ABSTRACT
Almost every cell in the human body extends a primary cilium. Defective cilia function leads to a set of disorders known as ciliopathies, which are characterised by debilitating developmental defects that affect many tissues. Here, we report a new role for regulator of calcineurin 2 (RCAN2) in primary cilia function. It localises to centrioles and the basal body and is required to maintain normal cilia length. RCAN2 was identified as the most strongly upregulated gene from a comparative RNAseq analysis of cells in which expression of the Golgi matrix protein giantin had been abolished by gene editing. In contrast to previous work where we showed that depletion of giantin by RNAi results in defects in ciliogenesis and in cilia length control, giantin knockout cells generate normal cilia after serum withdrawal. Furthermore, giantin knockout zebrafish show increased expression of RCAN2. Importantly, suppression of RCAN2 expression in giantin knockout cells results in the same defects in the control of cilia length that are seen upon RNAi of giantin itself. Together, these data define RCAN2 as a regulator of cilia function that can compensate for the loss of giantin function.
KEY WORDS: Cilia, RCAN2, Calcineurin, Giantin, Golgi
Summary: RCAN2 is a centriolar protein with roles in ciliary length control and can compensate for loss of function of giantin.
INTRODUCTION
Ciliogenesis, the emergence of a microtubule axoneme from the mother centriole, is fundamental for the ability of non-cycling cells to sense and respond to their environment (Lechtreck, 2015). Defects in the formation and/or function of the cilium lead to a cohort of human diseases known as ciliopathies (Braun and Hildebrandt, 2016), which affect multiple tissues leading to problems with respiratory, kidney and heart function, as well as vision, hearing and fertility. Cilia defects are also linked to obesity and diabetes. Movement of cargo and signalling complexes into and out of cilia requires intraflagellar transport (IFT), the process by which microtubule motors drive motility along the axoneme. The ability of cilia to act as confined signalling hubs directing key developmental and homeostatic pathways, such as those linked to sonic hedgehog (Shh) signalling (He et al., 2016) or mechanosensing, requires the trafficking of receptors and associated signalling molecules into cilia (He et al., 2016; Sung and Leroux, 2013). This includes ligand-gated receptors, G-protein-coupled receptors, Ca2+ channels and transcription factors. Considerable work supports a major role for Ca2+ in ciliary function (Delling et al., 2013). Resting cilium [Ca2+] is substantially higher than resting cytoplasmic [Ca2+], and ciliary [Ca2+] is important for cells to respond to Shh through activation of Gli transcription factors (Delling et al., 2013).
We have shown previously using RNAi that the coiled-coil Golgi protein giantin is required for ciliogenesis in vitro (Asante et al., 2013). This was linked to the accumulation of the dynein-2 motor complex around the ciliary base. Dynein-2 is the major motor driving retrograde IFT along the axoneme and is, among other functions, required for the transduction of the Shh signal (He et al., 2016). Consistent with our findings using RNAi in cultured cells, morpholino knockdown of giantin in zebrafish resulted in fewer but longer cilia in the neural tube (Bergen et al., 2017). In contrast, recent characterisation of giantin knockout (KO) zebrafish shows that, although they breed and develop similarly to wild-type fish, giantin KO fish show a significant decrease in body length (more notable in young adults) and have defects in extracellular matrix, cartilage and bone formation, but have only minor cilia defects (Bergen et al., 2017).
We recently generated a KO cell line that no longer expresses giantin (Stevenson et al., 2017). Here, we show that in contrast to depletion of giantin, complete knockout of the gene does not prevent cells from generating cilia on serum withdrawal. We hypothesised that compensatory mechanisms might enable KO cells to produce normal cilia, whereas acute suppression of expression using RNAi did not allow such compensation. Such mechanisms would also explain the relatively mild manifestation of ciliary defects in giantin mutant zebrafish compared with the effects of in vivo morpholino knockdown. RNAseq showed that RCAN2 (regulator of calcineurin-2; also called calcipressin-2 and ZAKI-4) is the most strongly upregulated gene in these giantin KO cells. Here, we show that RCAN2 localises to centrioles and its depletion affects ciliogenesis. Furthermore, removal of RCAN2 in giantin KO cells recapitulates the giantin RNAi ciliary length phenotype.
