these syndromes, although this association has not been comprehensively studied.
Predicted pathogenic variants in the DNA double-strand break repair genes ATM, BRCA2, and PALB2 have been identified in several families with hereditary diffuse gastric cancer.4, 10 However, given the rarity of these variants, the associated risk of diffuse gastric cancer is hard to quantify, and these variants are not used in routine clinical testing to aid management of these families.
We aimed to identify new candidate genes for predisposition to hereditary diffuse gastric cancer in families without pathogenic CDH1 variants.
Methods
Study design and participants
In this whole-exome sequencing study, we recruited 28 individuals diagnosed with diffuse gastric cancer and 11 unaffected relatives from 22 families with hereditary diffuse gastric cancer that had tested negative for CDH1 pathogenic germline mutations as part of the Familial Gastric Cancer study (MREC 97/5/32) and for whom blood and tumour samples were available. Families (including first-degree and second-degree relatives) were categorised as having hereditary diffuse gastric cancer on the basis of existing criteria.2, 3, 6
Whole-exome sequencing and variant filtering
DNA was extracted from blood or saliva and prepared for 125-bp paired-end whole-exome sequencing using the Nextera Rapid Capture Exome Enrichment Kit (Illumina, San Diego, CA, USA). Sequencing was done on HiSeq-4000 or HiSeq-2500 platforms (Illumina, San Diego, CA, USA). Variant Call Format files were generated with a standard pipeline following Genome Analysis Toolkit (GATK) Best Practices recommendations for whole-exome data (appendix). The dataset was filtered to select uncommon (allele frequency <0·05 in the 1000 Genomes Project European sample) protein-affecting variants, including loss-of-function variants (stop site gained, stop site lost, start site lost, splice acceptor, splice donor, or frameshift), deleterious (predicted with Sorting Intolerant From Tolerant version 5.2.2) and damaging (predicted with Polymorphism Phenotyping version 2.2.2) missense variants, and inframe indels, that were observed in at least one of the 28 affected individuals. These filters were chosen to remove variants that were least likely to affect predisposition to hereditary diffuse gastric cancer. We considered the 11 unaffected family members separately on a per-family basis as a control group on which we did segregation analysis for identified candidate variants. We determined the allele frequency of all candidate variants in healthy controls (with no history of cancer) and allele counts in affected and unaffected individuals within families, and predicted downstream effects on the protein product.
Variants were aggregated into unique genes, which were then filtered to select those that contained at least one loss-of-function variant. We also removed the top 1% most variable genes, which were identified by the number of rare, protein-affecting variants per gene. These genes typically possess many rare variants within the healthy population and, therefore, are unlikely to have a role in predisposition to hereditary diffuse gastric cancer or other diseases. Variant-filtering and gene-filtering steps are summarised in the appendix.
To analyse copy number variants, we applied the XHMM algorithm to the gene set, using principle component analysis to normalise read depth across exomes and a hidden Markov model to identify regions with variation in read depth.11 Around a 50% decrease or increase in read depth was required for the variant to be considered for further analysis. Copy number variants were further explored in selected individuals with a CytoScan 750K genotyping array (Affymetrix, Santa Clara, CA, USA), according to the clinical protocol.
The results published here are in whole or part based on data generated by The Cancer Genome Atlas (TCGA), managed by the National Cancer Institute and the National Human Genome Research Institute. Controlled access data was requested and downloaded for the TCGA-STAD dataset, of which a subset of data from 88 cases of diffuse gastric cancer were analysed to further validate the role of the identified candidate genes in predisposition to diffuse gastric cancer.
Gene-interaction network analysis
Gene-interaction network analysis was used to identify variant-enriched candidate genes with interacting protein products; non-antagonistic, physically interacting proteins might have a similar effect on cell function and, therefore, might produce a shared phenotype when mutated. The filtered genes were put through the GeneMANIA Cytoscape plugin version 3.4.1, which places physically interacting genes into clusters.12 A cluster was defined as a set of five or more physically interacting genes.
We used the PANTHER over-representation test (version 13.0) in the Gene Ontology Consortium enrichment analysis web-tool to assign Gene Ontology (GO) terms to clusters, applying the default Bonferroni correction for multiple testing.13 Of the significant terms highlighted in the analysis, the most significant term that encompassed between ten and 200 genes was selected, consistent with previous studies.14
Allelic counts of all filtered, loss-of-function variants within the selected GO terms (regardless of GeneMANIA Cytoscape clustering) were aggregated and contingency tables were drawn. Variants were also aggregated for each GO term over a comparably filtered set of genes from 503 European individuals in phase 3 of the 1000 Genomes study (appendix).15 We did a one-tailed Fisher's exact test using the R Stats package version 3.3.3 to test for enrichment of loss-of-function variants within each selected GO term in the families with hereditary diffuse gastric cancer compared with the 1000 Genomes European dataset. For this test, only one occurrence of a variant was counted per affected family. A link to the custom R scripts used for this analysis can be found in the appendix.
Validation by Sanger sequencing
Candidate variants were validated by Sanger sequencing. Germline DNA from blood and extracted tumour DNA were quantified with the Qubit dsDNA HS Kit (Invitrogen, Carlsbad, CA, USA), and custom flanking primers were designed for each variant (primer sequences are shown in the appendix). DNA fragments were amplified by PCR and the products were sequenced on an ABI Genetic Analyser (Applied Biosystems Foster City, CA, USA) with BigDye Terminator version 3.1 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions.
Tumour immunohistochemistry and microsatellite instability analysis
We used the Ventana MMR IHC Panel (Roche, Indianapolis, IN, USA) to do immunohistochemistry analysis of known mismatch repair genes in available tumours from individuals in which variants in mismatch repair genes were identified. The panel includes antibodies against MLH1, PMS2, MSH2, and MSH6.
