Abstract
Background
Limited information regarding the clonality of circulating E. coli strains in tertiary care hospitals in low and middle-income countries is available. The purpose of this study was to determine the serotypes, antimicrobial resistance and virulence genes. Further, we carried out a phylogenetic tree reconstruction to determine relatedness of E. coli isolated from patients in a tertiary care hospital in Tanzania.
Methods
E. coli isolates from inpatients admitted at Kilimanjaro Christian Medical Centre between August 2013 and August 2015 were fully genome-sequenced at KCMC hospital. Sequence analysis was done for identification of resistance genes, Multi-Locus Sequence Typing, serotyping, and virulence genes. Phylogeny reconstruction using CSI Phylogeny was done to ascertain E. coli relatedness. Stata 13 (College Station, Texas 77,845 USA) was used to determine Cohen’s kappa coefficient of agreement between the phenotypically tested and whole genome sequence predicted antimicrobial resistance.
Results
Out of 38 E. coli isolates, 21 different sequence types (ST) were observed. Eight (21.1%) isolates belonged to ST131; of which 7 (87.5.%) were serotype O25:H4. Ten (18.4%) isolates belonged to ST10 clonal complex; of these, four (40.0%) were ST617 with serotype O89:H10. Twenty-eight (73.7%) isolates carried genes encoding beta-lactam resistance enzymes. On average, agreement across all drugs tested was 83.9%. Trimethoprim/sulphamethoxazole (co-trimoxazole) showed moderate agreement: 45.8%, kappa =15% and p = 0.08. Amoxicillin-clavulanate showed strongest agreement: 87.5%, kappa = 74% and p = 0.0001. Twenty-two (57.9%) isolates carried virulence factors for host cells adherence and 25 (65.7%) for factors that promote E. coli immune evasion by increasing survival in serum. The phylogeny analysis showed that ST131 clustering close together whereas ST10 clonal complex had a very clear segregation of the ST617 and a mix of the rest STs.
Conclusion
There is a high diversity of E. coli isolated from patients admitted to a tertiary care hospital in Tanzania. This underscores the necessity to routinely screen all bacterial isolates of clinical importance in tertiary health care facilities. WGS use for laboratory-based surveillance can be an effective early warning system for emerging pathogens and resistance mechanisms in LMICs.
Electronic supplementary material
The online version of this article (10.1186/s13756-018-0361-x) contains supplementary material, which is available to authorized users.
Keywords: E. coli, Multi-locus sequence typing, Serotyping, And virulence, Whole genome sequencing, Tanzania
Background
Escherichia coli are Gram-negative bacterial commensals that naturally inhabit the human gastrointestinal tract (GIT). Through horizontal transfer and other mechanisms, commensal E. coli regularly acquire virulence, pathogenicity and multi-drug resistance properties from pathogenic E. coli. Consequently, E. coli is an important causative agent for a range of nosocomial and opportunistic infections including neonatal meningitis, diarrhoea, septicaemia, urinary tract and wound infections [1–5]. The global emergence and spread of multidrug resistant (MDR) E. coli in both community and as nosocomial infections, as well as in animals, warrants public health concerns [6–9]. Furthermore, virulent E. coli strains share resistance, virulence and pathogenic factors with avirulent or less virulent strains, enabling them to cause overlapping pathogenesis beyond their classical capacities [10]. Several E. coli outbreaks leading to serious health, social and economic impacts have been reported in high income countries (HICs) including the Netherlands [11], the UK [12], Norway, and Georgia [10].
Africa is highly burdened by diarrhoea and urinary tract infections (UTIs) that are E. coli related. For instance, in rural Kenya it was reported that 64.5% of UTI or bacteriuria was E. coli related [13]. In north-western Tanzania, 41.2 and 70% of UTI or bacteriuria was E. coli related among under-fives and febrile children, respectively [14, 15]. Similarly, in north-eastern part of Tanzania, E. coli accounted for 56.1% of all UTI cases among children [5] and E. coli accounted for 79 and 75% of UTI in non-malaria febrile children and adults, respectively [16]. In Dar es Salaam, two previous reports showed that, E. coli accounted for 51.1% of UTI in a survey done in 2010 [17] whereas as high as 64.0% of hospital- and community-acquired UTI was accounted for by E. coli in 2004 [18].
Finally, we recently showed that in Moshi, north-eastern Tanzania, E. coli was one of the most common bacterial pathogen isolates from a range of clinical manifestations [1]. Advanced molecular diagnostics such as whole genome sequencing (WGS) have revealed the emergence of a fatal diarrhoea-causing E. coli strain that combines virulence factors (VFs) from two E. coli strains [4]. The VFs are important properties that do enable an infectious agent to effectively and efficiently establish itself on or within its host by enhancing its potential to cause harm or disease. Some of the factors include those for host cell adherence, immune evasion, toxins and protease production for disrupting host cell pathways and scavenging minerals like iron in order to increase their survival. Certain E. coli strains have VFs that been linked to a serious outbreaks in China in 1999, where it caused 177 deaths [19], and in Germany in 2011, where it claimed 54 lives [20].
Molecular typing studies to evaluate E. coli related infections in low and middle-income countries (LMICs) are rare and little information is available regarding the specific subtypes causing infections in Tanzania or whether the high prevalence is due to sporadic infectious or clonal transmission between patients. The purpose of this study was to determine the serotypes, antimicrobial resistance and virulence genes. Further, we carried out a phylogenetic tree reconstruction to determine relatedness of E. coli isolated from patients in a tertiary care hospital in Tanzania using the WGS-based diagnostic platform installed at KCMC hospital in Moshi, Tanzania.
Methods
Study design, participants and specimen collection
A hospital based prospective cross-sectional study was conducted at KCMC between August 2013 and August 2015. Part of the study’s methods has been described in details by Kumburu et al. [1]. KCMC is located in Moshi municipality, north-eastern Tanzania and serves as a zonal referral hospital for a catchment area of around 15 million people. The hospital has a bed capacity of 650 with approximately 500 outpatients seeking medical services daily. This study was granted ethical approval by the KCMC Research Ethics Committee and the National Institute for Medical Research. A written informed consent was obtained from each participant or from parents or guardians of children before enrolment into the study. The study involved patients admitted in medical, surgical and paediatrics wards who were suspected to have bacterial infection. Specimens collected for bacterial culture were sputum, wound or pus swab, stool and blood. Bacteria culture, isolation, and identification were performed according to in-house standard operating procedures and the Clinical and Laboratory Standards Institute (CLSI) guidelines as described by Kumburu et al. [1]. Over a 2-year period, 590 samples were collected without apriori knowledge of the infecting agent. A total of 377 bacterial strains were isolated, and whole genome sequenced. A number of isolates from this collection were randomly selected for antimicrobial susceptibility testing. A total of 38 E. coli collected sequentially were included in this study; among which 24 E. coli isolates had antimicrobial susceptibility results.
