Abstract
Common genome-editing strategies are either based on non-homologous end joining (NHEJ) or, in the presence of a template DNA, based on homologous recombination with long (homology-directed repair [HDR]) or short (microhomology-mediated end joining [MMEJ]) homologous sequences. In the current study, we aim to develop a model system to test the activity of MMEJ after CRISPR/Cas9-mediated cleavage in cell culture. Following successful proof of concept in an episomally based reporter system, we tested template plasmids containing a promoter-less luciferase gene flanked by microhomologous sequences (mhs) of different length (5, 10, 15, 20, 30, and 50 bp) that are complementary to the mouse retinitis pigmentosa GTPase regulator (RPGR)-ORF15, which is under the control of a CMV promoter stably integrated into a HEK293 cell line. Luciferase signal appearance represented successful recombination events and was highest when the mhs were 5 bp long, while longer mhs revealed lower luciferase signal. In addition, presence of Csy4 RNase was shown to increase luciferase signaling. The luciferase reporter system is a valuable tool to study the input of the different DNA repair mechanisms in the replacement of large DNA sequences by mhs.
Keywords: genome editing, CRISPR-Cas, MMEJ, XLRP, Csy4
Introduction
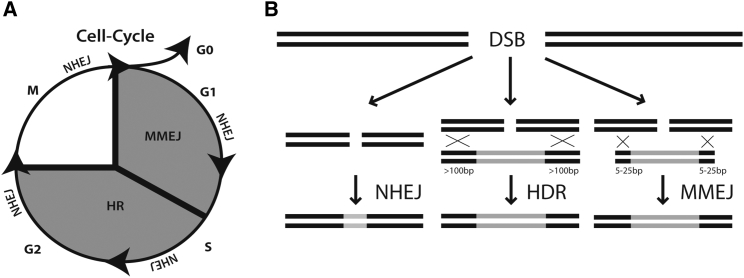
Programmable nucleases, such as TALENs and the CRISPR/Cas system, enable specific genome engineering. A cleavage at the desired genome position activates the endogenous cell repair machinery, which generates various gene modifications.1, 2, 3, 4 Two main repair pathways are well characterized: non-homologous end joining (NHEJ), which is an error-prone mechanism to induce knockouts; and homology-directed repair (HDR) with a defined DNA donor template, which is mainly used for gene knockins and specific gene modifications.5, 6, 7, 8 Gene therapy strategies that are aimed at correcting the disease-causing mutations are currently based on HDR events. While physiologically, the sister chromatid serves as a repair template for HDR during the G2 cell-cycle stage, a targeting vector as a template is used for therapeutic gene-editing strategies. HDR events with exogenous templates have been reported in several cell lines and organisms, with differences concerning their efficiencies and generally being at much lower levels than NHEJ events.9, 10, 11 One of the major challenges in advancing genome editing based on HDR is that this repair pathway is active almost exclusively during the late S/G2 phase, whereas NHEJ, which needs to be avoided in such strategies, is active throughout the cell cycle12 (Figure 1A).
Figure 1.
Double-Strand Break Repair Events
(A) Scheme showing the three preferred repair mechanisms during the cell cycle after DNA double-strand break. The error-prone NHEJ is active during the whole cell cycle. HDR leads to correct repair in late S/G2 phases. The MMEJ repair is active in early M and S phases, and can be used for targeted insertions with small homolog sequences. (B) Scheme of the different repair events highlighting the importance and length of the homologous sequences on the donor template DNA. For HDR, homologous regions of more than 100 bp up to several kbp are needed to edit a given sequence, depending on the size of the sequence to be replaced. For MMEJ, the length of the homologous sequences is about 5–25 bp.
More recently, an alternative NHEJ (A-NHEJ) pathway has been described, which was re-named microhomology-mediated end joining (MMEJ). Short microhomologies (currently considered to be within a 5–25 bp range) flanking a double strand break (DSB) can recombine to each other through this process, resulting in deletions in between the homology arms13, 14 (Figure 1B). MMEJ in combination with targeting vectors can also be used as a powerful technology for precise gene knockins.15, 16 The efficiency is strongly dependent on the length of the microhomologous sequences (mhs); 15 bp and longer was reported to result in a significant number of integration events.17 MMEJ seems to be most active during the M and early S phases in dividing cells,12, 18 and is associated with the presence of HDR- and NHEJ-independent proteins.19, 20 Because this repair pathway is mostly active in a different cell-cycle stage compared with HDR and is mechanistically associated with proteins not involved in HDR or NHEJ, this pathway may represent an alternative approach for developing therapeutic genome-editing applications.
The terminal exon ORF15 of the retinitis pigmentosa GTPase regulator (RPGR) gene, in which over 300 different mutations cause X-linked retinitis pigmentosa (XLRP),21, 22 may represent an ideal target locus to replace a mutant exon by the respective wild-type exon. In a first step, we developed a universal reporter-gene-based system to study MMEJ mechanisms in order to optimize the replacement efficiency of defined DNA sequences within the ORF15 exon. Exploiting the advantages of the CRISPR/Cas system, we were able to induce several DSBs simultaneously to trigger MMEJ.23, 24 Besides this, our reporter system may provide a tool in the future to gain more insights into cell-specific repair efficiencies in combination with different gene therapy strategies.