To analyse microsatellite instability, 5-μm formalin-fixed, paraffin-embedded tumour sections were mounted on glass slides for dewaxing and manual microdissection. DNA was extracted with the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). We assessed the DNA for five standard microsatellite markers (BAT25, BAT26, NR21, NR24, and MONO27) using the Promega MSI Analysis System, version 1.2 (Promega, Madison, WI, USA). Poorly and moderately differentiated gastric tissue was compared with adjacent tumour-free tissue.
Analysis of PALB2 and BRCA2 variants in published studies
We searched PubMed without language restrictions between Jan 1, 2015, and Dec 31, 2017, using the term “hereditary diffuse gastric cancer” to identify sequencing studies reporting loss-of-function variants in PALB2 and BRCA2 in hereditary diffuse gastric cancer probands with no detected pathogenic CDH1 variants. We included only publications released after the initial report4 in 2015 of PALB2 and BRCA2 mutations associated with hereditary diffuse gastric cancer. For each of the four identified publications4, 10, 16, 17 and this study, we aggregated the allele counts of loss-of-function PALB2 variants. The same counts were done across the 503 European samples from the 1000 Genomes Project and the 27 173 non-Finnish European individuals not in the TCGA from the Exome Aggregation Consortium (ExAC) control datasets.18 We removed the well characterised, non-pathogenic BRCA2 polymorphic stop codon in c.9976A→T from all datasets. We did a one-tailed Fisher's exact test using the R Stats package version 3.3.3 to test for enrichment of loss-of-function PALB2 or BRCA2 variants in the families with hereditary diffuse gastric cancer compared with either control dataset.
Role of the funding source
The funders had no role in study design, data collection, data analysis, data interpretation, or writing of the Article. EF, JR, and MT had access to the raw data. The corresponding author had full access to all of the data and the final responsibility for the decision to submit for publication.
Results
A whole-exome sequencing dataset of 39 individuals from 22 families with hereditary diffuse gastric cancer without pathogenic CDH1 variants (table 1) was filtered to select 3973 uncommon, protein-affecting variants that were aggregated into 2847 genes. Exclusion of the top 1% of highly variable genes, and retention of genes with at least one loss-of-function variant in affected individuals, resulted in a set of 732 genes (1228 variants). Eight highly variable genes were excluded, including ANKRD36, CDC27, HLA-DRB1, HLA-DRB5, MUC3A, MUC4, MUC6, and OR4C5. Additionally, the presence of affected and unaffected family members within our dataset allowed for selection of variants that segregated with phenotype on a per-family basis.
Table 1.
Characteristics of 28 affected individuals in 22 families with hereditary diffuse gastric cancer without CDH1 pathogenic variants
|
Number of samples sequenced* |
Age of proband at diagnosis (years) | Cancers diagnosed in relatives of probands† | Candidate gene identified in proband | ||
|---|---|---|---|---|---|
| Affected | Unaffected | ||||
| 1 | 2 | 0 | 41 | Diffuse gastric cancer (44)‡, gastric cancer (57) | None |
| 2 | 2 | 4 | 27 | Peritoneal cancer, ovarian cancer (22), diffuse gastric cancer (24)‡, diffuse gastric cancer (28) | None |
| 3 | 1 | 0 | 40 | Gastric cancer (28), diffuse gastric cancer (48) | None |
| 4 | 1 | 2 | 55 | Breast cancer, lung cancer, laryngeal cancer, gastric cancer, and diffuse gastric cancer (44, 52) | PALB2 |
| 5 | 1 | 0 | 36§ | Diffuse gastric cancer (37), lung cancer (54), colorectal cancer (57), breast cancer (50), diffuse gastric cancer (61), diffuse gastric cancer (79), lung cancer (83) | None |
| 6 | 1 | 0 | 37 | Breast cancer, gastric cancer (63), gastric cancer (64) | RECQL5 |
| 7 | 2 | 0 | 36 | Colorectal cancer, breast cancer (43), diffuse gastric cancer (55)‡ | None |
| 8 | 1 | 0 | 47¶ | Diffuse gastric cancer (44) | MSH2‖ |
| 9 | 1 | 0 | 44 | Diffuse gastric cancer (28) | None |
| 10 | 1 | 2 | 28 | Breast cancer, gastric cancer (44), gastric cancer (47) | None |
| 11 | 4 | 1 | 28 | Signet-ring cells‡, Signet-ring cells‡, breast cancer (40s), diffuse gastric cancer (45)‡, prostate cancer (60s), colorectal cancer (75) | ATR, NBN |
| 12 | 1 | 0 | 68 | Lung cancer, gastric cancer (49), gastric cancer (50), gastric cancer (76) | MSH2‖ |
| 13 | 1 | 0 | 47 | Gastric cancer, gastric cancer (50s), gastric cancer (60s) | None |
| 14 | 1 | 0 | 23 | Diffuse gastric cancer (40s), diffuse gastric cancer (46), thyroid cancer (30) | None |
| 15 | 1 | 0 | 53 | Gastric cancer (49), gastric cancer (67), gastric cancer (71) | None |
| 16 | 1 | 0 | 37 | Gastric cancer, breast cancer (54), breast cancer (65), colorectal cancer (66) | None |
| 17 | 1 | 0 | 45 | Diffuse gastric cancer (42) | None |
| 18 | 1 | 0 | 48 | Gastric cancer (44), gastric cancer (54) | None |
| 19 | 1 | 1 | 35 | Lung cancer, uterine cancer (65) | None |
| 20 | 1 | 0 | 55 | Gastric cancer (51), colorectal cancer (76) | None |
| 21 | 1 | 1 | 28 | Gastric cancer (53), breast cancer (76), gastric cancer (80) | RECQL5 |
| 22 | 1 | 0 | 30 | Gastric cancer, diffuse gastric cancer (67) | None |
In total, 39 individuals were sequenced, including 11 unaffected relatives. Numbers in parentheses indicate age in years at diagnosis.
