Abstract
RAG-1 and RAG-2 initiate V(D)J recombination through synapsis and cleavage of a 12/23 pair of V(D)J recombination signal sequences (RSS). RAG-RSS complex assembly and activity in vitro is promoted by high mobility group proteins of the “HMG-box” family, exemplified by HMGB1. How HMGB1 stimulates the DNA binding and cleavage activity of the RAG complex remains unclear. HMGB1 contains two homologous HMG-box DNA binding domains, termed A and B, linked by a stretch of basic residues to a highly acidic C-terminal tail. To identify determinants of HMGB1 required for stimulation of RAG-mediated RSS binding and cleavage, we prepared an extensive panel of mutant HMGB1 proteins and tested their ability to augment RAG-mediated RSS binding and cleavage activity. Using a combination of mobility shift and in-gel cleavage assays, we find that HMGB1 promotes RAG-mediated cleavage largely through the activity of box B, but optimal stimulation requires a functional A box tethered in the correct orientation. Box A or B mutants fail to promote RAG synaptic complex formation, but this defect is alleviated when the acidic tail is removed from these mutants.
During lymphocyte development, antigen receptor genes undergo a series of DNA rearrangements to generate functional exons encoding the antigen binding domains of these receptors (1). This rearrangement process, termed V(D)J recombination, is initiated when two lymphoid cell-specific proteins, called recombination-activating gene (RAG)1-1 and RAG-2, bring two gene segments into close proximity and then introduce a DNA double-strand break (DSB) at the end of each coding segment. Adjacent to each coding segment lies a recombination signal sequence (RSS) that serves as the binding site for the RAG-1/2 protein complex (hereafter termed the “RAG complex”) and directs the location of cleavage. The RSS contains a conserved heptamer and nonamer motif separated by either 12 or 23 bp of relatively nonconserved sequence (12-RSS and 23-RSS, respectively). Productive exon assembly is promoted by the 12/23 rule, a restriction that limits rearrangement to RSSs whose spacing between the heptamer and nonamer is different. RAG-mediated cleavage of RSS pairs produces four DNA ends: two blunt, 5′-phosphorylated signal ends and two coding ends terminating in DNA hairpin structures (2–4). These reaction products originate from a two-step cleavage reaction in which the RSS is first nicked at its 5′-end, and then the resulting 3′-OH is covalently linked to the bottom strand by direct transesterification (5, 6). After DNA cleavage, signal ends are generally ligated together to form precise signal joints, but coding ends, being sealed as DNA hairpins, are first resolved and then processed and joined to create coding joints in which nucleotides are frequently gained or lost at the junction. DNA hairpin opening is most likely catalyzed by a complex containing Artemis and the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) (7). Signal and coding joint formation is mediated by ubiquitously expressed proteins that comprise the non-homologous end-joining pathway (NHEJ) of DNA double-strand break repair, including Ku70, Ku80, XRCC4, and DNA ligase IV (1).
Early studies of RAG protein biochemistry demonstrated that both proteins were necessary and sufficient to cleave a DNA substrate containing a single RSS in vitro (5). Subsequent studies revealed that RAG complex binding to isolated recombination signals (especially a 23-RSS), and RAG-mediated syn-apsis and cleavage of RSS pairs according to the 12/23 rule are promoted by “architectural” DNA-binding proteins of the HMG-box family of high mobility group (HMG) proteins (8). In this respect, the RAG complex is part of a growing group of recombinases and transcription factors that are known to associate with, and have their activity augmented by, HMG-box proteins (9, 10). How these proteins stimulate the DNA binding and cleavage activity of the RAG complex remains poorly understood. Here, we investigate the mechanisms by which a prototypical HMG-box protein, HMGB1, facilitates RAG-RSS complex assembly and activity.
Mammalian members of the HMG-box family (including HMGB1 and HMGB2) contain tandem homologous DNA binding domains, termed HMG-box A and B, each of which consist of about 80 amino acids folded into three α-helices that adopt a characteristic L-shaped structure. The two HMG-box domains are followed by a short linker rich in basic amino acid residues and a C-terminal acidic tail comprised of about 30 consecutive aspartate and glutamate residues (10). Structural and biochemical studies of the individual HMG-box domains suggest both interact with the minor groove of DNA, but display different DNA binding properties: box A exhibits a preference for binding distorted DNA structures such as four-way junctions (11) and DNA interstrand cross-links (12), whereas box B does not selectively bind such DNA structures, but can itself introduce a severe bend into linear DNA when flanking sequences are present (13, 14). (The A domain lacks this ability.) The acidic tail of HMG-box proteins has been shown to mediate interactions with the HMG-boxes (15), as well as with core histones, thereby promoting association of the HMG-boxes with nucleosome linker DNA (16). In addition, the acidic tail plays a role in stimulating nucleosome sliding by chromatin remodeling factors (17). The distinct functional properties of the HMG-boxes and the acidic tail cause us to speculate that these regions may play separable roles in the assembly and activity of RAG-RSS complexes.
To explore this possibility, we prepared an extensive panel of truncated and mutant HMGB1 proteins and systematically tested their ability to promote RAG complex binding and cleavage of single and paired RSS substrates using a combination of mobility shift and in-gel cleavage assays. We find that, like full-length HMGB1, individual HMG-box domains supershift RAG-RSS complexes. Forms of full-length HMGB1 bearing alanine substitutions at residues thought to mediate important protein-DNA contacts in either or both HMG-boxes also super-shift RAG-RSS complexes. Interestingly, however, despite being incorporated into RAG-RSS complexes, individual HMG-box domain proteins and full-length HMGB1 bearing mutations in box B fail to stimulate RAG-mediated DNA cleavage of single RSS substrates. Mutations in box A reduce, but do not eliminate the ability of full-length HMGB1 to enhance RAG-mediated cleavage. However, the box A mutant is less effective at promoting site-specific cleavage of a 23-RSS, and stimulates 12/23-regulated cleavage poorly. Removal of the basic linker and/or the acidic tail from HMGB1 do not appreciably reduce its ability to stimulate RAG-mediated cleavage, but increases the nonspecific DNA binding activity of HMGB1, and enables mutant forms of HMGB1 to promote assembly of RAG synaptic complexes, which otherwise is impaired when these mutants contain the acidic tail. Interestingly, a tailless form of HMGB1 in which the positions of HMG-box A and B are reversed stimulates RAG-mediated synapsis and cleavage only slightly less effectively than its normally oriented counterpart. Taken together, these data argue that box B, likely by promoting DNA bending through interactions with RAG-1, plays a critical role in promoting RAG-mediated cleavage, but is not sufficient by itself to correctly position the recombinase active site in a 23-RSS, which requires the activity of a functional tethered box A. Moreover, the acidic tail may help enforce the 12/23 rule by reducing nonspecific protein-DNA interactions.
EXPERIMENTAL PROCEDURES
Expression Vectors
Eukaryotic expression constructs encoding core RAG-1 or core RAG-2, each fused at the amino terminus to MBP without additional sequence tags (pcMR1 and pcMR2, respectively), have been described previously (18). The prokaryotic expression construct pET11d-hHMGB1, encoding full-length human HMGB1 tagged at the amino terminus with polyhistidine, has been described elsewhere (19). Derivatives of this construct encoding forms of hHMGB1 that are truncated or contain alanine substitutions, as depicted in Fig. 1, were generated as described under supplemental materials.
