Abstract
Emerging evidence indicates that many microRNAs (miRNAs) are indispensable regulators of osteoblast differentiation and bone formation. However, the role of miRNAs in mechanotransduction of osteoblasts remains to be elucidated. This study aimed to identify a mechanosensitive miRNA that regulates Activin A receptor type I (ACVR1)-induced osteogenic differentiation. After 4 weeks of hindlimb unloading (HLU) suspension of 6-month-old male C57BL/6J mice, femurs and tibias were harvested to extract total bone RNAs. Elevated levels of miR-208a-3p correlated with a lower degree of bone formation in whole-bone samples of HLU mice. However, in vitro overexpression of miR-208a-3p inhibited osteoblast differentiation, whereas silencing of miR-208a-3p by antagomiR-208a-3p promoted expression of osteoblast activity, bone formation marker genes, and matrix mineralization under mechanical unloading condition. Bioinformatics analysis and a luciferase assay revealed that ACVR1 is a target gene of miR-208a-3p that negatively regulates osteoblast differentiation under mechanical unloading environment. Further, this study also demonstrates that in vivo pre-treatment with antagomiR-208a-3p led to an increase in bone formation and trabecular microarchitecture and partly rescued the bone loss caused by mechanical unloading. Collectively, these results suggest that in vivo, inhibition of miRNA-208a-3p by antagomiR-208a-3p may be a potential therapeutic strategy for ameliorating bone loss.
Keywords: microRNAs, osteoblast differentiation, ACVR1, hindlimb unloading, miR-208a-3p, bone loss
Introduction
The remodeling of bones is a process that takes place due to activity of bone forming and bone resorbing cells.1, 2, 3 Osteoblasts are the cells that are principally responsible for the bone post-natal formation.4 Their differentiation is a vital step in the procurement of bone mass, skeletal development, and ameliorating bone disorders. Osteoblast differentiation is triggered via mechanical stimulation by inducing the secretion of hormones and growth factors, thus affecting differentiation and proliferation potential of osteoblasts.5, 6, 7 Decreased mechanical loading owing to extended bed rest or exposure to microgravity (such as that experienced by astronauts) may result in significant bone loss of weight-bearing bones and a subsequent rapid progression of osteoporosis.8, 9, 10, 11 Additionally, microRNAs (miRNAs) play a substantial coordinating role in osteoblast differentiation and bone formation.12 Notably, miRNAs have potential roles in bone dynamics and metastatic bone diseases.13, 14
miRNAs are ∼22 nucleotide fragments of noncoding RNAs that are involved in various steps of regulatory mechanisms responsible for target gene expression in several biological responses.15, 16, 17, 18 Recently, it has been reported that miR-103a play a crucial role in osteoblast differentiation. This study further showed that in mechanical stress-regulated bone formation, miR-103a targets mainly Runx2.19 Similarly, the in vitro study stated that miR-33-5p suppresses osteoblast differentiation by inhibiting expression of Hmga2 under different mechanical environments.20 Moreover, mechanical loading-sensitive miR-153 downregulates osteoblast differentiation by directly targeting BMPR2.21 Various miRNAs are essential for osteoblast differentiation by targeting principal transcription factors and signaling pathways involved in the differentiation process,22, 23, 24 as well as influencing the function of mesenchymal stem cells (MSCs).25, 26 However, many of these miRNAs have only been identified in vitro. Biomolecules such as growth factors, cytokines (TGF-β1, BMP), signal transduction proteins (Wnt), and transcription factors (Runx2) are thought to be involved in the regulation of miRNA expression and their function in osteoblast differentiation.
The type I bone morphogenetic protein receptor ACVR1 (Activin A receptor, type I), which is also called ALK2, functions as the key receptor for bone morphogenic protein 7.27 In humans, the pathological role of ACVR1 is fairly well described: point mutations in the ACVR1 gene have been linked to the hereditary autosomal-dominant disorder of fibrodysplasia ossificans progressiva (FOP), which is characterized by excessive extra-skeletal bone formation (so-called heterotopic ossifications).28, 29, 30 However, the physiological role of ACVR1 and in particular its down- and upstream (miRNA) regulators are still partly unclear.
During adipocyte differentiation, it has been validated that ACVR1 is repressed by binding of miR-30c to its 3′ UTR.31 miR-130a was described as another regulator of ACVR1 in iron-deficient mouse liver.32 For bone, evidence from a knockout study on the endogenous role on ACVR1 in osteoblasts suggests a new regulatory BMP7-ACVR1-SOST/DKK1 axis. The authors found that a conditional disruption of ACVR1 in bone cells (including immature osteoblasts, mature osteoblasts, and osteocytes) led to an increase in endogenous bone mass during postnatal development33 via suppression of Wnt inhibitors sclerostin (SOST) and dickkopf-1 (DKK1).34 Another study also described that SOST play a central role in regulating bone’s response to mechanical loading.35
In the present study, we are interested in identifying a set of candidate miRNAs (mechanosensitive) that are expressed during hindlimb unloading (HLU) in mice with a special focus on those miRNAs that directly target ACVR1 and to test these miRNAs and their antagomiRs for their osteogenic potential. For this purpose, we screened differentially expressed miRNAs from HLU mouse bone and found that miR-208a-3p was negatively correlated with osteogenesis. This study demonstrated that inhibition of miRNA-208a-3p by antagomiR-208a-3p may be a potential therapeutic strategy for minimizing unloading-dependent bone loss in vivo. Further, our study shows that miR-208a-3p has a complementary role in osteoblast differentiation where it suppresses osteogenesis.
Results
Overexpression of miR-208a-3p in an HLU Mouse Model
To determine the inhibitory effects of mechanical unloading on the formation of bone, we used a HLU mice model (Figure S1A). This technique is extensively used for the purpose of studying numerous aspects of musculoskeletal unloading.19, 36, 37 Body weight did not differ significantly (total number of mice in each group = 8) among base line, control, and HLU groups at the end of the experimental period (Figure S2A). Additionally, serum and urine calcium levels of HLU groups were found to be higher compared to control (Figure S2B). Muscle weight (soleus and gastrocnemius) and mechanical properties (area moment of inertia; bending energy absorption; elastic modulus; maximum force and stiffness coefficient) of bone were also analyzed (Figures S2C and S2D).
Quantitative micro-computed tomography (μCT) was used to analyze the structure and parameters of trabecular bone of distal femurs, including trabecular bone volume (BV/TV), trabecular number (Tb.N), trabecular separation (Tb.Sp), and trabecular thickness (Tb.Th) in baseline, control, and HLU mice. These results showed that HLU led to bone resorption in mouse femur after 28 days of hindlimb suspension (Figure S1B). Moreover, 3D images showed the trabecular bone distance and separation, indicating bone loss in HLU mice (Figure S1C). Further significant changes were observed in bone mineral density (BMD) and bone mineral content (BMC) of the HLU model as compared to control mouse bones (Figure S1D). Bone histomorphometric analysis revealed that the bone formation-related parameters (Ob.S/BS, MAR, BFR, and N.Ob/B) were significantly lower in HLU mice (Figure S1E). Consistent with this μCT observation, HLU was accompanied by a decrease in bone formation marker genes ALP, Col1a1, and OCN when compared to base line and control group (Figure S1F).
