ABSTRACT
Proneural basic Helix-Loop-Helix (bHLH) proteins are required for neuronal determination and the differentiation of most neural precursor cells. These transcription factors are expressed in vastly divergent organisms, ranging from sponges to primates. Here, we review proneural bHLH gene evolution and function in the Drosophila and vertebrate nervous systems, arguing that the Drosophila gene atonal provides a useful platform for understanding proneural gene structure and regulation. We also discuss how functional equivalency experiments using distinct proneural genes can reveal how proneural gene duplication and divergence are interwoven with neuronal complexity.
KEY WORDS: bHLH gene, Neural development, Neurogenesis, Neuronal diversity, Proneural gene
Summary: This Review discusses how gene duplication and divergence are interwoven with neuronal complexity in Drosophila and vertebrates, highlighting atonal as a platform for understanding proneural gene structure and regulation.
Introduction
The function of the nervous system relies on a large number of neurons with diverse functions. Even the simple nervous system of the nematode C. elegans has more than 100 classes of neurons, and this number is vastly larger in more complex metazoans (White et al., 1986). The specification and differentiation of most neurons rely on a class of transcription factors known as proneural basic Helix-Loop-Helix (bHLH) transcription factors (Huang et al., 2014), which are also important in attempts to achieve neuronal regeneration (Guillemot and Hassan, 2017). There are ∼125 bHLH genes in the human genome, as compared with 59 in Drosophila (Ledent et al., 2002; Simionato et al., 2007). Vertebrate bHLH factors are further categorized into subfamilies, including the Atoh (Atonal homolog), Ascl (Achaete-Scute complex-like), Neurogenin, Neurod and Olig factors (Fig. 1), whose members act during neurogenesis, neuronal differentiation and/or gliogenesis. These related genes probably arose from common ancestors by gene or genome duplication during evolution. This raises the question of whether genetic complexity might contribute to neuronal diversity. Here, we discuss the evolution and function of complex proneural networks. We highlight how gene replacement studies, some made feasible by new genome editing technologies, can help evaluate evolutionary changes in proneural bHLH gene functions and clarify the extent to which neuronal diversity depends on increasing genetic complexity or on other factors.
Fig. 1.
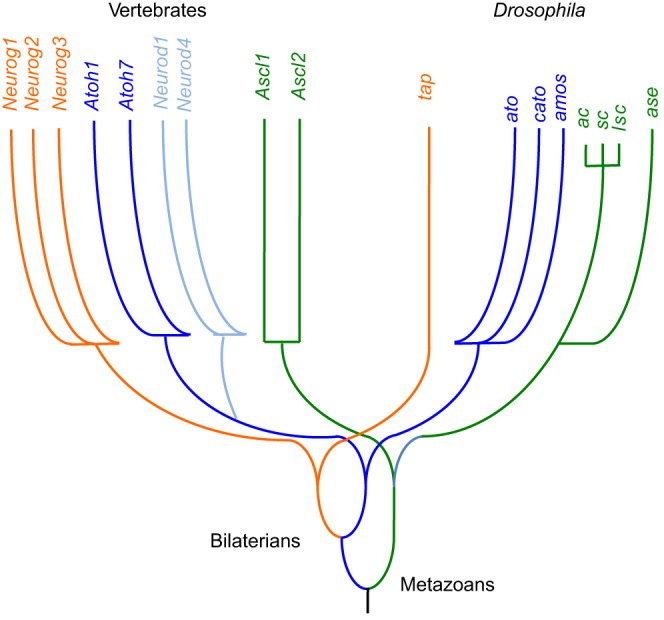
Schematic phylogeny of proneural bHLH genes. The relationships and ancestry of extant proneural bHLH genes in vertebrates and Drosophila (Simionato et al., 2007). Each color represents a related subfamily. The most ancient divergence is thought to be between Achaete/Scute-like genes and Atonal/Neurogenin/Neurod-like genes. The divergence of Atonal/Neurod-like and Neurog-like genes predates the vertebrate-insect divide, but the Atonal/Neurod family diverged within the chordate lineage. Achaete/Scute-like genes diversified within insects, including a recent triplication in the lineage leading to Drosophila (Negre and Simpson, 2009).
The Achaete-Scute gene complex and the proneural gene concept
The proneural concept was developed through the discovery and characterization of four Drosophila genes – achaete (ac), scute (sc), lethal of scute [lsc, or l(1)sc] and asense (ase) – that are responsible for development of much of the Drosophila CNS and PNS (Cubas et al., 1991; Garcia-Bellido and de Celis, 2009). Expression of these proneural genes defines regions of ectoderm with neurogenic competence, such that their default fate will be that of neural precursors unless diverted to another fate, for example by Notch signaling (Knust and Campos-Ortega, 1989; Simpson, 1990). ac, sc and lsc are proneural genes, conferring proneural competence that may or may not lead to neuronal determination in every cell, whereas ase is a neural precursor gene, expressed after the neural fate decision has been made. It has been suggested that the vertebrate homologs of these genes are expressed in ectoderm with previously acquired neural character, and therefore are not true proneural genes (Bertrand et al., 2002). However, we see this as a minor distinction because the same could be said for some proneural regions in Drosophila, and because it may not apply to some vertebrate tissues (Jarman and Groves, 2013). Like vertebrate proneural genes, ac, sc and lsc show a variety of overlapping and distinct expression patterns, reflecting both separate and redundant functions in the specification of different neural precursor cells, and exhibit cross-regulatory interactions whereby one gene functions to regulate the expression of another. Unlike most vertebrate proneural genes, however, ac, sc, lsc and ase are closely linked within a 100 kb segment of the X-chromosome, constituting the Achaete-Scute gene complex (AS-C) (Garcia-Bellido and de Celis, 2009).
The structure of the AS-C suggested that multiple proneural genes might be necessary to encode neuronal diversity. However, this idea has to be revisited, as it is now believed that the multigene AS-C of D. melanogaster is a recent evolutionary invention. Indeed, although proneural bHLH genes are found as far back as coelenterates, an AS-C of four linked genes is not found even in other dipteran insects. For example, the basal mosquito species Anopheles gambiae has only a single proneural gene of the AS-C class as well as a single neural progenitor gene related to ase. This simple arrangement seems to be the ancestral condition for insects and is also seen in the red flour beetle Tribolium castaneum, the honey bee Apis mellifera and the wasp Nasonia vitripennis (Negre and Simpson, 2009). Since the nervous systems of mosquitoes, honey bees or wasps does not appear to be significantly less complex than that of D. melanogaster, it can be argued that the multiple bHLH proteins in the AS-C are not really necessary to make complex neural structures. In addition, the potential contributions of multiple genes in other proneural gene families, such as the ato, cato and amos genes that diverged earlier in the lineage leading to Drosophila, or the Atoh genes that diverged in vertebrates (Fig. 1), merit consideration in light of this finding.
Drosophila atonal as a platform for understanding proneural gene structure and regulation
Ideally, a discussion of how proneural gene families arose by gene duplication and diversification would begin with the original progenitor genes. We currently lack access to those ancient genes, however, and therefore propose here that the Drosophila gene atonal (ato) provides a useful model because it is not a member of a gene complex and often acts alone, i.e. without coexpression of other proneural genes. Accordingly, a description and analysis of ato regulation and function in Drosophila provides a useful basis for comparison with other proneural genes, and a starting point for understanding proneural gene evolution.
ato is a homolog of vertebrate Atoh genes (Jarman and Groves, 2013). In Drosophila, ato functions to specify the retina, the chordotonal organs (stretch receptors of the PNS) and some of the olfactory sense organs, all without assistance from the AS-C. The two other Ato family genes in Drosophila are unlinked and show limited overlap in function with one another; amos is required for specification of the remaining olfactory sensillae and for specifying some dendritic neurons, while cato acts as a neural precursor gene in the chordotonal sensory lineage (Maung and Jarman, 2007; zur Lage and Jarman, 2010).