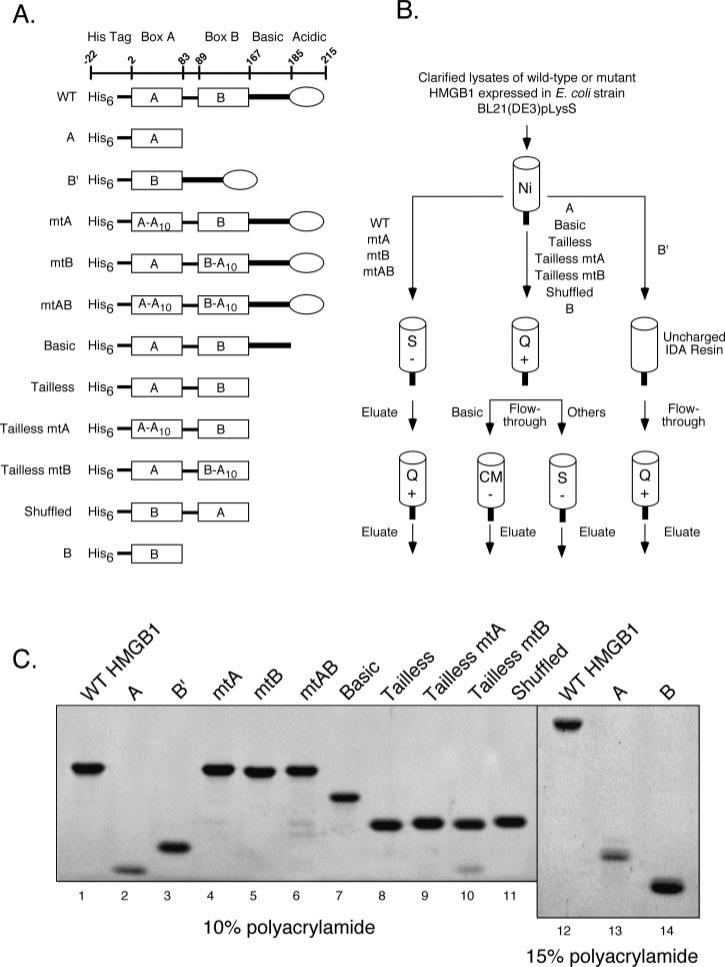
Fig. 1. Purification of HMGB1 proteins.
A, diagrams of HMGB1 proteins used in this study. The amino acid residues encompassing the amino-terminal polyhistidine tag, HMG boxes A and B (rectangles), the basic linker (heavy line), and acidic tail (oval) are indicated at the top. Various wild-type, truncated, and mutant forms of HMGB1 are shown, with designations at the left. Ten consecutive alanine substitutions (A10) have been introduced in either or both HMG-boxes, starting at residue 18 of box A and residue 102 of box B. B, purification strategy of HMGB1 proteins. All proteins were initially purified using IMAC (Ni), with eluates subsequently subjected to chromatography using strongly acidic (S), strongly basic (Q), or weakly acidic (CM) ion exchange supports as indicated. C, purified HMGB1 proteins were fractionated by SDS-PAGE using 10% (lanes 1–11) or 15% (lanes 12–14) polyacrylamide resolving gels, and stained with SYPRO Orange. The identity of purified full-length HMGB1 was confirmed by mass spectroscopy (data not shown).
Protein Expression and Purification
RAG-1 and RAG-2 fusion proteins were coexpressed in 293 cells and purified as described elsewhere (20). Wild-type, truncated, and mutant forms of HMGB1 were expressed in Escherichia coli strain BL21(DE3)pLysS and initially purified by immobilized metal affinity chromatography (IMAC) using iminodiacetate (IDA)-coupled Sepharose charged with Ni2+. Subsequent purification steps involved ion exchange chromatography (IEC) using strongly acidic (High S), weakly acidic (CM), or strongly basic (High Q) ion exchange supports as schematically diagrammed in Fig. 1. The purification procedures are described in detail under supplemental materials. We consistently observed lower overall yields of HMGB1 bearing alanine substitutions (either HMG-box, but particularly mtA), as these forms of HMGB1 are prone to degradation (not shown).
Oligonucleotide Binding Assays
Intact 12- and 23-RSS substrates were assembled and purified as described previously (18, 21). Electrophoretic mobility shift assays (EMSA) were performed under the same conditions as described previously (18, 22). The amounts of HMGB1 used in analytical EMSAs are noted in the figures or figure legends. Protein-DNA complexes were visualized from dried gels by autoradiography using a Molecular Dynamics Storm 860 Gel and Blot Imaging System.
In-gel Cleavage Assays
Preparative scale binding reactions were assembled in the presence of Ca2+, fractionated by EMSA, and protein-DNA complexes assayed for cleavage activity using a previously described in-gel cleavage assay (18, 23).
RESULTS
Preparation of Wild-type, Truncated, and Mutant Forms of HMGB1
In a previous study, the activities of HMGB1 and HMGB2 were directly compared in RAG-RSS binding and cleavage assays (24). In that study, HMGB1 and HMGB2 were found to comparably supershift RAG-RSS complexes containing either a 12- or 23-RSS, and stimulate RAG-mediated cleavage of an isolated 23-RSS, but not a 12-RSS, using an in-gel cleavage assay. In a subsequent study, HMGB1 was found to be required for RAG-mediated synapsis and cleavage of intact RSS pairs according to the 12/23 rule, and 12/23-regulated cleavage of nicked RSS pairs, despite not being rigorously required for synapsis of nicked substrates (18). In addition, HMGB1 was found to promote formation of RAG complexes with cleaved signal ends, but once formed, the RAG proteins did not require HMGB1 to mediate transposition of signal ends into a plasmid target substrate (18). To extend these studies, we wished to identify the structural features of HMGB1 responsible for stimulating RAG-mediated binding and cleavage of an isolated 23-RSS, and synapsis and 12/23-regulated cleavage of intact RSS pairs.
Sequence, structural, and biochemical characterization of HMGB1 and related HMG-box proteins suggest that HMG box A, box B, the basic linker, and the acidic tail of HMGB1 are encompassed within the amino acid residues shown in Fig. 1A (10). To systematically test how the various regions of HMGB1 affect RAG binding and cleavage activity, we prepared an extensive panel of bacterially expressed truncated and mutant forms of HMGB1 (Fig. 1A, see supplemental materials). To disrupt the DNA binding activity of HMGB1, we elected to replace ten consecutive residues with alanine at comparable positions in helix 1 of either or both HMG-boxes, starting at residue 18 of box A and residue 102 of box B, using ligation-assisted recombination PCR mutagenesis (LAR-PCR, see supplemental materials and supplemental Fig. 1). These residues were targeted for mutagenesis based on sequence and structural analysis indicating that they comprise part of a “hydrophobic wedge” in their respective HMG-box domains that mediates contact with the minor groove of DNA (10). Phenylalanine 103, for example, is thought to intercalate between DNA base pairs, promoting DNA bending; mutation of this residue impairs the DNA supercoiling activity of box B (25). Mutagenesis also replaces a highly conserved arginine residue present in both HMG-boxes (Arg24 in box A, and Arg110 in box B); substitution of this residue in either HMG-box reduces the DNA binding activity of the domain (25, 26). To test whether the orientation of the two HMG-box domains relative to one another is an important determinant for the ability of HMGB1 to stimulate RAG complex binding and cleavage activity, we also prepared a version of HMGB1 in which the positions of the two HMG-box domains are inverted or “shuffled.” Purifying the various HMGB1 proteins required different strategies, which are schematically diagrammed in Fig. 1B and described in detail under supplemental materials. Using these purification strategies, we obtained highly purified truncated and mutant HMGB1 proteins estimated to be between 90 and 95% pure (Fig. 1C).