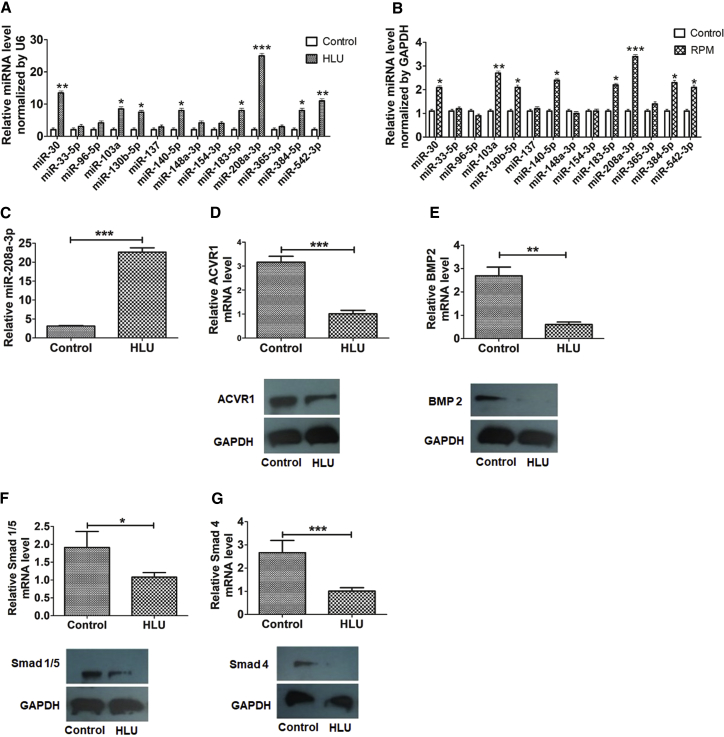
In order to decipher additional information regarding miRNA-208a-3p, we predict possible targets of miRNA-208a-3p with the help of TargetScan,38 miRanda,39 miRecords,40 and miRBase41 databases. The predicted targets included ACVR1, which has a binding site for miRNA-208a-3p in its 3' UTR. ACVR1 mRNA has a significantly long 3' UTR (1,101 nucleotides) based on RNA blotting and the sequence of cDNA clones (Figures S4A and S4B). miRNAs were demonstrated to possess a complicated role in the process of mechanical transduction.19, 42 In order to explore the response of miRNAs under the mechanical unloading condition in bone tissue, initially we studied miRNAs obtained from HLU mice bone, (Table S1) which, according to previous studies, have been highlighted to take part in the process of osteogenic differentiation.12, 43 In the beginning, we carefully chose 14 miRNAs involved in osteogenesis, including miR-30, miR-33-5p, miR-96-5p, miR-103a, miR-130b-5p, miR-137, miR-140-5p, miR-148a-3p, miR-154-3p, miR-183-5p, miR-208a-3p, miR-365-3p, miR-384-5p, and miR-542-3p; all were examined as a potential regulators of the ACVR1 3′ UTR through multiple binding sites. In our experiment, we observed significant differential expression of eight miRNAs from this list, including miR-30, miR-103a, miR-130b-5p, miR-140-5p, miR-183-5p, miR-208a-3p, miR-384-5p, and miR-542-3p from control and HLU mice bone (Figure 1A).
Figure 1.
Overexpression of miR-208a-3p in Hindlimb Unloading Mouse Model
(A) qPCR analysis of miRNAs extracted from tibia of control and HLU mice. (B) qPCR analysis of miRNAs extracted from MC3T3-E1 cell under RPM unloading condition. (C) Relative expression pattern of miR-208a-3p in HLU bone. (D) qPCR analysis of ACVR1 mRNA and western blot analysis of ACVR1 protein in control and HLU mice. (E) qPCR analysis of BMP2 mRNA level and western blot analysis of BMP2 protein in control and HLU mice. (F) qPCR analysis of Smad 1/5 mRNA level and western blot analysis of Smad 1/5 protein in control and HLU mice. (G) qPCR analysis of Smad 4 mRNA level and western blot analysis of Smad 4 protein in control and HLU mice. All data were expressed as means ± SD. Significance is noted at these thresholds: *p < 0.05, **p < 0.01, ***p < 0.001. One-way ANOVA with a post hoc test was performed. Statistical differences between two groups were determined by Student’s t test. n = 8 mice in each group.
qPCR analysis highlighted significant upregulation of miR-208a-3p expression compared with other miRNAs during HLU (Figure 1A), displaying a similar variation pattern as in random positioning machine (RPM; provide simulated microgravity), which may lead to the bone loss (Figure 1B). It was found that miR-208a-3p expression was more overexpressed than the other differentially expressed miRNAs in in vivo HLU and in in vitro RPM unloading conditions (Figures 1A and 1B).
Relative expression of miR-208a-3p was analyzed from control and HLU mouse bone by qPCR (Figure 1C). Consistently, we found that overexpression of miR-208a-3p lead to downregulation of ACVR1 mRNA in the HLU group, and further western blot analysis showed lower expression of ACVR1 protein in HLU mice as compared to the control group (Figure 1D). To gain more insight into the cellular functions possibly affected by miR-208a-3p, predicted target genes were analyzed. We hypothesized that if miRNA target genes are physiologically relevant, then they should produce significant interaction networks in the pathway analysis. Concurrently, we also investigated BMP2 and Smad expression. Western blot and qPCR analysis have shown that BMP2, Smad1/5, and Smad4 band intensity and their mRNA level were decreased in the HLU group as compared to control (Figures 1E–1G).
miR-208a-3p Inhibits Osteoblast Differentiation and Matrix Mineralization In Vitro
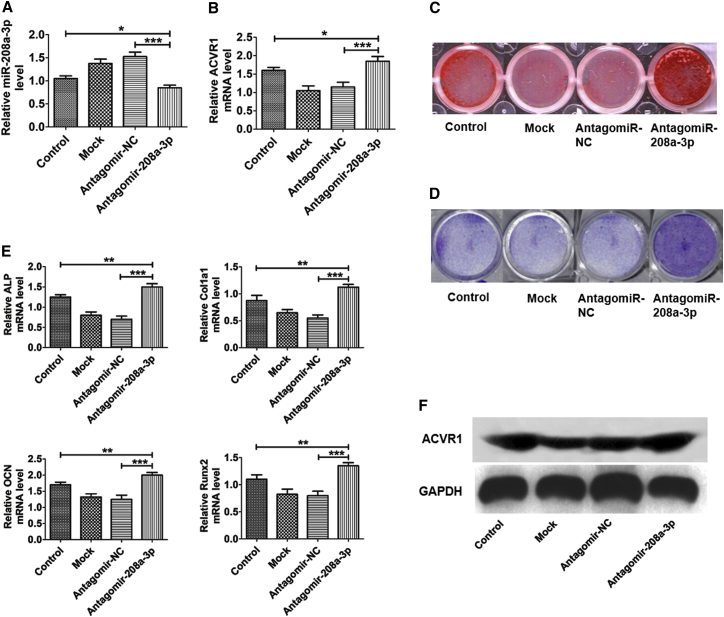
To determine whether miR-208a-3p could influence osteoblast differentiation, MC3T3-E1 cells were subjected to mock (Lipofectamine 2000 only), antagomiR-negative control (NC), and antagomiR-208a-3p transfection. We found that the level of miR-208a-3p was downregulated, while ACVR1 expression was upregulated in osteoblasts treated with antagomiR-208a-3p (MC3T3-E1 + antagomiR-208a-3p) compared to osteoblasts treated with mock (MC3T3-E1 + mock) and negative control (MC3T3-E1 + antagomiR-NC) (Figures 2A and 2B).
Figure 2.
miR-208a-3p Inhibits Osteoblast Activity and Matrix Mineralization In Vitro
(A) Relative expression of miR-208a-3p in MC3T3-E1 cells after transfection with mock, antagomiR-NC, and antagomiR-208a-3p. (B) Relative ACVR1 mRNA level in MC3T3-E1 cells after transfection with mock antagomiR-NC and antagomiR-208a-3p. (C) Staining of calcium deposition by alizarin red in MC3T3-E1 cells after transfection with mock, antagomiR-NC, and antagomiR-208a-3p in osteogenic medium for 24 days. (D) ALP staining of MC3T3-E1 cells after transfection with mock, antagomiR-NC, and antagomiR-208a-3p for 24 hr. (E) Effects of antagomiR-208a-3p transfection on expression of osteoblast differentiation marker genes ALP, Col1a1, OCN, and Runx2. (F) Western blot examination of ACVR1 protein expression in osteoblasts was enhanced after antagomiR-208a-3p transfection. All data were expressed as means ± SD. Significance is noted at these thresholds: *p < 0.05, **p < 0.01, ***p < 0.001. One-way ANOVA with a post hoc test was performed. Statistical differences between two groups were determined by Student’s t test.