Within the Drosophila eye, the role of ato is to specify the fate of R8 class photoreceptor neurons (Jarman et al., 1994). These are the founders of ommatidia – the individual units of the insect compound eye. Each R8 cell coordinates the induction of additional photoreceptor neurons from neighboring cells by a mechanism dependent on receptor tyrosine kinase signaling (Treisman, 2013). In the absence of ato no retinal neurogenesis takes place owing to the absence of the crucial R8 founder cell. The photoreceptor neurons of the eye differ markedly in structure and physiology from the other ato-dependent neurons, and are surrounded by very different support cells (Jarman and Groves, 2013). Clearly, distinct neural structures do not require distinct proneural genes, as ato exemplifies a single gene that defines multiple classes of neuron, albeit in combination with other genes that provide specific/unique contexts for ato function (Kiefer et al., 2005). This is also abundantly clear in vertebrates, where Atoh1 has diverse roles in the CNS, PNS and gut (reviewed by Huang et al., 2014).
An early step in neural fate determination is the transcription of ato. Detailed studies show that ato expression can best be understood in terms of distinct mechanisms of initiation and then of maintenance (Fig. 2, Box 1). There is no R8-specific transcription factor capable of initiating ato transcription in only these cells. Instead, ato transcription initiates in all undifferentiated retinal cells at a particular stage. Because the Drosophila eye develops progressively, with a wave of differentiation initiating at the posterior eye margin, a corresponding wave of ato transcription crosses the eye primordium just in advance of neurogenesis (Fig. 2) (Jarman et al., 1994). This spatiotemporal pattern is induced by the morphogens Hedgehog (Hh) and Decapentaplegic (Dpp). This eye-specific response to these morphogens requires a particular code of pre-existing transcription factors that includes eyeless (Drosophila Pax6) and sine oculis (a Drosophila homolog of the vertebrate Six genes), which are already expressed in the eye primordium (Zhang et al., 2006; Baker and Firth, 2011).
Fig. 2.
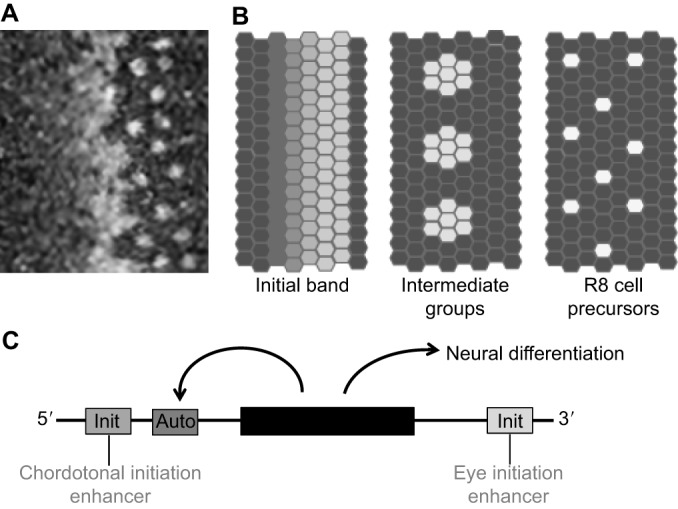
Regulation of Atonal expression and transcription in the Drosophila eye. (A) Confocal image of Atonal (Ato) protein expression at the leading edge of the Drosophila retina differentiation wave. Towards the anterior (left), Ato expression progressively accumulates in all the cells, then rapidly evolves through a pattern of intermediate groups of cells that transiently maintain Ato protein while the surrounding cells lose expression, resolving to only isolated single cells that maintain Ato expression posteriorly (right). These are now the committed and postmitotic R8 photoreceptor precursors. The wave of differentiation advances anteriorly at a rate of one column every 90-100 min. (B) The complete Ato expression pattern can be dissected into at least three temporally overlapping phases (Jarman et al., 1995). First, Ato expression accumulates uniformly, reaching higher and higher levels more posteriorly until this uniform expression abruptly ceases. Replacing the uniform expression, and just overlapping with it in time, is a transient phase of expression in ‘intermediate groups’ of up to ten cells. The intermediate groups are proneural preclusters that will all develop as R8 photoreceptor neurons unless Notch signaling is activated (Baker et al., 1996; Dokucu et al., 1996). Then, each intermediate group is refined to a single Ato-expressing cell that maintains expression for three to four ommatidial columns (∼6 h). (C) Uniform initiation of Ato expression depends on an eye initiation enhancer that is downstream of the coding region of the ato gene (black box), but is independent of functional Ato protein and of the 5′ autoregulatory enhancer. By contrast, expression in the intermediate groups of up to ∼10 cells is autoregulatory and depends on the 5′ autoregulatory enhancer because the 3′ initiation enhancer becomes inactive at this stage. Single cells escape Notch activity to maintain autoregulatory Ato expression and become determined as R8 photoreceptor precursor cells. They maintain Ato expression for ∼6 h, then Ato protein undergoes inhibitory phosphorylation, leading to the loss of expression. This loss is permanent because initiation is no longer active. Other neural regions that express Ato (e.g. chordotonal organs) rely on distinct enhancers (Baker et al., 1996; Sun et al., 1998; Quan et al., 2016).
Box 1. Gene initiation and maintenance programs.
The existence of both initiation and maintenance programs for gene expression reflects fundamental features of cell fate determination. Most developmental fates are initially induced by extrinsic signals and only remain stable through subsequent development by becoming independent of them. In fact, classical embryology defined cell determination as ‘acquired independence from inducing signals’, for example as revealed after explanting from the embryo or after experimental transplantation to a new embryonic location (Slack, 1983). Maintenance of key transcription factors by autoregulatory mechanisms is one way that this independence is achieved (Baker, 2004). The cell-cell signals that induce cell fates during development usually belong to a small number of conserved developmental signaling pathways (including the TGFβ, Hedgehog, Notch, Receptor tyrosine kinase and Wnt pathways), each of which has many functions in different tissues and developmental stages (Gerhart, 1999; Pires-daSilva and Sommer, 2003). Reuse of these signaling pathways is made possible because they initiate cell fates but are not generally required to maintain them.
Once initiated, the pattern of ato transcription (and protein) evolves rapidly, narrowing down first to proneural clusters of ∼10 cells and then to single cells that maintain expression for several hours and thereby acquire R8 photoreceptor fate (Fig. 2) (Jarman et al., 1994). In common with other proneural genes, this narrowing of ato expression is regulated by Notch, which establishes the final pattern of neuronal precursor cells. This refinement serves as a model for the Notch-dependent lateral inhibition that fine-tunes neural fate specification in numerous vertebrate and invertebrate developmental contexts (Fortini, 2009). During lateral inhibition, ato transcription no longer responds to Hh and Dpp. Instead, ato transcription is maintained by autoregulation, and the refinement of its expression by Notch reflects interruption of the autoregulatory loop in progressively more and more cells (Baker et al., 1996). Notch acts through transcriptional repressor proteins [E(spl) in Drosophila], which are sequence-specific DNA-binding proteins that can also be recruited to autoregulatory enhancers by protein-protein interactions with proneural proteins (Giagtzoglou et al., 2003). Blocking autoregulation in this way is an effective means of permanently extinguishing gene expression and might be exploited by Notch signaling in many situations (Baker, 2004). In addition, there is increasing evidence that Notch signaling affects proneural protein stability, which would also negatively impact autoregulation (Kiparaki et al., 2015; Weinberger et al., 2017).
The initiation and maintenance of ato gene expression segregate to distinct enhancers (Sun et al., 1998) (Fig. 2). Transgenes containing eye initiation enhancers drive reporter gene expression only in the initial uniform expression domain, just overlapping the first hints of spatially restricted expression. By contrast, transgenes containing the eye autoregulatory enhancer drive reporter gene expression only in the periodic clusters of cells and the single R8 precursors that derive from them. It makes sense that input from the initiation enhancer ceases before lateral inhibition, otherwise inhibiting autoregulation would not extinguish gene expression. Conversely, the autoregulatory enhancer must become active when initiation ceases, or Ato protein would decay before gene expression could be maintained. The basis of temporal coordination between these two regulatory programs is only partially known (Chen and Chien, 1999; Lim and Choi, 2004; Baker et al., 2009).