RESULTS AND DISCUSSION
Giantin KO cells show no gross defects in ciliogenesis
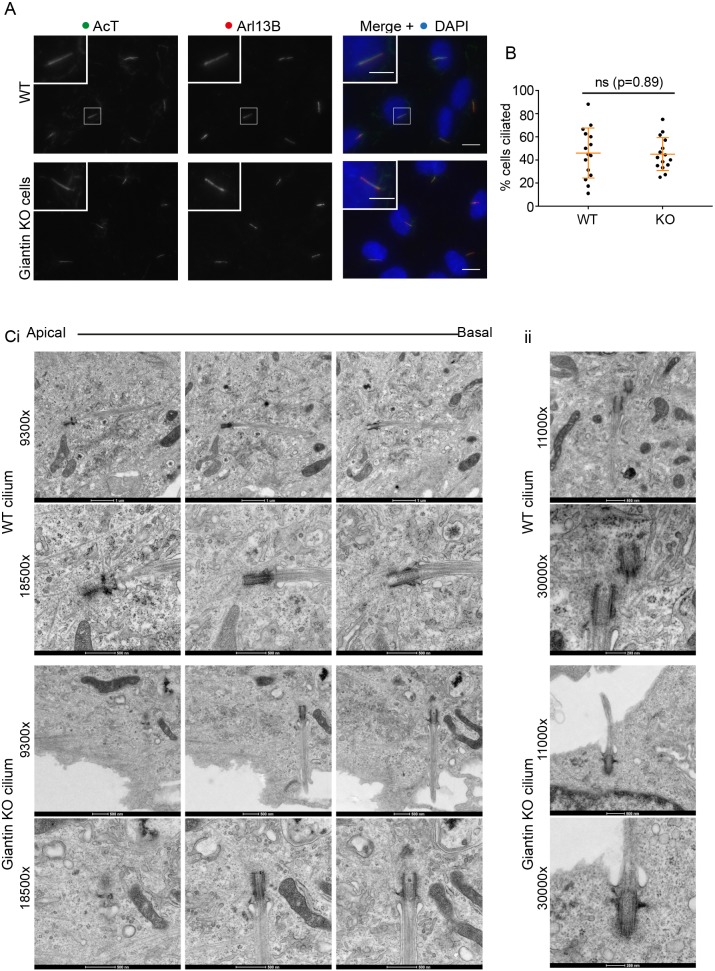
Using RNAi, we have shown previously that a reduction in the expression of giantin is associated with a ciliogenesis defect in cells (Asante et al., 2013). Giantin-depleted cells produce fewer cilia and those that remain are longer, suggesting a defect in length control. In contrast, analysis of giantin KO RPE-1 cells (Stevenson et al., 2017) shows no gross defects in ciliogenesis at the level of light microscopy (Fig. 1A, quantified in B) or any significant change in cilia length (Fig. 1A, quantified fully in later experiments). Using serial section transmission electron microscopy (TEM), we detected no obvious abnormalities in the structure of the basal body, appendages or ciliary pocket (Fig. 1C, enlarged in Cii). Furthermore, Golgi structure appears normal (see Stevenson et al., 2017). This phenotypic discrepancy between knockdown and KO cells is consistent with our observations in zebrafish (Bergen et al., 2017) and led us to hypothesise that altered expression of another gene or genes compensates for the loss of giantin. A caveat to these experiments is that we were only able to derive one knockout line for giantin in RPE1 cells.
Fig. 1.
Giantin knockout RPE1 cells have no obvious defects in ciliogenesis. (A) Representative images of RPE1 WT and giantin KO cells fixed after 24 h of serum starvation and labelled for Arl13B (red) and acetylated tubulin (green); nuclei are stained with DAPI (blue). Scale bars: 10 μm and 5 μm (inset). (B) Quantification of the experiment represented in A (n=3). The mean percentage of cells producing cilia does not change upon KO of giantin. Error bars represent s.d., Mann-Whitney U-test. (C) TEM images of (i) serial and (ii) single plane 70 nm sections through representative WT and giantin KO cilia at two different magnifications.
RNAseq of triplicate RNA samples from these and control cells followed by pairwise statistical comparison identified 1025 genes whose expression level is decreased >4-fold. These data are described in Stevenson et al. (2017) and the raw data are available via the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-5618. The most highly upregulated gene, and therefore the most likely to underpin any compensation for loss of giantin, is RCAN2. RCAN2 is a negative regulator of calcineurin and modulates Ca2+-dependent signalling. Calcineurin is required for transcriptional signalling; RCAN2 acts as a negative regulator of nuclear factor of activated T-cells (NFAT) activation and therefore of NFAT-dependent transcription. RNAseq showed that in giantin KO cells, RCAN2 is >256× upregulated compared with levels in the parental cell line (Stevenson et al., 2017), implicating RCAN2 in cell adaptation to the lack of giantin.
To determine whether RCAN2 upregulation is a conserved response to loss of giantin, we examined expression of RCAN2 in two giantin KO zebrafish lines (Bergen et al., 2017). Using cDNA derived from young adult dissected jaw and skull bone and cartilage elements, qPCR expression analyses showed that zebrafish rcan2 is 3.9- and 2.5-fold up-regulated in giantin X3078 and Q2948X mutant lines, respectively, when normalised to β-actin (actb1) as a control (Fig. 2A). Expression levels of hypoxanthine-guanine phosphoribosyl transferase 1 (hprt1) and glyceraldehyde-3-phosphate dehydrogenase (gapdh) were not significantly changed. This shows that upregulation of RCAN2 following loss of giantin is conserved and is therefore consistent with the hypothesis that the limited phenotypes seen in both giantin KO models are due to compensation. This also reduces concerns arising from the analysis of only one giantin knockout RPE1 cell line. We postulated that if this dramatic increase in expression is responsible for the limited phenotypes of giantin knockouts, then RCAN2 should have a key role in ciliogenesis.
Fig. 2.