To investigate the function of miR-208a-3p during extracellular matrix mineralization, cells were transfected with antagomiR-208a-3p. To maintain stable and constant expression, antagomiR-208a-3p, mock, and negative control were supplemented every 3 days. After 24 days, we found less mineral deposition in mock- and antagomiR-NC-transfected cells as compared to antagomiR-208a-3p-transfected cells (Figure 2C). Consistently, overexpression of miR-208a-3p reduced ALP activity and staining, whereas knockdown of miR-208a-3p with antagomiR-208a-3p transfection enhanced ALP activity and staining. Similarly, reduction in ALP activity and staining were also observed in mock and negative control groups (Figure 2D). However, in vitro transfection effects were further assessed by osteoblast activity, bone formation marker genes (ALP, Col1a1, OCN, and Runx2), and matrix mineralization. Cells were harvested for 48 hr after transfection, mRNA expression was assessed with qPCR. Bone formation marker genes were significantly upregulated after antagomiR-208a-3p transfection as compared to mock and antagomiR-NC (Figure 2E). Furthermore, western blot analysis indicates that after antagomiR-208a-3p transfection, ACVR1 protein expression was enhanced (Figure 2F). Together, our results suggest that miR-208a-3p plays a negative regulatory role in osteoblast differentiation and subsequently suppresses mineralization and ALP activity.
miR-208a-3p Directly Targets ACVR1 and Is Negatively Correlated with ACVR1
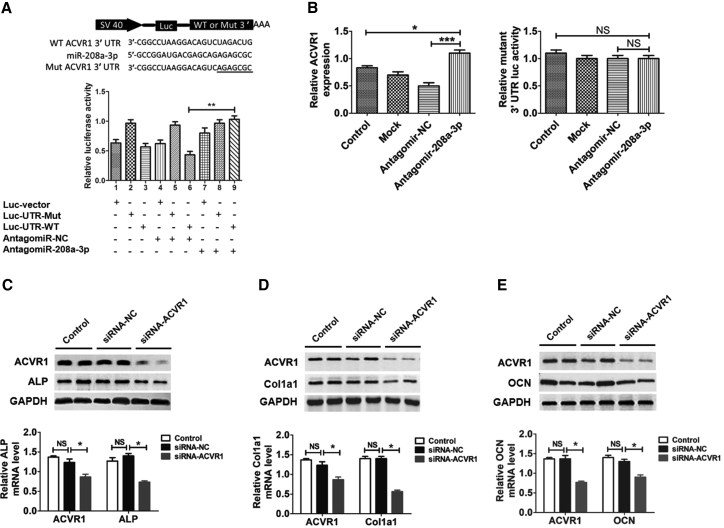
For the purpose of additional clarification of the connection existing between miR-208a-3p and ACVR1, the degree of expression of miR-208a-3p and ACVR1 was thoroughly examined in osteoblast differentiation. We observed that there was a significant upregulation of miR-208a-3p during the initial phase of osteoblast differentiation (day 1 to day 7), which slowly reduced afterwards (day 7 to day 28). But the ACVR1 mRNA expression appeared to be having a differing modification tendency to miR-208a-3p through the mineralization phase, except for day 14 to day 17 when mineral began to deposit (Figure S3A). During osteoblast mineralization phase, miR-208a-3p levels decreased gradually, and the amount of ACVR1 protein increased. To further identify the miR-208a-3p target region in the ACVR1 mRNA, we constructed luciferase reporter vectors. To test whether miR-208a-3p directly targets ACVR1, we constructed luciferase reporters that had either a wild-type (WT) ACVR1 3′ UTR or an ACVR1 3′ UTR containing mutant sequences in miR-208a-3p binding sites (Figure 3A). Further, we have co-transfected MC3T3-E1 cells with luciferase reporter constructs and miR-208a-3p oligonucleotides. Luciferase activity of ACVR1 with a mutated 3′ UTR was not repressed by antagomiR-208a-3p. Luciferase activity of WT ACVR1 3′ UTR and activity of the mutant ACVR1 3′ UTR reporter was significantly increased after treating MC3T3-E1 cells with antagomiR-208a-3p, and activity of mutant ACVR1 3′ UTR reporter was also affected (Figure 3B). To find out that osteoblast differentiation is ACVR1 dependent, we constructed ACVR1 siRNA and examined its effect on osteoblast differentiation. PCR analysis of ALP, OCN, and Col1-a1 mRNA levels was performed in MC3T3-E1 cells. The results have shown that suppression of siRNA-ACVR1 has downregulated the expression of ALP, Col1-a1, and OCN. These changes were also observed at the protein level, as indicated by western blot analysis of ALP, OCN, and Col1a1. (Figures 3C–3E). All together, our results demonstrate that miR-208a-3p directly targets ACVR1 through interaction with its 3′ UTR during osteoblast differentiation.
Figure 3.
miR-208a-3p Targets ACVR1 to Functionally Inhibit Osteoblast Activity In Vitro
(A) Effect of endogenous miR-208a-3p on the luciferase activity of WT ACVR1 3′ UTR (luc-UTR), ACVR1 3′ UTR mutant (luc-UTR-Mut), or the luciferase vector control (luc-vector) after treatment with antagomiR-208a-3p in MC3T3-E1 cells. (B) Effect of endogenous miR-208a-3p on the luciferase activity of WT ACVR1 3′ UTR (luc-UTR), the ACVR1 3′ UTR mutant (luc-UTR-Mut) after treatment with antagomiR-208a-3p or its negative control (antagomiR-NC) in MC3T3-E1 cells. (C–E) qPCR and western blot analysis of ALP (C), Col1a1 (D), and OCN (E) protein expression in MC3T3-E1 cells in which ACVR1 expression was inhibited by siRNA. All data were expressed as means ± SD. Significance is noted at these thresholds: *p < 0.05, **p < 0.01, ***p < 0.001. One-way ANOVA with a post hoc test was performed. Statistical differences between two groups were determined by Student’s t test. NS, not significant; NC, negative control; Rluc, Renilla luciferase.
AntagomiR-208a-3p Resumes the Reduction of Osteoblast Differentiation Induced by RPM Unloading
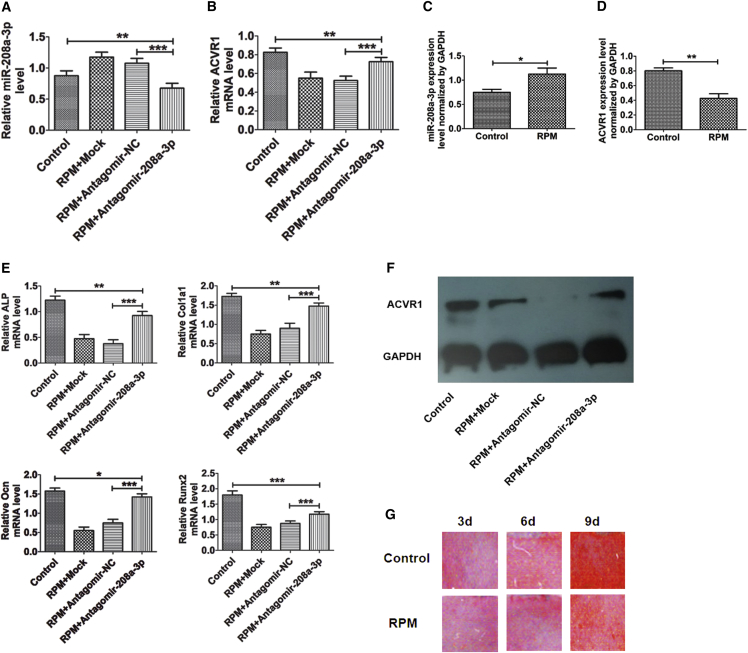
In order to determine the loss-of-function of miR-208a-3p on differentiation of osteoblasts under RPM conditions in vitro, a culture of MC3T3-E1 cells was set up with antagomiR-208a-3p transfection. The levels of miR-208a-3p in the antagomiR-208a-3p group was significantly reduced after unloading for 48 hr under RPM conditions as compared to that in the antagomiR-NC group (Figure 4A). Moreover, antagomiR-208a-3p resumed the ACVR1 expression that had been reduced due to RPM unloading (Figure 4B). Additional confirmation of these findings was established in MC3T3-E1 cells that had been placed under RPM exposure but not transfected with antagomiR-208a-3p (Figures 4C and 4D). Besides, antagomiR-208a-3p was able to restore the RPM unloading dependent reduced expression of genes involved in osteogenesis, such as ALP, Col1al, OCN, and Runx2 mRNA, compared to control and antagomiR-NC (Figure 4E).