Genomic DNA isolation, whole genome sequencing, and analysis
For all E. coli isolates genomic DNA (gDNA) was purified and its concentration determined using the Easy-DNA extraction kit (Invitrogen®) and the Qubit dsDNA Assay Kit (Invitrogen®), respectively. The gDNA library preparation was performed following Nextera® XT DNA Sample Preparation Guide [21]. In brief, each gDNA was tagmented (tagged and fragmented) by the Nextera® XT transposome. The transposome simultaneously fragments the input DNA and adds adapter sequences to the fragment ends. Then followed a limited-cycle PCR amplification whereby indexes required for cluster formation were added to each DNA piece. Then each gDNA library was normalized to ensure equal representation during sequencing. Equal volumes of the normalized library were combined, diluted in hybridization buffer, and heat denatured prior to sequencing on the Illumina MiSeq platform (Illumina Inc.). The sequencer output was analysed using the standard WGS pipeline at KCRI, which is based on local implementations of the bioinformatics services available at https://cge.cbs.dtu.dk/services/. Quality control of the reads was performed using FastQC 0.11.4 [22]. De novo assembly was performed with SPAdes 3.11.1 [23], and quality assessed using QUAST 4.5 [24]. For this article’s purpose the analyses included: resistance genes identification using ResFinder 2.1 [25], Multi-Locus Sequence Typing (MLST) determination using MLST 1.8 [26], serotyping using SeroTypeFinder 1.1 [27], and virulence genes determination using VirulenceFinder 1.4 [28]. Phylogeny reconstruction was done using CSI Phylogeny [29] (with reference strain EC958, NZ_HG941718.1). The 38 assembled E. coli genomes of the present study have been submitted to the European Nucleotide Archive with project accession number PRJEB23541. The phylogenetic analyses included 6 more E. coli genomes previously isolated from animals in Mwanza, north-western Tanzania, by Seni et al. [30]. The raw sequence data of the E. coli of animal origin were downloaded from the European Nucleotide Archive (ENA) under the project number PRJEB12335. Stata 13 (College Station, Texas 77,845 USA) was used to determine Cohen’s kappa coefficient of agreement between the phenotypically tested and whole genome sequence predicted antimicrobial resistance.
Results
Specimens and isolates
A total of 38 non-duplicate E. coli were isolated, of which 18 (47.4%) were from wound (or pus) swabs, 13 (34.2%) from diarrhoeal stool, 4 (10.5%) from sputum and 3 (7.9%) from blood. Twenty-three (60.5%) were isolated from patients in medical wards, 9 (23.7%) from patients in surgical wards, and 6 (15.8%) from patients in intensive care unit (ICU). Out of these six E. coli isolates, four were from surgical ICU and 2 from medical ICU. Thirteen (34.2%) E. coli were isolated in 2013, 16 (42.1%) in 2014 and nine (23.7%) in 2015 (Table 1).
Table 1.
Origin and drug susceptibility results of 38 clinical E. coli isolates
| Origin of isolate | Drug susceptibility resultsd | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | wardb | specimenc | yeara | AMC | AM | CZ | CAZ | CRO | C | CIP | GM | NA | SXT |
| 4 | MW | Stool | 2013 | S | R | S | S | S | S | S | S | S | R |
| 5 | MW | Stool | 2013 | S | S | S | S | S | S | S | S | S | S |
| 6 | MW | Stool | 2013 | S | S | S | S | S | S | S | S | S | S |
| 10 | SW | Swab | 2013 | R | R | S | S | S | S | R | S | R | S |
| 21 | MW | Stool | 2013 | R | S | S | S | S | S | S | S | S | R |
| 22 | MW | Stool | 2013 | N | N | N | N | N | N | N | N | N | N |
| 30 | SW | Swab | 2013 | S | S | S | S | S | S | S | S | S | S |
| 44 | SICU | Swab | 2013 | S | R | R | R | R | S | R | R | R | R |
| 70 | MW | Sputum | 2013 | R | R | R | R | R | S | R | S | R | R |
| 73 | MW | Stool | 2013 | R | R | R | R | R | S | R | S | R | R |
| 82 | MW | Stool | 2013 | R | R | R | S | R | S | R | S | R | R |
| 97 | MW | Stool | 2014 | N | N | N | N | N | N | N | N | N | N |
| 115 | SW | Swab | 2014 | S | R | R | R | R | S | S | S | S | R |
| 199 | MW | Blood | 2014 | R | R | R | R | R | R | R | R | R | R |
| 203 | MW | Stool | 2014 | R | R | S | S | S | S | R | S | R | R |
| 210 | MW | Blood | 2014 | R | R | R | R | R | S | R | R | R | R |
| 244 | MW | Sputum | 2014 | R | R | R | R | R | S | R | R | R | R |
| 245 | MW | Swab | 2014 | R | R | R | R | R | S | R | R | R | R |
| 247 | MW | Blood | 2014 | R | R | R | S | R | S | R | S | R | R |
| 298 | MW | Stool | 2014 | N | N | N | N | N | N | N | N | N | N |
| 365 | SICU | Swab | 2014 | R | R | R | R | R | S | R | S | R | R |
| 393 | MICU | Stool | 2015 | R | R | R | R | R | S | R | R | R | R |
| 521 | MW | Sputum | 2014 | S | S | S | S | S | S | S | S | R | R |
| 538 | SW | Swab | 2015 | N | N | N | N | N | N | N | N | N | N |
| 554 | MW | Stool | 2015 | N | N | N | N | N | N | N | N | N | N |
| 587 | SW | Swab | 2015 | N | N | N | N | N | N | N | N | N | N |
| 603 | MICU | Swab | 2015 | N | N | N | N | N | N | N | N | N | N |
| 118A | SW | Swab | 2014 | S | R | S | S | S | S | S | S | S | S |
| 119EC | MW | Stool | 2014 | S | S | R | R | R | R | S | S | S | S |
| 163A | SW | Swab | 2014 | S | S | S | S | S | S | S | S | S | S |
| 237C | SW | Swab | 2014 | N | N | N | N | N | N | N | N | N | N |
| 340B | MW | Sputum | 2014 | S | S | S | S | S | S | S | S | S | R |
| 431D | SICU | Swab | 2015 | N | N | N | N | N | N | N | N | N | N |
| 567B | SW | Swab | 2015 | N | N | N | N | N | N | N | N | N | N |
| 598A | MW | Swab | 2015 | N | N | N | N | N | N | N | N | N | N |
| 598B | MW | Swab | 2015 | N | N | N | N | N | N | N | N | N | N |
| 71E | MW | Swab | 2013 | R | R | R | R | R | S | R | R | R | R |
| 77E | SICU | Swab | 2013 | R | R | R | R | R | S | R | S | R | S |
ayear of collection
b MW Medical ward, SW surgical ward, MICU medical ICU, SICU surgical ICU
cWound or pus swab, diarrhoea or stool
d S Susceptible, R Resistant, N Not tested, AMC Amoxicillin-Clavulanate, AM ampicillin, CZ cefazoline, CAZ ceftazidime, CRO ceftriaxone, C chloramphenicol, CIP ciprofloxacin, GM gentamycin, NA Nalidixic acid, SXT trimethoprim sulphamethoxazole
Multi-locus sequence typing and serotyping
Out of 38 E. coli isolates, 21 different STs were observed. Eight (21.1%) E. coli belonged to ST131; 3 isolated in 2013, 2 in 2014 and 3 in 2015 (Table 1). Two of the E. coli ST131 were isolated from patients in surgical wards, and six from patients in medical wards, including one from medical ICU. Of these eight, seven (87.5%) E. coli had serotype O25:H4 and one had serotype O15:H1. Out of 38 isolates, 10 (18.4%) belonged to ST10 clonal complex; of these, four (40.0%) were ST617 with serotype O89:H10, of which 3 were isolated in 2014 and 1 in 2015 and three (30.0%) were ST10, of which two had serotype O89:H10 and one had serotype O89:H9 (Table 2). These ST10 were all isolated in 2014 from medical wards. We noted that, out of the three ST410, one had serotype O8:H21, and the others had unknown O-group but belonged to H9.