RCAN2 localises to centrioles. (A) qPCR of two giantin zebrafish mutant lines (Q2948X and X3078) compared with wild-type sibling controls shows that rcan2 is strongly upregulated relative to the housekeeping gene actb1. In contrast, hprt1 and gapdh are not upregulated. (B) Endogenous RCAN2 (red) localises to centrioles labelled with γ-tubulin (green) in hTERT-RPE1 cells. Scale bar: 10 μm. (C) RCAN2 is frequently (25±11%, n=3 independent experiments) found concentrated at the mother centriole (from which the cilium extends) as shown by acetylated tubulin labelling of the ciliary axoneme (arrowhead). Where only one puncta of RCAN2 labelling is found (asterisk; 7±3% of cells, n=3 independent experiments), a ciliary axoneme is always found extending from this centriole. (D) The distal appendage protein CEP170 is associated with the brighter of the two RCAN2-positive centrioles (arrowhead) as well as with single centrioles positive for CEP170 (asterisk). Boxes are 5×5 µm; >50 cells were analysed in each of the independent experiments in B-D.
The localisation of RCAN2 has not been reported. Immunofluorescence of RCAN2 in RPE-1 cells showed that it localised to centrioles (Fig. 2B) and occasionally showed enhanced localisation to the mother centriole (25±11% of cells, n=3 independent experiments) from which the primary cilium extends (shown by acetylated tubulin labelling in Fig. 2C). This was confirmed by labelling with the distal appendage protein CEP170 (Fig. 2D) which can be used as a marker of the mother centriole (Huang et al., 2017). In those cases where only one centriole is evident from RCAN2 labelling (7±3% of cells; n=3 independent experiments), we always see the axoneme extending from a centriole and CEP170 associated with that centriole.
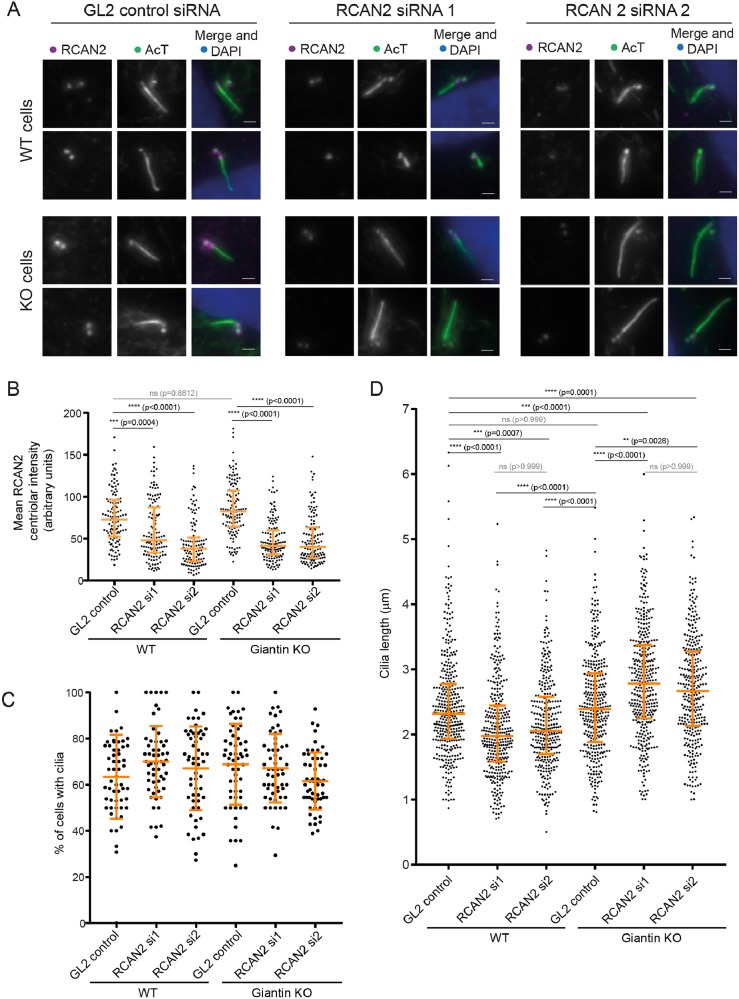
To determine whether RCAN2 was compensating for giantin depletion with respect to cilium length, we depleted RCAN2 using two individual siRNA duplexes (Fig. 3) as well as a pool of 4 duplexes (Fig. S1). This experiment validated the specificity of the antibody used since centriolar labelling was lost following RNAi suppression of RCAN2 expression (Fig. 3A). The efficacy of suppression was measured as a function of the intensity of RCAN2 labelling at the centrioles (Fig. 3B). Unfortunately, the antibody was not suitable for immunoblotting. Depletion of RCAN2 did not affect the proportion of cells that could generate cilia following serum withdrawal (Fig. 3C). Quantification of these data showed that in wild-type (WT) RPE-1 cells, suppression of RCAN2 resulted in shorter cilia (Fig. 3D). In contrast, depletion of RCAN2 in giantin KO cells resulted in longer cilia, even when compared with control cilia lengths. Our hypothesis was that RCAN2 could be suppressing the phenotypes seen following acute loss of giantin in RNAi experiments, namely an increase in cilia length. Consistent with this, we see that reducing the levels of RCAN2 in giantin KO cells recapitulates the longer cilia phenotypes seen upon giantin knockdown in WT cells (Asante et al., 2013). Depletion of RCAN2 has no effect on the integrity of the Golgi or on targeting of the glycosyltransferase galactosyltransferase T (GalT) (Fig. S1B versus Fig. S1A). The efficacy of depletion of RCAN2 using a pool of 4 siRNA duplexes is also shown (Fig. S1D). It was not possible to study the effects of RCAN2 overexpression in these experiments as transfection with myc-RCAN2 blocked ciliogenesis in >80% of transfected cells. Attempts to titrate expression levels were unsuccessful.