Figure 4.
Inhibition of miR-208a-3p Resumes Mechanical Unloading Reduced ACVR1 and Osteoblast Differentiation
(A) qPCR analysis of miR-208a-3p level in MC3T3-E1 cells after treatment with mock, antagomir-NC, and antagomir-208a-3p for 48 hr under RPM unloading condition. (B) Effect of mock, negative control, and antagomiR-208a-3p transfection for 48 hr on ACVR1 mRNA levels in osteoblast cells under RPM unloading conditions. (C) miR-208a-3p expression in osteoblasts for 48 hr under RPM unloading conditions without antagomiR-208a-3p transfection. (D) ACVR1 expression in osteoblasts for 48 hr under RPM unloading conditions without antagomiR-208a-3p transfection. (E) Effect of antagomir-208a-3p on ALP, Col Ia1 Ocn, and Runx2 mRNA levels in MC3T3-E1 cells for 48 hr under RPM unloading conditions. (F) Western blot analysis of ACVR1 protein in MC3T3-E1 cells after transfection with mock, negative control, and antagomiR-208a-3p for 48 hr under RPM unloading conditions. (G) Calcium deposition staining and area of mineralization with alizarin red in MC3T3-E1 cells under RPM for different time periods (the cells were seeded on coverslips). All data were expressed as means ± SD. Significance is noted at these thresholds: *p < 0.05, **p < 0.01, ***p < 0.001. One-way ANOVA with a post hoc test was performed. Statistical differences between two groups were determined by Student’s t test.
Consistent with the ability of antagomiR-208a-3p to silence miR-208a-3p, we found mock- and antagomiR-NC-transfected cells had lower levels of ACVR1 protein than control and antagomiR-208a-3p-transfected cells under RPM (Figure 4F). Moreover, alizarin red staining showed significantly more mineral deposition in control cells as compared to RPM (Figure 4G). Overall, our findings are conclusive with regards to the sensitivity of miR-208a-3p to mechanical unloading. It is highly plausible that miR-208a-3p is mechanosensitive because, during the process of osteoblast differentiation, it is negatively regulated.
Therapeutic Inhibition of miR-208a-3p Counteracts the Decrease of Bone Formation in HLU Mouse Model
In view of our observations that the decrease in osteoblast differentiation takes place under RPM unloading conditions and is accompanied by an increase in miR-208a-3p levels (Figure 4) and whole-bone (femur) after 28 days of HLU (Figure S1; Figure 2), we speculated that decreased bone formation could be restored by antagonizing miR-208a-3p in the HLU mouse model. To conclusively answer this question, the mice in the HLU group were separately administered 3 successive injections of PBS, antagomiR-NC, and antogmiR-208a-3p via the caudal tail vain36 (Figure 5B). To maintain the effect of miR-208a-3p, mice received more antagomiR-208a-3p injections as described above on days 1 to 3 during the third week of HLU (Figure 5B).
Figure 5.
Therapeutic Inhibition of miR-208a-3p with Bone-Targeting Delivery System Counteracts Decrease in Bone Formation of Hindlimb-Unloaded Mice
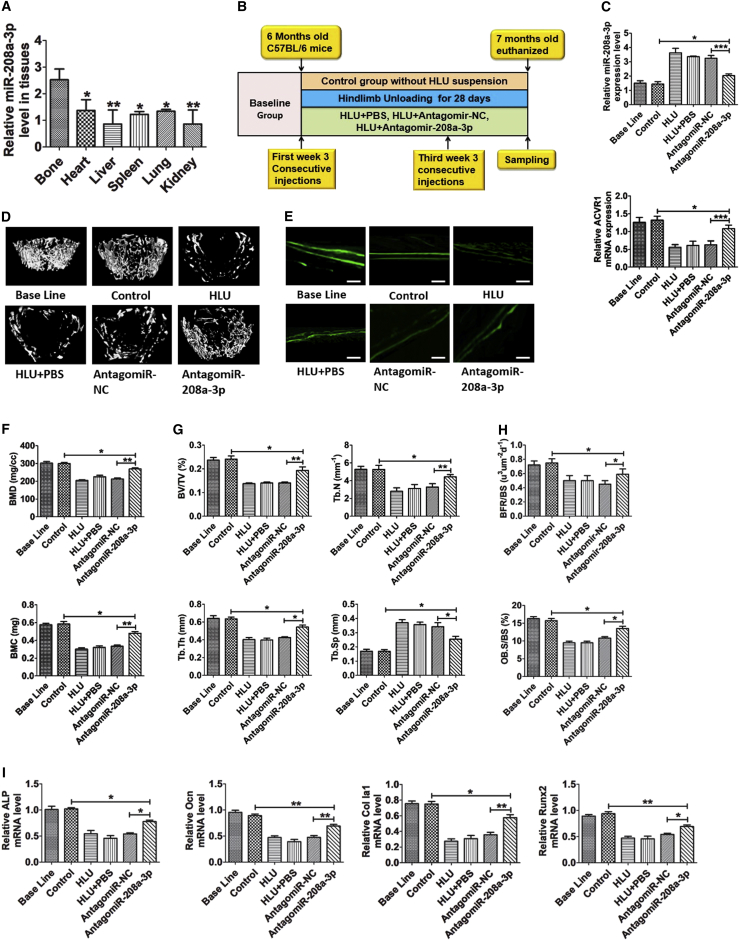
(A) qPCR analysis of miR-208a-3p levels in HLU bone and other tissues from C57BL/6J mice. (B) A schematic diagram illustrating experimental design. (C) Relative expression of miR-208a-3p and ACVR1 in femurs collected from baseline, control, HLU, HLU+PBS, HLU+antagomiR-NC, and HLU+antagomiR-208a-3p groups. (D) Representative μCT reconstructive images of distal femurs collected from baseline, control, HLU, HLU+PBS, HLU+antagomiR-NC, and HLU+antagomiR-208a-3p mice. (E) Representative images showing new bone formation assessed by double calcein labeling in mouse femurs collected from baseline, control, HLU, HLU+PBS, HLU+antagomiR-NC, and HLU+antagomiR-208a-3p. Scale bars, 10 μm. (F) BMD and BMC measurements in the femurs collected from baseline, control, HLU, HLU+PBS, HLU+antagomiR-NC, and HLU+antagomiR-208a-3p mice. (G) Trabecular bone parameters of bone volume/tissue volume ratio (BV/TV), trabecular separation (Tb.Sp), trabecular number (TB.N), and trabecular thickness (Tb.Th) in baseline, control, HLU, HLU+PBS, HLU+antagomiR-NC, and HLU+antagomiR-208a-3p mice. (H) Histomorphometric analysis of bone-formation-related parameters BFR and Ob.S/BS in HLU, HLU+PBS, HLU+antagomiR-NC, and HLU+antagomiR-208a-3p mice. (I) qPCR analysis expression of osteogenic genes ALP, Col Ia1, Ocn, and Runx2 in six group. Each experimental group was compared to the others. All data were expressed as means ± SD. Significance is noted at these thresholds: *p < 0.05, **p < 0.01, ***p < 0.001. One-way ANOVA with a post hoc test was performed. Statistical differences between two groups were determined by Student’s t test. n = 8 mice in each group.