All probands were sequenced in this study.
Includes both first-degree and second-degree relatives; age in years at diagnosis is in parentheses when known.
Sequenced in this study.
This proband was also diganosed with colorectal cancer at age 47 years.
This proband was also diagnosed with lobular breast cancer at age 36 years.
No microsatellite instability was detected in tumour.
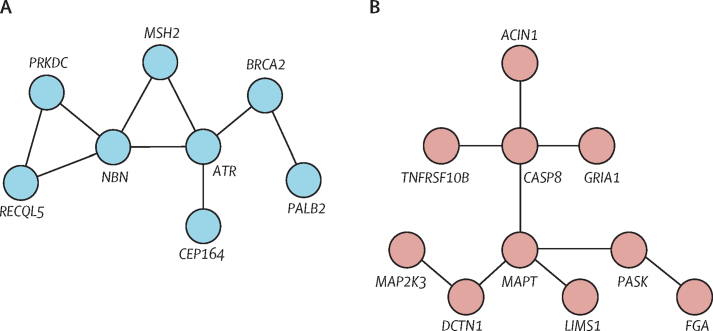
Gene-interaction network analysis of the 732 filtered genes identified two physical interaction clusters, to which GO terms were applied (figure 1). A cluster of eight genes was associated with the GO term double-strand break repair (GO:0006302; p<0·0001; figure 1A). A second cluster of ten genes was associated with the GO term negative regulation of extrinsic apoptotic signalling pathway via death domain receptors (GO:1902042; p=0·00517; figure 1B).
Figure 1.
Gene clusters identified via gene interaction analysis
Lines indicate a physical interaction, as assigned by the GeneMANIA plugin for Cytoscape.12 (A) Gene cluster to which the double-strand break repair GO term (GO:0006302) was assigned. (B) Gene cluster to which the negative regulation of extrinsic apoptotic signalling pathway via death domain receptors GO term (GO:1902042) was assigned. GO=Gene Ontology.
Loss-of-function variants within the filtered set of 1228 variants were aggregated under these two GO terms, including genes related to the GO terms that were not initially clustered by GeneMANIA. The double-strand break repair term was significantly enriched in families with hereditary diffuse gastric cancer compared with the 1000 Genomes European set (p=0·00051). By contrast, the apoptotic signalling pathway term was not enriched in families with hereditary diffuse gastric cancer (p=0·186), suggesting that the differences between the datasets in allele counts of DNA double-strand break repair genes cannot entirely be explained by technical differences that arise when using an externally produced control dataset.
Genes in the double-strand break repair GO term included PALB2, MSH2, RECQL5, ATR, and NBN, all of which were shown to be physically interacting in GeneMANIA (figure 1A). BRCA2 was also a part of this set, but was disregarded from further study because it contained the well characterised, benign polymorphic stop codon c.9976A→T.19
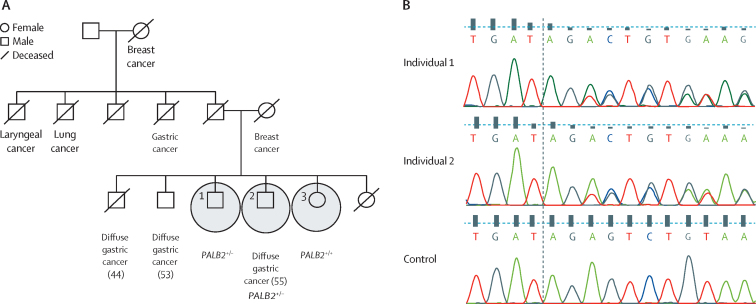
Table 2 summarises the candidate variants. A heterozygous 2 bp frameshift deletion was identified in PALB2 (c.757-758TAG→T [rs180177092, NM_024675.3]) in a patient from family 4 who was diagnosed with diffuse gastric cancer at age 55 years (figure 2). This loss-of-function variant at aminoacid position 253 is predicted to result in an early stop codon seven aminoacids downstream of the variant. Family 4 has a history of breast, lung, laryngeal, and diffuse gastric cancer (table 1, figure 2). Exome sequencing was also done on two unaffected siblings, one of whom also had the PALB2 (c.757-758TAG→T [rs180177092, NM_024675.3]) variant. The affected proband had previously received treatment for Helicobactor pylori infection, but had tested negative at subsequent endoscopies.
Table 2.
Candidate variants in six families with hereditary diffuse gastric cancer without CDH1 pathogenic variants
| Number of sequenced individuals | Gene | Variant | Consequence | Protein change |
Minimum allele frequency |
SIFT | Polymorphism Phenotyping | ||
|---|---|---|---|---|---|---|---|---|---|
| 1000 Genomes European sample | ExAC non-TCGA European sample | ||||||||
| 4 | 3 | PALB2 | c.757-758TAG→T | Frameshift deletion | Leu253fs | 0 | 0 | NA | NA |
| 6 | 1 | RECQL5 | c.2806-2T→C | Splice-acceptor variant | NA | 0 | 0 | NA | NA |
| 8 | 1 | MSH2 | c.967-968T→TCTCA | Frameshift insertion | Ser323fs | 0 | 0 | NA | NA |
| 11 | 5 | ATR | c.6075A→T | Stop site gain | Tyr2025X | 0 | 0 | NA | NA |
| 11 | 5 | NBN | c.1124+1G→C | Splice-donor variant | NA | 0 | 0 | NA | NA |
| 12 | 1 | MSH2 | c.1A→C | Start site loss | Met1?* | 0 | 0 | Deleterious | Benign |
| 21 | 2 | RECQL5 | c.2828C→T | Missense variant | Arg943His | 0·002 | 0·014332 | Deleterious | Probably damaging |
SIFT=Sorting Intolerant From Tolerant. ExAC=Exome Aggregation Consortium. TCGA=The Cancer Genome Atlas. fs=frameshift. NA=not applicable.