Fig. 3.
RCAN2 compensates for loss of function of giantin in cilia length control. (A) Depletion of RCAN2 using two different siRNA duplexes (1 and 2) is effective as judged by reduced centriolar labelling compared with cells transfected with control siRNA (GL2). Two examples are shown in each case. Scale bars: 1 µm. (B) Quantification of experiment shown in A. Depletion of RCAN2 results in shorter cilia in WT RPE-1 cells but longer cilia in giantin KO cells compared with GL2-transfected controls. (C) Depletion of RCAN2 does not affect the ability of cells to produce cilia. (D) Quantification of cilia length in RCAN2-depleted WT and giantin KO RPE-1 cells. Depletion of RCAN2 in wild-type RPE-1 cells results in a decrease in cilium length. In contrast, depletion of RCAN2 in giantin KO RPE-1 cells increases cilia length, even when compared with WT cells. Statistical significance was tested using one-way, non-parametric ANOVA (Kruskal-Wallis test) with Dunn's multiple comparisons test. N≥35 cilia were measured in each case from 3 independent biological replicates; ns, not significant.
Our data strongly suggest that upregulation of RCAN2 ameliorates defects in cilia length control. The phenotypes we observe following acute and partial depletion of giantin in mammalian cells (Asante et al., 2013) and zebrafish embryos (Bergen et al., 2017) are not seen following permanent loss of function of giantin. The fact that RCAN2 depletion leads to shorter cilia in WT cells and longer cilia in giantin KO cells probably reflects a general loss of length control rather than a linear relationship whereby increasing RCAN2 levels leads to reducing cilium lengths. In WT cells depleted of RCAN2, cilia are unable to maintain their length and so become shorter, whereas in giantin KO cells, the loss of length regulation means the effects of giantin loss become dominant and cilia become longer.
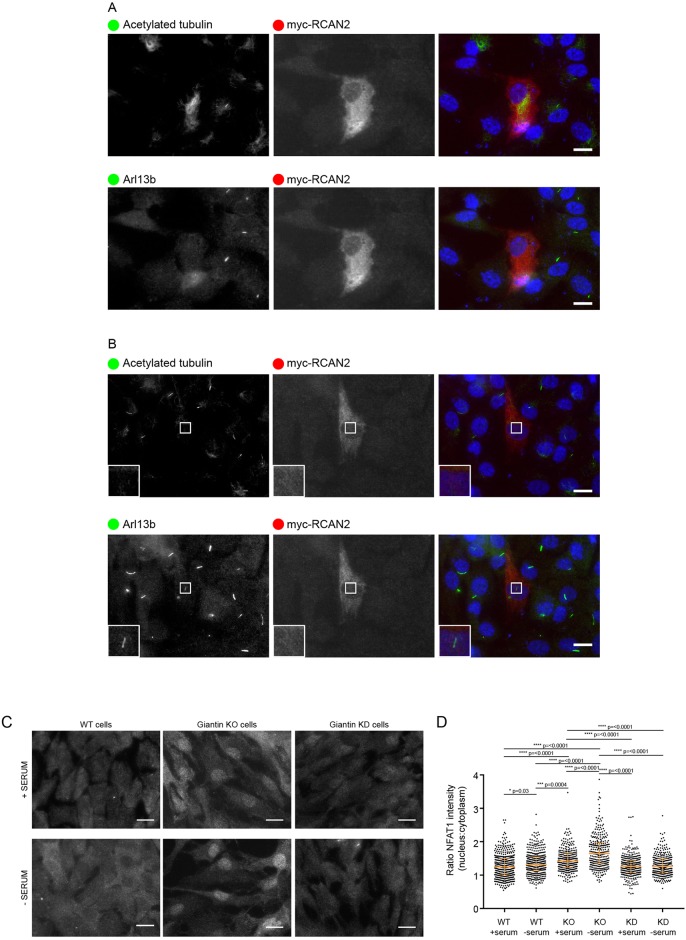
It is interesting that our data implicate cilium length control by compensatory pathways in vitro in proliferating cells. The importance of cilia to cell growth in culture is poorly understood and we cannot therefore predict whether defective ciliogenesis alone would provide sufficient adaptive pressure for upregulation of RCAN2. It is possible, however, that loss of giantin results in a pro-ciliary phenotype that is compensated for by upregulation of RCAN2. In support of this, we found that overexpression of myc-RCAN2 results in far fewer cells (<20%) that produce cilia (Fig. 4A, n=100 cells) and there were also a few incidences (∼10% of myc-RCAN2-expressing cells) where we could detect Arl13b-positive structures devoid of acetylated-tubulin labelling (Fig. 4B). These are similar in appearance to the ‘decapitated’ cilia that have been described by others to drive cilia resorption and cell cycle progression (Phua et al., 2017). In addition, RCAN2 may be acting as a regulator of gene transcription through NFAT signalling to correct other deficiencies in the KO cells. For example, we have shown previously that the glycosyltransferase content of the Golgi is altered in giantin KO cells, suggesting that glycosylation defects arising from loss of giantin function might have been corrected to generate bioequivalent glycans (Stevenson et al., 2017).