The mice were sacrificed after 28 days of HLU and analysis was performed. We found the expression profile of miR-208a-3p in different tissues of C57BL/6J mice. The results clearly showed that miR-208a-3p expression was much higher in HLU femur bone than in other tissues, indicating that miR-208a-3p may play a crucial role in bone remodeling (Figure 5A). On additional investigation, it was revealed that, in the femur bone of HLU mice treated with antagomiR-208a-3p (HLU+antagomiR-208a-3p), miR-208a-3p expression was downregulated and expression of ACVR1 was upregulated in comparison to HLU mice that were treated with negative control antagomiR (HLU+antaomiR-NC) (Figure 5C). Our results showed a significant decrease in BMD and BMC of HLU, HLU+PBS, and HLU+antagomiR-NC mice, whereas these parameters were significantly increased in HLU+antagomiR-208a-3p-treated mice (Figure 5F). Moreover, a schematic representation of 3D femur bone images showed bone loss in HLU, HLU+PBS, and HLU+antagomiR-NC groups, but the bone loss caused by mechanical unloading was partly rescued by antagomiR-208a-3p (Figure 5D). Calcein labeling indicated an increase in bone formation in the antagomiR-208a-3p-treated HLU group compared to other HLU groups (Figure 5E). Trabecular bone mass was markedly lost (BV/TV), and trabecular architecture was significantly impaired (lower in Tb.Th and Tb.N and higher in Tb.Sp) in HLU, HLU+PBS, and HLU+antagomiR-NC mice, whereas this decrease was prevented by treatment with antagomiR-208a-3p (Figure 5G). Furthermore, bone histomorphometric analysis revealed that bone formation-related parameters Ob.S/BS and BFR were increased in HLU+antagomiR-208a-3p mice as compared to HLU, HLU+PBS, and HLU+antagomiR-NC-treated mice (Figure 5H). Moreover, HLU mice in comparison to controls were shown to have low expression levels of osteogenic genes, such as ALP, Col Iα1, Runx2, and Ocn, while the HLU+antagomiR-208-3p group showed that the reduced mRNA levels of these genes were partially counteracted (Figure 5I). These observations highlight that antagonizing miR-208a-3p results in partial counteraction of low ACVR1 expression and a reduction in bone formation induced by mechanical unloading.
Discussion
The present study demonstrated that miR-208a-3p is an important inhibitor of bone formation under mechanical unloading conditions. Our results suggest that therapeutic inhibition of miR-208a-3p in osteoblasts may promote bone formation. By rescuing the trabecular bone, which has been shown to be lost in mechanical unloading conditions, inhibition of miR-208a-3p seems to be a potential promising therapeutic agent for mechanically unloaded bone loss. However, future studies have to elucidate that how much miR-208a-3p is dysregulated in bone loss under mechanical unloading conditions.
It has been shown that mechanical unloading leads to a decrease in the expression of osteogenic genes along with a reduction in bone mass and mineral deposition.44, 45, 46 Many researchers have reported the modulatory effects of mechanical unloading, including simulated microgravity, clinostat, and HLU on the miRNAs’ expression levels.19, 47, 48 Bin Zuo et al.19 has described that mechanosensitive miR-103a inhibit bone formation by targeting Runx2, which is a key regulator of osteogenesis. It has also been reported that Cav1.2 expression in simulated microgravity is suppressed by miR-103, leading to a decrease in osteoblast proliferation.48 In the current study, we have demonstrated findings similar to the ones mentioned before, highlighting the involvement of mechanosensitive miR-208a-3p in osteoblast differentiation.
Our present study has shown that under mechanical unloading, ACVR1 mRNA levels decreased with an expression profile concurrent to miR-208a-3p, whereas downregulation of ACVR1 downregulated BMP2 (Figure 1). This inhibition of BMP2 further downregulates SMAD1/5 and SMAD4, leading to the suppression of osteoblast-specific gene transcription (Figure 1). To understand the relation of miR-208a-3p with the mechanical unloading condition in regards to osteoblast differentiation and bone formation, we looked for possible targets of miR-208-3p. Our search revealed interesting data highlighting that the 3'UTR of ACVR1 has an 8-nt perfect match site to the miR-208a-3p seed region (Figure 3; Figure S4), and a decrease in miR-208a-3p levels as a consequence of transfection of antagomiR-208a-3p leads to enhancement of ACVR1 expression (Figure 2). It has been confirmed by Luciferase reporter experiment that miR-208a-3p directly targets the ACVR1 3′ UTR (Figure 3). Additionally, inverse association exists between expression levels of ACVR1 and miR-208a-3p. These findings indicate the possibility of ACVR1 being a physiological target of miR-208a-3p and the existence of negative feedback regulation between the two.
Regulation of gene expression plays vital regulatory roles in diverse cellular processes by various mechanisms at different levels. A number of recent studies have focused on miRNA’s role in post transcriptional regulation of gene expression in pathophysiological conditions.49 A recent study reported that miR-148a may directly downregulate ACVR1 by targeting the 3′ UTR of ACVR1 mRNA.50
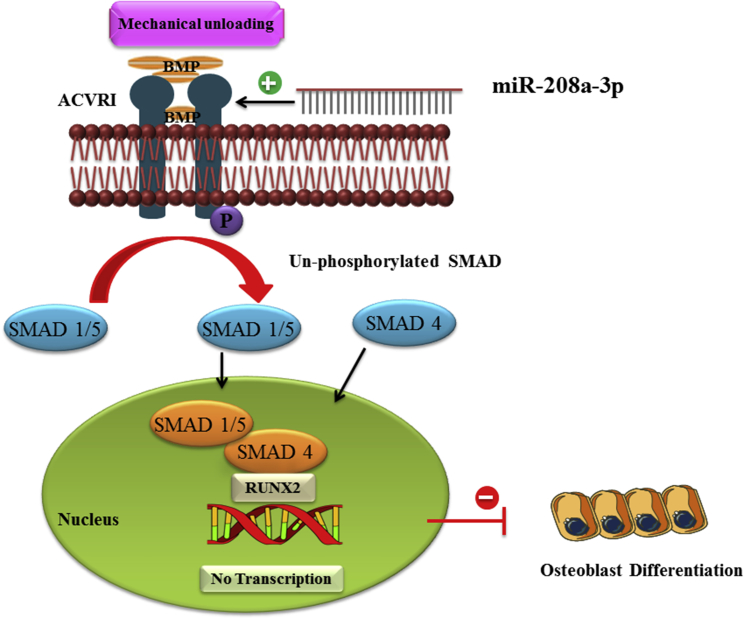
Moreover, we demonstrated that antagonizing miR-208-3p by antagomiR-208a-3p significantly increases the expression of genes related to osteogenesis along with ALP activity and ACVR1 expression under RPM mechanical unloading conditions (Figure 4). Similarly, our findings in vivo demonstrated that antagonizing miR-208a-3p leads to the counteraction of an HLU-induced ACVR1 expression alleviating a reduction in bone formation (Figure 5). These results suggest that miR-208a-3p inhibits bone formation through a negative feedback mechanism between miR-208a-3p and ACVR1 under mechanical unloading conditions. We have proposed a model for miR-208a-3p-mediated suppression of osteoblast differentiation targeting by ACVR1 (Figure 6). Interestingly, an in vivo study found that conditional interruption of ACVR1 activity favors bone formation by promoting osteoblast-specific gene expression. In addition, ACVR1 protein controls the growth and development of bones and muscles, including the gradual replacement of cartilage by bone.33 Likewise, a recently published study reported that ACVR1 mutant mice or cells have an important regulatory role in osteogenic differentiation of MSCs.51
Figure 6.
A Proposed Model of miR-208a-3p-Mediated Suppression of Osteoblast Differentiation
miR-208a-3p is expressed in bone and reduces ACVR1 translation, thereby decreasing BMP2 and its downstream target Smad 1/5 and Smad 4; subsequently, bone formation is suppressed.