Truncated and Mutant Forms of HMGB1 Differentially Supershift RAG-RSS Complexes
HMGB1 and HMGB2 are known from previous studies to become incorporated into RAG-RSS complexes, causing the protein-DNA complex to migrate more slowly in an EMSA (24). To compare the ability of full-length, truncated and mutant forms of HMGB1 to supershift RAG-RSS complexes, purified RAG proteins were incubated with a radiolabeled 23-RSS substrate in the presence of increasing concentrations of the various forms of HMGB1, and protein-DNA complex formation analyzed by EMSA (Fig. 2). All forms of HMGB1 tested, except B′, were found to supershift RAG-RSS complexes in a concentration-dependent manner. Based on these binding titrations, the concentration of each form of HMGB1 (except B′) needed to visibly supershift the RAG-RSS complexes was used in an EMSA to compare the mobility of supershifted protein-DNA complexes side-by-side, both in the absence and presence of the RAG proteins (Fig. 3).
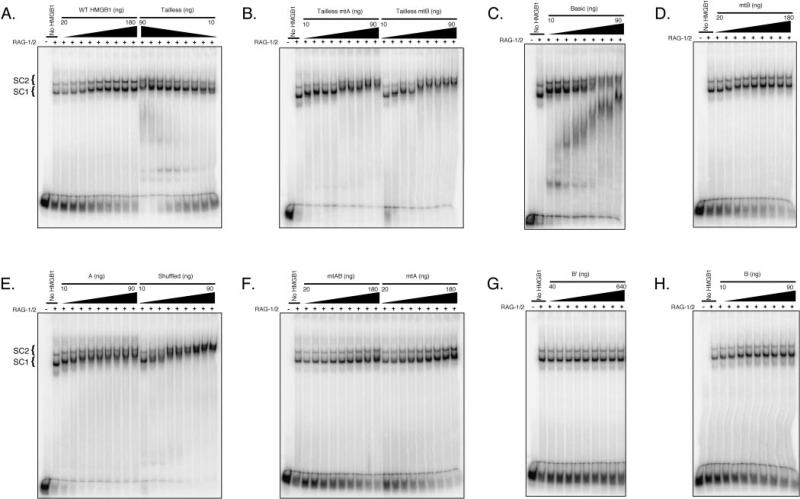
Fig. 2. Truncated and mutant forms of HMGB1 exhibit differential capacity to supershift RAG-RSS complexes.
Coexpressed MBP-RAG-1 and MBP-RAG-2 (RAG-1/2) was incubated with a radiolabeled intact 23-RSS substrate in the absence or presence of increasing amounts of the following forms of HMGB1 (as indicated above the gel): wild-type or tailless (A), tailless mtA or tailless mtB (B), basic (C), mtB (D), box A or shuffled (E), mtAB or mtA (F), box B′ (G), or box B (H). Protein-DNA complexes were fractionated by EMSA, and visualized from dried gels using a phosphorimager.
Fig. 3. Comparative binding of various HMGB1 proteins to a 23-RSS in the absence or presence of the RAG complex.
Purified forms of HMGB1 were incubated with a radiolabeled intact 23-RSS substrate in the absence or presence of the RAG complex (as indicated above the gels), and protein-DNA complex formation was analyzed using EMSA. A, EMSA of individual HMG-boxes and full-length forms of HMGB1. B, EMSA of basic, tailless, and shuffled forms of HMGB1. Based on the titrations performed in Fig. 2, the following amounts of each form of HMGB1 were used in the binding reaction: 150 ng of wild type, mtA, mtB, or mtAB; 50 ng of box A, basic, tailless, tailless mtA, tailless mtB, or shuffled. Because B′ failed to supershift RAG-RSS complexes at the concentrations tested (see Fig. 2), we used 150 ng in this assay for comparison to other forms of HMGB1.
In the absence of HMGB1, the RAG proteins form two distinct protein-DNA complexes when assembled on substrates containing a single RSS. We have previously shown that these two RAG-RSS complexes, called SC1 and SC2, both contain a RAG-1 dimer, but differ in the stoichiometry of RAG-2, with SC1 containing monomeric RAG-2 and SC2 containing two RAG-2 molecules (18). Both RAG-RSS complexes are super-shifted in the presence WT HMGB1 and individual HMGB boxes A and B, but not box B′, presumably because the negative charge associated with the acidic tail interferes with DNA binding (Figs. 2 and 3). Mixing HMG box A with either form of box B promotes formation of supershifted RAG-RSS complexes that comigrate with those formed in the presence of box A alone (Fig. 3). HMGB1 bearing mutations in box B supershift RAG-RSS complexes comparably to WT HMGB1, regardless of the presence of the acidic tail (Fig. 2). However, slightly more (~2-fold) mtA HMGB1 is required to reach a comparable shift in the mobility of the RAG-RSS complexes relative to WT or mtB HMGB1, although this difference is diminished if the acidic tail is removed (Fig. 2). Interestingly, mtAB HMGB1 exhibits a subtle and selective defect in its ability to supershift SC1, but supershifts SC2 comparably to mtA and mtB HMGB1 (Fig. 3). The observation that both mtA and mtB HMGB1 bind an isolated RSS and also supershift the SC1 and SC2 RAG complexes suggests that the introduced mutations do not severely disrupt the global folding of the protein, although some loss of α-helical content is observed in mtA and mtB HMGB1 compared with WT HMGB1, as indicated by circular dichroism spectroscopy (data not shown). Consistent with previous observations (27), tailless forms of HMGB1 exhibit greater DNA binding activity than full-length HMGB1, and are prone to forming higher order oligomeric complexes with DNA as a function of protein concentration, both in the absence and presence of the RAG proteins (Figs. 2 and 3). Notably, about 5–10-fold more full-length HMGB1 is required to visibly super-shift the RAG-RSS complexes compared with its tailless counterparts (Fig. 2). This effect is largely attributed to the presence of the acidic tail, as similar concentrations of basic and tailless HMGB1 are required to supershift the RAG-RSS complexes (Fig. 2). Perhaps surprisingly, “shuffled” HMGB1 also super-shifts SC1 and SC2, exhibiting a concentration dependence similar to that observed for tailless HMGB1.
Separable Roles for the HMG-box Domains in Facilitating RAG-mediated Cleavage of an Intact 23-RSS
One limitation of the mobility shift experiments described in the previous section is that they do not provide information about the functional significance of HMGB1 protein association with the RAG-RSS complex. To address this issue, we evaluated the ability of the various HMGB1 proteins to stimulate RAG-mediated cleavage of an intact 23-RSS within the context of the supershifted SC1 RAG-RSS complex using a previously described in-gel cleavage assay that enables a direct comparison of cleavage activity between multiple, discrete RAG-RSS complexes fractionated on a single nondenaturing polyacrylamide gel (23) (Fig. 4). As a negative control, the activity of a catalytically defective RAG complex (RAG-1 D600A) was analyzed in parallel with protein-DNA complexes assembled with WT RAG proteins. In these assays, we intentionally used the lowest concentration of the various forms of HMGB1 that yield a detectable supershift of the RAG complex to reduce the possibility of nonspecific binding by the HMGB1 proteins, while concomitantly retaining the ability to unambiguously determine whether the ability to supershift the RAG-RSS complex is the sole determinant for altering its activity.