Table 2.
Sequence types, Serotypes and Virulence factors of 38 clinical E. coli isolates
| Isolate | STe | Serotype | Virulence Factors | ||||
|---|---|---|---|---|---|---|---|
| Adherence | Toxin | Protease | Evasiong | Type IIIh | |||
| 70 | ST-131 | O25:H4 | Iha,nfaE | sat | iss | ||
| 73 | ST-131 | O25:H4 | Iha,nfaE | sat | iss | ||
| 199 | ST-131 | O25:H4 | Iha | astA,senB | sat | iss | |
| 210 | ST-131 | O25:H4 | Iha,nfaE | senB | sat | iss | |
| 587 | ST-131 | O25:H4 | Iha | sat | iss | ||
| 603 | ST-131 | O15:H1 | iha,nfaE lpfA,eilA | senB | sat | iss | air |
| 567B | ST-131 | O25:H4 | Iha | sat | iss | ||
| 71E | ST-131 | O25:H4 | iha,nfaE | senB | sat | iss | |
| 244 | ST-617f | O89:H10 | iss | ||||
| 245 | ST-617 f | O89:H10 | iss | ||||
| 538 | ST-617 f | O89:H10 | astA,senB | iss | capU | ||
| 237C | ST-617 f | O89:H10 | iss | ||||
| 393 | ST-405 | O102:H6 | eilA | air | |||
| 10 | ST-410 | O??:H9 | lpfA | ||||
| 22 | ST-410 | O??:H9 | lpfA | ||||
| 598B | ST-410 | O8:H21 | lpfA | iss | |||
| 97 | ST-10 f | O89:H10 | |||||
| 203 | ST-10 f | O89:H10 | |||||
| 340B | ST-10 f | O89:H9 | iss | ||||
| 44 | ST-167 f | O89:H21 | |||||
| 247 | ST-167 f | O89:H9 | senB | iss | capU | ||
| 5a | ST-226 | O40:H19 | bfpA,eae | espA,espF,espJ | nleB,nleC | ||
| 6a | ST-226 | O40:H19 | bfpA,eae | espA,espF,espJ | nleB,nleC | ||
| 118Ad | ST-73 | O6:H1 | Iha | pic | sat,vat | iss | mchB,mchC,mchF,mcmA |
| 21 | ST-942 | O39:H28 | lpfA | iss | |||
| 554c | ST-95 | O2(50):H4 | iss | mchF | |||
| 431D | ST-44 f | O89:H4 | |||||
| 119EC | ST-4959 | O154:H4 | |||||
| 521b | ST-504 | O166:H7 | Iha | vat | iss | mchB,mchC,mchF,mcmA, sigA, capU | |
| 4 | ST-2332 | O128:H45 | cfaC, lngA | eatA | |||
| 30 | ST-355 | O150:H5 | pic | vat | iss | ||
| 115 | ST-361 | O9:H30 | |||||
| 163A | ST-372 | O83:H31 | vat | iss | |||
| 298 | ST-38 | O86:H18 | Iha,nfaE eilA | senB | sat | iss | air |
| 365 | ST-224 | O8:H23 | lpfA | iss | |||
| 82 | ST-156 | O61:H34 | lpfA | iss | |||
| 598A | ST-1193 | O75:H5 | Iha | senB | sat,vat | ||
| 77E | ST-1284 | O89:H21 | iss | ||||
aHas virulence factor tir
bHas virulence factor iroN
cHas virulence factors ireA,iroN
dHas virulence factor ireA
eSequence Type (ST)
fST-10 clonal complex
gImmune evasion
hType III translocated protein
Virulence factors
Table 2 as well show the distribution of virulence factors (VFs) among the sequenced E. coli isolates. A total of 22 (57.9%) E. coli isolates carried VFs for host cells adherence. These included 12 (31.6%) for iha, 7 (18.4%) for lpfA and 6 (15.8%) for each of aafC and nfaE. The prevalence of eilA was 3 (7.9%) and 2 (5.3%) for each of eae and bfpA. A total of 9 (23.7%) E. coli isolates had VFs that promote toxin production. The distribution of VFs that promote toxin was 7 (18.4%) for senB and 2 (5.3%) for each of astA and pic. The VFs that promote E. coli protease production were detected in 18 (47.4%) E. coli isolates. The prevalent protease VFs were sat and vat with 13 (34.2%) and 5 (13.2%), respectively. A total of 25 (65.7%) E. coli isolates had iss, a factor that promotes E. coli immune evasion by increasing serum survival.
Acquired antimicrobial resistance
A total of 28 (73.7%) E. coli carried genes encoding beta-lactam resistance enzymes (Table 3). The blaCTX-M-15 was harboured by 17 (44.7%) isolates, blaOXA-1 harboured by 19 (50%) and blaTEM-1B by 12 (31.6%). The overall proportion of dfrA genes encoding trimethoprim resistance enzymes was 28 (73.7%). The dfrA17 was the most abundant gene with a proportion of 16 (42.1%) followed by 6 (15%) for dfrA14 genes, and 2 (5.3%) for each of dfrA5, dfrA7 and dfrA12.
Table 3.