Previously, it has been reported that miR-208a-3p is highly expressed in hepatocellular carcinoma, cardiac remodeling, and human esophageal squamous cell carcinoma, indicating that miR-208a-3p participates in carcinogenesis and metastasis as well.52, 53, 54 However, there have been no data available on the role of miR-208a-3p in osteoblast differentiation. In this study, we proposed a mechanism for how miR-208a-3p is involved in regulation of ACVR1 protein in bone. Several studies indicated that cyclic mechanical stretching could modulate the expression of a panel of miRNAs, which are involved in cellular response to mechanical force in different cell lines.19, 55, 56, 57 A few years back, studies reported that multiple miRNAs were shown to be important regulators of bone-remodeling-related gene expression on a post-transcriptional level and could further contribute to human skeletal disorders such as osteopenia or osteoporosis.36, 58 It is an established phenomenon that miRNAs can regulate the mRNA levels of their targets, and pharmacological silencing of miRNAs using antagomiRs might therefore lead to the regulation of many mRNAs.59 However, the function of specific mechanosensitive miRNAs in mechanical unloading-induced osteoblast differentiation have not been characterized or understood clearly.
Our data demonstrate that an elevation in miR-208a-3p expression during mechanical unloading results in downregulation of ACVR1 protein in MC3T3-E1 cells. To investigate the molecular mechanism by which miR-208a-3p inhibits osteogenic differentiation, we evaluated the target gene of miR-208a-3p. Our data show that a change in miR-208a-3p levels under mechanical conditions results in an alteration of the amount of ACVR1 protein. Upregulation of ACVR1 activity favors bone formation by promoting expression of its downstream osteoblast-specific marker genes, whereas in the HLU mouse model, therapeutic inhibition of miR-208a-3p in vivo may promote bone formation by exerting an anabolic effect under pathological mechanical conditions.
Conclusions
Importantly, we not only found that inhibition of miR-208a-3p by antagomiR-208a-3p increased osteoblast differentiation in cell line MC3T3-E1, but also confirmed a positive effect in HLU-induced bone loss. AntagomiR-208a-3p treatment reversed the bone loss induced by mechanical unloading and partially restored trabecular and cortical areas of bone. Furthermore, antagomiR-208a-3p treatment enhanced ACVR1 expression and bone formation marker genes and restored biomechanical properties of bone, demonstrating that bone architecture was also preserved. This robust response suggests that inhibition of miR-208a-3p and enhanced ACVR1 signaling by antagomiR-208a-3p represents a promising unique therapeutic approach for the treatment of bone-related disorders.
Materials and Methods
HLU Mice
Six-month-old male C57BL/6J mice were purchased from the University of Veterinary and Animal Sciences, Lahore, Pakistan. All mice were kept individually in clean plastic cages under standard controlled conditions (12-hr light/12-hr dark cycle, 25°C). Control animals were singly housed to mimic the increased stress of singly housed HLU animals (number of animals in each group = 8) at the University of Health Sciences, Lahore, Pakistan. Hindlimb of HLU mice were kept suspended for 4 weeks. The hindlimb suspension was based on the procedure described previously by Morey-Holton and Globus.60 In short, a strip of sterile adhesive tape (15 cm × 0.5 cm) was adhered laterally along the animal’s tail, which was suspended by sticking the tape to a chain that turns on a pivot attached to the top of the cage. This maneuver permitted only the forelimbs to contact the cage floor, and the animals had ad libitum access to food and water.
The height of the mice was adjusted to prevent the hindlimbs from contact with any supporting surface, and care was taken to maintain a suspension angle below 30°. The animal’s overall appearance, drinking/eating habits, and tail were monitored three times a day. The procedure did not occlude blood flow at the distal end of the tail (i.e., the tail remained pink). After euthanasia, HLU bones (femurs and tibias) were harvested and distal femurs were processed for microCT (μCT) examination and qPCR analysis. The research was conducted in accordance with the internationally accepted principles for laboratory animal use and care as found in the European Community guidelines (EEC Directive of 1986; 86/609/EEC) and the US guidelines (NIH publication #85-23, revised in 1985).
Physical and Biochemical Analysis
Body Weight
The body weight of the mice was monitored during a 28-day experimental period. Body weight of baseline, control, and HLU groups did not differ significantly during the experimental period (Figure S2A). The mice were weighed twice per week throughout the experiment, control animals showed normal growth, and HLU body weight decreased in the first week of suspension but gradually increased during the next 3 weeks of suspension. This showed significant growth during the experiment and suggested that suspension was not a significant source of stress.
Blood and Urine Collection
Blood and urine samples were collected before and after HLU suspension for determination of calcium metabolism. Baseline studies were conducted 1 week before suspension (Figure S2B). Total calcium was measured in serum and urine with a Perkin-Elmer 4000 atomic absorption spectrophotometer (Perkin-Elmer, Norwalk, MA, USA).
Mechanical Testing
A load-deformation test was conducted, and results were recorded with deformation as a function of the applied load. Mechanical strength of bone was examined by measuring the area moment of inertia, elastic modulus, bending energy absorption, maximum stress, and stiffness coefficient of femurs mid-diaphysis (Figure S2D) according to the guideline of Jepsen et al.61 Femurs were prepared for biomechanical testing by wrapping in normal saline-soaked gauze and freezing at −20°C and mechanically tested with three-point bending. Fresh-frozen femurs were thawed at room temperature then centered longitudinally with the anterior surface on the two lower support points spaced 10 mm apart. A material testing system (SANS CMT 5304 China) was used to apply a flexion moment in the anteroposterior plane of the surface at a constant displacement rate of 1.2 mm/min until fracture occurred by three-point bending. Mechanical data were acquired at 30 Hz and used to determine bending energy absorption (N.mm), elastic modulus (GPa), maximum stress (MPa), and stiffness coefficient (mm.N−1).
μCT Analyses
Distal femur from each mouse was scanned ex vivo using a μCT system GE eXplore Locus μCT 29 μm resolution scanner to produce images, and these images were then analyzed with Micview V2.1.2 evaluation software for segmentation, three-dimensional morphometric analysis, density, and distance parameters (GE, USA). Soleus and gastrocnemius muscles were separated and weighted (Figure S2C). In brief, femurs and tibias were harvested from euthanized mice. Soft tissues were cleaned, and the femur bones were fixed in ethanol for μCT analysis and tibias were preserved in RNA Later at −80°C for miRNA extraction using PCR. Three-dimensional structural parameters including: Tb.N, BV/TV per tissue volume, Tb.Th, Tb.Sp, average cortical thickness (Ct.Th), cortical area fraction (Ct.Ar/Tt.Ar), cortical bone area (Ct.Ar), total cross-sectional area (Tt.Ar), and analysis of BMD, tissue mineral density (TMD), and BMC were analyzed. To assess dynamic indices of bone formation, mice received subcutaneous injections of green-fluorescent calcein (Sigma; 5 mg.kg−1 body weight) on day 8 of the experiment and 2 days before euthanasia.
