Abstract
We report the development of the SEVENS database, which contains information on G-protein coupled receptor (GPCR) genes that are identified with high confidence levels (A, B, C, and D) from various eukaryotic genomes, by using a pipeline comprising bioinformatics softwares, including a gene finder, a sequence alignment tool, a motif and domain assignment tool, and a transmembrane helix predictor.
SEVENS compiles detailed information on GPCR genes, such as chromosomal mapping position, phylogenetic tree, sequence similarity to known genes, and protein function described by motif/domain and transmembrane helices. They are presented in a user-friendly interface. Because of the comprehensive gene findings from genomes, SEVENS contains a larger data set than that of previous databases and enables the performance of a genome-scale overview of all the GPCR genes. We surveyed the complete genomes of 68 eukaryotes, and found that there were between 6 and 3,470 GPCR genes for each genome (Level A data). Within these genes, the number of receptors for various molecules, including biological amines, peptides, and lipids, were conserved in mammals, birds, and fishes, whereas the numbers of odorant receptors and pheromone receptors were highly diverse in mammals. SEVENS is freely available at http://sevens.cbrc.jp or http://sevens.chem.aoyama.ac.jp.
Keywords: G-protein coupled receptor, gene finding, bioinformatics, comparative genome analysis, functional annotation
Significance.
This paper introduces the SEVENS database, which contains information on G-protein coupled receptor (GPCR) genes identified from the genome of 68 eukaryotic species by using a bioinformatics approach. The coordinates of the GPCR gene on the genome and functional annotation information of the genes are methodically displayed. SEVENS is useful for the comparative genomic analysis of GPCRs.
G-protein coupled receptors (GPCRs), characterized by seven transmembrane (TM) helices, bind to various kinds of ligands, such as biological amines, peptides, hormones and odorant substances, from the extracellular side. This induces signal transduction to the inner cell through the activation of G-proteins. GPCRs exist in most types of cells and abnormalities in the signal transduction pathways are related to a variety of serious conditions, including heart trouble, cancer, and high blood pressure. As approximately 30% of medicines used worldwide have been designed to control this receptor system [1,2], both academic and industrial researchers have attempted to elucidate the functional mechanisms of GPCRs. For several decades the experimental complexity of structure determination and gene expression process prevented the researchers from understanding the functional mechanisms of GPCRs. However, the recent reporting of large amount of genomic information of many species and several new structures of GPCRs has made it possible to obtain an overview of the structure and sequence space to understand the GPCR functions through bioinformatic methodologies. Many useful GPCR databases are already available for the collection and overview of GPCR sequences, such as GPCRDB [3], IUPHAR (GPCR database) [4], GPCR-PD™ [5], and ORDB [6]. GPCRDB, the most popular database, provides known GPCR sequences gathered from UniProt and GenBank. IUPHAR and GPCR-PD™ accumulate literature information and known GPCR sequences. ORDB specifically focuses on the olfactory receptors, a subfamily of GPCRs. These databases collate known GPCR sequences obtained from experiments and are well organized for the analysis of known GPCR genes. However, to achieve an overview of the “GPCR proteome”, it is necessary to consider the comprehensive dataset that includes not only known GPCR sequences, but also novel sequences that cannot be detected by in vivo experiments, despite their confirmed existence in genomic DNA sequences. If GPCR genes can be comprehensively collected, including the novel genes, the general rules of function common to all GPCR genes will be understood, rather than biased rules common to limited families. Furthermore, if these novel GPCR genes can be predicted in advance from the genome sequences, they present new pathways for drug discovery. For this purpose, we have developed a bioinformatics pipeline for the comprehensive identification of GPCR genes in the genome sequences of various species and have stored information on GPCR families in an integrated database, known as SEVENS. The contents of the database are updated as the number of known genomes of eukaryotic species increases. For example, in 2005, we reported the analysis of GPCR genes from seven eukaryotic genomes [7], whereas in 2009, we analyzed GPCRs from 34 eukaryotic genomes [8], and we have discussed the usefulness of SEVENS for reverse genetic analysis. Furthermore, the statistical analysis of GPCR genes from 57 eukaryotic genomes confirmed their variations from the perspective of amino acid conservation and gene position on the genome [9].