Results
18-bp Microhomology Is Sufficient to Trigger MMEJ in an Episomal System
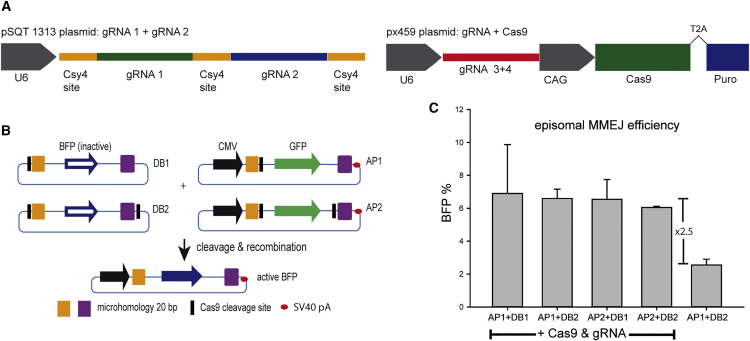
We detected MMEJ events in HEK293 cells in an episomal-based assay (Figure 2). The acceptor plasmid (AP) in the reporter system consists of a GFP gene under the control of a CMV promoter and an SV40 polyadenylation signal flanked by two different 18-bp mhs (Figure 2B). The donor plasmid (DB) harbors identical mhs flanking a blue fluorescent protein (BFP) gene, which is inactive due to the absence of a promoter sequence. Next to the mhs, one (DB1, AP1) or two (DB2, AP2) Cas9 sites are incorporated for cleavage (Figure 2B). These target sites represent two different but therapeutically relevant sequences within the ORF15 of the RPGR gene (Table 1). The guide RNAs (gRNAs 1 and 2) and the Cas9 (px459) sequences were provided as separate plasmids (Figure 2A). The episomal-based assay showed significantly higher BFP levels compared with control samples without induced Cas9 cleavage (Figure 2C). Interestingly, the number of BFP-positive cells was about 6%–7%, regardless of whether one or two Cas9 target sites were included in each vector, indicating that a pathway other than NHEJ was active. If NHEJ would have resulted in the edited gene, the BFP signaling would differ between the combinations AP1+DB1 and AP1+DB2, as the former would have the SV40 polyadenylation signal not in close proximity to the BFP gene following recombination, because of the integration of the entire plasmid backbone (Figure 2B).
Figure 2.
Episomal-Based MMEJ Assay
(A) The plasmids used in this assay. pSQT1313 harbors gRNAs from therapeutic relevant target sequences. The px459 plasmid encodes only the Cas9 protein in the episomal-based assays. For the genomic editing experiments it harbors gRNA3 resp. gRNA4. (B) Schematic overview of plasmid combinations to induce MMEJ recombination events. The donor plasmid (DB) harbors a complete BFP plasmid without promotor. BFP is flanked by 18-bp microhomology regions (orange and purple), and the plasmid can be linearized (DB1) or the insert can be cleaved out (DB2). The AP was generated following the same principle. The Cas9 cleavage occurs in a PAMout manner (cf. Figure 3). (C) Episomal gene-editing efficiencies measure by quadruple transfections of the reporter plasmids. Depicted is the number of BFP-positive cells after DNA repair. Error bars indicate the SD of the data set.
Table 1.
Guide RNA Sequences
| Guide RNA | Sequence 5′→3′ |
|---|---|
| gRNA used in episomal-based assay | TCGAACTTTGGGAAAAATCTGG |
| gRNA used in episomal-based assay and gRNA1 | GTCAGGGATACCAGAGGAGCAGG |
| gRNA g2 | TCCAGAATCGTTCGGAGCCTAGG |
| gRNA g3 | GAGCGCCACCATGGTGAGCAAGG |
| gRNA g4 | GGCCACAAGTTCAGCGTGTCCGG |
List of the gRNA sequences that have been used during the experimentation. Underlined sequences represent the PAM (protospacer adjacent motif) site.