For bone static histomorphometric analysis, distal femurs were dehydrated in graded ethanol and embedded without decalcification in methyl methacrylate (MMA).36 Frontal sections of trabecular bone were obtained from distal femur (at a thickness of 15 μm) for osteoblast surface/bone surface and osteoblast number/bone perimeter, (Ob.S/BS and Ob.N/B.Pm) and bone dynamic histomorphometric analysis for mineral apposition rate and bone formation rate/bone surface (MAR and BFR/BS) were performed using professional image analysis software (ImageJ, NIH, USA) under fluorescence microscopy (Leica image analysis system, Q500MC). Bone histomorphometric parameters were calculated and expressed according to the standardized nomenclature for bone histomorphometry.
miRNA Target Site Prediction
Several databases were utilized to search possible target genes for differentially expressed miRNAs: PicTar (http://pictar.mdc-berlin.de/), TargetScan (http://www.targetscan.org), miRBase (http://www.mirbase.org), and miRanda (http://www.microrna.org). Another database, miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2) was used for the prediction and validation of miRNAs targets. Computational target predictions were ranked according to the total context score of the conserved sites.
miRNA Extraction and qPCR
Bones from control and experimental animal groups were harvested and soft tissues and growth plates were removed and preserved in RNA Later (QIAGEN) until extraction of miRNAs. All samples were stored at −80°C. Total RNA was extracted using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. For detection and purification of miRNA from bone tissue and MC3T3-E1 cells, a TaKaRa miRNA reverse transcription-PCR kit was used for cDNA construction. For the validation of miRNAs, cDNA samples were used as a template for qPCR. For qPCR validation of miRNAs, cDNA were obtained according to the protocol of the one-step PrimeScript miRNA cDNA synthesis kit. All samples were analyzed twice, and equal amounts of cDNA were amplified with SYBR Premix Ex TaqII (TaKaRa, Japan). After the reverse transcription reaction, qPCR analysis was performed by the Thermal Cycler C-1000 Touch system (Bio-Rad CFX Manager, USA). All amplifications were normalized to GAPDH mRNA and U6 small nuclear RNA (snRNA). Data were analyzed using the comparative Ct method (2−ΔΔCt) and expressed as fold change compared to their respective control. The miRNA primer sequences used in this study are provided in Table S1. For the quantitative determination, SYBR green reagents were used, and primer sequence for ALP, OCN, and Type I Collagen mRNA are is provided in Table S2.
Cell Culture and Transfection
The mouse osteoblast cell line MC3T3-E1 was kindly donated by the University of Veterinary and Animals Sciences, Lahore, Pakistan. The MC3T3-E1 cell line was cultured in complete DMEM (Hyclone) supplemented with 10% fetal bovine serum (FBS) (Gibco), 1% penicillin and streptomycin (Hyclone), and 0.3 mg/mL G418 (Sigma). Cell cultures were incubated and maintained at 37°C with 5% CO2 at a relative humidity of approximately 95% and were not used beyond passage 10. When necessary, cells were detached using 0.25% trypsin/10 mM EDTA (Hyclone), resuspended in antibiotic-free growth medium, and seeded in 24-well plates at cell densities of 2.0 × 105 cells per well.
Transfection of antagomiR-208a-3p and antagomiR-NC (Sangon Biotech Shanghai) at a concentration of 200 μM was carried out with Lipofectamine TM 2000 (Invitrogen, USA) according to the manufacturer’s instructions.
For transfection of miRNAs or siRNA oligos, Lipofectamine 2000 (Invitrogen, USA) was used according to manufacturer’s instructions. AntagomiR-208a-3p (Sangon Biotech Shanghai) was transfected at a concentration of 200 μM, and ACVR1 siRNA (Sangon Biotech Shanghai) was transfected at a concentration of 50 nM.
Alkaline Phosphatase Staining
The detailed procedure of cell culture was explained above. Alkaline phosphatase (ALP) staining was performed with BCIP/NBT alkaline phosphatase color development kit (Beyotime, China) according to the manufacturer’s instructions.62, 63 In brief, cells were carefully rinsed with PBS and fixed with 10% neutral buffered formalin to cover the cellular monolayer at room temperature for 15 min. After fixation, cells were rinsed in washing buffer and then incubated in BCIP/NBT liquid substrate for 1–24 hr. Sample preparation and incubation were performed at room temperature while protected from exposure to light. Color change was monitored (osteoblast cells will stain blue/purple) to avoid non-specific staining and cells were observed under a CCD microscope, and the stained cell cultures were imaged by a scanner (BenQ 5560B, Taiwan, China). All the staining data were confirmed by three repeated tests.
Alizarin Red Staining
The detailed procedure of cell culture was explained above. To induce osteoblast mineralization, MC3T3-E1 cells were cultured in 24-well plates with osteogenic medium containing 100 nM dexamethasone, 50 μM ascorbic acid, and 10 mM β-glycerophosphate (Sigma). Cells were fixed with 1 mL of 10% formalin per well, and plates were incubated at room temperature for 15 min. Cells were rinsed with 1 mL of 1× PBS and stained with 500 μL 0.5% alizarin red stain solution, pH 4.0, for 15 min at room temperature. After incubation, cells were rinsed with 1 mL of ddH2O on an orbital shaker for 5 min. The mineralized nodules were stained as red spots after removal of unincorporated excess dye with ddH2O. Finally, plates were observed under a CCD microscope and scanned with scanner (BenQ 5560B, Taiwan, China). All the staining data were confirmed by three repeated tests.
ACVR1 3′ UTR Cloning and Luciferase Reporter Assay
The mouse ACVR1 3′ UTR containing the miR-208a-3p binding sequence for the ACVR1 gene (gene ID 11477) was amplified by PCR from mouse genomic DNA. The PCR product was then subcloned into the XbaI site downstream of the stop codon in a pGL3 vector (Promega). Binding-region mutations were achieved using a Quik Change Site-Directed Mutagenesis Kit (Stratagene) by following the manufacturer’s instructions. Transient transfection of MC3T3-E1 cells (1 × 106 cells per well) was carried out in six-well plates with Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. The cells were co-transfected with 200 ng of luciferase constructs and 50 ng of the pRL-TK (Promega) Renilla luciferase plasmid, and luciferase assays were performed with the dual-luciferase reporter assay system (Promega) according to the manufacturer’s instructions. Luminescence was quantified with a luminometer (Glomax, Promega), and each value from the firefly luciferase construct was normalized to the Renilla luciferase signal.
RPM
The mechanical unloading condition for cell culture was simulated as described previously by making use of a desktop RPM (The Center for Space Science and Applied Research of Chinese Academy of Sciences).64, 65 RPM was placed inside an incubator at 37°C, and vessels used for cell culture were secured to the inner frame of RPM. For the purpose of studying mechanical unloading, 2.0 × 105 cells/cm2 of MC3T3-E1 cells were seeded on glass coverslips placed in a 90 mm dish and incubated at 37°C. Cells were allowed to reach confluence and then transfected as described earlier (without vitamin C and β-glycerophosphate) and kept in DMEM containing 10% FBS (Gibco) with 1% penicillin and streptomycin (Hyclone). The RPM culture flasks containing growth medium were firmly capped (air bubbles avoided) and fixed on the inner frame of the RPM. It was operated for 48 hr using a random mode for speed (0–8 rpm) and direction, including both inner and outer frames.66 The static control group cells were also cultured inside the same incubator at 37°C but without rotation. All experiments were repeated three times.
Western Blot Analysis
Western blot cells were harvested and then lysed on ice using cell lysis buffer (Beyotime, China) with 1% protease inhibitor Cocktail Set III (Merck, Germany). The collection of protein fractions was done by 5 min centrifugation at 15,000 g, 4°C and then subjected to SDS-PAGE. Afterward, the protein fractions were transferred onto an NC membrane (PALL, USA). After treatment with 5% skim milk, the membrane was incubated with specific antibodies at 4°C overnight. After that, a secondary antibody was added labeled with HRP (CoWin Bioscience, Beijing, China). Visualization was performed by using a chemiluminescence detection system (Progma, USA) as recommended by the manufacturer. The primary antibodies that were used included MACF1 rabbit pAb (1:500, Abcam, USA), β-catenin rabbit pAb (1:1,000, Cell Signaling Technology, USA), and GAPDH mouse mAb (1:1,000, Calbiochem, USA). All experiments were repeated three times.