Human Genome Variation Society nomenclature to indicate loss of a start site without experimental evidence of a new start site.20
Figure 2.
Pedigree and cancer history for family 4
(A) Whole-exome sequencing was done on the three circled individuals; age at diagnosis of cancer is shown in parentheses when known. (B) Chromatograms showing the PALB2 frameshift variant (c.757-758TAG→T) in DNA from individuals 1 and 2 compared with control DNA.
Two heterozygous loss-of-function variants were identified in the mismatch repair gene MSH2: a start site loss (c.1A→C [rs267607911, NM_000251.2]) in a patient from family 12 and a frameshift insertion of 4 bp (c.967-968T→TCTCA [NM_000251.2]) in a patient from family 8 (table 2; appendix). Both families had a strong history of gastric cancer; however, only DNA from the probands was available for sequencing, so segregation analysis could not be done. Heterozygosity of both variants was maintained in tumour DNA from the patients, as confirmed by Sanger sequencing. Tumours from both probands showed normal expression of MSH2 and other mismatch repair proteins by immunohistochemistry compared with adjacent tumour-free tissue (appendix), and neither tumour showed evidence of microsatellite instability (appendix). Both probands with MSH2 variants had previously tested negative for H pylori.
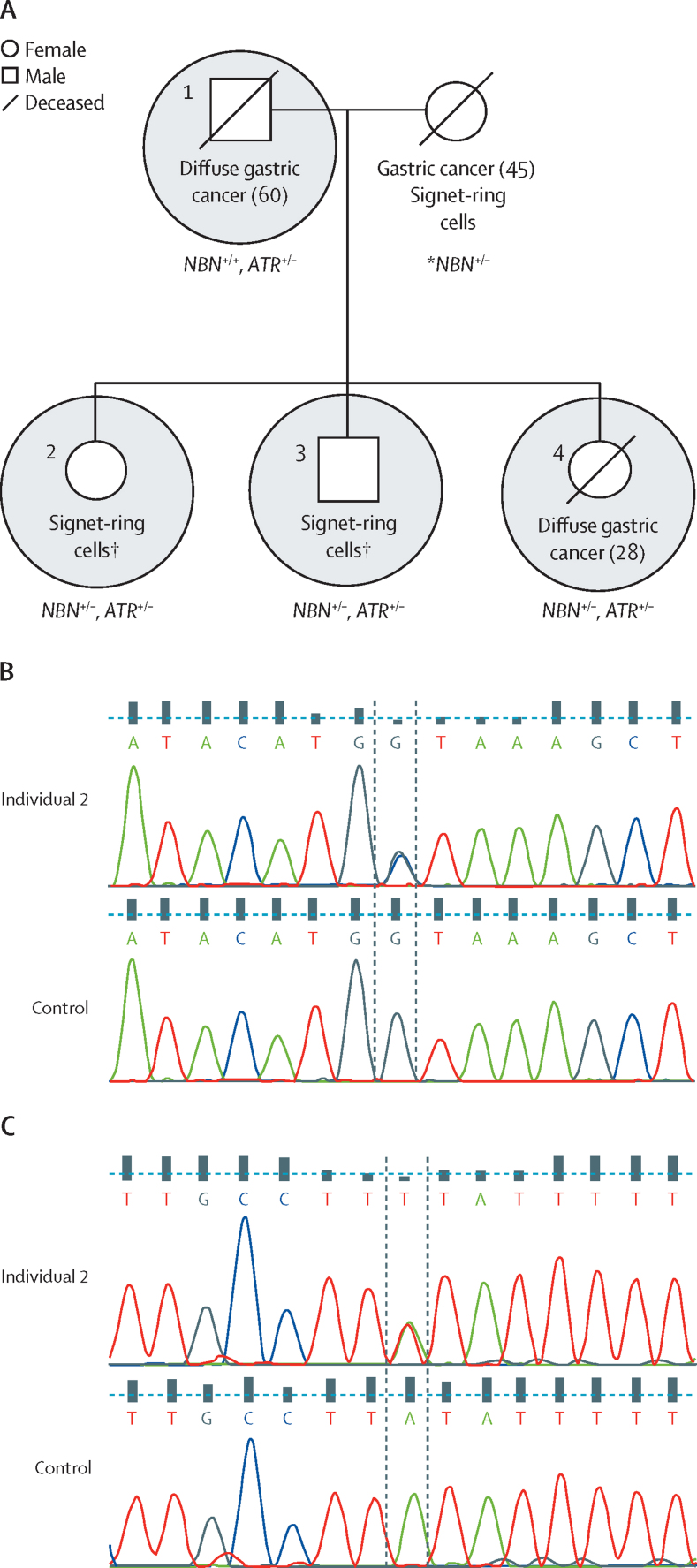
Heterozygous variants in the DNA repair genes ATR and NBN—a splice-donor variant (c.1124+1G→C [NM_002485.4]) in NBN and a predicted stop site-gain variant (c.6075A→T [NM_001184.3]) in ATR—were identified in the proband from family 11, who was diagnosed with diffuse gastric cancer at age 28 years (figure 3, table 2). Two siblings underwent risk-reducing gastrectomies, and subsequent pathological analysis of gastric tissue revealed the presence of signet-ring cells in both individuals. As such, these individuals were considered to be affected family members in this analysis. The father of the proband was diagnosed with diffuse gastric cancer at age 60 years and the mother had metastatic disease characterised by signet-ring cells, which suggests that she had primary gastric cancer. All individuals in this family whose DNA was sequenced tested negative for H pylori. The proband and both siblings were heterozygous for loss-of-function variants in both ATR and NBN. The splice-donor variant in NBN was not seen in the father, and so was presumed to have been inherited maternally (DNA was only available for the father). The ATR variant was identified in the father, in whom no clinically relevant copy number variants were found. An unaffected, second-degree relative in family 11 did not have either variant.