Application to the RPGR ORF15 Exon
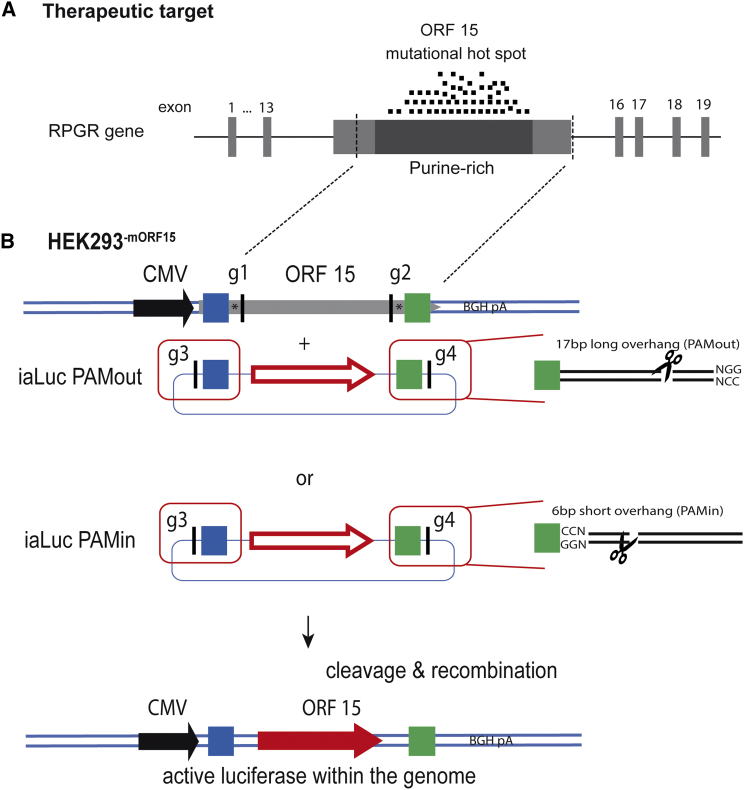
The RPGR gene produces two major isoforms: one being a constitutively expressed form containing exons 1–19; and a second form expressing exons 1–13 and a terminal exon, commonly referred to as ORF15. The ORF15 exon contains 80% of all disease-causing mutations found in XLRP (Figure 3A). Therefore, the replacement of most of the ORF15 sequence would represent a single therapeutic approach for a large number of patients. To study the MMEJ mechanism at the genomic level, we generated a stable HEK293 cell line containing the murine RPGR-ORF15 sequence under the control of a CMV promoter. The DNA sequence, which would serve as mhs, flanking the ORF15 was chosen with a distance of 30 bp to the Cas9 cleavage site. This distance was chosen to avoid indels formation within the mhs, which could be potentially induced by NHEJ activities following DSB induction (Figure 3B). The donor vector contains the luciferase gene lacking promotor sequences flanked by the mhs (length 5, 10, 15, 20, 30, and 50 bp) (Figure 6A). The Cas9 target sites flank the mhs in a way that either a short (6 bp, PAMin) or a long overhang (17 bp, PAMout) results after cleavage (Figure 3B).
Figure 3.
DNA Repair Reporter System in Stable HEK293-mORF15 Cells
(A) X-linked retinitis pigmentosa is associated with mutations in the RPGR gene. The terminal ORF15 exon with its repetitive sequences is a mutational hotspot with almost 300 known disease-causing mutations. A large part from ORF15 has been cloned into HEK293 cells as indicated by dashed lines. (B) Schematic representation of the MMEJ model system. The ORF15 sequence was almost entirely cloned between a CMV promoter and a BGH polyadenylation signal and stable integrated into the HEK293 cell line. Blue and green squares indicate the microhomologous regions (mhs). Vertical black lines indicate the guide RNA target sites g1 to g2. Stars indicate a buffer zone of 30 bp between the gRNA sites and the mhs. The template plasmid contains the luciferase cDNA flanked by two mhs and gRNA target sites 3 and 4. The orientation is either PAMout, when the polyspacer adjacent motif (PAM) is 17 bp away from the gRNA target site, or PAMin, when the PAM is only 6 bp away. iaLuc PAMout, inactive luciferase template PAMout; iaLuc PAMin, inactive luciferase template PAMin. Upon cleavage and recombination, the luciferase cDNA is integrated at the ORF15 locus, and the CMV promoter drives transgene expression.
Figure 6.
Efficacy of Luciferase Integration in Dependence of the Length of the Microhomologous Sequences in the Template Plasmid
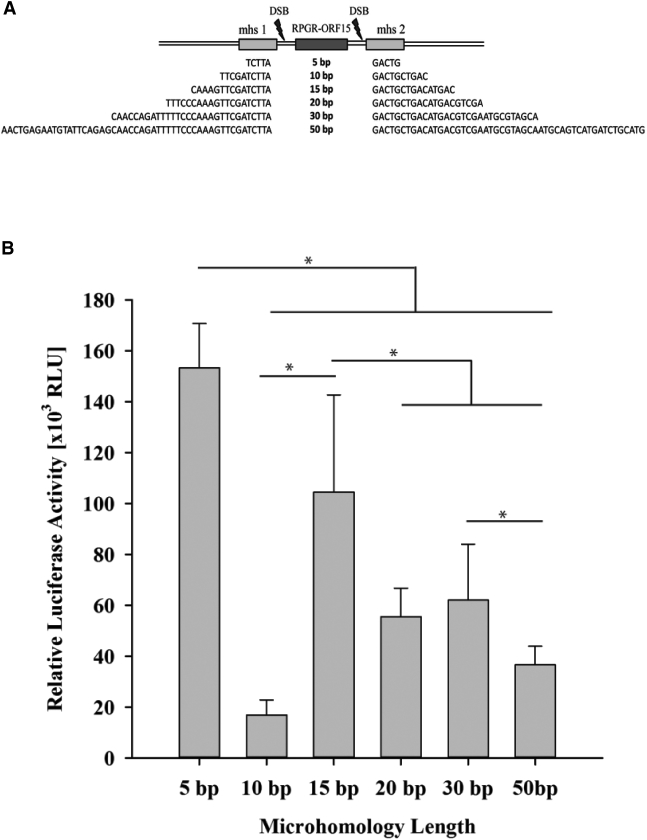
(A) Schematic representation of the different mhs sequences supplied with the luciferase donor templates iaPAMout. (B) Quantification of luciferase integration following mhs-mediated DSB repair. Different mhs length results in significantly different RLU levels, with 5 bp mhs being by far the most efficient variant. *p < 0.001 (Holm-Sidak pairwise multiple comparison). Error bars indicate the SD of the data set.