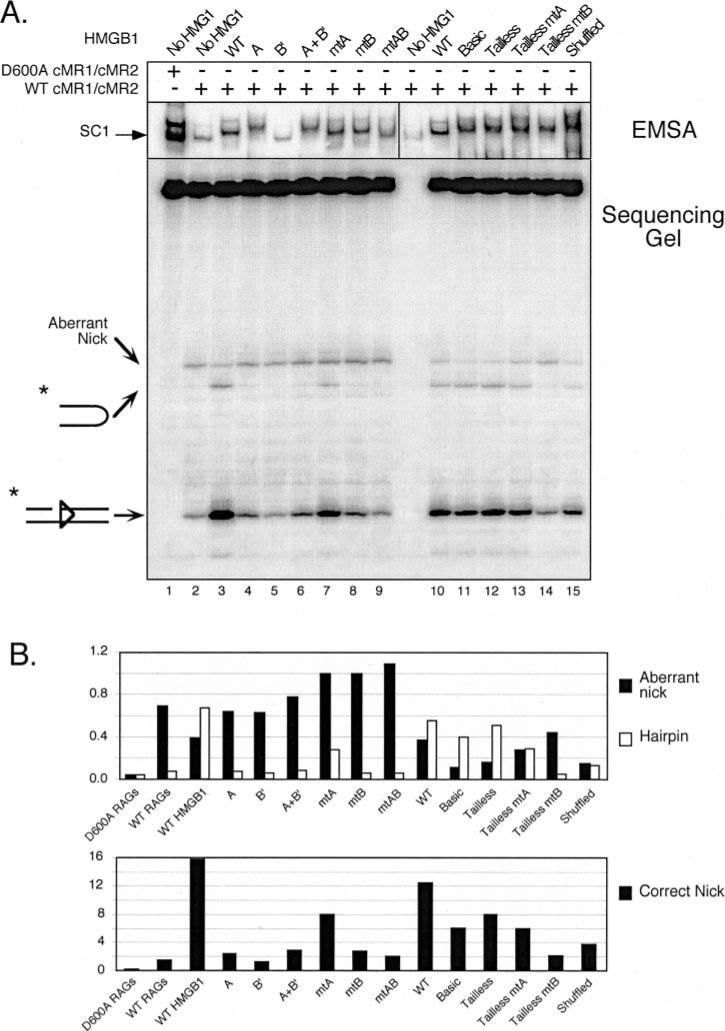
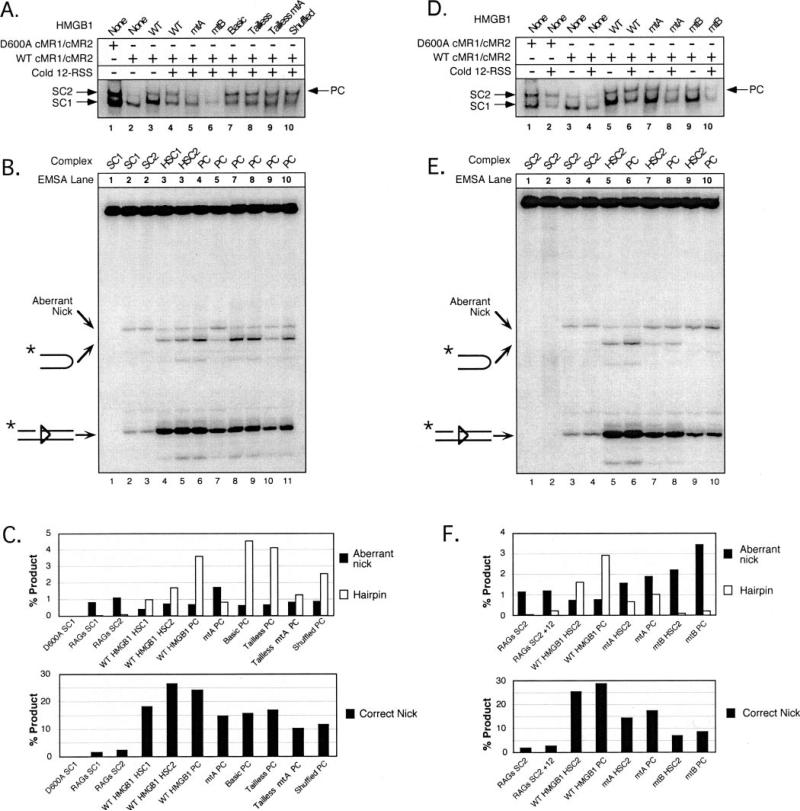
Fig. 4. Stimulation of RAG-mediated 23-RSS cleavage by HMGB1 is primarily mediated by box B, but requires a tethered box A.
A, in-gel cleavage assay of RAG-RSS complexes super-shifted by different forms of HMGB1. A wild-type or inactive RAG complex (WT cMR1/cMR2 and D600A cMR1/cMR2, respectively) was incubated with radiolabeled intact 23-RSS either alone or with various forms of HMGB1 (indicated above the gel) in preparative binding reactions containing Ca2+ and protein-DNA complexes were fractionated by EMSA. After electrophoresis, gels were submerged in buffer containing Mg2+ for 1 h at 37 °C to initiate cleavage and then the DNA was transferred to DEAE cellulose paper. Reaction products recovered from complexes of interest (identified by autoradiography; see exposure at the top) were then analyzed on a 15% polyacrylamide sequencing gel (bottom). Positions of nicked, hairpin, and aberrantly nicked products are indicated at the left. Note that samples from lanes 1–9 and 10 –15 were obtained from different in-gel cleavage assays (given space limitations for the EMSA), but all reaction products were fractionated on the same sequencing gel. B, quantification of reaction products. The abundance of aberrant nicks and hairpins (top bar graph) or correctly positioned nicks (bottom bar graph) are shown for each lane depicted in A and presented in the same order. The data are representative of independent experiments.
Consistent with previous results (24), a RAG-RSS SC1 complex supershifted by wild-type, full-length HMGB1 catalyzes more site-specific nicking and hairpin formation, while exhibiting less aberrant nicking in the spacer region of the 23-RSS, than an unshifted SC1 complex. Despite its ability to supershift the SC1 complex, box A alone does not appreciably stimulate nicking or hairpin formation when present in this complex. Because box B′ containing the acidic tail does not supershift the SC1 complex, it is perhaps not surprising that the complex formed in the presence of this form of HMGB1 does not stimulate cleavage. Interestingly, the supershifted SC1 complex formed in the presence of both box A and box B′ proteins exhibits no more activity than the SC1 complex supershifted by box A alone. This observation holds true regardless of whether or not the acidic tail is present on box B (data not shown).
When incorporated into the SC1 complex, mtA HMGB1 domain stimulates nicking and hairpin formation, albeit to lower levels than WT HMGB1. However, in contrast to WT HMGB1, there is a small increase in the abundance of the aberrantly nicked product. Perhaps more importantly, and in marked contrast with the mtA HMGB1, an SC1 complex supershifted with mtB HMGB1 is no more active than an unshifted SC1 complex in the catalysis of appropriately sited nicks and DNA hairpins. However, similarly to mtA HMGB1, a modest increase in aberrant nicks is reproducibly observed in this complex. This effect is also observed when both HMG-box domains are mutated.
We next compared the activity of SC1 complexes super-shifted by wild-type or mutant forms of HMGB1 lacking the basic linker and/or the acidic tail. We find that basic HMGB1 stimulates RAG-mediated cleavage of a 23-RSS as well as WT HMGB1. Further truncation of the basic linker does not significantly alter the activity of HMGB1 in this regard. We find that tailless mtA promotes more RAG-mediated cleavage and less aberrant nicking than its full-length counterpart in this assay, but tailless and full-length mtB HMGB1 are comparably defective in stimulating 23-RSS cleavage by the RAG complex. Interestingly, shuffled HMGB1 stimulates RAG-mediated nicking and hairpin formation about half as well as tailless HMGB1, suggesting that the orientation of the HMG boxes relative to one another is not a critical factor for HMGB1 to stimulate RAG-mediated cleavage of a single RSS.