Figure 3.
Pedigree and cancer history of family 11
(A) Whole-exome sequencing was done on the three circled individuals; the proband was patient 4. The presence of the variants in NBN (c.1124+1G→C) and ATR (c.6075A→T) among the four affected family members are shown. Age at diagnosis of cancer is shown in parentheses when known. *DNA was not available for the mother, and so the mother's genotype was assumed on the basis of the genotypes of the father and children. †These individuals underwent risk-reducing gastrectomies. (B) Chromatograms showing the NBN variants in DNA from individual 2 compared with control DNA. (C) Chromatograms showing the ATR variants in DNA from individual 2 compared with control DNA.
Two variants were identified in the helicase gene RECQL5 in different families. One was a missense variant (c.2828C→T [rs200535477, NM_004259.6]) in the proband from family 21, who was diagnosed with diffuse gastric cancer at age 28 years, and the other was a loss-of–function, splice-acceptor variant (c.2806-2T→C [rs201841487, NM_004259.6]) in the proband from family 6, who was diagnosed with diffuse gastric cancer at age 37 years (table 2; appendix). Both of these families included individuals across three generations who were diagnosed with gastric cancer and breast cancer. The proband of family 6 tested negative for H pylori, and the H pylori status of the proband from family 21 was unknown. DNA from the father of the proband in family 21 was sequenced, and the missense variant in RECQL5 was not found.
We did not explore variants in other genes within the double-strand break repair cluster because the variants did not segregate with the disease in families containing affected and unaffected members.
We analysed data from previous studies4, 10, 16, 17 to estimate enrichment of loss-of-function variants in PALB2 and BRCA2 in families with hereditary diffuse gastric cancer. A loss-of-function variant (c.1438A→T [rs1057520653, NM_024675.3]) was identified and reported to us by collaborators (Teixeira MR, unpublished), however this variant was not included in the analysis because it did not fit our search criteria. Five (2%) of the 329 probands tested in these studies (including the present study) had loss-of-function PALB2 variants (table 3). By contrast, PALB2 variants were identified in 26 (<1%) of 27 173 individuals in the non-TCGA, non-Finnish European ExAC database (p<0·0001) and in one (<1%) of 503 individuals in the 1000 Genomes Project European database (p=0·039). Loss-of-function BRCA2 variants were not enriched in families with hereditary diffuse gastric cancer compared with ExAC (p=0·47) or 1000 Genomes Project (p=1·00) individuals. No loss-of-function PALB2 variants were identified in a set of 88 cases with sporadic diffuse gastric cancer from the TCGA.
Table 3.
PALB2 variants identified in hereditary diffuse gastric cancer sequencing studies
| Race | Patient ID | Diagnosis of proband (age at diagnosis in years) | Variant | Consequence | Protein change | |
|---|---|---|---|---|---|---|
| Hansford et al (2015)4 | European | P124 | Diffuse gastric cancer (45) | c.1193AC→A | fs deletion | Val398fs |
| Sahasrabudhe et al (2017)10 | European | CG-12 | Intestinal gastric cancer (69) | c.1240C→T | Stop-site gain | Arg414Ter |
| Sahasrabudhe et al (2017)10 | European | CG-008 | Diffuse gastric cancer (48) | c.1240C→T | Stop-site gain | Arg414Ter |
| Sahasrabudhe et al (2017)10 | European | GM037589 | Gastric cancer (46) | c.1240C→T | Stop-site gain | Arg414Ter |
| Sahasrabudhe et al (2017)10 | European | CG-05 | Diffuse gastric cancer (50) | c.3201+1G→T | Splice-site variant | NA |
| Sahasrabudhe et al (2017)10 | European | CG-039 | Diffuse gastric cancer (47) | c.1882_1890delAAGTCCTGC | In-frame deletion | Lys628_Cys630del |
| Sahasrabudhe et al (2017)10 | Latin American | CG-028 | Intestinal gastric cancer (81) | c.1882_1890delAAGTCCTGC | In-frame deletion | Lys628_Cys630del |
| Sahasrabudhe et al (2017)10 | Latin American | 3CG-103 | Mixed (79) | c.2753C→A | Missense | Pro918Gln |
| Fewings et al (this study) | European | GST_172_301 | Diffuse gastric cancer (55) | c.757_758TAG→T | fs deletion | Leu253fs |
| Teixeira (unpublished) | European | GM048157 | Diffuse gastric cancer (56) | c.1438A→T | Stop-site gain | Lys480Ter |
None of the identified PALB2 variants appeared in the 1000 Genomes Project European samples or in the Exome Aggregation Consortium European datasets. fs=frameshift. NA=not applicable.