After Ato expression is stable and Ato protein is active as a transcription factor, R8 photoreceptor fate can be specified. The eye-specific targets of Ato might be defined combinatorially with other, eye-specific factors. Some individual eye targets are known, such as the zinc-finger transcription factor Senseless (sens, a homolog of Gfi1) and Fasciclin 2 (Fas2), an adhesion molecule that is elevated transiently by Ato function (Frankfort et al., 2001; Acar et al., 2006; Quan et al., 2016). Studies of transcription in a variety of systems indicate that proneural genes activate many transcriptional targets including other transcription factors, structural proteins, receptors and channel proteins (Portman and Emmons, 2004; Reeves and Posakony, 2005; Cachero et al., 2011). In addition, it has been shown that both the levels and duration of Ato expression in R8 cells are important for proper development (White and Jarman, 2000). If Ato levels are too high, the subsequent recruitment of other photoreceptors by receptor tyrosine kinase signaling becomes overactive (White and Jarman, 2000). Conversely, failure to maintain full expression levels results in inadequate recruitment of other ommatidial cells (White and Jarman, 2000).
Ato expression is ultimately downregulated in the R8 neurons during their terminal differentiation (Jarman and Groves, 2013), a feature that is generally shared by vertebrate proneural bHLHs. In normal development, Ato is active in R8 precursors during only a short time window of 6-8 h, before transcription fades. Recent studies show that Ato protein itself persists for longer but becomes inactivated by phosphorylation so that autoregulatory ato transcription is not maintained, and the protein subsequently decays. Mutation of the phosphorylation site leads to reduced recruitment of other photoreceptors, apparently owing to persistently high levels of the cell adhesion protein Fas2 (Quan et al., 2016). Thus, a second, Notch-independent process ultimately interrupts autoregulation to terminate Ato expression.
The possible origins of proneural gene complexes by gene duplication
If the single AS-C progenitor gene resembled ato in its regulation, then the complexity of the AS-C in Drosophila might have arisen from duplications of this ancestral gene (Fig. 3) (Taylor and Raes, 2004). Gene duplicates are expected to begin life identical in sequence, and it is easy to see that natural selection need not maintain both identical copies. Mutational loss or inactivation of one copy is thought to be a frequent outcome. When the duplicated gene has multiple functional elements that may be lost independently, a process of subfunctionalization can occur. For example, when an important feature of the expression pattern is lost from one copy, its retention by the other copy becomes essential. If both copies lose distinct subsets of the original gene's function, selection must maintain both partially defective copies to retain the full complement of functions. For this reason, subfunctionalization is thought to be a widespread feature of duplicated gene families and the underlying cause for the retention of both copies during evolution (Force et al., 1999; Taylor and Raes, 2004; Conant and Wolfe, 2008).
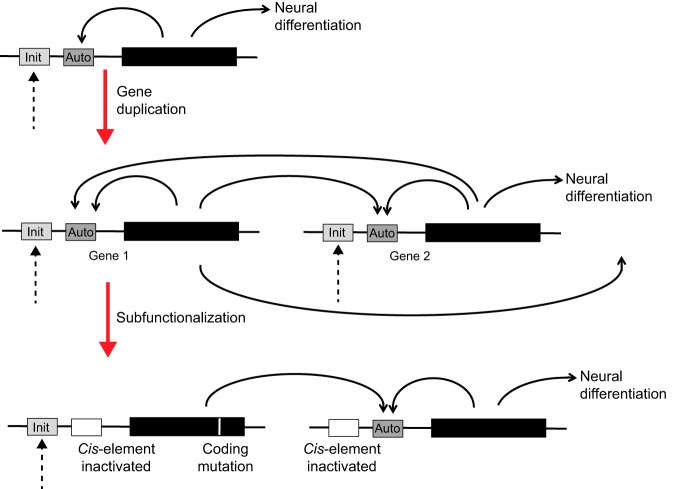
Fig. 3.
Illustration of subfunctionalization. A hypothetical proneural gene with separate enhancers controlling initiation versus autoregulatory transcriptional activities is shown (top). The complete duplication of this gene (middle) produces two functionally redundant, identical genes (genes 1 and 2) that are expected to positively regulate one another through inheritance of the autoregulatory machinery. However, natural selection does not have to maintain redundant functions and can give rise to subfunctionalization (bottom). In the example shown, protein 1 has lost the capacity to specify neural differentiation because of a missense mutation in the coding region and has also lost the ability to autoregulate through divergence of its regulatory sequences, but remains functional as long as protein 1 cross-activates gene 2 and protein 2 then specifies neuronal differentiation. In this example, it would not be necessary for gene 2 to retain regulatory sequences for initiation, since this subfunction could be performed by gene 1. Duplicated genes 1 and 2 would now appear to have temporally distinct roles, although together they fulfill the function of a single ancestral gene. In principle, many other interdependent network topologies can arise through other patterns of subfunctionalization (Taylor and Raes, 2004; Conant and Wolfe, 2008).
Considering duplication of a gene such as ato, with separate initiation and maintenance enhancers, one imagines that both descendant genes should have been capable not only of autoregulation but also of cross-regulation (Fig. 3). This situation would not have to be retained and many scenarios are possible as the enhancer and protein sequences drift during evolution. It is easy to imagine a variety of complex network topologies that could arise via subfunctionalization, with each network retaining the full function of the original progenitor gene (Fig. 3) (Taylor and Raes, 2004; Conant and Wolfe, 2008). In many respects, the regulation of the AS-C looks like one possible outcome of this process. A variety of ‘prepattern’ pathways activate one or more AS-C genes in different proneural regions. Thereafter, the autoregulation and cross-regulation of AS-C genes play important roles in neural fate determination (Gómez-Skarmeta et al., 1995). These features of gene regulation could have been inherited from an autoactivating progenitor (Culi and Modolell, 1998).
In the simplest case, subfunctionalization can maintain multiple members of a gene family even when they have not acquired new functions compared with their common progenitor. However, it is also possible that when multiple functions divide among separate descendant genes, selection then optimizes each now-independent function to a greater degree, leading to enhanced, more specialized functions of the descendant family members. Finally, new functions sometimes arise in duplicated genes, although this is thought to occur less frequently (Taylor and Raes, 2004; Conant and Wolfe, 2008). Functional distinctions have been demonstrated between individual AS-C genes in transgenic assays (Chien et al., 1996), consistent with differences that contribute to neuronal diversity (Huang et al., 2014). Could individual genes have thus evolved new functions, or has each retained subsets of the ancestral functions?
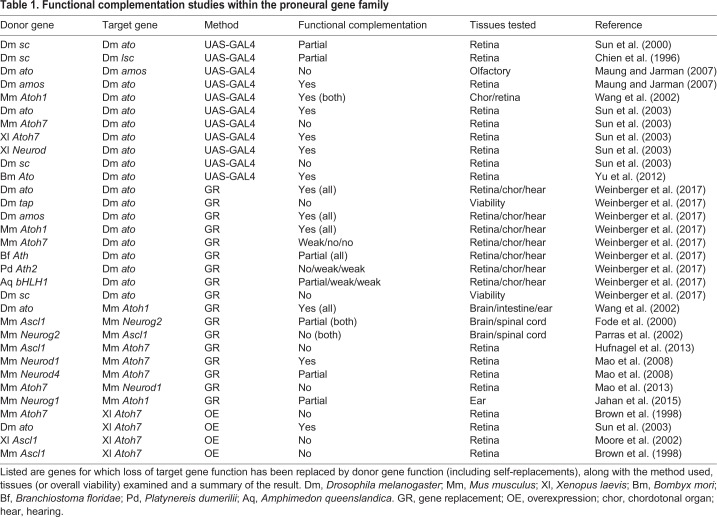
Gene replacement to compare proneural gene function
In principle, the origins of functional distinctions between AS-C genes could be addressed in an experiment that tests whether the single homolog of the proneural AS-C genes from honey bees, wasps or mosquitoes could rescue any or all of the Drosophila AS-C genes. The AS-C structure would make this difficult, but a recent study has made use of gene knock-in methods to replace the ato coding region at the endogenous Drosophila locus with sequences encoding other proneural proteins, an assay that should test the functional attributes of various proteins precisely (Table 1) (Weinberger et al., 2017). These studies showed that Amos is almost indistinguishable from its paralog Ato in terms of function when expressed from the ato locus, whereas previous transgenic assays had suggested distinctions between these proteins (Maung and Jarman, 2007; Weinberger et al., 2017). Thus, the Ato family paralogs in Drosophila might have diverged primarily in expression pattern not protein function, although further studies are required to confirm this, including studies of the olfactory sensilla subtypes that depend on either ato or amos.
Table 1.