In this paper, we have introduced the most up-to-date version of the SEVENS database (http://sevens.cbrc.jp and http://sevens.chem.aoyama.ac.jp), which contains an exhaustive collection of GPCR genes from the genomes of 68 eukaryotic species.
Method
Computational identification of GPCR genes from genome sequences
We identified GPCR genes from 68 eukaryotic genome sequences by using our computational gene discovery pipeline, which is composed of the following two stages: (a) GPCR gene finding stage and (b) the GPCR gene screening stage.
The genome sequences are obtained from the ftp sites of NCBI (http://www.ncbi.nih.gov/), UCSC (http://genome.ucsu.edu), Ensembl (http://ensemblgenomes.org/), the Broad Institute (https://www.broadinstitute.org/), Baylor College of Medicine (https://www.bcm.edu/), dictyBase (http://www.dictybase.org/), and IRGSP (http://rgp.dna.affrc.go.jp/J/IRGSP/).
(a) Gene finding stage
To maximize the total number of gene candidates, we selected two types of sequence set from the genome sequences. The first sequence set is termed the “6f-sequences”, which were all the possible combinations between the initial and stop codons in six reading frames, by using the rule that most upstream ATG in the same sequence will be adopted. The second sequence set is termed the “ALN-sequences”. The genomic regions where at least partial exon regions of the known GPCR sequences were a hit with a significant score in TBLASTN [10] are listed. Around these exon regions, the full gene structure was built by using ALN [11], which performs dynamic alignment of known GPCR and genome sequence through consideration of the exon/intron boundary. These candidate sequence datasets still contain several redundancies: (1) Perfect matches or overlaps at the same genomic position: we regarded these as the same gene and adjusted the duplicate count; (2) multiple sequence copies in different genomic positions: we considered these to represent different genes; and (3) separate sequence fragments linked by a known protein sequence: These originate from an erroneous prediction by a gene finding program, so were merged by using the linker sequence (the known protein sequence). These redundancies were detected by the following clustering method: first, Smith-Waterman sequence alignment [12] was applied to the candidate sequences in an all-against-all fashion. Sequences were then linked together if the hit was more than 50 amino acids and with over 95% sequence identity, and shared the same chromosome number and overlapping genomic position. If chromosome numbers were unknown for (either/both) sequences, more than 99% sequence identity was required for the linking. After computing the transitive closures of the links, each of the known GPCR sequences from the Swiss-Prot database (http://www.uniprot.org/) was aligned against all the candidate sequences. All clusters for which the hit was more than 50 amino acids and with more than 99% identity were merged. Finally, in each cluster, the longest sequence was selected as representative. Short amino acid sequence less than 150 residues were eliminated; because GPCRs have seven transmembrane helices of approximately 20 residues each, the sequence length must be more than 140 residues.
In a previous work [7], we also reported another dataset called GD-sequences. However, as all GD-sequences were included in the ALN sequence regions and are finally integrated into the same gene by the above procedures, we decided not to use this sequence set here.
(b) GPCR gene screening stage
The identified amino acid sequences were analyzed by using gapped BLAST [13] for a similarity search to known GPCRs, HMMER to assign the Pfam domain [14], an in-house program for assigning PROSITE [15] patterns, and SOSUI [16] and our original tool (TMWindows) for predicting transmembrane helix (TMH) regions. TMwindows assigns the Engelman-Steitz-Goldman hydropathy [17] index to amino acid sequences and calculates the average hydrophobicity within a pre-determined window of 19 to 27 residues in length. After a comparison of all indices in the AAindex database [18], this index was selected as the most powerful for the discrimination membrane proteins from other proteins by using the total average hydrophobicity: if the average hydrophobicity within each window exceeds 2.5, the region is regarded as a transmembrane helix.