Discussion
We found predicted pathogenic (protein-affecting) germline variants in known cancer-predisposing DNA repair genes (including PALB2, MSH2, ATR, NBN, and RECQL5) in six (27%) of 22 families with hereditary diffuse gastric cancer. This finding reflects the increasing number of cancer phenotypes found to be associated with existing cancer-predisposing genes as genomic analyses extend to rarer cancer subtypes. For example, mismatch repair genes were initially associated with increased risk of colorectal cancer, but were subsequently associated with risk of developing gastric and pancreatic cancers, among others.21, 22
Simply identifying predicted pathogenic variants in known cancer-predisposing genes does not imply causality. For example, pathogenic germline variants in MSH2 were not accompanied by altered expression of DNA mismatch repair proteins in tumour tissue in our study. To investigate causality, larger studies with matched controls are required, which is not typically feasible for rare diseases. The generation of large control datasets, such as ExAC and 1000 Genomes, can be used to strengthen possible associations, as we have shown in the case of PALB2. When combining our data with those from previously published relevant studies,4, 10, 16, 17 including those in which no PALB2 variants were found, we saw a significant over-representation of PALB2 (but not BRCA2) pathogenic variants in families with hereditary diffuse gastric cancer compared with ExAC and 1000 Genomes controls; however, this finding was much less significant when comparing with the 503 Europeans in the 1000 Genomes Project set than when comparing with the 27 173 individuals from the non-Finnish, non-TCGA ExAC dataset.15
In several of the cases described by Sahasrabudhe and colleagues,10 tumour molecular profiling was done and showed that carriers of PALB2 mutations had mutational signatures indicative of defects in homologous recombination. PALB2 has an important role in homologous recombination during double-stranded DNA break repair through recruitment of BRCA2 and RAD51 to DNA breaks. Mutations in this gene are associated with an increased risk of breast and pancreatic cancers.23, 24, 25 Even within families carrying PALB2 mutations, cases of diffuse gastric cancer are likely to be rare and could be masked by a larger number of sporadic gastric adenocarcinomas, which means that associations with certain cancer subtypes might be missed in epidemiological studies of these families unless the pathology of all reported cancers is known. For example, a recent study26 revealed that a rare serous subtype of endometrial cancer is over-represented in carriers of BRCA1 variants, identifying a novel cancer association with a gene that has been intensively studied for more than 20 years.
ATR and NBN are also involved in initiating the response to double-strand DNA breaks. The NBN gene product (NBS1) associates with MRE11 and RAD50 to form a complex involved in the activation of the ataxia proteins ATM and ATR, which have roles in the recruitment of damage repair proteins, cell cycle regulation, and apoptosis. We identified single loss-of-function variants in NBN or ATR in the parents of the proband in family 11, both of whom had, or were suspected to have, diffuse gastric cancer. These variants were co-inherited in all three siblings in the family. Expression of either one of these variants might predispose family members to diffuse gastric cancer, although no other incidences of gastric cancer, and only one instance of late-onset prostate cancer, were noted in an extensive maternal and paternal family history. Slavin and colleagues17 also identified a stop site-gain variant in ATR in an individual with intestinal-type adenocarcinoma and a strong family history of gastric cancer.
The unusual cancer pattern seen in family 11 (with all immediate family members [siblings and parents], but no extended family members, of the proband affected) might be attributed to multi-locus, inherited neoplasia alleles syndrome, in which inheritance of pathogenic mutations in multiple cancer-predisposing genes leads to an atypical or severe phenotype.27 The close functional relationship between NBN and ATR in double-stranded DNA break repair could indicate a potential combinatorial effect of variants in these genes, as potentially suggested by the young age of diagnoses or the presence of signet-ring cells in the siblings carrying these variants. By contrast, double heterozygosity of mutations in the DNA repair genes BRCA1 and BRCA2 in patients with breast cancer was found to be no more deleterious than a single heterozygous mutation.28 Nevertheless, such double heterozygosity might have implications for genetic counselling that should be considered.
Genetic variants in genes involved in the mismatch DNA repair pathway are also associated with Lynch syndrome. A variant similar to the start-site loss variant that we identified in MSH2 (c.1A→G) has previously been shown to have only a mild effect on protein function;29 thus, this variant should not be treated as a typical loss-of-function variant. This attenuated effect on protein function could be due to the presence of an alternative start codon and a second non-mutated MSH2 allele. Increased microsatellite instability, a measure of decreased MSH2 function, has been shown in patients and tumours with the c.1A→G start site-loss variant.29 However, in tumours from both cases analysed here, MSH2 expression (detected by immunohistochemistry staining) was normal and no microsatellite instability was found. Although it is most likely that tumorigenesis was not caused by mismatch repair deficiency, we cannot rule out the possibility of a novel, non-mismatch repairmediated mechanism of carcinogenesis driven by variants in MSH2.
The helicase RECQL5 is important for prevention of aberrant homologous recombination and accumulation of double-strand DNA breaks, and thus for preservation of genome stability.30 A missense RECQL5 variant was identified in the proband from family 21, and was not found in the proband's unaffected father. A splice-acceptor variant in RECQL5 was identified in an individual in family 6 who was diagnosed with diffuse gastric cancer at age 37 years. Both family 6 and family 21 had a history of breast and gastric cancer.
Previous studies have explored the role of known cancer predisposition genes in individuals with hereditary diffuse gastric cancer who do not have known CDH1 mutations. Sahasrabudhe and colleagues10 identified germline variants in PALB2, BRCA1, and RAD51C in families with diffuse gastric cancer. Hansford and colleagues4 described variants in ATM, BRCA2, MSR1, and STK11, as well as a frameshift deletion in PALB2. This group has also uncovered a role for the CDH1-related adhesion gene CTNNA1. Although we did not find any variants of interest in ATM, BRCA1, BRCA2, CTNNA1, MSR1, RAD51C, or STK11, an exploration of PALB2 variants in all families with hereditary diffuse gastric cancer sequenced in recent studies showed enrichment of loss-of-function variants in these families compared with control datasets. This finding makes a case for inclusion of PALB2 in genetic testing for families with hereditary gastric cancer without CDH1 mutations, and it is possible that individuals who carry PALB2 mutations might benefit from platinum-based chemotherapy and treatment with PARP inhibitors.31 However, the evidence is not yet sufficient to recommend surveillance of diffuse gastric cancer in carriers of PALB2 mutations because the absolute risk is likely to be low in the absence of a family history.