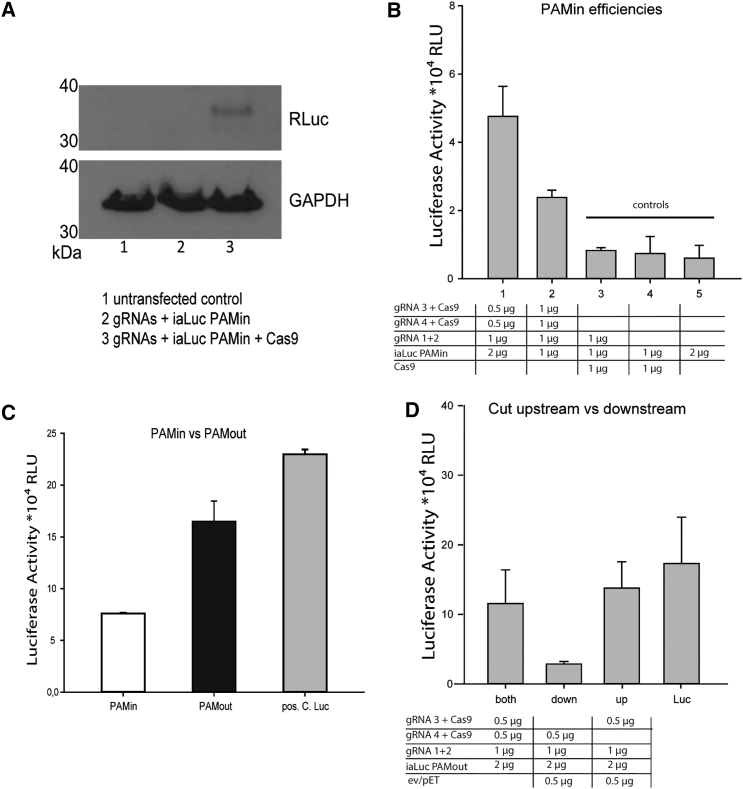
The luciferase expression was verified 48 hr following transfection (Figure 4A). A band at the expected size was visible only in the lane with cell lysate that was transfected with gRNAs, template, and Cas9 protein. By using the PAMin variant, 20-bp mhs length, and different ratios of the plasmids, we were able to show five times higher luciferase signal compared with control samples (Figure 4B), indicating that DSB repair takes place at the site. Interestingly, our data show that a DSB in the donor template is a prerequisite for recombination events to take place, because the control experiment with donor vector, Cas9, and gRNAs 1 and 2 (only cutting within the target genome) did not show any increased signal compared with donor template transfection alone. This could indicate NHEJ being the major repair mechanism. Following this, we compared the PAMin variant with the highest luciferase signal to the respective PAMout variant, again with 20-bp mhs length. We observed that the luciferase signal increased by 100% (Figure 4C). This observation, in turn, indicates that NHEJ is not the major repair mechanism because luciferase signal strength would not have been dependent on the position of cleavage.
Figure 4.
Quantification of the Genome-Editing Activity by Using the Relative Luciferase Activity Measurement
(A) Luciferase expression was verified by western blot with a luciferase-specific antibody. Only in lane 3 with gRNA, template, and Cas9 protein expression, a light luciferase band was visible. (B) RLU measured as a function of different combinations of gRNAs, Cas9 expression plasmid, and PAMin template DNA. Controls lack one of the necessary plasmids. (C) Comparison of PAMin versus PAMout template plasmid. The PAMout template plasmid results in 100% higher luciferase signal. (D) Influence of the DSB introduction either upstream, downstream, or at both sides of the template plasmid. The DSB induced by g3 RNA is more important than the one induced by the g4 RNA. All experiments were performed at least in triplicates. Error bars indicate the SD of the data set.
The 5′ Cleavage Is More Important Than the 3′ Cleavage to Trigger MMEJ
In order to investigate which repair mechanism is active in the model system, we analyzed whether a double cleavage or a linearization of the donor vector propagates high genome-editing efficiency. A cleavage upstream (5′ end) of the luciferase gene resulted in a similar luciferase signal compared with a cut at both sides of the gene, while a single cut downstream (3′ end) of the gene did not result in a significant amount of luciferase signal (Figure 4D). The observation that only one cut in the donor template upstream of the luciferase gene is necessary to generate high luciferase signal suggests that MMEJ takes place. If NHEJ would have occurred after the single cut, the resulting luciferase signal would be much lower because of the incorporation of the entire plasmid backbone, disrupting the connection between luciferase gene and bovine growth hormone (BGH) polyadenylation signal.