Functional complementation studies within the proneural gene family
By contrast, neither sc nor the Drosophila Neurogenin-like gene tap could replace ato, reflecting the much earlier evolutionary divergence of the Achaete/Scute-like, Neurog and Ato families (Fig. 1). On the other hand, a sponge proneural gene equally resembling the Neurog and Ato gene families could partially substitute for ato, consistent with the idea that once-shared features of the Neurog and Ato genes might have been lost from at least the Neurog family gene tap. Interestingly, this study also suggested that the degrees of partial rescue by diverged family members, including mouse Atoh1 and Atoh7 and Atoh genes from cephalochordates and annelids, might largely reflect quantitative differences in protein stability, in particular destabilization by Notch signaling, rather than qualitative functional differences (Table 1) (Weinberger et al., 2017).
The expression, regulation and function of proneural genes in the vertebrate nervous system
To what extent are these studies of Drosophila proneural genes, and of the Ato gene family in particular, relevant to the more complex network of vertebrate proneural genes (Huang et al., 2014)? Based on amino acid sequence conservation within their bHLH domains, it is thought that the vertebrate Atoh, Neurog and Neurod subfamilies evolved from a common ancestor with ato, cato, amos and tap, whereas Drosophila AS-C genes are most related to the vertebrate Ascl1 and Ascl2 genes (Fig. 1). However, the vertebrate genes differ functionally from one another, and from the Drosophila genes, in a variety of ways. Ascl2 is not expressed in the nervous system and is not relevant here. Others are expressed in, and required for, the development of distinct neural cell populations.
The expression domains of vertebrate Ascl and Atoh gene families have been comprehensively described (see Huang et al., 2014). In the vertebrate retina, for example, Atoh7 (also known as Ath5) function is crucial for retinal ganglion cell (RGC) formation (Kanekar et al., 1997; Brown et al., 2001; Kay et al., 2001; Wang et al., 2001). By contrast, mutations in mouse Neurog2 delay the temporal progression of retinal neurogenesis (Hufnagel et al., 2010). Neurod1 is required for photoreceptor and amacrine cell differentiation (Morrow et al., 1999), while Ascl1 (also known as Mash1) maintains a progenitor pool for late fates and regulates rod photoreceptor and bipolar cell neurogenesis (Jasoni and Reh, 1996; Tomita et al., 1996b). Of the three members of the Neurogenin family, Neurog1 and Neurog2 are expressed in the developing forebrain, olfactory system, spinal cord, dorsal root ganglia, and cranial ganglia, whereas Neurog3 is expressed in the developing spinal cord, hypothalamus, and pancreatic endocrine progenitor cells. In addition to being required in the retina, Ascl1 is required in the ventral forebrain and dorsal spinal cord. Yet another distinction, exemplified by retinal bHLH factors, is temporally separable expression. Neurog2 and Ascl1 are present in actively mitotic progenitors, whereas Atoh7 is expressed by progenitors completing terminal mitosis, and Neurod1 and Neurod4 are only present in postmitotic cells.
There are also differences between related sets of genes. For instance, Atoh1 (also known as Ath1) and Atoh7 appeared during whole-genome duplications in basal vertebrate species (Fig. 1) but have non-overlapping, distinct expression domains, as well as a subdivision of ato sensory functions. Whereas Atoh7 is crucial for RGC neurogenesis, Atoh1 is required by inner ear sensory hair cells, dorsal spinal cord interneurons, Merkel cells and cerebellar granule cells (Bermingham et al., 1999, 2001; Ben-Arie et al., 2000). These two genes also exhibit separate expression domains in zebrafish, frog, chick and mouse embryos, although their complete functional separation can only be tested by gene replacement. Remarkably, the genetic hierarchy of retinal neurogenesis exhibits similarities in flies and mammals: in mammals, Pax6 (a mouse homolog of Drosophila Eyeless, which activates ato expression) activates multiple bHLH factors, while Shh signaling and Notch signaling refine spatiotemporal expression. In line with this, it has been shown that Atoh7 has a conserved Pax6 binding site in its 5′ primary enhancer, located ∼1.5 kb upstream from the ATG codon (Riesenberg et al., 2009; Willardsen et al., 2009). Although not formally tested, there are also consensus binding sites for Gli and Hes proteins, which are the transcriptional effectors of Hh and Notch signaling pathways. Together, these findings suggest that vertebrate Atoh7 performs a similar function in the retina to that performed by Drosophila ato in the fly eye. However, even though Atoh1 is not expressed in the vertebrate retina, it behaves more like the ato gene, since it positively autoregulates itself (via a 3′ enhancer) (Helms et al., 2000) and can partially substitute for ato in the fly eye (Table 1) (Wang et al., 2002; Weinberger et al., 2017). In addition, the mammalian Atoh7 gene differs in its acquisition of a second ‘shadow’ enhancer, the deletion of which in humans causes the agenesis of RGCs (Ghiasvand et al., 2011). Although there is a partial atoh7 shadow enhancer in bony fishes, it is located closer to the primary enhancer, and a comparison of mouse and human genomes shows that these two regulatory elements are increasingly farther apart (Ghiasvand et al., 2011). This is suggestive of an additional, mammalian-specific mode of regulation in the eye, whereby changing chromatin configurations or epigenetic factors might be needed to rapidly open or close the Atoh7 locus.
There are also examples of both positive autoregulation and cross-regulation among the vertebrate proneural genes. Mouse Atoh1 positively autoregulates its own expression in the dorsal neural tube, cerebellum, vestibular and auditory systems (Helms et al., 2000). By contrast, neither Atoh7 nor Neurog2 autoregulates in the mouse eye. However, Neurog2 appears first along the advancing retinal wavefront, and directly activates transcription through an evolutionarily conserved Atoh7 retinal enhancer (Hufnagel et al., 2007; Skowronska-Krawczyk et al., 2009), so Neurog2 cross-regulation of Atoh7 could be considered analogous to the Ato autoregulation, or AS-C cross-regulation, that is observed in the fly. Cross-regulation between Neurog2 and Atoh7 is potentially mammalian specific, however, because there is no Neurog2 homolog in zebrafish (Furlong and Graham, 2005), in which initiation of atoh7 expression depends at a minimum on Hh signaling (Neumann and Nuesslein-Volhard, 2000; Masai et al., 2005).
Could other mechanisms thus be used to replace autoregulation to maintain stable neural fate? Positive autoregulation has not been found in studies of some Drosophila neurons, indicating that although the segregation of initiation and autoregulation provides an important conceptual insight into mechanisms of fate specification, similar outcomes might also be achieved by other regulatory mechanisms (Holohan et al., 2006; Zhou et al., 2017). It is possible that transcription factors further downstream are responsible. The Ato-Sens hierarchy that functions in the fly eye is not found in the vertebrate retina, although the Sens homolog Gfi1 works with Atoh1 during vertebrate auditory and intestinal development (Shroyer et al., 2007; Kirjavainen et al., 2008). Instead, Atoh7 transcriptionally activates both Pou4f2 and Isl1, which act synergistically to lock in an RGC fate during vertebrate retinal neurogenesis (Wu et al., 2015). Immediately downstream of Pou4f2-Isl1 is a hierarchy of transcription factors [Onecut1 (Oc1), Oc2, Ebf1, 3, Irx2, 5, 8, Myt1] that control integral aspects of the RGC differentiation program (reviewed by Zagozewski et al., 2014). Each factor acts similarly in other parts of the CNS. Some of these genes have homologs that are also active in the fly eye, suggesting that there might be some deep conservation of mechanisms. Indeed, the autoregulatory expression of ato in the fly eye is transient, lasting only ∼8 h, and the neural fate of photoreceptor cells must thus be maintained by other genes thereafter.