Both HMG-box Domains and the Acidic Tail Collaborate to Promote 12/23-regulated Synapsis and Cleavage of RSS Pairs by the RAG Complex
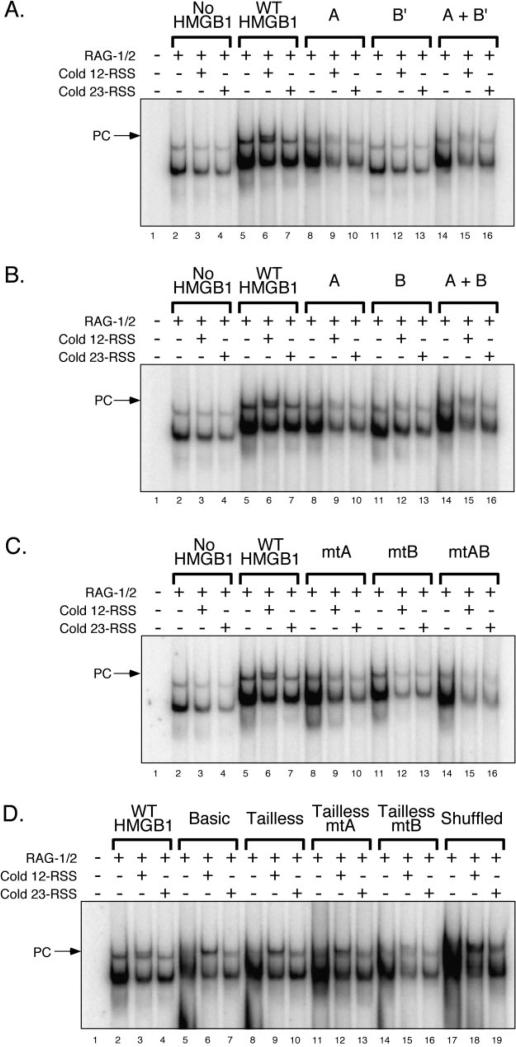
Having characterized the activity of the various truncated and mutant forms of HMGB1 in stimulating RAG binding and cleavage of an intact 23-RSS, we next wished to determine whether they promoted 12/23-regulated synapsis and cleavage of RSS pairs by the RAG complex. In the first set of experiments, the RAG complex was incubated with a radio-labeled 23-RSS substrate in the presence of various forms of HMGB1 with or without cold 12- or 23-RSS partner DNA, and RAG synaptic complex formation was analyzed by EMSA (see “Experimental Procedures”). As expected from previous studies (18), the addition of cold 12- or 23-RSS partner DNA to RAG complexes assembled on a radiolabeled 23-RSS in the absence of HMGB1 reduced the abundance of both SC1 and SC2 complexes. In the presence of WT HMGB1, however, addition of cold 12-RSS partner, but not 23-RSS partner, promoted formation of a RAG-RSS complex whose mobility is slightly retarded relative to the SC2 complex supershifted by WT HMGB1 alone (hereafter termed HSC2) (Fig. 5A). This higher order protein-DNA complex is designated a paired complex (PC) because its formation is regulated by the 12/23 rule. Interestingly, box A alone modestly promotes 12/23-regulated PC formation by the RAG complex, despite its inability to stimulate RAG-mediated cleavage of an isolated 23-RSS. In contrast, box B′ fails to stimulate PC formation, but box B, which lacks the acidic tail, is capable of promoting PC assembly similarly to box A (compare Fig. 5, A and B). Moreover, paired complexes assembled in the presence of box A mixed with either form of box B (tailed or tailless) resemble those formed in the presence of HMG-box A alone (compare Fig. 5, A and B).
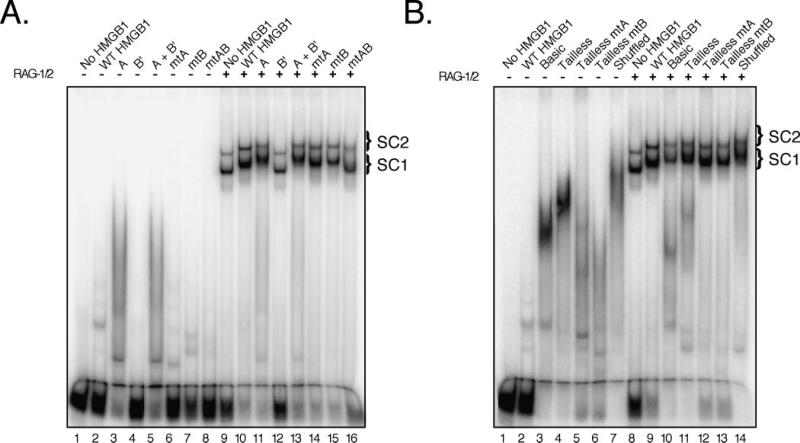
Fig. 5. The acidic tail of HMGB1 helps enforce 12/23-regulated synapsis of RSS pairs by the RAG complex.
A–D, RAG complex was incubated with radiolabeled intact 23-RSS in the absence or presence of various forms of HMGB1 (see Fig. 3 for amounts) with or without added cold 12- or 23-RSS partner DNA in the combinations indicated above the gel. Protein-DNA complexes were fractionated by EMSA and visualized using a phosphorimager.
Compared with WT HMGB1, mutant forms of full-length HMGB1 all failed to promote PC formation (Fig. 5C). Removal of the acidic tail with or without additional truncation of the basic linker facilitated 12/23-regulated PC formation (Fig. 5D). Interestingly, in contrast to their full-length counterparts, tailless mtA and mtB HMGB1 stimulate 12/23-regulated PC formation (compare Fig. 5, C and D). Whether shuffled HMGB1 supports 12/23-regulated PC formation remains somewhat ambiguous, as the higher order protein-DNA complexes formed in the presence of cold 12- or 23-RSS partner are of similar mobility and are not well-resolved from complexes formed in the absence of cold partner RSS. Nevertheless, like the PCs assembled in the presence of tailless HMGB1, the abundance of the PC-like complex is greater in reactions containing 12-RSS partner than in samples containing 23-RSS partner (Fig. 5D).
Using an in-gel cleavage assay, we next determined whether various forms of HMGB1 stimulate cleavage similar to WT HMGB1 in the context of the PC (Fig. 6). Because the individual HMG box domains, either alone or in combination, failed to stimulate RAG-mediated cleavage of a single 23-RSS in the context of the supershifted SC1 complex, we focused our efforts on characterizing the activity of PCs formed in the presence of tailless or mutant forms of HMGB1. One difficulty we encountered with analyzing the activity of the PC using the in-gel cleavage assay is that, unlike analytical EMSAs, the PC could not be unambiguously separated from the HSC2 complex after fractionation by preparative EMSA and subsequent electro-phoretic transfer to DEAE cellulose (Fig. 6A). Thus, the cleavage products observed may be contributed from both complexes, and any stimulation observed in the complex may therefore under-represent the true level of PC activity. This issue may be more important for the PC formed in the presence of full-length HMGB1 than for the truncated versions of HMGB1 analyzed in this assay, as little of the HSC2 complex is observed when the PC is assembled in the presence of the basic and tailless forms of HMGB1 (Fig. 5D). Despite these limitations, we readily detect enhanced cleavage activity in the PC formed in the presence of full-length HMGB1 relative to HSC2 (Fig. 6B). Consistent with previous results, the PC assembled with mtA and mtB HMGB1 failed to promote cleavage to levels observed with WT HMGB1, regardless of whether the acidic tail was present or absent (Fig. 6, B and E). The PCs formed with basic or tailless HMGB1 possessed similar to slightly higher cleavage activity than the PC assembled with full-length HMGB1. Given the ambiguity about whether shuffled HMGB1 supports 12/23-regulated PC formation, it is interesting to note that the complex formed with shuffled HMGB1 that comigrates with the PC formed in the presence of tailless HMGB1 exhibits greater cleavage activity than the HSC2 complex formed with full-length HMGB1, but less than the PCs formed with full-length or tailless versions of HMGB1. These data support the conclusion that shuffled HMGB1 can promote PC formation by the RAG complex. This conclusion is additionally strengthened by the observation that a PC assembled on a radiolabeled 23-RSS substrate in the presence of shuffled HMGB1 and cold 12-RSS partner is about 4-fold more active than its counterpart assembled in the presence of cold 23-RSS partner, as assessed using an in-gel cleavage assay (data not shown). This level of stimulation is similar to that observed with tailless HMGB1 when analyzed in the same gel, further suggesting that shuffled HMGB1 can promote RAG-mediated cleavage according to the 12/23 rule.