Addition of RNases Increases DNA Repair Efficacy
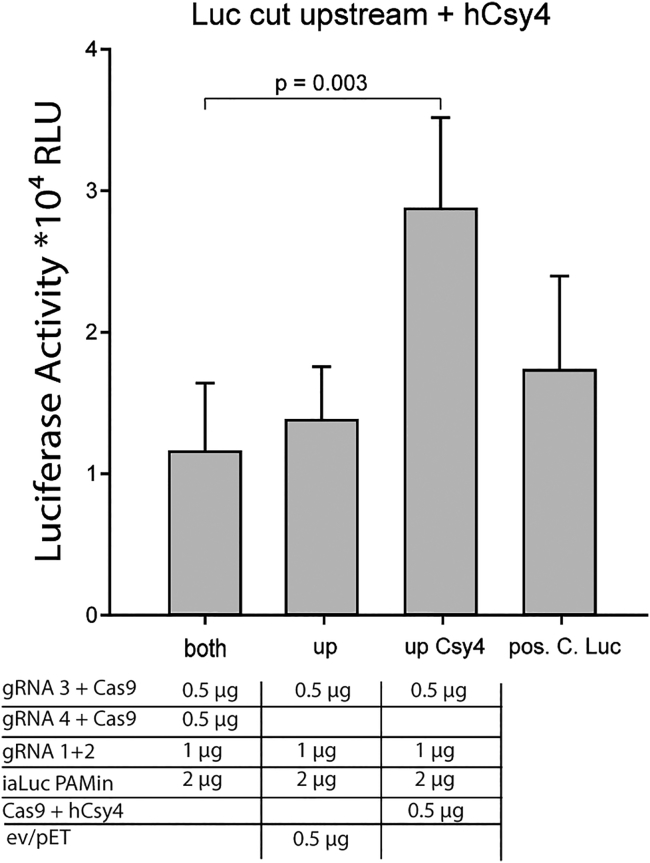
We also analyzed whether supplementary RNases would be superior for the processing of the RNA transcripts into gRNAs, which would be an important aspect in therapeutic applications. Because the plasmid pSQT1313 harboring the gRNA 1+2 contains Csy4 RNase sites in between the two RNA sequences (Figure 2A), we were able to study DNA repair activities with and without the addition of Csy4-expressing plasmids to the transfections.25 Interestingly, an addition of the enzyme further increased the DNA repair activity by 100% (Figure 5), indicating that even though eukaryotic RNases are able to process guide RNases, the efficiency of DSB repair events in a therapeutic setting may be improved by the addition of RNA-processing enzymes.
Figure 5.
Improvement of Guide RNA Processing in Multiplex CRISPR/Cas9 Applications
A plasmid coding the bacterial RNase Csy4 was added to the triple transfection to process the Cas9 guide RNAs (see also Figure 2A). Addition of Csy4 increased the Luciferase signal by 100%, indicating that cellular RNases are only partially capable of processing multiple guide RNAs in multiplex CRISPR/Cas9 applications. Error bars indicate the SD of the data set.
5-bp mhs Length Is Most Efficient to Trigger MMEJ
Finally, we studied whether the mhs length would have an impact on the luciferase signal intensity after MMEJ (Figure 6A). We found significantly higher luciferase signal with all different mhs (5, 10, 15, 20, 30, 50) tested compared with negative control (Figure 6B). The intensity of the RLU for the different microhomology pairs varied between 153.268 ± 17.387 and 16.874 ± 5.985 RLU. The 5-bp microhomology donor plasmid showed the maximum RLU followed by 15, 30, 20, 50, and 10 bp.
Discussion
In this study, we developed an in vitro model system to study MMEJ at the locus of the RPGR gene on chromosome X, in which mutations are associated with XLRP. We showed that homologous recombination can take place at the target site with mhs ranging from 5 to 50 bp in length. Interestingly, mhs with 5-bp length was most efficient.
Until recently, it was considered that high-fidelity genome editing is generally based on HDR events. Unfortunately, this major knockin strategy shows different success rates depending on locus, nuclease, cell types, and organisms.26, 27, 28 Sakuma and colleagues16 demonstrated that precise knockin experiments are possible with short homologies based on the MMEJ repair pathway. One advantage is that MMEJ donor templates do not need large homology regions, as this is the case for donor templates for HDR-based editing with very long homology arms (Figure 1). Even though single-stranded oligodeoxynucleotide (ssODN) can also be used as templates for HDR, this option is only used to edit the genome with small modifications (<100 bp) and is not suitable for the insertion of long DNA sequences, as it would be the case when replacing the ORF15 exon.29, 30, 31 The NHEJ pathway is also an option to incorporate exogenous sequences, but this pathway is error prone by inducing mutagenic in-dels. Therefore, NHEJ is most often used to generate knockouts, for which no exogenous template is needed.26, 32, 33 Consequently, MMEJ may represent an alternative way to HDR-based genome editing to replace larger DNA sequences of choice.
Although MMEJ and HDR are the repair pathways of choice to replace mutant DNA by wild-type DNA, it is often difficult to discern by which repair event a DSB was processed. In our model system, we consider the short homology arms as the key elements for MMEJ to take place, as it was reported in several studies that HDR for larger sequences to be replaced requires homology arms way beyond 100 bp of length.10, 11, 16 Of note, we observed an overall decline in replacement activity with longer mhs, which could well be due to a shift from MMEJ activity more toward an HDR-dependent repair mechanism. This question is currently addressed by the authors by knocking down/out respective repair proteins.
NHEJ and the two homology-dependent pathways are always competing on the DNA ends following a DSB. The processing of nucleotides (removal or adding) at the target site is a characteristic feature of the NHEJ mechanism. A few nucleotides in the luciferase donor vector with its Cas9 PAMin and PAMout sites can be deleted after Cas9 cleavage through NHEJ. However, the PAMin site has only a 6-bp distance between the Cas9 cleavage site and the mhs. Larger deletions will directly interfere with the mhs, which influences the MMEJ success rate, and alterations of a few nucleotides can have a huge impact on the MMEJ efficiency.17 This could be an explanation for the difference in the measured luciferase activity between the PAMin and PAMout donor vectors, and an indication that MMEJ is the primary repair process active here (Figure 4B). An integration of both constructs via NHEJ would lead to similar results because no microhomology is required.