Although proneural gene autoregulation may not be universal, mouse Atoh1 and Neurog2 proteins do exhibit the conserved phosphorylation event that terminates autoregulation of fly Ato. As mentioned above, the phosphorylation of Ato on a particular serine residue blocks protein-DNA binding, thereby halting stimulation of neurogenesis by all three bHLH proteins (Quan et al., 2016). Curiously, substitution of Ser with Ala in Atoh1 also appears to reduce function (Xie et al., 2017). The bHLH domain of mammalian Atoh7 also possesses the relevant serine, but the importance of this particular protein phosphorylation mechanism in the mouse retina remains unclear for both Neurog2 and Atoh7 (Brown et al., 1998). However, additional phosphorylation events that post-translationally regulate bHLH proneural activity have been described. In the frog retina, GSK3 phosphorylates a distinct C-terminal serine in Xenopus Neurod (Moore et al., 2002). This serine is not present in Xenopus Atoh7, thereby allowing GSK3 activity to behave as a toggle switch between amacrine and RGC fates. During motor neuron formation in mice, Neurog2 is phosphorylated at particular serine residues, enabling it to complex with the LIM-domain proteins Islet1 and Lhx3 and execute a motor neuron specification program in the ventral spinal cord (Ma et al., 2008). More generally among bHLH proteins, phosphorylation of multiple amino acid residues has increasingly been shown to control key aspects of differentiation (Hardwick and Philpott, 2015; Philpott, 2015; Wylie et al., 2015; Hardwick et al., 2016; Azzarelli et al., 2017; Krentz et al., 2017). Thus, post-translational regulation of multiple bHLH factors, rather than transcriptional cross-regulation, correlates with situations in which neurogenesis occurs rapidly and/or bHLH factors are co-expressed in the same progenitor cell.
Other types of transcriptional cross-regulation can also occur in vertebrates. For example, Neurog2 and Ascl1 cross-regulate one another in multiple contexts (Fode et al., 2000). In the developing forebrain, these factors are expressed in distinct dorsal (Neurog2) and ventral (Ascl1) domains and are thought to instruct separate forebrain identities via similar neural determination programs. Loss of Neurog2 causes ectopic dorsal Ascl1 expression, and dorsal misexpression of Ascl1 is sufficient for induction of ventral-specific markers and fate. Cross-regulation is negative and non-autonomous; the molecular mechanism involves bHLH factor activation of a Notch ligand, which signals to an adjacent cell to downregulate the other bHLH factor, rather than to itself as would be typical for classical lateral inhibition (Kageyama et al., 2008; Shimojo et al., 2008). Because Neurog2-mediated suppression of Ascl1 maintains dorsal identity, functional distinctions between Neurog2 and Ascl1 were expected, but surprisingly the ectopic expression of Neurog2 in ventral forebrain progenitors is unable to induce progenitors to adopt a dorsal fate, although it does generally induce neuronal differentiation (Parras et al., 2002).
In contrast to the forebrain, Ascl1 and Neurog2 act sequentially in spinal cord progenitor cells (Scardigli et al., 2001; Parras et al., 2002; Helms et al., 2005). Ascl1 is expressed by proliferative dorsal interneuron progenitor cells [(dI) 3 and 5 regions], where it is required for specification. Neurog2 expression follows that of Ascl1, and modulates the size of each cell population. Finally, in the ventral midbrain, Neurog2 and Ascl1 are co-expressed in dopaminergic neuronal precursors. The specification of these cells requires Neurog2, but not Ascl1 (Andersson et al., 2006), but when Ascl1 is used to replace Neurog2 in a gene replacement knock-in mouse model, midbrain dopaminergic neurogenesis is partially rescued (Table 1) (Parras et al., 2002). The overlapping Neurog2 and Ascl1 function in the ventral midbrain questions how distinct their functions are in the forebrain and spinal cord, where the similarities are as apparent as the differences.
How functionally distinct are vertebrate proneural genes?
If the multiplicity of vertebrate proneural genes contributes to the complexity of neural development then this may be revealed by functional differences between proteins. This indeed is sometimes evident. For example, bHLH domains from the AS-C and Ato families prefer to bind to slightly different DNA sequences (Powell et al., 2008). However, even though the Ascl, Atoh and Neurog families diverged long ago and are likely to have the most distinct properties, there is still evidence for interchangeable, and hence shared, proneural functions of ato and sc in the development of Drosophila chordotonal organs (zur Lage and Jarman, 2010). The best test of shared function is the capacity of one protein to substitute for another in knock-in experiments (Table 1). In the most dramatic example of such experiments, ato from Drosophila substitutes for Atoh1 in mouse, even in the mouse intestine despite the fact that the fly intestine does not express ato (Wang et al., 2002). This is a powerful demonstration that genes long separated by evolution can retain ancestral functions at the protein level, while their regulatory sequences can diverge to allow species-specific expression patterns. It also highlights that distinct expression patterns do not always indicate divergent protein function at the molecular level.
In contrast to Atoh1, mouse Atoh7 shows less rescue of ato eye phenotypes in Drosophila (Table 1) (Sun et al., 2003; Weinberger et al., 2017). This might reflect loss of ancestral functions by Atoh7 that are retained by Atoh1. However, mouse Atoh7 cannot even induce RGCs in the frog retina, whereas both Drosophila ato and Xenopus Atoh7 can (Brown et al., 1998; Sun et al., 2003). Because Xenopus Atoh7 does rescue the Drosophila ato eye phenotype partially (Brown et al., 1998; Sun et al., 2003) (Table 1), it appears that the mammalian Atoh7 gene has lost ancestral functions at some point after the initial duplication and divergence of the Atoh1 and Atoh7 genes. A potential test of whether Atoh7 protein has acquired novel functions would be to examine whether fly ato can rescue the mouse Atoh7 retinal defects, as it does for Atoh1.
Is it possible that other bHLH genes fulfill shared ancestral functions? Only Ascl1;Neurod4 double mutants completely lack bipolar neurons, suggesting that both genes contribute overlapping but distinct functions to define these cells. Since Ascl1 and Neurod4 are expressed successively, in principle it could be that their expression dynamics are the only reason neither alone is fully sufficient. It should be noted, however, that defining specificities for proneural genes by replacement studies may lead to complicated results. Substituting Atoh7 for Neurod1 reprograms retinal cells to adopt an erroneous RGC fate, even though Neurod1, together with Neurod4, rescues RGC genesis in the absence of Atoh7 (Mao et al., 2008, 2013) (Table 1). It is unclear whether it is protein expression levels and stability that normally distinguish the functions of Atoh7 and Neurod1, or local environmental cues or additional intrinsic factors modify the outcome of Neurod1 expression.
In summary, studies that have examined functional differences between vertebrate proneural genes provide evidence for both functional distinctions and for conserved, shared functions, but it is not yet clear to what extent proneural gene number itself contributes to neural diversity.
Conclusions
As we have highlighted (and as summarized in Table 1), functional complementation studies have revealed many differences between proneural bHLH genes but also provide examples of functional similarity. Particularly striking is the functional replacement of Drosophila ato and mouse Atoh1, despite their evolutionary separation (Ben-Arie et al., 2000; Wang et al. 2002), and the comparative evidence that the complexity of the AS-C might not contribute much to neuronal complexity in insects (Negre and Simpson, 2009). The complex expression and function of the AS-C could have evolved by duplication of progenitor genes similar to ato, as could vertebrate proneural gene networks that show some regulatory similarities. It should be noted, however, that whereas a positive outcome in gene replacement argues powerfully for shared functions between proneural proteins, negative outcomes can have multiple explanations. Some distinctions between proneural bHLH genes must be due to their interactions with other regulatory factors, which explains, for example, how genes such as ato and Atoh1 can be responsible for different neuronal cell types in distinct tissues (Kiefer et al., 2005; Huang et al., 2014). There is also increasing evidence that, in addition to transcriptional regulation, the regulation of bHLH protein stability is important (Qu et al., 2013; Kiparaki et al., 2015; Weinberger et al., 2017; Li and Baker, 2018). Also, genes under constraint by multiple mechanisms may behave less effectively in gene replacement experiments.
Although new functions may have been acquired during evolution to enable neuronal diversity, it is also likely that functional attributes that were already present in an evolutionary precursor have been selectively lost. For example, whereas a Drosophila Neurog protein lacks Ato function, a sponge gene possibly resembling the Neurog/Ato common ancestor exhibits partial rescue, suggesting that an ancestral function has been lost from the Drosophila Neurog protein (Weinberger et al., 2017) (Table 1). At present, only a small minority of possible replacements have been tested in endogenous gene replacement studies, which provide the most reliable results (Table 1), but the availability of new gene editing methods promises more such studies in the future. These studies might clarify the extent to which neuronal complexity relies on a multiplicity of proneural bHLH genes, and how much complexity could already have been encoded by ancient progenitor genes.