Fig. 4.
Giantin KO cells show higher levels of NFAT1 activation. (A) Expression of myc-RCAN2 (red in merge) prevents cilia formation as shown by acetylated tubulin or Arl13b (both pseudocoloured green in merge; images were obtained from cells triple labelled with Alexa Fluor 488, 568 and 647). 80% of myc-RCAN2-expressing cells failed to extend a cilium (n=100). (B) In 5% of these myc-RCAN2-expressing cells, we observed ‘decapitated’ cilia, positive for Arl13b but not for acetylated tubulin. (C) Representative images showing WT and giantin KO and knockdown (KD) cells grown in serum or serum deprived for 24 h and then immunolabelled for NFAT1. Scale bars: 10 μm. (D) Quantification of the ratio of nuclear to cytoplasmic NFAT1 staining intensity in experiments represented in A [n=3, bars represent median and interquartile range, P-values calculated using non-parametric ANOVA (Kruskal-Wallis test with Dunn's multiple comparisons test)].
Many studies have shown that RCAN2 regulates calcineurin-dependent activation of NFAT signalling, and subsequently, NFAT-dependent transcriptional programmes. To confirm that upregulation of RCAN2 in giantin KO cells is of functional importance, we examined localisation of NFAT1 in WT and giantin KO cells using immunofluorescence (Fig. 4C). As shown in Fig. 4D, the ratio of nuclear versus cytoplasmic levels of NFAT1 was higher in giantin KO cells compared with that in WT cells. This nuclear localisation was further enhanced in response to serum starvation. The distribution of NFAT1 in cells depleted of giantin by shRNA was similar to that in WT cells, supporting our argument that acute depletion of giantin does not induce adaptation within these timescales, hence the longer cilia. The higher level of RCAN2 expression seen in the giantin KO cells could thus result in greater activation of the calcineurin-NFAT signalling pathways. Although RCAN2 is commonly considered to be a negative regulator of calcineurin (Cao et al., 2002; Loh et al., 1996), other studies have found that it can positively promote NFAT signalling (Sanna et al., 2006), as appears to be the case here. Alternatively, enhanced RCAN2 expression could result from attempts to suppress upregulated calcineurin-based signalling. Activation of NFAT signalling has been described to occur downstream of α6β4 integrin activation in metastatic cancer cells (Jauliac et al., 2002) and RCAN1 can modulate cancer cell migration (Espinosa et al., 2009). Of note, both giantin KO cells and giantin KO fish have substantial defects in extracellular matrix-related functions (Bergen et al., 2017; Stevenson et al., 2017). Thus, we speculate that changes in matrix glycoproteins could be the trigger for compensatory changes in gene expression in giantin KO models.
Rodent knockouts for giantin (rat: Katayama et al., 2011 and mouse: Lan et al., 2016) exhibit defects in extracellular matrix secretion and assembly, cartilage and bone formation, and have syndromic cleft palate. RCAN2 KO mice are viable but show selective defects in osteoblast function (Bassett et al., 2012), in impaired intramembranous ossification, and in cortical bone formation. Indeed, loss of RCAN2 function is associated with reduced bone mass, whereas giantin mutant zebrafish (with increased RCAN2) show enhanced bone formation in the intervertebral discs (Stevenson et al., 2017). Evidence also exists that inhibition of calcineurin using cyclosporin is linked to cilia function, shown by the presence of abnormal basal bodies in proximal convoluted tubule cells from patients receiving Cyclosporin A (Kirwan, 1982). Ciliary dysfunction is frequently linked to skeletal defects. Indeed, the ‘skeletal ciliopathies’ are grouped according to phenotypic descriptions of skeletal defects (Huber and Cormier-Daire, 2012). These diseases include Jeune syndrome in which subunits of the dynein-2 motor are mutated. Intriguingly, our work has led from the biology of giantin to the dynein-2 intraflagellar transport motor (Asante et al., 2013) and on to the identification of RCAN2 as a key regulator of ciliary function.