Each analysis tool was first assessed to determine two threshold settings: the “best specificity threshold” and “best sensitivity threshold”, when compared with the reference dataset which are 1,242 known GPCR sequences and 73,493 non-GPCR sequences in the Swiss-Prot (version 41) database. Here the known GPCR sequences were chosen from the file of Swiss-Prot under the condition that there are seven places of “TRANSMEM” descriptions on the “FT” region or “G-protein coupled receptor” descriptions on the “KW” region. And we searched known GPCR sequences, as query, against this reference dataset using Gapped BLAST or HMMER. Here, the upper limit of the E-values of pairs on known GPCRs is defined as the “best sensitivity threshold”, and the lower limit of the E-value of pairs on known GPCR and non-GPCR is defined as the “best specificity threshold”. The “best specificity threshold” is intended to achieve, when applied to the reference dataset, almost 100% specificity and the minimum false-negatives. In contrast, the “best sensitivity threshold” is intended to achieve almost 100% sensitivity and the minimum false-positives. For example, for a gapped BLAST search, the “best specificity threshold” is determined as E value <10−80 and the “best sensitivity threshold” is E value <10−30.
Then, four confidence levels (A, B, C, and D) of the datasets were generated through several combination (see Table 1) of the “best specificity threshold” and the “best sensitivity threshold”. For example, Level A dataset are obtained by “AND” combination of the candidate sequences obtained by the “best specificity thresholds” of the sequence similarity search and domain assignments. To discover remote homologs of GPCRs, we further added candidates using the three-level (B, C, and D; see Table 1) thresholds of TMH prediction with the level A dataset. The current prediction performances of candidate sequences are expected to be equivalent to the sensitivity and specificity (Table 1) of the datasets gathered by using above rules from the reference dataset.
Table 1.
Thresholds used for GPCR discovery
| Level A | Level B | Level C | Level D | |
|---|---|---|---|---|
| Sequence search with BLASTP | E<10−80* | E<10−30** | E<10−30** | E<10−30** |
| Pfam domain assignment with HMMER | E<10−10* | E<1.0** | E<1.0** | E<1.0** |
| PROSITE motif assignment | Not used | Match | Match | Match |
| TMH Prediction | Not used | TMwindows (7) AND SOSUI (7) | TMwindows (7) AND SOSUI (6–8) | TMwindows (7) OR SOSUI (7) |
| Sensitivity | 99.40% | 99.80% | 99.90% | 99.90% |
| Specificity | 96.60% | 70.00% | 48.40% | 20.00% |
Best specificity threshold of BLAST and HMMER against the reference data set.
Best sensitivity threshold of BLAST and HMMER against the reference data set.
The number in the parentheses represents the predicted number of TMH.
Finally, the screened sequences which were hits to the known GPCRs in Swiss-Prot (http://www.uniprot.org/) with an E-value <10−30 in the gapped BLAST search were categorized as the same subfamily of the known GPCR. In each subfamily, we also classified the sequences as known products when they show hits with known sequences with 96% similarity for more than 100 residues of the alignment regions. For sequences that do not satisfy this condition, we define them as novel genes.
Results
Annotated subfamily distribution
Currently, SEVENS (ver. 1.72) stores 109,835 genes (including 23,114 pseudogenes) from 68 eukaryotes. Of these gene candidates, the number of genes at each level is 67,901 (including 19,672 pseudogenes), 86,919 (23,063), 93,878 (23,090), 109,835 (23,114) for Levels A, B, C, and D, respectively. Novel genes for 68 species were 34,184 (Level A), 44,625 (Level B), 50,450 (Level C) and 64,630 (Level D). Especially, human novel genes are 24 (Level A), 184 (Level B), 310 (Level C) and 710 (Level D). From 68 eukaryotes, we identified some GPCRs in yeasts, a dozen in plants, approximately two hundred in insects, several hundred in fishes and birds, and between several hundred and several thousands in mammals. When we divide these numbers into the subfamily classification (represented by different colors in the tree view), there are only several in the genomes of lower species, whereas the number of subfamilies started to rapidly increase from fish genomes, and reached approximately 60 in mammalian genomes.