Acknowledgements
We thank Jean Herbert and Francesca Pignoni for critical comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Neural research in N.E.B.’s lab is supported by National Institutes of Health (NIH) grant R01 GM047892. Research in N.L.B.’s lab is supported by NIH grant R01 EY013612. Deposited in PMC for release after 12 months.
References
- Acar M., Jafar-Nejad H., Giagtzoglou N., Yallampalli S., David G., He Y., Delidakis C. and Bellen H. J. (2006). Senseless physically interacts with proneural proteins and functions as a transcriptional co-activator. Development 133, 1979-1989. 10.1242/dev.02372 [DOI] [PubMed] [Google Scholar]
- Andersson E., Jensen J. B., Parmar M., Guillemot F. and Björklund A. (2006). Development of the mesencephalic dopaminergic neuron system is compromised in the absence of neurogenin 2. Development 133, 507-516. 10.1242/dev.02224 [DOI] [PubMed] [Google Scholar]
- Azzarelli R., Hurley C., Sznurkowska M. K., Rulands S., Hardwick L., Gamper I., Ali F., McCracken L., Hindley C., McDuff F. et al. (2017). Multi-site neurogenin3 phosphorylation controls pancreatic endocrine differentiation. Dev. Cell 41, 274-286 e275. 10.1016/j.devcel.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. E. (2004). Atonal points the way-protein-protein interactions and developmental biology. Dev. Cell 7, 632-634. 10.1016/j.devcel.2004.10.010 [DOI] [PubMed] [Google Scholar]
- Baker N. E. and Firth L. C. (2011). Retinal determination genes function along with cell-cell signals to regulate Drosophila eye development: examples of multi-layered regulation by master regulators. BioEssays 33, 538-546. 10.1002/bies.201000131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. E., Yu S. and Han D. (1996). Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr. Biol. 6, 1290-1302. 10.1016/S0960-9822(02)70715-X [DOI] [PubMed] [Google Scholar]
- Baker N. E., Bhattacharya A. and Firth L. C. (2009). Regulation of Hh signal transduction as Drosophila eye differentiation progresses. Dev. Biol. 335, 356-366. 10.1016/j.ydbio.2009.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Arie N., Hassan B. A., Bermingham N. A., Malicki D. M., Armstrong D., Matzuk M., Bellen H. J. and Zoghbi H. Y. (2000). Functional conservation of atonal and Math1 in the CNS and PNS. Development 127, 1039-1048. [DOI] [PubMed] [Google Scholar]
- Bermingham N. A., Hassan B. A., Price S. D., Vollrath M. A., Ben-Arie N., Eatock R. A., Bellen H. J., Lysakowski A. and Zoghbi H. Y. (1999). Math1: an essential gene for the generation of inner ear hair cells. Science 284, 1837-1841. 10.1126/science.284.5421.1837 [DOI] [PubMed] [Google Scholar]
- Bermingham N. A., Hassan B. A., Wang V. Y., Fernandez M., Banfi S., Bellen H. J., Fritzsch B. and Zoghbi H. Y. (2001). Proprioceptor pathway development is dependent on Math1. Neuron 30, 411-422. 10.1016/S0896-6273(01)00305-1 [DOI] [PubMed] [Google Scholar]
- Bertrand N., Castro D. S. and Guillemot F. (2002). Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517-530. 10.1038/nrn874 [DOI] [PubMed] [Google Scholar]
- Brown N. L., Kanekar S., Vetter M. L., Tucker P. K., Gemza D. L. and Glaser T. (1998). Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development 125, 4821-4833. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Patel S., Brzezinski J. and Glaser T. (2001). Math5 is required for retinal ganglion cell and optic nerve formation. Development 128, 2497-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachero S., Simpson T. I., Zur Lage P. I., Ma L., Newton F. G., Holohan E. E., Armstrong J. D. and Jarman A. P. (2011). The gene regulatory cascade linking proneural specification with differentiation in Drosophila sensory neurons. PLoS Biol. 9, e1000568 10.1371/journal.pbio.1000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-K. and Chien C.-T. (1999). Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors. Proc. Natl. Acad. Sci. USA 96, 5055-5060. 10.1073/pnas.96.9.5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C.-T., Hsiao C.-D., Jan L. Y. and Jan Y. N. (1996). Neuronal type information encoded in the basic-helix-loop-helix domain of proneural genes. Proc. Natl. Acad. Sci. USA 93, 13239-13244. 10.1073/pnas.93.23.13239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant G. C. and Wolfe K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938-950. 10.1038/nrg2482 [DOI] [PubMed] [Google Scholar]
- Cubas P., de Celis J.-F., Campuzano S. and Modolell J. (1991). Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal wing disc. Genes Dev. 5, 996-1008. 10.1101/gad.5.6.996 [DOI] [PubMed] [Google Scholar]
- Culi J. and Modolell J. (1998). Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 12, 2036-2047. 10.1101/gad.12.13.2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokucu M. E., Zipursky S. L. and Cagan R. L. (1996). Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development 122, 4139-4147. [DOI] [PubMed] [Google Scholar]
- Fode C., Ma Q., Casarosa S., Ang S. L., Anderson D. J. and Guillemot F. (2000). A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 14, 67-80. [PMC free article] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L. and Postlethwait J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini M. E. (2009). Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell 16, 633-647. 10.1016/j.devcel.2009.03.010 [DOI] [PubMed] [Google Scholar]
- Frankfort B. J., Nolo R., Zhang Z., Bellen H. and Mardon G. (2001). senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron 32, 403-414. 10.1016/S0896-6273(01)00480-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong R. F. and Graham A. (2005). Vertebrate neurogenin evolution: long-term maintenance of redundant duplicates. Dev. Genes Evol. 215, 639-644. 10.1007/s00427-005-0023-x [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A. and de Celis J. F. (2009). The complex tale of the achaete-scute complex: a paradigmatic case in the analysis of gene organization and function during development. Genetics 182, 631-639. 10.1534/genetics.109.104083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J. (1999). Warkany lecture: signaling pathways in development. Teratology 60, 226-239. [DOI] [PubMed] [Google Scholar]
- Ghiasvand N. M., Rudolph D. D., Mashayekhi M., Brzezinski J. A. t., Goldman D. and Glaser T. (2011). Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat. Neurosci. 14, 578-586. 10.1038/nn.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giagtzoglou N., Alifragis P., Koumbanakis K. A. and Delidakis C. (2003). Two modes of recruitment of E(spl) repressors onto target genes. Development 130, 259-270. 10.1242/dev.00206 [DOI] [PubMed] [Google Scholar]
- Gómez-Skarmeta J. L., Rodriguez I., Martinez C., Culi J., Ferres-Marco D., Beamonte D. and Modolell J. (1995). Cis-regulation of achaete and scute: shared enhancer-like elements drive their coexpression in proneural clusters of the imaginal discs. Genes Dev. 9, 1869-1882. 10.1101/gad.9.15.1869 [DOI] [PubMed] [Google Scholar]
- Guillemot F. and Hassan B. A. (2017). Beyond proneural: emerging functions and regulations of proneural proteins. Curr. Opin. Neurobiol. 42, 93-101. 10.1016/j.conb.2016.11.011 [DOI] [PubMed] [Google Scholar]
- Hardwick L. J. A. and Philpott A. (2015). Multi-site phosphorylation regulates NeuroD4 activity during primary neurogenesis: a conserved mechanism amongst proneural proteins. Neural Dev. 10, 15 10.1186/s13064-015-0044-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick L. J. A., Davies J. D. and Philpott A. (2016). MyoD phosphorylation on multiple C terminal sites regulates myogenic conversion activity. Biochem. Biophys. Res. Commun. 481, 97-103. 10.1016/j.bbrc.2016.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms A. W., Abney A. L., Ben-Arie N., Zoghbi H. Y. and Johnson J. E. (2000). Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127, 1185-1196. [DOI] [PubMed] [Google Scholar]