Sporadic stomach cancers have been analysed as part of the TCGA study,32 and an association was identified between truncating PALB2 mutations and sporadic stomach adenocarcinoma. Of the individuals with sporadic stomach adenocarcinoma and PALB2 mutations from the TCGA database, we selected 88 individuals with diffuse gastric cancer, as described by Bass and colleagues,33 among which we did not identify any truncating PALB2 variants. However, the average age at diagnosis for this cohort was 66 years, so this finding is perhaps not unexpected given the younger age of onset usually observed in hereditary cancers.
The rarity of patients with hereditary diffuse gastric cancer without pathogenic CDH1 variants makes the collection of large datasets challenging. We used gene-interaction network analysis to prioritise candidate gene variants that co-segregated with disease phenotype and were likely to be involved in predisposition to hereditary diffuse gastric cancer on the basis of knowledge of the biology of the disease. This approach did not overcome the problem of low statistical power due to a small sample size, which is often seen with rare cancer datasets, but it did allow for selection of the most plausible candidates from the available data.
We attempted an additional analysis of copy number variants within this dataset using the XHMM algorithm.11 Although this analysis did not suggest any plausible candidates, at present, copy number variant analysis of germline whole-exome sequencing data is not validated and, therefore, some causal copy number variants could have been missed.
In summary, we found that rare, protein-affecting variants in DNA damage repair genes were enriched in families with hereditary diffuse gastric cancer without pathogenic CDH1 variants compared with control datasets. Further studies of these genes in similar families are required to increase knowledge of the genetic basis of hereditary diffuse gastric cancer so that better informed decisions about risk reduction and management in affected family members can be made. Lastly, for many families with hereditary diffuse gastric cancer without pathogenic CDH1 variants, the underlying cause remains unexplained even after whole-exome sequencing, and although whole-genome sequencing might identify some additional candidates in regulatory elements or structural variants, it seems unlikely that high-impact genes other than CDH1 will be implicated in hereditary diffuse gastric cancer. Therefore, focusing on moderate-impact or low-impact cancer genes, such as PALB2, might be the way forward for future studies of genes associated with predisposition to disease in these patients.
Acknowledgments
Acknowledgments
This study was supported by UK Medical Research Council (Sackler programme), European Research Council (310018) under the European Union's Seventh Framework Programme (2007–13), National Institute for Health Research Cambridge Biomedical Research Centre, Experimental Cancer Medicine Centres, and Cancer Research UK. We thank the Human Research Tissue Bank, which is supported by the National Institute for Health Research Cambridge Biomedical Research Centre, Addenbrooke's Hospital, Cambridge, UK. We also thank Tara Clancy (Manchester Centre for Genomic Medicine, Manchester, UK), Cecilia Compton (Clinical Genetics Service, Guy's and St Thomas' NHS Foundation Trust, London, UK), Sarah Everest (Peninsula Clinical Genetics Service, Exeter, UK), Vicky Hunt (Peninsula Clinical Genetics Service), Emma Kivuva (Peninsula Clinical Genetics Service), Anna Lehmann (Clinical Genetics, St George's University Hospitals NHS Foundation Trust, London, UK), Mark Longmuir (West of Scotland Genetics Services, Glasgow, UK), Ana Peixoto (Department of Genetics, Portuguese Oncology Institute of Porto, Porto, Portugal), Peter Risby (Oxford Centre for Genomic Medicine, Oxford University Hospitals NHS Foundation Trust, Oxford, UK), and Sarah Rose (Clinical Genetics Service, Guy's and St Thomas' NHS Foundation Trust) for their help in recruiting families and the mutation analysis.
Contributors
MT, PP, CC, and RCF conceived and designed the study. EF, AL, MAG, JS, JR, GRC, JH, and S-FC did the sequencing and data analyses. CB, RD, IE, DGE, DH, LI, PM, VMcC, LV, MRT, MdP, and RH recruited patients. SR and RM were responsible for coordinating recruitment, consent, and sample handling. MO'D and OTG provided histopathology input and reviewed reports and tumour samples. EF, AL, and PP did the statistical analyses and data interpretation. EF and MT wrote the manuscript and all authors reviewed the final version. MT is the guarantor.
Declaration of interests
DGE declares personal fees from AstraZeneca outside the submitted work. All other authors declare no competing interests.