Within these subfamilies, the number of receptors for molecules such as biological amines, peptides, and lipids were conserved in mammals, birds, and fishes. In contrast, the number of receptors for chemical substances (such as odorants and pheromones) followed a unique distribution in different species. For example, the olfactory receptors extended to approximately 70% in GPCR genes and showed a large diversity in the numbers in mammals. Human and primate olfactory receptors constitute approximately 60% of their GPCR genes. The percentage of olfactory receptors is extremely high (more than 80%) in elephant, armadillo, sloth, cow, horse, and squirrel. It is interesting that the number of pheromone receptors increased only for limited species, such as tree shrew, rabbit, mouse, rat, platypus, and western clawed frog. In contrast, sea animals (e.g. dolphin) have a smaller number of olfactory receptors.
The current SEVENS version (ver. 1.72) has increased 12 kinds of species (7584 Level A GPCR genes including 2602 pseudo genes) than the 56 species of the previous version (1.70, [9]). These increases are Horse (Equuus Caballus), Sloth (Choepus hoffmanne), Nematode ( Caenorhabditis japonica) and subfamilies of Fruit fly ( Drosophila pseudoobscura, ananassae, erecta, grimshawi, mojavensis, persimilis, sechelliav, virilism and willistoni). These collections makes it possible to compare the genomes of 12 different kinds of Fruit Fly in the current SEVENS. They have 215, on average, Level A GPCR genes (from 184 to 240 genes). The difference in the number of genes was mainly due to the numbers of insect sensory receptors, Family 2 (B) receptors and Family 3 (C) receptors (metatropic glutamate and calcium receptors). Similarly, comparing the five Nematode genomes, there was large spread in the number of Level A GPCRs (from 349 to 889 genes). The difference in these numbers is largely due to Nematode chemosensory receptors, but it is also possible due to the qualities of the genome sequences.
Web representation
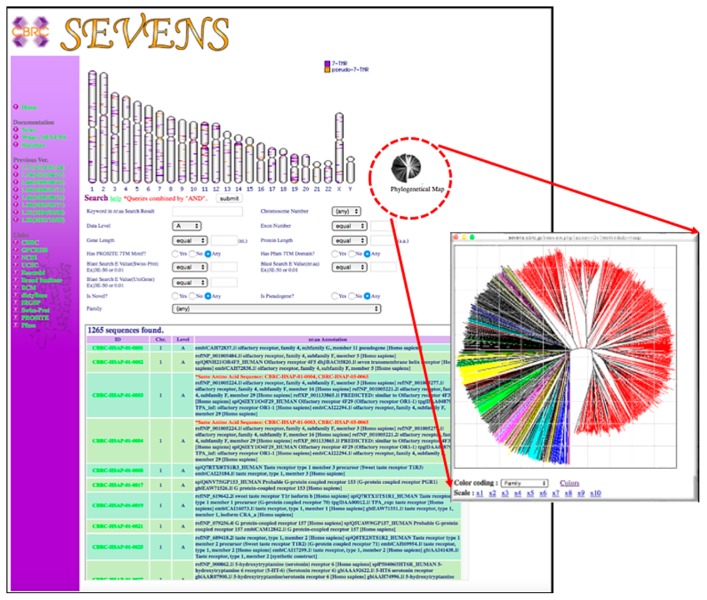
SEVENS is an integrated database in which various functional and structural information for each GPCR gene are visually presented and organized in hierarchical manner. The top page shows the table of the eukaryote genomes with information on the major GPCR subfamily of this genome. The selection of one species navigates to the next page for a content search (Fig. 1). Here, the chromosomal map, the phylogenetical icon, or the search condition entry forms are shown as the search items (Fig. 1). The chromosomal map shows the position of the GPCR genes, which are colored according to their status as actual genes or pseudogenes, and the selection of this leads to the gene annotation information page (Fig. 2). The phylogenetic icon in Figure 1 navigates to a GPCR tree viewer, where each leaf line can be colored according to the GPCR family or information on the chromosome number. The selection of a gene navigates to the gene annotation information page (Fig. 2). The search condition entry form in Figure 1 retrieves candidate GPCR genes by the “AND” combination of following information: keyword in nr.aa database search results; chromosome number; data level; predicted exon number; DNA and protein sequence length; E-value of sequence search against the Swiss-Prot or nr.aa database; and whether the query has GPCR-specific Pfam domains [14] and GPCR-specific PROSITE [15] motifs. These search results appear again in the chromosomal viewer and the lower table, which are linked to the “gene annotation information page” (Fig. 2).