- Helms A. W., Battiste J., Henke R. M., Nakada Y., Simplicio N., Guillemot F. and Johnson J. E. (2005). Sequential roles for Mash1 and Ngn2 in the generation of dorsal spinal cord interneurons. Development 132, 2709-2719. 10.1242/dev.01859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan E. E., zur Lage P. I. and Jarman A. P. (2006). Multiple enhancers contribute to spatial but not temporal complexity in the expression of the proneural gene, amos. BMC Dev. Biol. 6, 53 10.1186/1471-213X-6-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Chan J. A. and Schuurmans C. (2014). Proneural bHLH genes in development and disease. Curr. Top. Dev. Biol. 110, 75-127. 10.1016/B978-0-12-405943-6.00002-6 [DOI] [PubMed] [Google Scholar]
- Hufnagel R. B., Riesenberg A. N., Saul S. M. and Brown N. L. (2007). Conserved regulation of Math5 and Math1 revealed by Math5-GFP transgenes. Mol. Cell. Neurosci. 36, 435-448. 10.1016/j.mcn.2007.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufnagel R. B., Le T. T., Riesenberg A. L. and Brown N. L. (2010). Neurog2 controls the leading edge of neurogenesis in the mammalian retina. Dev. Biol. 340, 490-503. 10.1016/j.ydbio.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufnagel R. B., Riesenberg A. N., Quinn M., Brzezinski J. A. t., Glaser T. and Brown N. L. (2013). Heterochronic misexpression of Ascl1 in the Atoh7 retinal cell lineage blocks cell cycle exit. Mol. Cell. Neurosci. 54, 108-120. 10.1016/j.mcn.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I., Pan N., Kersigo J. and Fritzsch B. (2015). Neurog1 can partially substitute for Atoh1 function in hair cell differentiation and maintenance during organ of Corti development. Development 142, 2810-2821. 10.1242/dev.123091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman A. P. and Groves A. K. (2013). The role of Atonal transcription factors in the development of mechanosensitive cells. Semin. Cell Dev. Biol. 24, 438-447. 10.1016/j.semcdb.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman A. P., Grell E. H., Ackerman L., Jan L. Y. and Jan Y. N. (1994). Atonal is the proneural gene for Drosophila photoreceptors. Nature 369, 398-400. 10.1038/369398a0 [DOI] [PubMed] [Google Scholar]
- Jarman A. P., Sun Y., Jan L. Y. and Jan Y. N. (1995). Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development 121, 2019-2030. [DOI] [PubMed] [Google Scholar]
- Jasoni C. and Reh T. (1996). Temporal and spatial pattern of MASH-1 expression in the developing rat retina demonstrates progenitor cell heterogeneity. J. Comp. Neurol. 369, 319-327. [DOI] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Shimojo H. and Imayoshi I. (2008). Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat. Neurosci. 11, 1247-1251. 10.1038/nn.2208 [DOI] [PubMed] [Google Scholar]
- Kanekar S., Perron M., Dorsky R., Harris W. A., Jan L. Y., Jan Y. N. and Vetter M. L. (1997). Xath5 participates in a network of bHLH genes in the developing Xenopus retina. Neuron 19, 981-994. 10.1016/S0896-6273(00)80391-8 [DOI] [PubMed] [Google Scholar]
- Kay J. N., Finger-Baier K. C., Roeser T., Staub W. and Baier H. (2001). Retinal ganglion cell genesis requires lakritz, a zebrafish atonal Homolog. Neuron 30, 725-736. 10.1016/S0896-6273(01)00312-9 [DOI] [PubMed] [Google Scholar]
- Kiefer J. C., Jarman A. and Johnson J. (2005). Pro-neural factors and neurogenesis. Dev. Dyn. 234, 808-813. 10.1002/dvdy.20522 [DOI] [PubMed] [Google Scholar]
- Kiparaki M., Zarifi I. and Delidakis C. (2015). bHLH proteins involved in Drosophila neurogenesis are mutually regulated at the level of stability. Nucleic Acids Res. 43, 2543-2559. 10.1093/nar/gkv083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirjavainen A., Sulg M., Heyd F., Alitalo K., Ylä-Herttuala S., Möröy T., Petrova T. V. and Pirvola U. (2008). Prox1 interacts with Atoh1 and Gfi1, and regulates cellular differentiation in the inner ear sensory epithelia. Dev. Biol. 322, 33-45. 10.1016/j.ydbio.2008.07.004 [DOI] [PubMed] [Google Scholar]
- Knust E. and Campos-Ortega J. A. (1989). The molecular genetics of early neurogenesis in Drosophila melanogaster. BioEssays 11, 95-100. 10.1002/bies.950110405 [DOI] [PubMed] [Google Scholar]
- Krentz N. A. J., van Hoof D., Li Z., Watanabe A., Tang M., Nian C., German M. S. and Lynn F. C. (2017). Phosphorylation of NEUROG3 links endocrine differentiation to the cell cycle in pancreatic progenitors. Dev. Cell 41, 129-142 e126. 10.1016/j.devcel.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledent V., Paquet O. and Vervoort M. (2002). Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 3, RESEARCH0030 10.1186/gb-2002-3-6-research0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K. and Baker N. E. (2018). Regulation of the Drosophila ID protein Extra macrochaetae by proneural dimerization partners. Elife 7, e33967 10.7554/eLife.33967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. and Choi K. W. (2004). Induction and autoregulation of the anti-proneural gene Bar during retinal neurogenesis in Drosophila. Development 131, 5573-5580. 10.1242/dev.01426 [DOI] [PubMed] [Google Scholar]
- Ma Y.-C., Song M.-R., Park J. P., Henry Ho H.-Y., Hu L., Kurtev M. V., Zieg J., Ma Q., Pfaff S. L. and Greenberg M. E. (2008). Regulation of motor neuron specification by phosphorylation of neurogenin 2. Neuron 58, 65-77. 10.1016/j.neuron.2008.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C.-A., Wang S. W., Pan P. and Klein W. H. (2008). Rewiring the retinal ganglion cell gene regulatory network: Neurod1 promotes retinal ganglion cell fate in the absence of Math5. Development 135, 3379-3388. 10.1242/dev.024612 [DOI] [PubMed] [Google Scholar]
- Mao C.-A., Cho J.-H., Wang J., Gao Z., Pan P., Tsai W.-W., Frishman L. J. and Klein W. H. (2013). Reprogramming amacrine and photoreceptor progenitors into retinal ganglion cells by replacing Neurod1 with Atoh7. Development 140, 541-551. 10.1242/dev.085886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai I., Yamaguchi M., Tonou-Fujimori N., Komori A. and Okamoto H. (2005). The hedgehog-PKA pathway regulates two distinct steps of the differentiation of retinal ganglion cells: the cell-cycle exit of retinoblasts and their neuronal maturation. Development 132, 1539-1553. 10.1242/dev.01714 [DOI] [PubMed] [Google Scholar]
- Maung S. M. T. W. and Jarman A. P. (2007). Functional distinctness of closely related transcription factors: a comparison of the Atonal and Amos proneural factors. Mech. Dev. 124, 647-656. 10.1016/j.mod.2007.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K. B., Schneider M. L. and Vetter M. L. (2002). Posttranslational mechanisms control the timing of bHLH function and regulate retinal cell fate. Neuron 34, 183-195. 10.1016/S0896-6273(02)00666-9 [DOI] [PubMed] [Google Scholar]
- Morrow E. M., Furukawa T., Lee J. E. and Cepko C. L. (1999). NeuroD regulates multiple functions in the developing neural retina in rodent. Development 126, 23-36. [DOI] [PubMed] [Google Scholar]
- Negre B. and Simpson P. (2009). Evolution of the achaete-scute complex in insects: convergent duplication of proneural genes. Trends Genet. 25, 147-152. 10.1016/j.tig.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Neumann C. J. and Nuesslein-Volhard C. (2000). Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science 289, 2137-2139. 10.1126/science.289.5487.2137 [DOI] [PubMed] [Google Scholar]
- Parras C. M., Schuurmans C., Scardigli R., Kim J., Anderson D. J. and Guillemot F. (2002). Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 16, 324-338. 10.1101/gad.940902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott A. (2015). Multi-site phospho-regulation of proneural transcription factors controls proliferation versus differentiation in development and reprogramming. Neurogenesis 2, e1049733 10.1080/23262133.2015.1049733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires-daSilva A. and Sommer R. J. (2003). The evolution of signalling pathways in animal development. Nat. Rev. Genet. 4, 39-49. 10.1038/nrg977 [DOI] [PubMed] [Google Scholar]