Supplementary Material
References
- 1.Guilford P, Blair V, More H, Humar B. A short guide to hereditary diffuse gastric cancer. Hered Cancer Clin Pract. 2007;5:183–194. doi: 10.1186/1897-4287-5-4-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Post RS, Vogelaar IP, Carneiro F. Hereditary diffuse gastric cancer: updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. J Med Genet. 2015;52:361–374. doi: 10.1136/jmedgenet-2015-103094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caldas C, Carneiro F, Lynch HT. Familial gastric cancer: overview and guidelines for management. J Med Genet. 1999;36:873–880. [PMC free article] [PubMed] [Google Scholar]
- 4.Hansford S, Kaurah P, Li-Chang H. Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol. 2015;1:23–32. doi: 10.1001/jamaoncol.2014.168. [DOI] [PubMed] [Google Scholar]
- 5.Mi EZ, Mi EZ, di Pietro M. Comparative study of endoscopic surveillance in hereditary diffuse gastric cancer according to CDH1 mutation status. Gastrointest Endosc. 2018;87:408–418. doi: 10.1016/j.gie.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzgerald RC, Hardwick R, Huntsman D. Hereditary diffuse gastric cancer: updated consensus guidelines for clinical management and directions for future research. J Med Genet. 2010;47:436–444. doi: 10.1136/jmg.2009.074237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sereno M, Aguayo C, Guillén Ponce C. Gastric tumours in hereditary cancer syndromes: clinical features, molecular biology and strategies for prevention. Clin Transl Oncol. 2011;13:599–610. doi: 10.1007/s12094-011-0705-y. [DOI] [PubMed] [Google Scholar]
- 8.van Lier MGF, Westerman AM, Wagner A. High cancer risk and increased mortality in patients with Peutz-Jeghers syndrome. Gut. 2011;60:141–147. doi: 10.1136/gut.2010.223750. [DOI] [PubMed] [Google Scholar]
- 9.Masciari S, Dewanwala A, Stoffel EM. Gastric cancer in individuals with Li-Fraumeni syndrome. Genet Med. 2011;13:651–657. doi: 10.1097/GIM.0b013e31821628b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahasrabudhe R, Lott P, Bohorquez M. Germline mutations in PALB2, BRCA1, and RAD51C, which regulate DNA recombination repair, in patients with gastric cancer. Gastroenterology. 2017;152:983–986. doi: 10.1053/j.gastro.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fromer M, Purcell SM. Using XHMM software to detect copy number variation in whole-exome sequencing data. Curr Protoc Hum Genet. 2014;81:7.23.1–7.2321. doi: 10.1002/0471142905.hg0723s81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montojo J, Zuberi K, Rodriguez H. GeneMANIA cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blake JA, Christie KR, Dolan ME. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milne RL, Kuchenbaecker KB, Michailidou K. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat Genet. 2017;49:1767–1778. doi: 10.1038/ng.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auton A, Abecasis GR, Altshuler DM. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogelaar IP, van der Post RS, van Krieken JHJ. Unraveling genetic predisposition to familial or early onset gastric cancer using germline whole-exome sequencing. Eur J Hum Genet. 2017;25:1246–1252. doi: 10.1038/ejhg.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slavin T, Neuhausen SL, Rybak C. Genetic gastric cancer susceptibility in the International Clinical Cancer Genomics Community Research Network. Cancer Genet. 2017;216–17:111–119. doi: 10.1016/j.cancergen.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lek M, Karczewski KJ, Minikel EV. Analysis of protein-coding genetic variation in 60 706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgs JE, Harkness EF, Bowers NL. The BRCA2 polymorphic stop codon: stuff or nonsense? J Med Genet. 2015;52:642–645. doi: 10.1136/jmedgenet-2015-103206. [DOI] [PubMed] [Google Scholar]
- 20.Human Genome Variation Society Discussions regarding the description of sequence variants. http://www.hgvs.org/mutnomen/disc.html#Met
- 21.Oliveira C, Pinheiro H, Figueiredo J, Seruca R, Carneiro F. Familial gastric cancer: genetic susceptibility, pathology, and implications for management. Lancet Oncol. 2015;16:e60–e70. doi: 10.1016/S1470-2045(14)71016-2. [DOI] [PubMed] [Google Scholar]
- 22.Lynch HT, Voorhees GJ, Lanspa SJ, McGreevy PS, Lynch JF. Pancreatic carcinoma and hereditary nonpolyposis colorectal cancer: a family study. Br J Cancer. 1985;52:271–273. doi: 10.1038/bjc.1985.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antoniou AC, Casadei S, Heikkinen T. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pauty J, Rodrigue A, Couturier A, Buisson R, Masson J-Y. Exploring the roles of PALB2 at the crossroads of DNA repair and cancer. Biochem J. 2014;460:331–342. doi: 10.1042/BJ20140208. [DOI] [PubMed] [Google Scholar]
- 25.Easton DF, Pharoah PDP, Antoniou AC. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372:1–15. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saule C, Mouret-Fourme E, Briaux A. Risk of serous endometrial carcinoma in women with pathogenic BRCA1/2 variant after risk-reducing salpingo-oophorectomy. J Natl Cancer Inst. 2018;110:2017–2019. doi: 10.1093/jnci/djx159. [DOI] [PubMed] [Google Scholar]
- 27.Whitworth J, Skytte A-B, Sunde L. Multilocus inherited neoplasia alleles syndrome. JAMA Oncol. 2016;2:373. doi: 10.1001/jamaoncol.2015.4771. [DOI] [PubMed] [Google Scholar]
- 28.Leegte B. Phenotypic expression of double heterozygosity for BRCA1 and BRCA2 germline mutations. J Med Genet. 2005;42:e20. doi: 10.1136/jmg.2004.027243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kets CM, Hoogerbrugge N, van Krieken JHJM, Goossens M, Brunner HG, Ligtenberg MJL. Compound heterozygosity for two MSH2 mutations suggests mild consequences of the initiation codon variant c.1A>G of MSH2. Eur J Hum Genet. 2009;17:159–164. doi: 10.1038/ejhg.2008.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saponaro M, Kantidakis T, Mitter R. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell. 2014;157:1037–1049. doi: 10.1016/j.cell.2014.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ledermann JA, Kohn EC. PARP inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann Oncol. 2014;25:32–40. doi: 10.1093/annonc/mdt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu C, Xie M, Wendl MC. Patterns and functional implications of rare germline variants across 12 cancer types. Nat Commun. 2015;6:10086. doi: 10.1038/ncomms10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bass AJ, Thorsson V, Shmulevich I. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.