Acquired antimicrobial resistance genes of 38 clinical E. coli isolates
| Isolate | AMGa | BLb | FQAc | Macrolide | Phenicol | Quinolone | Sulphonamide | Tetracycline | Trimethoprim |
|---|---|---|---|---|---|---|---|---|---|
| 4 | strA strB | blaTEM-1B | sul2 | dfrA14 | |||||
| 5 | |||||||||
| 6 | |||||||||
| 10 | blaOXA-1 | aac(6’)Ib-cr | catB3 | tet(A) | |||||
| 21 | strA strB | sul2 | tet(A) | dfrA14 | |||||
| 22 | blaOXA-1 | aac(6’)Ib-cr | catB3 | tet(A) | |||||
| 30 | |||||||||
| 44 | aac(3)-IId aadA2 | blaCTX-M-15 blaTEM-1B | mph(A) | catA1 | QnrS1 qepA QnrD | sul1 | tet(A) | dfrA12 | |
| 70 | aadA5 | blaCTX-M-15 blaOXA-1 | aac(6’)Ib-cr | mph(A) | catB3 | sul1 | tet(A) | dfrA17 | |
| 73 | aadA5 | blaCTX-M-15 blaOXA-1 | aac(6’)Ib-cr | mph(A) | catB3 | sul1 | tet(A) | dfrA17 | |
| 82 | strA strB aadA2 | blaTEM-1B | mph(A) | sul2 sul1 | tet(B) tet(A) | dfrA12 | |||
| 97 | blaOXA-1 | aac(6’)Ib-cr | catB3 | tet(B) | |||||
| 115 | aadA1 aac(3)-IIa strB strA | blaCTX-M-15 blaTEM-1B blaOXA-1 | aac(6’)Ib-cr | catB3 catA1 | QnrB1 | sul2 | dfrA14 | ||
| 163A | |||||||||
| 199 | aadA1 strB strA aadA5 aadB | blaCTX-M-15 blaTEM-1B blaOXA-1 | aac(6’)Ib-cr | mph(A) | catB3 cmlA1 | sul1 sul2 | tet(A) | dfrA17 | |
| 203 | strB strA | blaTEM-1C blaOXA-1 | aac(6’)Ib-cr | catB3 | sul2 | tet(A) | dfrA14 | ||
| 210 | aadA5 aac(3)-IIa strB strA | blaCTX-M-15 blaOXA-1 | aac(6’)Ib-cr | mph(A) | catB3 | sul1 sul2 | tet(A) | dfrA17 | |
| 237C | aac(3)-Iia aadA5 strA strB | blaCTX-M-15 blaOXA-1 | aac(6’)Ib-cr | mph(A) | catB3 | sul1 sul2 | tet(B) | dfrA17 | |
| 244 | aac(3)-IIa aadA5 strA strB | blaCTX-M-15 blaOXA-1 | aac(6’)Ib-cr | mph(A) | catB3 | sul2 sul1 | tet(B) | dfrA17 | |
| 245 | strA aac(3)-IIa strB aadA5 | blaCTX-M-15 blaOXA-1 | aac(6’)Ib-cr | mph(A) | catB3 | sul1 sul2 | tet(B) | dfrA17 | |
| 247 | strA strB aadA5 | blaOXA-1 blaCTX-M-15 | aac(6’)Ib-cr | mph(A) | catB3 | sul2 sul1 | tet(A) | dfrA17 | |
| 298 | strB strA | blaTEM-1B | catA1 | sul2 | tet(D) | dfrA7 | |||
| 365 | aadA5 | blaCTX-M-15 blaOXA-1 | aac(6’)Ib-cr | mph(A) | catB3 | sul1 | tet(A) | dfrA17 | |
| 393 | aac(3)-Iia aadA5 | blaCTX-M-15 blaOXA-1 | aac(6’)Ib-cr | mph(A) | catB3 | sul1 | tet(B) | dfrA17 | |
| 431D | erm(B) strA strB | sul2 | |||||||
| 521 | sul2 sul1 | dfrA5 | |||||||
| 538 | aac(3)-Iia aadA5 strA strB | blaCTX-M-15 blaOXA-1 | aac(6’)Ib-cr | mph(A) | catB3 | sul1 sul2 | tet(A) tet(B) | dfrA17 | |
| 554 | strA strB | blaTEM-1B | sul2 | dfrA5 | |||||
| 567B | aadA5 | mph(A) | sul1 | dfrA17 | |||||
| 587 | aadA5 | blaCTX-M-15 | mph(A) | sul1 | dfrA17 | ||||
| 603 | strB aadA5 aac(3)-IId strA | blaTEM-1B | mph(A) | sul2 sul1 | dfrA17 | ||||
| 118A | strB strA | blaTEM-1B | sul1 sul2 | dfrA7 | |||||
| 119EC | |||||||||
| 340B | strB strA | sul2 | tet(A) | dfrA14 | |||||
| 598A | strB strA | blaTEM-1B | mph(A) | sul2 | tet(B) | dfrA17 | |||
| 598B | aac(3)-IIa strB strA | blaTEM-1B blaCTX-M-15 blaOXA-1 | aac(6’)Ib-cr | mph(A) | catB3 | sul2 | tet(A) | dfrA14 | |
| 71E | aadA5 aac(3)-IIa strB strA | blaCTX-M-15 blaOXA-1 | aac(6’)Ib-cr | mph(A) | catB3 | sul1 sul2 | tet(A) | dfrA17 | |
| 77E | strA strB | blaCTX-M-15 blaOXA-1 | aac(6’)Ib-cr | catB3 | sul2 | tet(B) | |||
a Aminoglycoside
b Beta-Lactam
c Fluoroquinolones and aminoglycoside
A total of 19 (50%) E. coli carried mph(A), a gene encoding macrolide resistance enzymes including towards erythromycin and azithromycin. The prevalence of genes encoding sulphonamides resistance enzyme for sul1 and sul2 were 19 (47.5%) and 24 (60.5%), respectively. The proportion of genes encoding tetracycline resistance enzymes for tet(A) and tet(B) were 16 (42.1%) and 9 (23.7%) respectively. The proportions of genes encoding chloramphenicol resistance enzymes were 19 (50%) for catB3 and 3 (7.9%) for catA1. A total of 22 (57.9%) isolates carried both strA and strB genes against aminoglycoside. The gene aadA5 was detected in 15 (39.5%), for each aadA2 and aadA1 in 2 (5.3%) and aadB in 1 (2.6%) E. coli isolates. Others included aac(3)-IIa in 9 (23.7%) and aac(3)-IId in 2 (5.3%) of the E. coli isolates.