Figure 1.
Content search page, which shows the chromosome map, phylogenetic icon, and search condition entry form. The chromosomal map in the upper region shows the position of GPCR genes colored according to their status as actual genes (purple) or pseudogenes (orange) and the selection of these positions leads to the result page. Selection of the phylogenetic icon indicates a GPCR tree viewer with each leaf line colored according to the GPCR family or chromosome number information. Selection of a gene navigates to the result page. The search condition entry form (in the middle of the figure) retrieves candidate GPCR genes through the “AND” combination of keyword in nr.aa (non-redundant amino acid) database search results, chromosome number, data level, predicted exon number, DNA and protein sequence length, E-value of sequence search against the Swiss-Prot or nr.aa database, and whether the query has GPCR-specific PROSITE motifs and GPCR-specific Pfam domains. The search results appear in the chromosomal viewer and the lower table, which are linked to the result page.
Figure 2.
a. Chromosomal coordinate information, together with the known information on regulatory regions (green bars). The board color represents the GC content of the genome sequence. The selected gene is colored red. In a table in the lower region of this figure, the information of the selected protein sequences (sequence search result against the Swiss-Prot/TrEMBL, nr.aa, and UniGene database using BLAST) is shown. Furthermore, information such as the gene expression pattern, binding ligand, the type of binding G-protein, and the composition of the amino acid sequence, are described in the bottom region. b. Structural information, such as predicted TM helix region (red bar) by SOSUI, PROSITE motif pattern regions (green bar), domain regions (blue bar), predicted disorder regions by DISOPRED (white bar), exon-intron boundary, pseudogene, novel genes, and regions of known structure (purple bar), can be observed. Each exon sequence appears when “EXON SEQUENCES” is clicked. The structures for class A GPCRs, determined by comparative modeling, are presented in Jmol 3D viewer. Based on these structures, the actual TM helix regions are displayed (yellow bar) on the structure board.
On the gene annotation information page (Fig. 2), information on the chromosomal coordinate, together with the information on the known regulatory regions appear on the board (Fig. 2a), with color changes according to the GC content of the genome sequence. This viewer has several functions: zoom in/out; centering; and position sliding. The selected gene is colored by red and this selection links to the information page of the selected protein sequence (Fig. 2a). Here, the table shows a sequence search result against the Swiss-Prot/TrEMBL, nr.aa, and UniGene databases by using gapped BLAST [13]. Furthermore, information as such as the gene expression pattern, the binding ligand, the type of binding G-protein, and composition of amino acid sequence is described.
In contrast, the structure board (Fig. 2b) shows the following information: the TM helix region predicted by SOSUI [16], PROSITE motif pattern regions, domain regions, disorder regions predicted by DISOPRED [19], exon-intron boundary, judgement of pseudogene, judgement of novel genes, and regions of known structure. Each exon sequence appears when the “EXON SEQUENCES” button is clicked.