These and other findings implicate RCAN2 and calcineurin in transcriptional regulation via cilia. This provides a potential point of integration at the nexus of Ca2+ and ciliary signalling. In support of this, tax-6, the C. elegans orthologue of calcineurin, regulates the trafficking of the key ciliary protein polycystin-2 (PKD2) (Hu et al., 2006). PKD2 is a Ca2+-permeable cation channel that is mutated in 15% of cases of autosomal dominant polycystic kidney disease. Of note here, the partner of PKD2, PKD1 (mutated in the other 85% of ADPKD cases) is understood in osteoblasts to be a mechanoreceptor transducing signals from the cilia to induce osteoblastic gene transcription by potentiating Ca2+-dependent calcineurin-NFAT signalling (Dalagiorgou et al., 2013). Its role in canonical NFAT signalling suggests a mechanism by which it could regulate coupling of ciliary Ca2+ to cellular functions through transcriptional control. Our findings also have significant potential for understanding the mechanisms of cilia-based Ca2+-dependent signal transduction, including modulation of Shh signalling through Gli (Delling et al., 2013). One possibility is that the concentration of RCAN2 at the base of the cilium acts to quench Ca2+-dependent calcineurin signalling, restricting it to the cilium itself. Understanding the role of RCAN2 in ciliary signalling will therefore have significant implications for diverse areas of fundamental importance to our understanding of cell function, as well as clinical relevance in bone formation, renal function, developmental signalling and mechanosensing.
MATERIALS AND METHODS
All reagents were purchased from Sigma-Aldrich unless stated otherwise.
Cell culture
Human telomerase-immortalised retinal pigment epithelial cells (hTERT-RPE-1, ATCC) were grown in DMEM-F12 supplemented with 10% FCS (Life Technologies, Paisley, UK). Cell lines were not authenticated after purchase other than confirming absence of mycoplasma contamination. Transfections were performed using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Myc-DDK-tagged-human RCAN2 (NCBI accession number NM_005822.2) was purchased from Generon (Maidenhead, UK).
Knockout models
Giantin KO RPE-1 cells have been described previously (Stevenson et al., 2017) and Giantin KO zebrafish are described in Bergen et al. (2017). London AB, Tupfel long fin (TL) and AB/TL zebrafish were used and maintained according to standard conditions (Westerfield, 2000), kept in 3.5 litre tanks in groups of ≤15 adult fish, and staged accordingly (Kimmel et al., 1995). Ethical approval was obtained from the University of Bristol Ethical Review Committee using the Home Office Project License number 30/2863.
RNAi
RCAN2 was depleted in RPE-1 cells using RNA interference. siRNA duplexes directed against RCAN2 were made by Dharmacon [MQ-020054-01-0002 siGENOME Human RCAN2 (10231), Dharmacon, GE Healthcare]. Transfections were carried out using either a control or RCAN2 siRNA. Four pooled siRNAs were used to deplete RCAN2, with the following target sequences, RCAN2 siRNA #1 (GGAUAGAGCUUCAUGAAAC), RCAN2 siRNA #2 (CGUAUAAACUUCAGCAAUC), RCAN2 siRNA #3 (CCACGCCAGUCCUCAACUA) and RCAN2 siRNA #4 (GCUCUACUUUGCACAGGUU). GL2 control siRNA (CGUACGCGGAAUACUUCGAUU) was used as a negative control. All siRNA reagents were prepared in 2 M CaCl2 and incubated for 5 min at room temperature before an equal volume of 2× BES buffered solution (BBS) was added to the solution and allowed to equilibrate for a further 30 min at room temperature. Final siRNA concentrations used were 5 μM. Confluent RPE-1 cells were grown on round 22 mm glass coverslips in Costar 6-well plates and incubated at 37°C, 3% CO2 for 24 h before replacing the growth medium. Cells were grown for an additional 24 h at 37°C, 5% CO2 then serum starved for ciliogenesis assays.
Antibodies, labelling and microscopy
Antibodies used: mouse monoclonal anti-giantin (full-length, Abcam, ab37266; 1:1000), rabbit polyclonal anti-giantin (N-terminus, Covance, PRB-114C; 1:1000), rabbit anti-RCAN2 (GTX31373, Insight Biotechnology, London UK; 1:500), anti-Arl13b (17711-1-AP, Proteintech, Manchester, UK; 1:1000), sheep anti-myc was a kind gift from Harry Mellor (University of Bristol) 1:1000, Fan et al., 2010 (PMID: 20233848), sheep anti-GRASP65 was a kind gift from Jon Lane (University of Bristol) Cheng et al., (2010) PMID: 21368855, anti-GalT (CB002, CellMab, Gothenberg, Sweden; 1:500), anti-NFAT1 (GTX127932, GeneTex, Insight Biotechnology, London, UK; 1:500).
For antibody labelling, cells were grown on autoclaved coverslips (Menzel #1.5, Fisher Scientific, Loughborough, UK), rinsed with PBS and fixed in methanol for 4 min at −20°C. Cells were then blocked in 3% BSA-PBS for 30 min and incubated with primary then secondary antibody for 1 h each, washing in between. Nuclei were stained with DAPI [4,6-diamidino-2-phenylindole (Life Technologies, Paisley, UK, D1306)] for 3 min and coverslips mounted in Mowiol (MSD, Hertfordshire, UK) or Prolong Diamond antifade (Thermo Fisher, Paisley, UK). Fixed cells were imaged using an Olympus IX70 microscope with 60×1.42 NA oil-immersion lens, Exfo 120 metal halide illumination with excitation, dichroic and emission filters (Semrock, Rochester, NY), and a Photometrics Coolsnap HQ2 CCD, controlled by Volocity 5.4.1 (Perkin Elmer, Seer Green, UK). Chromatic shifts in images were registration corrected using TetraSpek fluorescent beads (Thermo Fisher). Images were acquired as 0.2 µm z-stacks. Electron microscopy methods are descried fully in Stevenson et al. (2017).