- Portman D. S. and Emmons S. W. (2004). Identification of C. elegans sensory ray genes using whole-genome expression profiling. Dev. Biol. 270, 499-512. 10.1016/j.ydbio.2004.02.020 [DOI] [PubMed] [Google Scholar]
- Powell L. M., Deaton A. M., Wear M. A. and Jarman A. P. (2008). Specificity of Atonal and Scute bHLH factors: analysis of cognate E box binding sites and the influence of Senseless. Genes Cells 13, 915-929. 10.1111/j.1365-2443.2008.01217.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Afelik S., Jensen J. N., Bukys M. A., Kobberup S., Schmerr M., Xiao F., Nyeng P., Veronica Albertoni M., Grapin-Botton A. et al. (2013). Notch-mediated post-translational control of Ngn3 protein stability regulates pancreatic patterning and cell fate commitment. Dev. Biol. 376, 1-12. 10.1016/j.ydbio.2013.01.021 [DOI] [PubMed] [Google Scholar]
- Quan X.-J., Yuan L., Tiberi L., Claeys A., De Geest N., Yan J., van der Kant R., Xie W. R., Klisch T. J., Shymkowitz J. et al. (2016). Post-translational control of the temporal dynamics of transcription factor activity regulates neurogenesis. Cell 164, 460-475. 10.1016/j.cell.2015.12.048 [DOI] [PubMed] [Google Scholar]
- Reeves N. and Posakony J. W. (2005). Genetic programs activated by proneural proteins in the developing Drosophila PNS. Dev. Cell 8, 413-425. 10.1016/j.devcel.2005.01.020 [DOI] [PubMed] [Google Scholar]
- Riesenberg A. N., Le T. T., Willardsen M. I., Blackburn D. C., Vetter M. L. and Brown N. L. (2009). Pax6 regulation of Math5 during mouse retinal neurogenesis. Genesis 47, 175-187. 10.1002/dvg.20479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardigli R., Schuurmans C., Gradwohl G. and Guillemot F. (2001). Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron 31, 203-217. 10.1016/S0896-6273(01)00358-0 [DOI] [PubMed] [Google Scholar]
- Shimojo H., Ohtsuka T. and Kageyama R. (2008). Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58, 52-64. 10.1016/j.neuron.2008.02.014 [DOI] [PubMed] [Google Scholar]
- Shroyer N. F., Helmrath M. A., Wang V. Y., Antalffy B., Henning S. J. and Zoghbi H. Y. (2007). Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology 132, 2478-2488. 10.1053/j.gastro.2007.03.047 [DOI] [PubMed] [Google Scholar]
- Simionato E., Ledent V., Richards G., Thomas-Chollier M., Kerner P., Coornaert D., Degnan B. M. and Vervoort M. (2007). Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol. Biol. 7, 33 10.1186/1471-2148-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P. (1990). Notch and the choice of cell fate in Drosophila neuroepithelium. Trends Genet. 6, 343-345. 10.1016/0168-9525(90)90260-D [DOI] [PubMed] [Google Scholar]
- Skowronska-Krawczyk D., Chiodini F., Ebeling M., Alliod C., Kundzewicz A., Castro D., Ballivet M., Guillemot F., Matter-Sadzinski L. and Matter J.-M. (2009). Conserved regulatory sequences in Atoh7 mediate non-conserved regulatory responses in retina ontogenesis. Development 136, 3767-3777. 10.1242/dev.033449 [DOI] [PubMed] [Google Scholar]
- Slack J. M. W. (1983). From Egg to Embryo. Cambridge: Cambridge University Press. [Google Scholar]
- Sun Y., Jan L. Y. and Jan Y. N. (1998). Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development 125, 3731-3740. [DOI] [PubMed] [Google Scholar]
- Sun Y., Jan L. Y. and Jan Y. N. (2000). Ectopic scute induces Drosophila ommatidia development without R8 founder photoreceptors. Proc. Natl. Acad. Sci. USA 97, 6815-6819. 10.1073/pnas.110154497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Kanekar S. L., Vetter M. L., Gorski S., Jan Y.-N., Glaser T. and Brown N. L. (2003). Conserved and divergent functions of Drosophila atonal, amphibian, and mammalian Ath5 genes. Evol. Dev. 5, 532-541. 10.1046/j.1525-142X.2003.03058.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. S. and Raes J. (2004). Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 38, 615-643. 10.1146/annurev.genet.38.072902.092831 [DOI] [PubMed] [Google Scholar]
- Tomita K., Nakanishi S., Guillemot F. and Kageyama R. (1996b). Mash1 promotes neuronal differentiation in the retina. Genes Cells 1, 765-774. [DOI] [PubMed] [Google Scholar]
- Treisman J. E. (2013). Retinal differentiation in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 2, 545-557. 10.1002/wdev.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. W., Kim B. S., Ding K., Wang H., Sun D., Johnson R. L., Klein W. H. and Gan L. (2001). Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 15, 24-29. 10.1101/gad.855301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang V. Y., Hassan B. A., Bellen H. J. and Zoghbi H. Y. (2002). Drosophila atonal fully rescues the phenotype of Math1 null mice: new functions evolve in new cellular contexts. Curr. Biol. 12, 1611-1616. 10.1016/S0960-9822(02)01144-2 [DOI] [PubMed] [Google Scholar]
- Weinberger S., Topping M. P., Yan J., Claeys A., De Geest N., Ozbay D., Hassan T., He X., Albert J. T., Hassan B. A. et al. (2017). Evolutionary changes in transcription factor coding sequence quantitatively alter sensory organ development and function. Elife 6, e26402 10.7554/eLife.26402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N. M. and Jarman A. P. (2000). Drosophila Atonal controls photoreceptor R8-specific properties and modulates both Receptor Tyrosine Kinase and Hedgehog signalling. Development 127, 1681-1689. [DOI] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N. and Brenner S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1-340. 10.1098/rstb.1986.0056 [DOI] [PubMed] [Google Scholar]
- Willardsen M. I., Suli A., Pan Y., Marsh-Armstrong N., Chien C.-B., El-Hodiri H., Brown N. L., Moore K. B. and Vetter M. L. (2009). Temporal regulation of Ath5 gene expression during eye development. Dev. Biol. 326, 471-481. 10.1016/j.ydbio.2008.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Kaczynski T. J., Sethuramanujam S., Li R., Jain V., Slaughter M. and Mu X. (2015). Two transcription factors, Pou4f2 and Isl1, are sufficient to specify the retinal ganglion cell fate. Proc. Natl. Acad. Sci. USA 112, E1559-E1568. 10.1073/pnas.1421535112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie L. A., Hardwick L. J. A., Papkovskaia T. D., Thiele C. J. and Philpott A. (2015). Ascl1 phospho-status regulates neuronal differentiation in a Xenopus developmental model of neuroblastoma. Dis. Model. Mech. 8, 429-441. 10.1242/dmm.018630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W. R., Jen H.-I., Seymour M. L., Yeh S.-Y., Pereira F. A., Groves A. K., Klisch T. J. and Zoghbi H. Y. (2017). An Atoh1-S193A phospho-mutant allele causes hearing deficits and motor impairment. J. Neurosci. 37, 8583-8594. 10.1523/JNEUROSCI.0295-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Zhou Q., Zhang C. and Pignoni F. (2012). Identification of Bombyx atonal and functional comparison with the Drosophila atonal proneural factor in the developing fly eye. Genesis 50, 393-403. 10.1002/dvg.20816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagozewski J. L., Zhang Q. and Eisenstat D. D. (2014). Genetic regulation of vertebrate eye development. Clin. Genet. 86, 453-460. 10.1111/cge.12493 [DOI] [PubMed] [Google Scholar]
- Zhang T., Ranade S., Cai C. Q., Clouser C. and Pignoni F. (2006). Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development 133, 4881-4889. 10.1242/dev.02669 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Yu L., Friedrich M. and Pignoni F. (2017). Distinct regulation of atonal in a visual organ of Drosophila: organ-specific enhancer and lack of autoregulation in the larval eye. Dev. Biol. 421, 67-76. 10.1016/j.ydbio.2016.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Lage P. I. and Jarman A. P. (2010). The function and regulation of the bHLH gene, cato, in Drosophila neurogenesis. BMC Dev. Biol. 10, 34 10.1186/1471-213X-10-34 [DOI] [PMC free article] [PubMed] [Google Scholar]