As several three dimensional (3D) structures have already been elucidated the class A GPCR family, we were able to show the structures for all Class A GPCRs by the comparative modeling, which is presented by Jmol 3D viewer [20]. Based on these 3D structures, the actual TM helix regions are displayed on the structure board (Fig. 2b). The GPCR sequence regions that have a significant match (>80% sequence similarity) and a large coverage (>80%) of the PDB sequence are indicated with purple bars.
Discussion
From the perspective of the comprehensive collection of GPCRs, there are many databases available [3–6] in addition to SEVENS. With regard to comprehensive GPCR gene analysis, Fredriksson et al. [21,22] classified GPCR genes based on a new taxonomy, “GRAFS” of individual genomes, and many publications, including those on insects [23,24], plants [25], human, and mice [26] are available.
It seems that the number of GPCR genes collected in the Level A data set by our pipeline is in good agreement with the above studies, because the number of GPCR of these studies are based on Swiss-Prot data and most genes in Level A sequences show good hits to Swiss-Prot data. Each study has different criteria for collecting GPCRs, so it is impossible to compare, exactly, the number of GPCRs of SEVENS and that of other works with the same selection criteria. However, if we compare roughly the number of human GPCRs, it is 418 for GPCRdb, for example, 438 for SEVENS Level A dataset, which are in good agreement. (It is a comparison when olfactory receptor is excluded.)
The total number of genes in SEVENS are larger than those of the previous studies, because SEVENS also includes both predicted novel genes and pseudogenes. Naturally, the number of genes depends on how we combined each bioinformatic tools and how the thresholds are decided in the tools. Nevertheless, the accuracy of our results, at least for the Level A data, must be sufficient because our system can predict GPCR genes from known sequences with 99.4% sensitivity and 96.6% specificity; in case of the Levels B, C, and D datasets, although the specificity is smaller than that of Level A data, almost 100% sensitivity was achieved. Therefore, by using this pipeline, it is possible to cover the entire set of GPCR genes which is expected to include a larger number of GPCR genes than previous datasets.
The significance of collecting the entire set of GPCR genes is as follows.
Unbiased statistical analysis can be performed using all genes, not limited species and families. This will be useful for finding functional expression rules of universal GPCR across species and families. (2) The position of the GPCR gene on the genome sequence can be overlooked. By comparative genome analysis, we can see how the GPCR genes spread on genome sequences and function differentiated. This gives us new information that can not be gained by phylogenetic analysis of the sequence alone. Because of these advantages, new findings have been found [7,24,27,28] in the fields of structural analysis and genome analysis using SEVENS.
Concluding Remarks
SEVENS provides an infrastructure to access the general information of the GPCR universe. Additional information, such as expression data, tertiary structure constructed by modeling, functional data including as binding ligands or G-protein type, will be appended into the database with every update opportunity. Furthermore, we have a project in progress to gather GPCR signaling pathway data; in the near future, these data will also be compiled into SEVENS.
As this system is applicable to all species, our next project is to conduct comparative genomic studies of the GPCR genes among the available eukaryote genomes, the total of which is expected to increase rapidly. We hope that SEVENS will contribute to large-scale comparative genomic studies.
Acknowledgement
This research was partly supported by the Program for the Strategic Research Foundation at Private Universities, 2013–2017, from MEXT (Ministry of Education, Culture, Sports, Science and Technology) and partly supported by JSPS KAKENHI Grant Numbers 258056, 268041, and 15HP8050.
Footnotes
Conflicts of Interest
All authors declare that they have no conflict of interest.
Author Contributions
M. Suwa. directed the research. M. Ikeda. and M. Sugihara. helped to draft the manuscript. All authors discussed the database development, and critically reviewed and approved the final manuscript.