Fig. 6. Optimal 12/23-regulated RSS cleavage by the RAG complex requires both HMG-box domains in the correct orientation, but not the basic linker or acidic tail.
Preparative binding reactions containing Ca2+ and either a wild-type or inactive RAG complex (WT cMR1/cMR2 and D600A cMR1/cMR2, respectively) were incubated with radiolabeled intact 23-RSS in the absence or presence of various forms of HMGB1 with or without added cold 12- or 23-RSS partner DNA in the indicated combinations. Protein-DNA complexes were fractionated by EMSA, and their cleavage activity assessed using an in-gel cleavage assay as described in the legend to Fig. 4. A and D, autoradiographic exposure of DEAE paper after transfer of reaction products. B and E, reaction products fractionated on a sequencing gel. C and F, quantification of the three major cleavage products. Data are displayed as described in the legend to Fig. 4 and are representative of independent experiments.
DISCUSSION
HMGB1 Determinants for Stimulation of RAG-mediated RSS Binding and Cleavage
Earlier studies demonstrated that HMGB1 stimulates RAG-mediated binding and cleavage of isolated 23-RSS substrates, and promotes RAG-mediated synapsis and cleavage of RSS pairs according to the 12/23 rule (8, 18, 24). To determine what structural features of HMGB1 are necessary to promote RSS binding and cleavage by the RAG proteins, we prepared a large panel of truncated and mutant forms of HMGB1, and systematically characterized their activity in assays of RAG-RSS complex formation and function in vitro. In principle, the ability of HMGB1 to stimulate RAG-mediated RSS cleavage could be simply because of its capacity to promote RAG-RSS complex formation. To address this possibility, we have used in-gel cleavage assays rather than intube assays to specifically evaluate the catalytic activity of RAG-RSS complexes supershifted by incorporation of the various HMGB1 proteins prepared for this study. A summary of how the various HMGB1 proteins examined in this study modulate the binding and cleavage activity of the RAG complex is provided in Table I.
Table I.
Summary of activity of HMGB1 proteins in RAG binding and cleavage assays
| HMGB1 protein | Single RSS bindinga | Supershift of SC1 and/or SC2b | RSS cleavage (MT SC1/WT SC1)c |
||
|---|---|---|---|---|---|
| Nick | Hairpin | Ab nick | |||
| WT | +; 150 ng | +(SC1/SC2) | 1 | 1 | 1 |
| A | + + +; 50 ng | +(SC1/SC2) | 0.15 | 0.10 | 1.64 |
| B′ | –; 150 ng | – (up to 640 ng) | 0.07 | 0.09 | 1.62 |
| B | ND | +(SC1/SC2) | |||
| mtA | +; 150 ng | +(SC1/SC2) | 0.50 | 0.42 | 2.56 |
| mtB | +; 150 ng | +(SC1/SC2) | 0.17 | 0.09 | 2.56 |
| mtAB | –/+; 150 ng | –/+ (SC1), + (SC2) | 0.12 | 0.09 | 2.79 |
| Basic | + + +; 50 ng | +(SC1/SC2) | 0.30 | 0.73 | 0.48 |
| Tailless | + + +; 50 ng | +(SC1/SC2) | 0.62 | 0.93 | 0.43 |
| Tailless mtA | + +; 50 ng | +(SC1/SC2) | 0.48 | 0.53 | 0.76 |
| Tailless mtB | + +; 50 ng | +(SC1/SC2) | 0.16 | 0.09 | 1.19 |
| Shuffled | + + +; 50 ng | +(SC1/SC2) | 0.07 | 0.11 | 1.57 |
| HMGB1 protein | PC formationd | RSS cleavage (MT PC/WT PC)e |
||
|---|---|---|---|---|
| Nick | Hairpin | Ab nick | ||
| WT | + + | 1 | 1 | 1 |
| A | + (weak) | ND | ND | ND |
| B′ | – | ND | ND | ND |
| B | + (weak) | ND | ND | ND |
| mtA | – | 0.60 | 0.23 | 2.49 |
| mtB | – | 0.30 | 0.07 | 4.55 |
| mtAB | – | ND | ND | ND |
| Basic | + + | 0.64 | 1.26 | 0.91 |
| Tailless | + + | 0.7 | 1.15 | 0.96 |
| Tailless mtA | + + | 0.41 | 0.35 | 1.20 |
| Tailless mtB | + | ND | ND | ND |
| Shuffled | + | 0.47 | 0.71 | 1.26 |
Binding to 23-RSS in the absence of RAGs at indicated amounts of HMGB1 protein using EMSA (Fig. 3); –, no detectable binding; –/+, weak binding; +, binding evident, but little oligomerization; + +, moderate oligomerization; + + +, extensive oligomerization. ND, not done.
Ability to supershift SC1 and/or SC2 at amounts of HMGB1 protein used to detect single RSS binding (Fig. 3); –, no supershift detected (up to indicated amount of HMGB1 protein); –/+, impaired supershift; +, supershift comparable to WT.
Abundance of cleavage products isolated from SC1 complex using in-gel cleavage assay relative to WT HMGB1 (Fig. 4).
Ability to support PC formation detectable by EMSA (Fig. 5). –, no PC formation detected; +, PC formation detectable, but bands diffuse or weak; + +, PC clearly evident.
Abundance of cleavage products isolated from PC using in-gel cleavage assay relative to WT HMGB1 (Fig. 6). ND, not done.
We find that individual HMG-box domains (not B′) super-shift all RAG-RSS complexes and promote 12/23-regulated syn-apsis of RSS pairs, but fail to stimulate DNA cleavage when incorporated into RAG complexes assembled on an isolated 23-RSS (HSC1), even when both HMG box domains are present during RAG-RSS complex assembly. Thus, to stimulate RAG-mediated cleavage in these complexes, the HMG-box domains must be physically coupled to one another. This observation raises the possibility that the two HMG-box domains interact with separate portions of the RAG proteins and/or the RSS, bridging the two regions and bringing them into close proximity.