The upstream or downstream cleavage of the luciferase gene within the donor template also results in differences in repair efficiency. The downstream cleavage is linked to decreased luciferase activity levels, while the upstream cleavage results in similar luciferase activity levels as the double-cut experiment. In the case of NHEJ, both single-cut experiments would result in lower levels of luciferase activity, because the donor plasmid backbone would have been incorporated into the target site either between the CMV promotor and the luciferase gene or following the luciferase gene, but in front of the polyadenylation signal, both resulting in suboptimal mRNA processing. The reason for the downstream cleavage with lower levels of luciferase activity signaling remains unknown, but may potentially be associated with a negative influence of the very long donor plasmid backbone on the single-strand invasion during the homology-based DNA repair.
The cellular RNA processing machinery in eukaryotes is obviously capable of producing active gRNAs that allow the cut by Cas9 at the desired locus. This has been shown in many experiments before, and it is assumed that endogenous mammalian RNases assist in pre-crispr RNA (crRNA) maturation.24 However, whether the mammalian RNase system enables optimal processing or whether this feature is also a point where efficacy could be optimized was not clear. We showed that high amounts of Csy4, which cut the pre-crRNA between gRNA1 and gRNA2, increased the measured luciferase activity in our experiments. However, the increased amount of enzyme did not change the ratios within the experiments, which implies a pre-crRNA processing with endogenous RNases in HEK293 cells. The cleavage rates for g1 and g2 alone were analyzed in preliminary assays with the traffic light reporter system (TLR)34 to ensure their activity (data not shown). Therefore, if multiplex CRISPR/Cas systems are used in therapeutic settings, the addition of the cutting enzyme to process the different gRNAs may be useful.
All different mhs lengths (5, 15, 20, 30, and 50 bp) within the template DNA resulted in significant repair activity at the RPGR-ORF15 locus. The data are in line with the results reported earlier in budding yeast.17, 20 Interestingly, the plasmid donor with an mhs length of 5 bp was found to be the best among the ones tested, followed by the 15-bp mhs length. However, our results differed from that of Tadi and colleagues,20 who tested mhs of 5, 8, 10, 13, 16, 19, and 22 bp in DSB repair during mitochondrial DNA lesions and reported an enhanced efficiency of MMEJ with the increase in the length of the mhs. The reasons for the highest repair activity measured with the shortest mhs could be because of simple MMEJ being active at that particular site, as opposed to the DNA synthesis-dependent (SD)-MMEJ mechanisms that usually occur with the larger mhs.35, 36, 37 It could also be a mixture of MMEJ and NHEJ mechanisms, which is currently under intensive exploration by the authors. Knocking down key proteins in either pathway should shed some light on these correlations.
The stepwise development of the reporter system started with an episomal-based assay using fluorescent reporter proteins that was optimized further into a luciferase-based assay on the genomic level and proved to be promising as a sensitive method to measure DNA repair efficiencies in targeted gene-editing approaches. We used the system at the locus of the RPGR gene, but it can also be used with therapeutically relevant DNA sequences in any target cells assuming that DNA delivery is established. After establishing the best strategy for each disease model, the system may be further evaluated in vivo in newly generated animal models.
Materials and Methods
Cloning Vectors for Episomal-Based Assay
The GFP AP pEGFP-N1 is commercially available from Invitrogen. The BamHI/KpnI (upstream) and MfeI/XbaI (downstream) were used for cloning cassettes harboring the mhs. The donor plasmid with BFP is based on the pSQT1313 vector (Addgene plasmid 53370). BFP was incorporated via BamHI and XhoI cleavage sites. The sites NheI and BamHI upstream of BFP and the sites downstream XhoI and PciI are recruited for cloning the microhomology sequences with Cas9 recognitions sites. The two gRNAs for Cas9 are cloned into the pSQT1313 vector as described by Tsai et al.38 The sequences are listed in Table 1.
Transfection of Vectors for the Episomal-Based Assay
HEK293T cells were seeded in six-well plates at a density of 5 × 105 cells per well and cultured in DMEM supplemented with 10% fetal bovine serum, penicillin/streptomycin (PAN Biotech, Aidenbach, Germany), and l-glutamine (200 mM) (PAN Biotech). After 24 hr, cells were transfected using 200 μL of polyethylenimine (PEI) mix (pH 7; 0.1 g PEI/L) in addition with 100 μL of 150 mM NaCl with 600 ng of addressed target within the TLR3 system and 600 ng of each nuclease, and additionally 300 ng of gRNA in case of Cas9-FokI. The transfection reactions were filled up with pET-19b or circular pTOPO-TA empty vector (Novagen/Merck, USA/Thermo Fisher, Darmstadt, Germany) to 1.8 μg of DNA. After 6 hr, the medium was substituted, and cells were harvested 72 h posttransfection; the number of BFP-positive cells was determined by flow cytometry (FACS Canto Calibur; BD Biosciences, USA).