Phenotype and sequence based antimicrobial resistance comparison
Agreement between phenotype and whole genome sequence based antimicrobial resistance was done for 24 out of 38 E. coli isolates (Table 4). On average, agreement across all drugs tested was 83.9%. Overall, the phenotypically determined resistance was higher than sequence-based resistance. However, all but trimethoprim sulpha or co-trimoxazole showed strong agreement (81–100%) between phenotype and sequence-based resistance results. Trimethoprim/sulphamethoxazole (co-trimoxazole) showed moderate agreement: 45.8%, kappa =15% and p = 0.08. Sequence-based analysis predicted resistance in 4 (16.7%) isolates, whereas phenotypic testing revealed 17 (70.8%) isolates to be resistant. Amoxicillin-clavulanate showed strongest agreement: 87.5%, kappa = 74% and p = 0.0001. Sequence-based analysis predicted resistance to amoxicillin-clavulanate in 14 (58.3%) isolates, whereas 15 (62.5%) isolates were found to be resistant phenotypically.
Table 4.
Agreement between phenotypically tested and whole genome sequence predicted antimicrobial resistance
| Antibiotic name | DSTa | WGSb | Agreement | Kappa | P value |
|---|---|---|---|---|---|
| Amoxicillin-Clavulanate | 15 (62.5%) | 14 (58.3%) | 0.875 | 0.74 | 0.0001 |
| Ampicillin | 16 (66.7%) | 14 (58.3%) | 0.9167 | 0.82 | 0.00 |
| Ceftazidime | 13 (54.2%) | 13 (54.2%) | 0.9167 | 0.83 | 0.00 |
| Ceftriaxone | 15 (62.5) | 13 (54.2%) | 0.9167 | 0.84 | 0.00 |
| Chloramphenicol | 2 (8.3%) | 2 (8.3%) | 0.8333 | −0.09 | 0.672 |
| Ciprofloxacin | 15 (62.5%) | 14 (58.3%) | 0.875 | 0.74 | 0.0001 |
| Gentamycin | 7 (29.2%) | 7 (29.2%) | 0.9167 | 0.79 | 0.00 |
| Trimethoprim Sulpha | 17 (70.8%) | 4 (16.7%) | 0.4583 | 0.15 | 0.0799 |
a phenotype-based resistance
b whole genome sequence-based resistance
Phylogeny and genome comparison
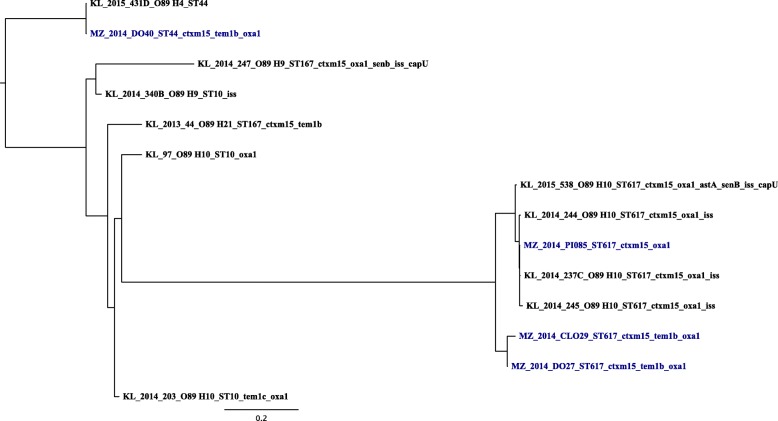
The observed minimum and maximum SNPs difference between one isolate and another in a pairwise genome comparison for ST131 were 234 and 10,425 respectively (Additional file 1: Table S1). The tree topology for ST131 showed one isolate segregating very distinctly from the rest (Fig. 1). For ST10 clonal complex the observed minimum and maximum SNPs difference between one isolate and another were 157 and 35,103 respectively (Additional file 2: Table S2). Looking at the tree for ST10 clonal complex (Fig. 2), limited pattern of the E. coli isolates was observed. In this tree, with the exception of ST617 in the middle clade of the ST10 clonal complex tree, all other STs making this complex occurred across the trees with little apparent segregation. Furthermore, the resistance and virulence genes were spread almost universally across both phylogenetic trees.
Fig. 1.
Phylogenetic analysis of 10 ST131 E.coli isolates showing serotypes, STs, resistance and virulence genes. KL stands for isolates sampled in the present study from patients hospitalised at a tertiary hospital in Kilimanjaro. MZ (blue colour) stands for isolates from companion and domesticated animals in Mwanza. The MZ sequences were downloaded as raw reads from European Nucleotide Archive (ENA) under the project number PRJEB12335. Number next to KL and MZ is date (year) of sampling or E. coli isolation. CTXM15 stands for blaCTX-M-15, OXA1 for blaOXA-1 and TEM1 for blaTEM-1
Fig. 2.
Phylogenetic analysis of 14 ST10 clonal complex (ST10, ST44, ST167, ST617) E.coli isolates showing serotypes, STs, resistance and virulence genes. KL stands for stands for isolates from patients hospitalised at a tertiary hospital in Kilimanjaro. MZ (blue colour) stands for isolates from companion and domesticated animals in Mwanza. The MZ sequences were downloaded as raw reads from European Nucleotide Archive (ENA) under the project number PRJEB12335. Number next to KL and MZ is date (year) of sampling or E. coli isolation. CTXM15 stands for blaCTX-M-15, OXA1 for blaOXA-1 and TEM1 for blaTEM-1
Discussion
The present study revealed a high diversity of E. coli strains circulating in KCMC hospital settings as measured by Multi-Locus Sequence Typing and Serotyping. However, strains belonging to O25:H4-ST131 and O89:H10-ST617 (ST10 clonal complex) were found to predominate. These findings are similar to the findings from other continents describing the spread and predominance of these endemic clones in health facilities [31–34]. The observed clonal diversity in this hospital may suggest sporadic introductions of diverse strains into the hospital from the community. To explore whether or not similar STs were clonally related, SNP difference between isolates and phylogenetic analysis suggested the existence of multiple clones of E. coli in these settings. A similar clonal diversity was observed when STs from the present study were compared to similar STs of E. coli isolates from companion and other domesticated animals in Mwanza, north western Tanzania.
Overall, levels of antimicrobial resistance in E. coli isolates were observed to be high. Trimethoprim/sulphamethoxazole (co-trimoxazole) resistance was observed to be lower (70%) than the one (93%) found by Seni et al. [30], but in the present study on average 54–60% resistance to amoxicillin-clavulanate, ampicillin, ceftazidime, ceftriaxone, and ciprofloxacin was higher than that reported by Seni et al. [30]. Strong agreement (81–100%) between sequence and phenotype-based resistance to all drugs tested was observed. However, trimethoprim/sulphamethoxazole revealed moderate agreement (45.8%) between the two methods. The phenotypically determined resistance to trimethoprim/sulphamethoxazole was higher than sequence-based resistance. The plausible explanations for the observed difference could be that our analysis used only known resistance genes and did not include point mutations. Also, resistance in gram negative bacteria including E. coli is multifactorial and not all genes involved in resistance mechanisms have been uncovered.