Reverse transcriptase PCR and quantitative real-time PCR
Young adult fish [60 and 63 days post fertilisation for Q2948X (F4) and X3078 (F3) lines respectively, 3 individuals from each genotype group] were lethally anaesthetised (<0.15% Tricaine) and their frontal skull and ventral jaw bone and cartilage elements were carefully dissected using a standard dissecting microscope. From these separate samples, total RNA was isolated using RNeasy mini kit (cat# 74104, Qiagen) and reverse transcriptase reaction was performed by using Superscript III (cat# 18080093, Thermo Fisher Scientific) according to the manufacturers' protocols. Quantitative Real-Time PCR (qPCR) reaction [with the following primers: rcan2 (ENSDART00000143379.2) F, 5′-CCAGGTGCAGAACCCAGTAT-3′ and R, 5′-CCTCAGACCCTGGTTGTGTT-3′; hprt1 F, 5′-ATGGACCGAACTGAACGTCT-3′ and R, 5′GGTCTGTATCCAACGCTCCT-3′; actb1 F, 5′TCAACACCCCTGCCATGTAT-3′ and R, 5′-CAGGAAGGAAGGCTGGAAGA-3′; gapdh F, 5′-TGTTCCAGTACGACTCCACC-3′ and R, 5′-GCCATACCAGTAAGCTTGCC-3′] was undertaken with DyNAmo HS SYBR green (F410L, Thermo Fisher Scientific) on 175 ng/µl cDNA libraries, cycling (40 times) at 95°C for 25 s, 57.5°C for 30 s and 70°C for 45 s followed by a standard melt curve (QuantStudio3, Applied Biosystems).
RNAseq
Triplicate samples of total RNA (three independent passages) from WT and giantin KO RPE-1 cells were analysed by RNAseq by the Earlham Institute (formerly The Genome Analysis Centre). The methodology and analysis is fully described elsewhere (Stevenson et al., 2017).
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism 7.00. The tests used and sample sizes are indicated in the figure legends, P-values are shown on the figures. All tests met standard assumptions and the variation between each group is shown. Sample sizes were chosen based on previous, similar experimental outcomes and based on standard assumptions. No samples were excluded. Randomisation and blinding were not used except where the genotype of zebrafish was determined after experimentation. Statistical significance was tested using one-way, non-parametric ANOVA (Kruskal-Wallis test) with multiple comparisons using Dunn's test.
Supplementary Material
Acknowledgements
We would like to thank the Earlham Institute for the RNAseq analysis. We also thank the MRC and Wolfson Foundation for establishing the Wolfson Bioimaging Facility, Roderick Skinner for his help with zebrafish-related work and the Wolfson Bioimaging Facility staff for help with microscopy.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: N.S., D.S.; Methodology: N.S., D.B., D.S.; Validation: N.S.; Formal analysis: N.S., D.B., A.X., E.W., F.H., D.S.; Investigation: N.S., D.B., A.X., E.W., F.H., J.M., L.V., D.S.; Resources: N.S.; Data curation: N.S., D.S.; Writing - original draft: N.S., D.B., J.M., L.V., C.H., D.S.; Writing - review & editing: N.S., D.S.; Visualization: N.S., D.B., A.X., E.W., F.H., D.S.; Supervision: N.S., J.M., L.V., C.H., D.S.; Project administration: C.H., D.S.; Funding acquisition: C.H., D.S.
Funding
This work was supported by the Biotechnology and Biological Sciences Research Council (BB/L014181/1 and BB/N000420/1), the Medical Research Council (MR/K018019/1), the Wellcome Trust (099848/Z/12/Z) and the University of Bristol. Deposited in PMC for release after 6 months.