References
- 1.Vauquelin G, Mentzer BG. Protein-coupled Receptors. John Wiley & Sons, Ltd; West Sussex, England: 2007. [Google Scholar]
- 2.Rosenbaum DM, Rasmaussen SGF, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horn F, Vriend G, Cohen FE. Collecting and harvesting biological data: the GPCRDB and NucleaRDB information systems. Nucleic Acids Res. 2001;29:346–349. doi: 10.1093/nar/29.1.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harmar AJ, Hills RA, Rosser EM, Jones M, Buneman OP, Dunbar DR, et al. IUPHAR-DB: the IU-PHAR database of G protein coupled receptors and ion channels. Nucleic Acids Res. 2009;37:D680–D685. doi: 10.1093/nar/gkn728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodges PE, Carrico PM, Hogan JD, O’Neill KE, Owen JJ, Mangan M, et al. Annotating the human proteome: the Human Proteome Survey Database (HumanPSDTM) and an in-depth target database for G protein-coupled receptors (GPCR-PDTM) from Incyte Genomics. Nucleic Acids Res. 2002;30:137–141. doi: 10.1093/nar/30.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crasto C, Marenco L, Miller P, Shepherd G. Olfactory Receptor Database: a metadata-driven automated population from sources of gene and protein sequences. Nucleic Acids Res. 2002;30:354–360. doi: 10.1093/nar/30.1.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ono Y, Fujibuchi W, Suwa M. Automatic gene collection system for genome-scale overview of G-protein coupled receptors in Eukaryotes. Gene. 2005;364:63–73. doi: 10.1016/j.gene.2005.05.038. [DOI] [PubMed] [Google Scholar]
- 8.Suwa M, Ono Y. Computational overview of GPCR gene universe to support reverse chemical genomics study. In: Koga H, editor. Rev Chem Genet, Methods Mol Biol. Vol. 577. Human Press, a part of Springer Sci; New Jersey: 2009. pp. 41–54. [DOI] [PubMed] [Google Scholar]
- 9.Suwa M, Sugihara M, Ono Y. Functional and structural overview of G-Protein-Coupled Receptors comprehensively obtained from genome sequences. Pharmaceuticals. 2011;4:652–664. [Google Scholar]
- 10.Gertz EM, Yu YK, Agarwala R, Schäffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: Improving the TBLASTN module of BLAST. BMC Biol. 2006;4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gotoh O. Homology-based gene structure prediction: simplified matching algorithm using a translated codon (tron) and improved accuracy by allowing for long gaps. Bioinformatics. 2000;16:190–202. doi: 10.1093/bioinformatics/16.3.190. [DOI] [PubMed] [Google Scholar]
- 12.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PAI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sigrist CJA, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, et al. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 17.Engelman DM, Steitz TA, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 18.Kawashima S, Kanehisa M. AAindex: amino acid index database. Nucleic Acids Res. 2000;28:374. doi: 10.1093/nar/28.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Janes TJ. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 20.Hanson RM. Jmol—a paradigm shift in crystallographic visualization. J Appl Crystal. 2010;43:1250–1260. [Google Scholar]
- 21.Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 22.Fredriksson R, Schioh HB. The repertoire of G-protein coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67:1414–1425. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- 23.Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, et al. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- 24.The International Silkworm Genome Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38:1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Josefsson LG. Evidence for kinship between diverse G-protein coupled receptors. Gene. 1999;239:333–340. doi: 10.1016/s0378-1119(99)00392-3. [DOI] [PubMed] [Google Scholar]
- 26.Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, et al. The G protein-coupled receptor repertoires of human and mouse. Proc Natl Acad Sci USA. 2003;100:4903–4908. doi: 10.1073/pnas.0230374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugihara M, Fujibuchi W, Suwa M. Structural elements of the signal propagation pathway in squid rhodopsin and bovine rhodopsin. J Phys Chem B. 2011;115:6172–6179. doi: 10.1021/jp1101785. [DOI] [PubMed] [Google Scholar]
- 28.Nagarathnam B, Kalaimathy S, Balakrishnan V, Sowdhamini R. Cross-Genome Clustering of Human and C. elegans G-Protein Coupled Receptors. Evol Bioinform Online. 2012;8:229–259. doi: 10.4137/EBO.S9405. [DOI] [PMC free article] [PubMed] [Google Scholar]