Following analysis of resistance genes, over 70% of the E. coli strains carried genes encoding beta-lactamases, with blaOXA-1 being predominant (50%) followed by blaCTX-M-15 (44.7%) and blaTEM-1B (31.6%). The prevalence of blaCTX-M-15 found in the present study contrasts with Manyahi et al. [18] who found blaCTX-M-15 as the most prevalent gene (90.6%) in a tertiary hospital in Dar es Salaam. Nonetheless, similar to the present study, O25:H4-ST131 was reported to be a major cause of MDR E. coli infections [35]. The proportion (70%) of E. coli strains carrying genes encoding beta-lactamases, correlates in this study with a high (50%) presence of aac(6’)Ib-cr encoding ciprofloxacin resistance enzymes. Although aac(6’)Ib-cr encodes low level ciprofloxacin resistance by itself (as well as aminoglycoside resistance), and usually requires additional mutations (e.g. in chromosomal gyrA or parC) to confer high level resistance, it is a threat to ciprofloxacin which in our settings is one of the most prescribed drugs. Other studies in the US [36, 37], Brazil [38] and Korea [34] have documented similar findings to the present study of the co-carriage of aac(6’)Ib-cr, ESBL genes and the existence of ciprofloxacin resistance in E. coli ST131. Further, an agreement between the presence or absence of aac(6’)Ib-cr and phenotypic resistance results in particular ciprofloxacin was noted by Madoshi et al. [39] who characterised E. coli isolates from healthy cattle and cattle attendants in Morogoro, Tanzania [39]. Among other fluoroquinolones, ciprofloxacin is one of the most prescribed antibiotics in Tanzania. Plausibly this explains the observed linkage between ciprofloxacin resistance and carriage of aac(6’)Ib-cr and CTX-M ESBL [31]. The linkage between quinolone resistance and sub-lineages of ST131 has been recently explored by Zakour et al. [40] whereby this work suggested that quinolone use is associated with the acquisition of virulence and fluoroquinolone resistance determinants and expansion of clade C2/H30-Rx of quinolone-resistant ST131. Also a gene (mph(A)) conferring resistance to macrolides including erythromycin and azithromycin was detected in a high proportion. The presence of the mph(A) in E. coli isolates of the current study was 50%, and thus, higher than the 13% that was found in E. coli from 5 countries from 4 continents by Nguyen et al. [41]. High exposures to erythromycin and azithromycin could be one possible reason leading to emergence of resistance to macrolides [42].
Furthermore, the present study noted a high proportion (73.7%) of dfrA genes encoding trimethoprim resistance enzymes similar to findings by Madoshi et al. [39] in Morogoro, Tanzania. Nonetheless, the present study proportion of dfrA genes was relatively higher than that (43%) found in E. coli isolates from healthy college students in Ghana and Nigeria in 2005 and 2009 [43]. The observed proportion difference could be explained by the fact that resistance in hospital-based studies is relatively higher than in community-based ones. Another reason could be that the present study was hospital-based whereas the West African study population were healthy individuals.
The present study also characterised VFs in all E. coli isolates. The VFs that facilitate E. coli and host cells or E. coli and E. coli adherence including long polar fimbriae (lpfA), adhesin (iha) and intimin (eae) were predominant among the circulating E. coli O25:H4-ST131 strains. These VFs have been identified as suggestive of virulent serotypes and may be used as reliable markers for the identification of pathogenic E. coli [44]. Another prevalent group of VFs this study noted were those responsible for E. coli immune evasion by increasing serum survival (iss). Although the predominance of iss in the present study was from E. coli isolated from hospitalised patients, in Mwanza north-western Tanzania, Msahana et al. [45] noted a similar pattern of iss in E. coli that were community-acquired. In this study we observed no clear correlation or pattern of VFs with the clinical findings or patient’s outcomes. However, colonisation events rather than infections were common as wound or pus swab and stool specimens constituted the majority of strains studied. The observed existence of multiple VFs further underlines the exceptional ability of E. coli in colonising or causing infections to a wide range of hosts and niches.
Potential clinical implications for the obtained results are that caution should be taken when interpreting and utilising microbiology results especially when E. coli is isolated in LMICs as most often E. coli is regarded of low threat. Contrary to that notion, these findings highlight that E. coli should not be regarded non-pathogenic until pathogenic and antimicrobial resistance determinants have been truly confirmed absent. Additionally, the WGS-based findings hint on the existence of nosocomial transmissions in the hospital thus prompting the formulation of pragmatic antimicrobial stewardships and infection prevention and control initiatives.
We understand and acknowledge the limitations of the present study. First, the analysis was performed on a small number of E. coli isolates, which may limit generalisation of the findings. With such a small number of the isolates analysed, it is important to point out that another limitation that this work was likely to suffer from was the lack of deeper statistical analysis to correlate the isolates resistance and virulence findings with patients characteristics (age, gender, ward, room, specimen) Third, the existence of genes encoding different resistance and virulence factors is only indicative of the genes present in the isolates. On the other hand, RNA sequencing which is an avenue for future studies could have given a strong evidence of expression levels of genes encoding different resistance and virulence factors.
Conclusion
The observed high levels of E. coli diversity in terms of their antimicrobial resistance genes, serotypes and virulence genes underlines the necessity for concerted efforts to routinely screen all bacterial isolates of clinical importance especially in tertiary health care facilities. WGS use for laboratory-based surveillance can be an effective early warning system for emerging pathogens and resistance mechanisms in LMICs. The information generated in this study will not only provide updates on levels of virulence and antimicrobial resistance at hospital level but will also be used as a basis in formulating pragmatic antimicrobial stewardships and infection prevention and control initiatives.
Additional files
Table S1. A pairwise genome comparison matrix of SNP differences for E. coli ST131. (XLS 34 kb)
Table S2. A pairwise genome comparison matrix of SNP differences for E. coli ST10 clonal complex. (XLS 29 kb)
Acknowledgements
We thank the management of Kilimanjaro Christian Medical Centre and all patients who consented to participate in this study.
Funding
This study was supported by DANIDA through Danida Fellowship Centre award number DFC No. 12-007DTU.
Availability of data and materials
Data are available on request to the authors.
Authors’ contributions
TS conceived the initial idea; FA, OL, MA, BM and GK refined the idea. TS and HK performed laboratory analyses. TS and MZ analysed data and prepared manuscript draft. All authors read, revised and approved the final manuscript.
Ethics approval and consent to participate
This study was granted ethical approval by the KCMC Research Ethics Committee and the National Institute for Medical Research with approval numbers 893 and NIMR/HQ/R.8a/Vol.IX/2080 respectively. A written informed consent was obtained from each participant or from parents or guardians of children before enrolment into the study.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13756-018-0361-x) contains supplementary material, which is available to authorized users.