We also show that forms of full-length HMGB1 bearing multiple alanine substitutions in HMG-box A or B (or both) super-shift RAG-RSS complexes assembled with an individual 23-RSS, but exhibit either a partial impairment (mtA) or almost a complete loss (mtB or mtAB) of their ability to stimulate RAG-mediated 23-RSS cleavage. Because the B domain is capable of introducing a severe bend in linear DNA, whereas the A domain lacks this ability (10, 14), the evidence presented here strongly suggests that the main mechanism by which HMGB1 facilitates RAG-mediated nicking and hairpin formation is by promoting or stabilizing a bent DNA structure in the 23-RSSRAG complex. The bending activity mediated by the B domain may enable the active site of the RAG complex to remain stably associated with the 23-RSS at the heptamer-coding junction after nicking. However, while bending of the 23-RSS may be sufficient to promote enhanced RAG-mediated cleavage, the efficiency of this process is clearly perturbed by mutations in box A. A plausible explanation for this observation, given other studies suggesting that box A preferentially recognizes distorted DNA structures (13), is that conformational changes introduced into the heptamer-coding junction by the RAG proteins are recognized and stabilized by box A, possibly involving protein-protein interactions with the RAGs near the active site. Alternatively, or in addition, the protein-DNA interactions between box-A and the heptamer-coding junction may further promote DNA flexure imposed through box B and the RAG complex (14). These data, taken together with evidence from a previous study suggesting RAG-mediated RSS cleavage occurs in trans (23), and pull-down assays suggesting that both HMG-box domains associate with the nonamer binding domain of RAG-1 (NBD; using residues 377–477) (28), raises the intriguing possibility that HMG-box A and B may interact with the NBD of RAG-1 bound to the nonamer of one RSS, with the A domain further interacting with the active site domain bound to heptamer-coding junction of the other RSS (for a speculative model, see Ref. 29). Tethering of the two HMG-box domains may be required to provide the necessary “pull” between the active site and the NBD, with their associated protein-DNA interactions, to guide proper active site placement, and enforce conformational changes necessary to promote 23-RSS cleavage by the RAG complex. The finding that mtA, mtB, and mtAB HMGB1, unlike WT HMGB1, all modestly promote aberrant nicking by the RAG complex is not inconsistent with this model, as increased aberrant nicking could be attributed to either poor bending that impairs the correct positioning of the heptamer-coding junction proximal to the RAG active site (e.g. mtB), or the loss of protein-protein and/or protein-DNA interactions near the heptamer-coding junction that stabilize the association of the catalytic core of the RAG complex at the appropriate cleavage site (e.g. mtA).
Full-length box A and box B mutants have a profound defect in promoting 12/23-regulated synapsis of RSS pairs by the RAG complex, but are nevertheless capable of modestly stimulating aberrant nicking (mtA and mtB) or cleavage (mtA) of a 23-RSS in RAG-RSS complexes assembled in the presence of cold 12-RSS partner. Surprisingly, box A and box B mutants lacking the acidic tail retain the ability to promote 12/23-regulated synapsis of RSS pairs by the RAG complex. Although the PC formed with tailless mtA exhibits slightly greater cleavage activity and less aberrant nicking than the PC assembled with its full-length counterpart, we cannot determine whether this enhanced activity represents an intrinsic property of the tailless mtA PC, or whether it rather reflects a greater abundance of the synaptic complex relative to the contaminating unshifted HSC2 complex in the PC, which cannot be uniquely separated using the in-gel cleavage assay. The observation that the loss of the acidic tail promotes oligomerization on RAG-RSS complexes and enables mtA and mtB HMGB1 to bypass the normal constraints that inhibit their ability to mediate 12/23-regulated synapsis suggests that the acidic tail may play an active role in enforcing the 12/23 rule, perhaps either by generally reducing nonspecific protein-DNA interactions through charge repulsion of the DNA, or by specifically promoting and/or stabilizing interactions that facilitate 12- and 23-RSS discrimination by the RAG complex through contacts between the acidic tail and the HMG box domains (15, 30). The acidic tail of HMGB1 may play additional roles in regulating RSS accessibility by promoting nucleosome sliding by chromatin remodeling factors (16, 17), which may help reposition nucleosomes away from the RSS where they may preferentially reside (31, 32).
A surprising finding is that shuffled HMGB1 stimulates RAG-mediated cleavage of isolated and paired RSS substrates almost as well as its normally oriented counterpart, despite poor resolution of the protein-DNA complexes. This finding might seem to contradict the model proposed above in which the two HMG-box domains are suggested to interact with different parts of the RSS, because it is hard to imagine how the normally oriented and inverted di-domain HMGB1 proteins could similarly interact with the RSS in these RAG complexes. However, at least three factors complicate this interpretation. First, the stoichiometry of HMGB1 in these complexes is unknown; thus, there may be enough conformational flexibility within a multimeric HMGB1-RAG complex for a given HMG-box domain to function appropriately. Second, RAG-1 binds the RSS as a dimer (18, 33); thus, interactions with a given RSS by HMGB1 could potentially be facilitated through protein-protein contacts with either one of the RAG-1 subunits. Third, the orientation of the RAG-1 subunits relative to one another when bound to DNA, and their conformational flexibility within such complexes, remains unclear. These shortcomings, taken together with the known propensity of the RAG complex to bend DNA (28), raises the possibility that although HMGB1 would seem incapable of bridging heptamer and nonamer in a linear model of the RSS, the two elements may be close enough in space, particularly when a trans mode of RSS cleavage is considered, for normally oriented and inverted HMGB1 di-domains to function similarly in these assays. Nevertheless, we cannot completely rule out an alternative possibility that the two HMG-box domains are functionally independent and essentially equivalent in their ability to promote RAG activity as long as they are linked together. In this model, the protein-protein and protein-DNA interactions mediated by each of the two HMG-boxes in the RAG-RSS-HMGB1 complex can be complemented by one another, at least partially, and that the stimulation of RAG activity achieved through these interactions is largely driven by avidity. However, one reason we disfavor this idea is that shuffled HMGB1 does not support 12/23-regulated synapsis and cleavage as well as its normally oriented counterpart, which provides evidence against domain redundancy and illuminates a direct role for HMG-box domain organization in mediating RSS discrimination and cleavage by the RAG complex. Ultimately, structural studies will be required to resolve these issues.
Comparative Analysis of HMGB1 Function in V(D)J Recombination, Sleeping Beauty Transposition, and Adeno-associated Viral DNA Integration
HMGB1 is an abundant non-histone architectural DNA-binding protein that has been identified as an important cofactor in the assembly of nucleoprotein complexes involved in DNA transcription, replication, recombination, and repair (9, 10). In many cases, HMGB1 appears to promote these cellular processes by directly interacting with the requisite sequence-specific DNA-binding protein. This mode of interaction has been described for p53 (34), the TATA-binding protein (19), and progesterone receptor (35, 36), among others. However, in other cases, most notably the ZEBRA and RTA viral transactivators, HMGB1 acts indirectly to promote nucleoprotein complex formation, most likely by mediating DNA bending without requiring stable association with a sequence-specific DNA-binding protein (26, 37). In addition to facilitating assembly of nucleoprotein complexes, HMGB1 is also known to stimulate the activity of enzymes that catalyze DNA strand cleavage and strand transfer reactions. Three known examples include the RAG complex involved in V(D)J recombination (8, 18, 24, 28, 38), the engineered Tc1-like transposase that mediates Sleeping Beauty transposition (SB transposase) (39), and the Rep protein that initiates site-specific integration of adeno-associated virus DNA (40). In all three cases, the available evidence suggests that HMGB1 is capable of directly interacting with the sequence-specific DNA-binding protein(s), stimulating the DNA binding activity of the involved protein(s), and becoming incorporated into ternary protein-DNA complexes containing the sequence-specific DNA-binding protein(s). However, whereas ternary complexes formed with Rep or the RAG proteins are stable to electrophoresis, as indicated by the appearance of one or more supershifted protein-DNA complexes using an EMSA, ternary complexes containing the SB transposase are not, despite their detection by immunoprecipitation. Moreover, HMGB1 has been shown to stimulate the activity of all three proteins in vivo (8, 18, 24, 28, 38).