Generation of the HEK293-mORF15 Stable Cell Line
The ORF15 exon from the mouse RPGR gene [AL671042 (gb), Riken cDNA clone (ImaGenes)], containing four point mutations that are unrelated to this study, was cloned into the pcDNA 3.1 (−) vector, under the control of a CMV promoter (Invitrogen/Thermo Fisher Scientific, USA). Four micrograms of the vector was linearized with EcoRI and then transfected into HEK293 cells in a six-well plate as mentioned earlier. 48 h following the transfection, 0.5 μg/mL Geneticin (Life Technologies) was added to the medium for selection. Surviving cell foci were picked and transferred to 24-well plates. The selection process was repeated twice. The integration was analyzed using automated DNA sequencing by Sanger methods. Multiple integrations of RPGR-ORF15, if any, were not analyzed.
Vectors for Transfection in Stable HEK293-mORF15
The px459 vector [pSpCas9(BB)-2A-Puro (PX459) V2.0; Addgene plasmid 62988] was used to cleave the donor plasmid containing the luciferase gene. The gRNA was cloned into the vector as described by Ran et al.39 The gRNAs (g1 and g2) targeting the RPGR-mORF15 sequence within the genome were cloned into the pSQT1313 vector as described by Tsai et al.38 All Cas9 target sites are listed in Table 1, with the polyspacer adjacent motif (PAM) sites being underlined. The donor plasmid is based on sequences that have homology to the RPGR-mORF15 and are between the length of 5 and 50 bp.
Transfection and Lysis of HEK293-mORF15 Cells, Western Blot, and Measurement of Luciferase Activity
HEK293-mORF15 cells were seeded in six-well plates at a density of 5 × 105 cells per well with DMEM and Geneticin (0.5 μg/mL), and the quadruple transfection with PEI was performed as described earlier. Different plasmid concentrations were used and are listed in the Results. Cells were harvested 48–72 hr posttransfection using the cell lysis buffer for luciferase assays (Promega, Mannheim, Germany). Additionally, the cell lysates were snap chilled twice before pipetting 20 μL of the cleared lysate (17,000 rpm; 5 min; 4°C) into COSTAR Lumiplates Flat White (Corning, Berlin, Germany).
Protein samples were processed for western blot application. Forty micrograms of total protein was loaded onto two identical SDS-PAGE gels and migrated along their molecular weight. Primary antibodies against luciferase (rabbit anti-RLuc, ab187338; Abcam, Cambridge, UK) and GAPDH (rabbit anti-glycerine aldehyde 3-phosphate dehydrogenase [GAPDH]; Cell Signaling, Frankfurt, Germany) were used, following the secondary antibody fused to HR peroxidase (goat anti-rabbit IgG-HRP; Sigma, Darmstadt, Germany). Chemiluminescence detection assay was performed with the ECL peroxidase detection kit (Amersham Bioscience, Buckinghamshire, UK).
The luciferase activity was measured after adding the substrate h-Coelenterazine (NanoLight, Pinetop USA) in 500 μL of NanoLight-Fuel Solvent and diluted in 1:100 in 1× PBS. 100 μL of the Renilla luciferase substrate solution was used for each well, and the measurements were recorded using Tecan Infinite M1000 Pro plate reader (Tecan, Groeding, Austria) with open filters and an integration time of 1 s to collect photons produced by the luciferase (photons/s RLU).
Statistical Analysis
All of the experiments were performed and measured at least in triplicate; Student’s paired t test and the Holm-Sidak method were used to compare the results. The p value for each dataset is fixed independently, as mentioned in the respective bar graphs/figures.
Author Contributions
M.Y. designed and performed experiments, analysed data, and wrote and edited the paper. S.P.G.P. designed the experiments, interpreted data, and wrote the paper. T.W., L.T., C.M., I.G., J.G., A.J., and J.D. performed experiments and interpreted data. W.W. and B.L. interpreted data, and wrote and edited the paper. K.S. designed the experiments, analyzed/interpreted data, and edited the paper.
Acknowledgments
The authors thank Prof. Hackstein, Dr. Nelli Baal, and Gaby Michel for performing the FACS analysis. We thank Dr. Müller and Dr. Lopez for helpful discussions concerning the experimental approach. Dr. Ponnam acknowledges the financial support received, as a DBT Overseas associate, from the Department of Biotechnology, Government of India. This work was supported by the European Research Council (starting grant 311244 to K.S.).