Contributor Information
Tolbert Sonda, Email: t.sonda@kcri.ac.tz.
Happiness Kumburu, Email: h.kumburu@kcri.ac.tz.
Marco van Zwetselaar, Email: zwets@kcri.ac.tz.
Michael Alifrangis, Email: micali@sund.ku.dk.
Blandina T. Mmbaga, Email: b.mmbaga@kcri.ac.tz
Frank M. Aarestrup, Email: fmaa@food.dtu.dk
Gibson Kibiki, Email: g.kibiki@gmail.com.
Ole Lund, Email: lund@cbs.dtu.dk.
References
- 1.Kumburu HH, Sonda T, Mmbaga BT, Alifrangis M, Lund O, Kibiki G, Aarestrup FM. Patterns of infections, aetiological agents and antimicrobial resistance at a tertiary care hospital in northern Tanzania. Trop Med Int Health. 2017;22:454–464. doi: 10.1111/tmi.12836. [DOI] [PubMed] [Google Scholar]
- 2.Moissenet D, Salauze B, Clermont O, Bingen E, Arlet G, Denamur E, Mérens A, Mitanchez D, Vu-Thien H. Meningitis caused by Escherichia coli producing TEM-52 extended-spectrum ??-lactamase within an extensive outbreak in a neonatal ward: epidemiological investigation and characterization of the strain. J Clin Microbiol. 2010;48:2459–2463. doi: 10.1128/JCM.00529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afset JE, Bevanger L, Romundstad P, Bergh K. Association of atypical enteropathogenic Escherichia coli (EPEC) with prolonged diarrhoea. J Med Microbiol. 2004;53:1137–1144. doi: 10.1099/jmm.0.45719-0. [DOI] [PubMed] [Google Scholar]
- 4.Olesen B, Scheutz F, Andersen RL, Menard M, Boisen N, Johnston B, Hansen DS, Krogfelt KA, Nataro JP, Johnson JR. Enteroaggregative Escherichia coli O78:H10, the cause of an outbreak of urinary tract infection. J Clin Microbiol. 2012;50:3703–3711. doi: 10.1128/JCM.01909-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahende C, Ngasala B, Lusingu J, Butichi A, Lushino P, Lemnge M, Premji Z. Aetiology of acute febrile episodes in children attending Korogwe District hospital in North-Eastern Tanzania. PLoS One. 2014;9:e104197. doi: 10.1371/journal.pone.0104197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berman H, Barberino MG, Moreira ED, Riley L, Reis JN. Distribution of strain type and antimicrobial susceptibility of Escherichia coli isolates causing meningitis in a large urban setting in Brazil. J Clin Microbiol. 2014;52:1418–1422. doi: 10.1128/JCM.03104-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabal A, García-Castillo M, Cantón R, Gortázar C, Domínguez L, Álvarez J. Prevalence of Escherichia coli virulence genes in patients with diarrhea and a subpopulation of healthy volunteers in Madrid, Spain. Front Microbiol. 2016;7:1–6. doi: 10.3389/fmicb.2016.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shahrani M, Dehkordi FS, Momtaz H. Characterization of Escherichia coli virulence genes, pathotypes and antibiotic resistance properties in diarrheic calves in Iran. Biol Res. 2014;47:28. doi: 10.1186/0717-6287-47-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imuta N, Ooka T, Seto K, Kawahara R, Koriyama T, Kojyo T, Iguchi A, Tokuda K, Kawamura H, Yoshiie K, Ogura Y, Hayashi T, Nishi J. Phylogenetic analysis of enteroaggregative Escherichia coli (eaec) isolates from Japan reveals emergence of ctx-m-14-producing eaec o25:h4 clones related to sequence type 131. J Clin Microbiol. 2016;54:2128–2134. doi: 10.1128/JCM.00711-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beutin L, Hammerl JA, Strauch E, Reetz J, Dieckmann R, Kelner-Burgos Y, Martin A, Miko A, Strockbine NA, Lindstedt BA, Horn D, Monse H, Huettel B, Muller I, Stuber K, Reinhardt R. Spread of a distinct Stx2-encoding phage prototype among Escherichia coli O104:H4 strains from outbreaks in Germany, Norway, and Georgia. J Virol. 2012;86:10444–10455. doi: 10.1128/JVI.00986-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferdous M, Zhou K, de Boer RF, Friedrich AW, Kooistra-Smid AMD, Rossen JWA. Comprehensive characterization of Escherichia coli O104:H4 isolated from patients in the Netherlands. Front Microbiol. 2015;6:1–9. doi: 10.3389/fmicb.2015.01348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dallman TJ, Chattaway MA, Cowley LA, Doumith M, Tewolde R, Wooldridge DJ, Underwood A, Ready D, Wain J, Foster K, Grant KA, Jenkins C. An investigation of the diversity of strains of enteroaggregative escherichia coli isolated from cases associated with a large multi-pathogen foodborne outbreak in the UK. PLoS One. 2014;9:e98103. doi: 10.1371/journal.pone.0098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masika WG, O’Meara WP, Holland TL, Armstrong J. Contribution of urinary tract infection to the burden of febrile illnesses in young children in rural Kenya. PLoS One. 2017;12:e0174199. doi: 10.1371/journal.pone.0174199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Msaki BP, Mshana SE, Hokororo A, Mazigo HD, Morona D. Prevalence and predictors of urinary tract infection and severe malaria among febrile children attending Makongoro health Centre in Mwanza city, north-western Tanzania. Archives of Public Health. 2012;70:4. doi: 10.1186/0778-7367-70-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed M, Moremi N, Mirambo MM, Hokororo A, Mushi MF, Seni J, Kamugisha E, Mshana SE. Multi-resistant gram negative enteric bacteria causing urinary tract infection among malnourished underfives admitted at a tertiary hospital, northwestern, Tanzania. Ital J Pediatr. 2015;41:1–5. doi: 10.1186/s13052-015-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hildenwall H, Amos B, Mtove G, Muro F, Cederlund K, Reyburn H. Causes of non-malarial febrile illness in outpatients in Tanzania. Trop Med Int Health. 2016;21:149–156. doi: 10.1111/tmi.12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moyo SJ, Aboud S, Kasubi M, Lyamuya EF, Maselle SY. Antimicrobial resistance among producers and non-producers of extended spectrum beta-lactamases in urinary isolates at a tertiary Hospital in Tanzania. BMC research notes. 2010;3:348. doi: 10.1186/1756-0500-3-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manyahi J, Moyo SJ, Tellevik MG, Ndugulile F, Urassa W, Blomberg B, Langeland N. Detection of CTX-M-15 beta-lactamases in Enterobacteriaceae causing hospital- and community-acquired urinary tract infections as early as 2004, in Dar es salaam, Tanzania. BMC Infect Dis. 2017;17:282. doi: 10.1186/s12879-017-2395-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong Y, Wang P, Lan R, Ye C, Wang H, Ren J, Jing H, Wang Y, Zhou Z, Bai X, Cui Z, Luo X, Zhao A, Wang Y, Zhang S, Sun H, Wang L, Xu J. A novel escherichia coli O157:H7 clone causing a major hemolytic uremic syndrome outbreak in China. PLoS One. 2012;7:1–10. doi: 10.1371/annotation/a5edef40-e46d-4810-9008-dbda429ccc2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, Bernard H, Fruth A, Prager R, Spode A, Wadl M, Zoufaly A, Jordan S, Kemper MJ, Follin P, Müller L, King LA, Rosner B, Buchholz U, Stark K, Krause G. Epidemic profile of Shiga-toxin–producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med. 2011;365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 21.Nextera® XT Library Prep Reference Guide. http://support.illumina.com/downloads/nextera_xt_sample_preparation_guide_15031942.html.