In vitro assays of strand cleavage and/or strand transfer conducted without fractionation of protein-DNA complexes demonstrate that HMGB1 augments the activity of the RAG complex, Rep, and the SB transposase (8, 39, 40). In principle, the enhanced activity observed under these conditions could be attributed solely to an increased abundance of active protein-DNA complexes, whose assembly is facilitated by HMGB1. Alternatively, protein-DNA complexes containing HMGB1 may be intrinsically more active than comparable counterparts lacking HMGB1. In the case of Rep and the SB transposase, whether protein-DNA complexes formed in the presence of HMGB1, supershifted or otherwise, are more active in the catalysis of strand cleavage than counterparts assembled in the absence of HMGB1 remains largely untested. Using in-gel cleavage assays, we have shown in previous studies (18, 24), and again in this work, that RAG-RSS complexes supershifted with HMGB1 (or HMGB2) are more active than comparable complexes lacking HMGB1. It is tempting to speculate that, like the RAG complex, HMGB1 may play a direct role in facilitating DNA cleavage by the Rep protein and the SB transposase that is independent of its ability to promote DNA binding by these proteins.
Supplementary Material
Acknowledgments
We thank Sandor Lovas for assistance in performing circular dichroism and mass spectroscopy experiments, which were conducted with support from the INBRE Program of the National Institutes of Health National Center for Research Resources (1P20RR16469). This investigation was conducted in a facility constructed with support from the Research Facilities Improvement Program of the National Institutes of Health National Center for Research Resources (C06 RR17417-01).
Footnotes
This work was supported by Grant RSG-01-020-01-CCE and Grant R01 AI055599 from the National Institutes of Health (to P. C. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental materials.
The abbreviations used are: RAG, recombination-activating gene; DSB, double-strand break; RSS, recombination signal sequence; DNAPKcs, DNA-dependent protein kinase catalytic subunit; NHEJ, nonhomologous end-joining; HMG, high mobility group; MBP, maltose-binding protein; IMAC, immobilized metal affinity chromatography; EMSA, electrophoretic mobility shift assay; LAR-PCR, ligation-assisted recombination polymerase chain reaction; IDA, iminodiacetate; IEC, ion exchange chromatography; WT, wild type; SB, sleeping beauty; PC, paired complex.
REFERENCES
- 1.Bassing CH, Swat W, Alt FW. Cell. 2002;109(suppl.):S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 2.Roth DB, Zhu C, Gellert M. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10788–10792. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roth DB, Menetski JP, Nakajima PB, Bosma MJ, Gellert M. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 4.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 5.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 6.van Gent DC, Mizuuchi K, Gellert M. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 7.Ma Y, Pannicke U, Schwarz K, Lieber MR. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 8.van Gent DC, Hiom K, Paull TT, Gellert M. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bustin M. Mol. Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas JO, Travers AA. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 11.Webb M, Thomas JO. J. Mol. Biol. 1999;294:373–387. doi: 10.1006/jmbi.1999.3150. [DOI] [PubMed] [Google Scholar]
- 12.Kasparkova J, Delalande O, Stros M, Elizondo-Riojas MA, Vojtiskova M, Kozelka J, Brabec V. Biochemistry. 2003;42:1234–1244. doi: 10.1021/bi026695t. [DOI] [PubMed] [Google Scholar]
- 13.Teo SH, Grasser KD, Thomas JO. Eur. J. Biochem. 1995;230:943–950. doi: 10.1111/j.1432-1033.1995.tb20640.x. [DOI] [PubMed] [Google Scholar]
- 14.Stros M. J. Biol. Chem. 1998;273:10355–10361. [PubMed] [Google Scholar]
- 15.Knapp S, Muller S, Digilio G, Bonaldi T, Bianchi ME, Musco G. Biochemistry. 2004;43:11992–11997. doi: 10.1021/bi049364k. [DOI] [PubMed] [Google Scholar]
- 16.Ueda T, Chou H, Kawase T, Shirakawa H, Yoshida M. Biochemistry. 2004;43:9901–9908. doi: 10.1021/bi035975l. [DOI] [PubMed] [Google Scholar]
- 17.Bonaldi T, Langst G, Strohner R, Becker PB, Bianchi ME. EMBO J. 2002;21:6865–6873. doi: 10.1093/emboj/cdf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swanson PC. Mol. Cell. Biol. 2002;22:7790–7801. doi: 10.1128/MCB.22.22.7790-7801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge H, Roeder RG. J. Biol. Chem. 1994;269:17136–17140. [PubMed] [Google Scholar]
- 20.Swanson PC, Volkmer D, Wang L. J. Biol. Chem. 2004;279:4034–4044. doi: 10.1074/jbc.M311100200. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Swanson P, Desiderio S. Mol. Cell. Biol. 1997;17:6932–6939. doi: 10.1128/mcb.17.12.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swanson PC, Desiderio S. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 23.Swanson PC. Mol. Cell. Biol. 2001;21:449–458. doi: 10.1128/MCB.21.2.449-458.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swanson PC. Mol. Cell. Biol. 2002;22:1340–1351. doi: 10.1128/mcb.22.5.1340-1351.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stros M, Muselikova E. J. Biol. Chem. 2000;275:35699–35707. doi: 10.1074/jbc.M007167200. [DOI] [PubMed] [Google Scholar]
- 26.Mitsouras K, Wong B, Arayata C, Johnson RC, Carey M. Mol. Cell Biol. 2002;22:4390–4401. doi: 10.1128/MCB.22.12.4390-4401.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee KB, Thomas JO. J. Mol. Biol. 2000;304:135–149. doi: 10.1006/jmbi.2000.4206. [DOI] [PubMed] [Google Scholar]
- 28.Aidinis V, Bonaldi T, Beltrame M, Santagata S, Bianchi ME, Spanopoulou E. Mol. Cell Biol. 1999;19:6532–6542. doi: 10.1128/mcb.19.10.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swanson PC. Immunol. Rev. 2004;200:90–114. doi: 10.1111/j.0105-2896.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- 30.Ramstein J, Locker D, Bianchi ME, Leng M. Eur. J. Biochem. 1999;260:692–700. doi: 10.1046/j.1432-1327.1999.00185.x. [DOI] [PubMed] [Google Scholar]
- 31.Baumann M, Mamais A, McBlane F, Xiao H, Boyes J. EMBO J. 2003;22:5197–5207. doi: 10.1093/emboj/cdg487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon J, Imbalzano AN, Matthews A, Oettinger MA. Mol. Cell. 1998;2:829–839. doi: 10.1016/s1097-2765(00)80297-x. [DOI] [PubMed] [Google Scholar]
- 33.Swanson PC, Desiderio S. Mol. Cell Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jayaraman L, Moorthy NC, Murthy KG, Manley JL, Bustin M, Prives C. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi ME, Taraseviciene L, Nordeen SK, Allegretto EA, Edwards DP. Mol. Cell Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Onate SA, Prendergast P, Wagner JP, Nissen M, Reeves R, Pettijohn DE, Edwards DP. Mol. Cell Biol. 1994;14:3376–3391. doi: 10.1128/mcb.14.5.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellwood KB, Yen YM, Johnson RC, Carey M. Mol. Cell Biol. 2000;20:4359–4370. doi: 10.1128/mcb.20.12.4359-4370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mo X, Bailin T, Noggle S, Sadofsky MJ. Nucleic Acids Res. 2000;28:1228–1236. doi: 10.1093/nar/28.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zayed H, Izsvak Z, Khare D, Heinemann U, Ivics Z. Nucleic Acids Res. 2003;31:2313–2322. doi: 10.1093/nar/gkg341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costello E, Saudan P, Winocour E, Pizer L, Beard P. EMBO J. 1997;16:5943–5954. doi: 10.1093/emboj/16.19.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.