Data availability
Raw RNAseq data are available via the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-5618.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.212258.supplemental
References
- Asante D., Maccarthy-Morrogh L., Townley A. K., Weiss M. A., Katayama K., Palmer K. J., Suzuki H., Westlake C. J. and Stephens D. J. (2013). A role for the Golgi matrix protein giantin in ciliogenesis through control of the localization of dynein-2. J. Cell Sci. 126, 5189-5197. 10.1242/jcs.131664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett J. H. D., Logan J. G., Boyde A., Cheung M. S., Evans H., Croucher P., Sun X. Y., Xu S., Murata Y. and Williams G. R. (2012). Mice lacking the calcineurin inhibitor Rcan2 have an isolated defect of osteoblast function. Endocrinol. 153, 3537-3548. 10.1210/en.2011-1814 [DOI] [PubMed] [Google Scholar]
- Bergen D. J. M., Stevenson N. L., Skinner R. E. H., Stephens D. J. and Hammond C. L. (2017). The Golgi matrix protein giantin is required for normal cilia function in zebrafish. Biol. Open 6, 1180-1189. 10.1242/bio.025502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun D. A. and Hildebrandt F. (2016). Ciliopathies. Cold Spring Harbor Perspect. Biol. 9, a028191, 10.1101/cshperspect.a028191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Kambe F., Miyazaki T., Sarkar D., Ohmori S. and Seo H. (2002). Novel human ZAKI-4 isoforms: hormonal and tissue-specific regulation and function as calcineurin inhibitors. Biochem. J. 367, 459-466. 10.1042/bj20011797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalagiorgou G., Piperi C., Georgopoulou U., Adamopoulos C., Basdra E. K. and Papavassiliou A. G. (2013). Mechanical stimulation of polycystin-1 induces human osteoblastic gene expression via potentiation of the calcineurin/NFAT signaling axis. Cell. Mol. Life Sci. 70, 167-180. 10.1007/s00018-012-1164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling M., DeCaen P. G., Doerner J. F., Febvay S. and Clapham D. E. (2013). Primary cilia are specialized calcium signalling organelles. Nature 504, 311-314. 10.1038/nature12833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa A. V., Shinohara M., Porchia L. M., Chung Y. J., McCarty S., Saji M. and Ringel M. D. (2009). Regulator of calcineurin 1 modulates cancer cell migration in vitro. Clin. Exp. Metastasis 26, 517-526. 10.1007/s10585-009-9251-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Agbu S. and Anderson K. V. (2016). Microtubule motors drive hedgehog signaling in primary cilia. Trends Cell Biol. 27, 110-125. 10.1016/j.tcb.2016.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Bae Y. K., Knobel K. M. and Barr M. M. (2006). Casein kinase II and calcineurin modulate TRPP function and ciliary localization. Mol. Biol. Cell 17, 2200-2211. 10.1091/mbc.E05-10-0935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N., Xia Y., Zhang D., Wang S., Bao Y., He R., Teng J. and Chen J. (2017). Hierarchical assembly of centriole subdistal appendages via centrosome binding proteins CCDC120 and CCDC68. Nat. Commun. 8, 15057 10.1038/ncomms15057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber C. and Cormier-Daire V. (2012). Ciliary disorder of the skeleton. Am. J. Med. Genet. C Semin. Med. Genet. 160C, 165-174. 10.1002/ajmg.c.31336 [DOI] [PubMed] [Google Scholar]
- Jauliac S., López-Rodriguez C., Shaw L. M., Brown L. F., Rao A. and Toker A. (2002). The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat. Cell Biol. 4, 540-544. 10.1038/ncb816 [DOI] [PubMed] [Google Scholar]
- Katayama K., Sasaki T., Goto S., Ogasawara K., Maru H., Suzuki K. and Suzuki H. (2011). Insertional mutation in the Golgb1 gene is associated with osteochondrodysplasia and systemic edema in the OCD rat. Bone 49, 1027-1036. 10.1016/j.bone.2011.08.001 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. and Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kirwan P. D. (1982). Giant mitochondria and multiple cilia in proximal convoluted tubules of renal transplant patients receiving cyclosporin a immunosuppression. Micron 13, 353-354. 10.1016/0047-7206(82)90055-3 [DOI] [Google Scholar]
- Lan Y., Zhang N., Liu H., Xu J. and Jiang R. (2016). Golgb1 regulates protein glycosylation and is crucial for mammalian palate development. Development 143, 2344-2355. 10.1242/dev.134577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K. F. (2015). IFT-cargo interactions and protein transport in cilia. Trends Biochem. Sci. 40, 765-778. 10.1016/j.tibs.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh C., Shaw K. T.-Y., Carew J., Viola J. P. B., Luo C., Perrino B. A. and Rao A. (1996). Calcineurin binds the transcription factor NFAT1 and reversibly regulates its activity. J. Biol. Chem. 271, 10884-10891. 10.1074/jbc.271.18.10884 [DOI] [PubMed] [Google Scholar]
- Phua S. C., Chiba S., Suzuki M., Su E., Roberson E. C., Pusapati G. V., Setou M., Rohatgi R., Reiter J. F., Ikegami K. et al. (2017). Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell 168, 264-279.e15. 10.1016/j.cell.2016.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna B., Brandt E. B., Kaiser R. A., Pfluger P., Witt S. A., Kimball T. R., van Rooij E., De Windt L. J., Rothenberg M. E., Tschop M. H. et al. (2006). Modulatory calcineurin-interacting proteins 1 and 2 function as calcineurin facilitators in vivo. Proc. Natl. Acad. Sci. USA 103, 7327-7332. 10.1073/pnas.0509340103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson N. L., Bergen D. J. M., Skinner R. E. H., Kague E., Martin-Silverstone E., Robson Brown K. A., Hammond C. L. and Stephens D. J. (2017). Giantin-knockout models reveal a feedback loop between Golgi function and glycosyltransferase expression. J. Cell Sci. 130, 4132-4143. 10.1242/jcs.212308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung C.-H. and Leroux M. R. (2013). The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat. Cell Biol. 15, 1387-1397. 10.1038/ncb2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). 4th ed Eugene: Univ. of Oregon Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.