- 22.FastQC A Quality Control Tool for High Throughput Sequence Data. www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 23.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joensen KG, Tetzschner AMM, Iguchi A, Aarestrup FM, Scheutz F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol. 2015;53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014;52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One. 2014;9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seni J, Falgenhauer L, Simeo N, Mirambo MM, Imirzalioglu C, Matee M, Rweyemamu M, Chakraborty T, Mshana SE. Multiple ESBL-producing Escherichia coli sequence types carrying quinolone and aminoglycoside resistance genes circulating in companion and domestic farm animals in Mwanza, Tanzania, harbor commonly occurring plasmids. Front Microbiol. 2016;7:1–8. doi: 10.3389/fmicb.2016.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valenza G, Nickel S, Pfeifer Y, Pietsch M, Voigtländer E, Lehner-Reindl V, Höller C. Prevalence and genetic diversity of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli in nursing homes in Bavaria, Germany. Vet Microbiol. 2017;200:138–141. doi: 10.1016/j.vetmic.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 32.Aibinu I, Odugbemi T, Koenig W, Ghebremedhin B. Sequence Type ST131 and ST10 Complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin Microbiol Infect. 2012;18:E49–E51. doi: 10.1111/j.1469-0691.2011.03730.x. [DOI] [PubMed] [Google Scholar]
- 33.Oteo J, Diestra K, Juan C, Bautista V, Novais Â, Pérez-Vázquez M, Moyá B, Miró E, Coque TM, Oliver A, Cantón R, Navarro F, Campos J. Extended-spectrum β-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/a, ST23 complex/a and ST131/B2. Int J Antimicrob Agents. 2009;34:173–176. doi: 10.1016/j.ijantimicag.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Cha MK, Kang CI, Kim SH, Cho SY, Ha YE, Wi YM, Chung DR, Peck KR, Song JH, Son JS, Moon SY, Kim CK, Lee SS, Lee JA, Kim YS, Sohn KM, Rhee JY, Jung SI, Park KH, Kang SJ, Kim SW, Chang HH, Ryu SY, Kim HA, Kwon KT, Lim MH. Comparison of the microbiological characteristics and virulence factors of ST131 and non-ST131 clones among extended-spectrum β-lactamase-producing Escherichia coli causing bacteremia. Diagn Microbiol Infect Dis. 2016;84:102–104. doi: 10.1016/j.diagmicrobio.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 35.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. Escherichia coli sequence type ST131 as the major cause of serious multidrug-Resistant E. Coli infections in the United States. Clin Infect Dis. 2010;51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 36.Peirano G, Costello M, Pitout JDD. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli from the Chicago area: high prevalence of ST131 producing CTX-M-15 in community hospitals. Int J Antimicrob Agents. 2010;36:19–23. doi: 10.1016/j.ijantimicag.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 37.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother. 2006;50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peirano G, Asensi MD, Pitondo-Silva A, Pitout JDD. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli from Rio de Janeiro, Brazil. Clin Microbiol Infect. 2011;17:1039–1043. doi: 10.1111/j.1469-0691.2010.03440.x. [DOI] [PubMed] [Google Scholar]
- 39.Madoshi BP, Kudirkiene E, Mtambo MMA, Muhairwa AP, Lupindu AM, Olsen JE. Characterisation of commensal Escherichia coli isolated from apparently healthy cattle and their attendants in Tanzania. PLoS One. 2016;11:e0168160. doi: 10.1371/journal.pone.0168160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, Khanh Nhu NT, Roberts LW, Stanton-Cook M, Schembri MA, Beatson SA. Sequential Acquisition of Virulence and Fluoroquinolone Resistance has Shaped the evolution of Escherichia coli ST131. mBio. 2016;7:e00347-16. doi: 10.1128/mBio.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phuc Nguyen MC, Woerther P-L, Bouvet M, Andremont A, Leclercq R, Canu A. Escherichia coli as reservoir for macrolide resistance genes. Emerg Infect Dis. 2009;15:1648–1650. doi: 10.3201/eid1510.090696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noguchi N, Takada K, Katayama J, Emura A, Sasatsu M. Regulation of transcription of the mph(a) gene for macrolide 2′-phosphotransferase I in Escherichia coli: characterization of the regulatory gene mphR(a) J Bacteriol. 2000;182:5052–5058. doi: 10.1128/JB.182.18.5052-5058.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labar AS, Millman JS, Ruebush E, Opintan JA, Bishar RA, Aboderin AO, Newman MJ, Lamikanra A, Okeke IN. Regional dissemination of a trimethoprim-resistance gene cassette via a successful transposable element. PLoS One. 2012;7:e38142. doi: 10.1371/journal.pone.0038142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torres AG, Blanco M, Valenzuela P, Slater TM, Patel SD, Dahbi G, López C, Barriga XF, Blanco JE, Gomes TAT, Vidal R, Blanco J. Genes related to long polar fimbriae of pathogenic Escherichia coli strains as reliable markers to identify virulent isolates. J Clin Microbiol. 2009;47:2442–2451. doi: 10.1128/JCM.00566-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mshana SE, Falgenhauer L, Mirambo MM, Mushi MF, Moremi N, Julius R, Seni J, Imirzalioglu C, Matee M, Chakraborty T. Predictors of blaCTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC Infectious Diseases. 2016;16:1–9. doi: 10.1186/s12879-016-1527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. A pairwise genome comparison matrix of SNP differences for E. coli ST131. (XLS 34 kb)
Table S2. A pairwise genome comparison matrix of SNP differences for E. coli ST10 clonal complex. (XLS 29 kb)
Data Availability Statement
Data are available on request to the authors.