References
- 1.Gaj T., Gersbach C.A., Barbas C.F., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mussolino C., Mlambo T., Cathomen T. Proven and novel strategies for efficient editing of the human genome. Curr. Opin. Pharmacol. 2015;24:105–112. doi: 10.1016/j.coph.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Yanik M., Alzubi J., Lahaye T., Cathomen T., Pingoud A., Wende W. TALE-PvuII fusion proteins—novel tools for gene targeting. PLoS ONE. 2013;8:e82539. doi: 10.1371/journal.pone.0082539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibikova M., Golic M., Golic K.G., Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H.J., Kim E., Kim J.S. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20:81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee H.J., Kweon J., Kim E., Kim S., Kim J.S. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 2012;22:539–548. doi: 10.1101/gr.129635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heyer W.D., Ehmsen K.T., Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C., Katibah G.E., Amora R., Boydston E.A., Zeitler B. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hockemeyer D., Wang H., Kiani S., Lai C.S., Gao Q., Cassady J.P., Cost G.J., Zhang L., Santiago Y., Miller J.C. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sommer D., Peters A., Wirtz T., Mai M., Ackermann J., Thabet Y., Schmidt J., Weighardt H., Wunderlich F.T., Degen J. Efficient genome engineering by targeted homologous recombination in mouse embryos using transcription activator-like effector nucleases. Nat. Commun. 2014;5:3045. doi: 10.1038/ncomms4045. [DOI] [PubMed] [Google Scholar]
- 12.Lieber M.R. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 13.Sinha S., Villarreal D., Shim E.Y., Lee S.E. Risky business: microhomology-mediated end joining. Mutat. Res. 2016;788:17–24. doi: 10.1016/j.mrfmmm.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng S.K., Gibb B., de Almeida M.J., Greene E.C., Symington L.S. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014;21:405–412. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakade S., Tsubota T., Sakane Y., Kume S., Sakamoto N., Obara M., Daimon T., Sezutsu H., Yamamoto T., Sakuma T., Suzuki K.T. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014;5:5560. doi: 10.1038/ncomms6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakuma T., Nakade S., Sakane Y., Suzuki K.T., Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protoc. 2016;11:118–133. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- 17.Villarreal D.D., Lee K., Deem A., Shim E.Y., Malkova A., Lee S.E. Microhomology directs diverse DNA break repair pathways and chromosomal translocations. PLoS Genet. 2012;8:e1003026. doi: 10.1371/journal.pgen.1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taleei R., Nikjoo H. Biochemical DSB-repair model for mammalian cells in G1 and early S phases of the cell cycle. Mutat. Res. 2013;756:206–212. doi: 10.1016/j.mrgentox.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Sharma S., Javadekar S.M., Pandey M., Srivastava M., Kumari R., Raghavan S.C. Homology and enzymatic requirements of microhomology-dependent alternative end joining. Cell Death Dis. 2015;6:e1697. doi: 10.1038/cddis.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tadi S.K., Sebastian R., Dahal S., Babu R.K., Choudhary B., Raghavan S.C. Microhomology-mediated end joining is the principal mediator of double-strand break repair during mitochondrial DNA lesions. Mol. Biol. Cell. 2016;27:223–235. doi: 10.1091/mbc.E15-05-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vervoort R., Lennon A., Bird A.C., Tulloch B., Axton R., Miano M.G., Meindl A., Meitinger T., Ciccodicola A., Wright A.F. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat. Genet. 2000;25:462–466. doi: 10.1038/78182. [DOI] [PubMed] [Google Scholar]
- 22.Vervoort R., Wright A.F. Mutations of RPGR in X-linked retinitis pigmentosa (RP3) Hum. Mutat. 2002;19:486–500. doi: 10.1002/humu.10057. [DOI] [PubMed] [Google Scholar]
- 23.Horvath P., Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 24.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haurwitz R.E., Jinek M., Wiedenheft B., Zhou K., Doudna J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cristea S., Freyvert Y., Santiago Y., Holmes M.C., Urnov F.D., Gregory P.D., Cost G.J. In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol. Bioeng. 2013;110:871–880. doi: 10.1002/bit.24733. [DOI] [PubMed] [Google Scholar]
- 27.Li J., Zhang B., Bu J., Du J. Intron-based genomic editing: a highly efficient method for generating knockin zebrafish. Oncotarget. 2015;6:17891–17894. doi: 10.18632/oncotarget.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyaoka Y., Berman J.R., Cooper S.B., Mayerl S.J., Chan A.H., Zhang B., Karlin-Neumann G.A., Conklin B.R. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Sci. Rep. 2016;6:23549. doi: 10.1038/srep23549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bedell V.M., Wang Y., Campbell J.M., Poshusta T.L., Starker C.G., Krug R.G., 2nd, Tan W., Penheiter S.G., Ma A.C., Leung A.Y. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen F., Pruett-Miller S.M., Huang Y., Gjoka M., Duda K., Taunton J., Collingwood T.N., Frodin M., Davis G.D. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer M., Ortiz O., Hrabé de Angelis M., Wurst W., Kühn R. Modeling disease mutations by gene targeting in one-cell mouse embryos. Proc. Natl. Acad. Sci. USA. 2012;109:9354–9359. doi: 10.1073/pnas.1121203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auer T.O., Duroure K., De Cian A., Concordet J.P., Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014;24:142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orlando S.J., Santiago Y., DeKelver R.C., Freyvert Y., Boydston E.A., Moehle E.A., Choi V.M., Gopalan S.M., Lou J.F., Li J. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res. 2010;38:e152. doi: 10.1093/nar/gkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Certo M.T., Ryu B.Y., Annis J.E., Garibov M., Jarjour J., Rawlings D.J., Scharenberg A.M. Tracking genome engineering outcome at individual DNA breakpoints. Nat. Methods. 2011;8:671–676. doi: 10.1038/nmeth.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Truong L.N., Li Y., Shi L.Z., Hwang P.Y., He J., Wang H., Razavian N., Berns M.W., Wu X. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. USA. 2013;110:7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu A.M., McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010;38:5706–5717. doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Y., Caron P., Legube G., Paull T.T. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014;42:e19. doi: 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D., Goodwin M.J., Aryee M.J., Joung J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]