Therapeutic Inhibition of miR-208a-3p in HLU Mice
Six-month-old C57BL/6J mice received daily tail-vein injections of antagomiR-208a-3p (HLU+antagomiR-208a-3p), negative control (HLU+antagomiR-NC), and (HLU+PBS) at a dose of 80 mg kg−1 body weight (0.2 mL per injection), for three consecutive days before HLU suspension.36 AntagomiR was purchased from Sangon Biotech Shanghai. For in vivo transfection, liposomal nanoparticle (DSS)6-liposome delivery system was used. The mice were then subjected to HLU through tail suspension for 28 days. To maintain the effect of the treatment, HLU mice received another three-treatment regimen of consecutive daily injections of antagomiR-208a-3p after 3 weeks of HLU suspension. All the mice received subcutaneous injections of green-fluorescent calcein (Sigma; 5 mg kg−1 body weight) on day 8 of the experiment and 2 days before sacrifice. On completion of experimental period, mice were euthanized, bones and tissues were harvested, and miR-208a-3p levels in bone tissue were measured.
Statistical Analyses
All numerical data is expressed as the mean ± SD. Statistical differences among groups were analyzed by one-way ANOVA with a post hoc test (after normalization to baseline in HLU study) to determine group differences in the study parameters. Statistical differences between two groups were determined by using Student’s t test analysis. All statistical analyses were performed with GraphPad Prism software, version 5.0. *p < 0.05 was considered statistically significant.
Author Contributions
Y.A., M.S., and K.M. conceived and designed the experiments Y.A., M.A.R.B., M.S., K.M., N.M., and H.M. performed the experiments. Y.A., M.A.R.B., and M.S. analyzed the data. Y.A., M.A.R.B., M.S., and K.M. contributed reagents/materials/analysis tools. N.M. and H.M. performed statistical analyses. Y.A. and K.M. wrote the paper. Y.A. edited the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
This work was supported by funds from the Post-doctoral Science Foundation of China (2016M592831).
Footnotes
Supplemental Information includes four figures and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.11.009.
Contributor Information
Yasir Arfat, Email: yasir@nwu.edu.cn.
Muhammad Shahzad, Email: shahzad912@hotmail.com.
Supplemental Information
References
- 1.Karsenty G. The complexities of skeletal biology. Nature. 2003;423:316–318. doi: 10.1038/nature01654. [DOI] [PubMed] [Google Scholar]
- 2.Marx J. Coming to grips with bone loss. Science. 2004;305:1420–1422. doi: 10.1126/science.305.5689.1420. [DOI] [PubMed] [Google Scholar]
- 3.Alliston T., Derynck R. Medicine: interfering with bone remodelling. Nature. 2002;416:686–687. doi: 10.1038/416686a. [DOI] [PubMed] [Google Scholar]
- 4.Ducy P., Schinke T., Karsenty G. The osteoblast: a sophisticated fibroblast under central surveillance. Science. 2000;289:1501–1504. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- 5.Bikle D.D. Integrins, insulin like growth factors, and the skeletal response to load. Osteoporos. Int. 2008;19:1237–1246. doi: 10.1007/s00198-008-0597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papadopoulou A.K., Papachristou D.J., Chatzopoulos S.A., Pirttiniemi P., Papavassiliou A.G., Basdra E.K. Load application induces changes in the expression levels of Sox-9, FGFR-3 and VEGF in condylar chondrocytes. FEBS Lett. 2007;581:2041–2046. doi: 10.1016/j.febslet.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 7.Reijnders C.M., Bravenboer N., Tromp A.M., Blankenstein M.A., Lips P. Effect of mechanical loading on insulin-like growth factor-I gene expression in rat tibia. J. Endocrinol. 2007;192:131–140. doi: 10.1677/joe.1.06880. [DOI] [PubMed] [Google Scholar]
- 8.Rittweger J., Frost H.M., Schiessl H., Ohshima H., Alkner B., Tesch P., Felsenberg D. Muscle atrophy and bone loss after 90 days’ bed rest and the effects of flywheel resistive exercise and pamidronate: results from the LTBR study. Bone. 2005;36:1019–1029. doi: 10.1016/j.bone.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Bucaro M.A., Fertala J., Adams C.S., Steinbeck M., Ayyaswamy P., Mukundakrishnan K., Shapiro I.M., Risbud M.V. Bone cell survival in microgravity: evidence that modeled microgravity increases osteoblast sensitivity to apoptogens. Ann. N Y Acad. Sci. 2004;1027:64–73. doi: 10.1196/annals.1324.007. [DOI] [PubMed] [Google Scholar]
- 10.Zwart S.R., Pierson D., Mehta S., Gonda S., Smith S.M. Capacity of omega-3 fatty acids or eicosapentaenoic acid to counteract weightlessness-induced bone loss by inhibiting NF-kappaB activation: from cells to bed rest to astronauts. J. Bone Miner. Res. 2010;25:1049–1057. doi: 10.1359/jbmr.091041. [DOI] [PubMed] [Google Scholar]
- 11.Trudel G., Payne M., Madler B., Ramachandran N., Lecompte M., Wade C., Biolo G., Blanc S., Hughson R., Bear L. Bone marrow fat accumulation after 60 days of bed rest persisted 1 year after activities were resumed along with hemopoietic stimulation: the Women International Space Simulation for Exploration study. J. Appl. Physiol. 2009;107(1985):540–548. doi: 10.1152/japplphysiol.91530.2008. [DOI] [PubMed] [Google Scholar]
- 12.Inose H., Ochi H., Kimura A., Fujita K., Xu R., Sato S., Iwasaki M., Sunamura S., Takeuchi Y., Fukumoto S. A microRNA regulatory mechanism of osteoblast differentiation. Proc. Natl. Acad. Sci. USA. 2009;106:20794–20799. doi: 10.1073/pnas.0909311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taipaleenmäki H., Browne G., Akech J., Zustin J., van Wijnen A.J., Stein J.L., Hesse E., Stein G.S., Lian J.B. Targeting of Runx2 by miR-135 and miR-203 impairs progression of breast cancer and metastatic bone disease. Cancer Res. 2015;75:1433–1444. doi: 10.1158/0008-5472.CAN-14-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heilmeier U., Hackl M., Skalicky S., Weilner S., Schroeder F., Vierlinger K., Patsch J.M., Baum T., Oberbauer E., Lobach I. Serum microRNAs are indicative of skeletal fractures in postmenopausal women with and without type 2 diabetes and influence osteogenic and adipogenic differentiation of adipose-tissue derived mesenchymal stem cells in vitro. J. Bone Miner. Res. 2016;31:2173–2192. doi: 10.1002/jbmr.2897. [DOI] [PubMed] [Google Scholar]
- 15.Rigoutsos I., Furnari F. Gene-expression forum: decoy for microRNAs. Nature. 2010;465:1016–1017. doi: 10.1038/4651016a. [DOI] [PubMed] [Google Scholar]
- 16.Chitwood D.H., Timmermans M.C. Small RNAs are on the move. Nature. 2010;467:415–419. doi: 10.1038/nature09351. [DOI] [PubMed] [Google Scholar]
- 17.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 18.Kosik K.S. MicroRNAs and cellular phenotypy. Cell. 2010;143:21–26. doi: 10.1016/j.cell.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Zuo B., Zhu J., Li J., Wang C., Zhao X., Cai G., Li Z., Peng J., Wang P., Shen C. microRNA-103a functions as a mechanosensitive microRNA to inhibit bone formation through targeting Runx2. J. Bone Miner. Res. 2015;30:330–345. doi: 10.1002/jbmr.2352. [DOI] [PubMed] [Google Scholar]
- 20.Wang H., Sun Z., Wang Y., Hu Z., Zhou H., Zhang L., Hong B., Zhang S., Cao X. miR-33-5p, a novel mechano-sensitive microRNA promotes osteoblast differentiation by targeting Hmga2. Sci. Rep. 2016;6:23170. doi: 10.1038/srep23170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Y., Lv Q., Lv C. MicroRNA-153 suppresses the osteogenic differentiation of human mesenchymal stem cells by targeting bone morphogenetic protein receptor type II. Int. J. Mol. Med. 2015;36:760–766. doi: 10.3892/ijmm.2015.2275. [DOI] [PubMed] [Google Scholar]
- 22.Li Z., Hassan M.Q., Jafferji M., Aqeilan R.I., Garzon R., Croce C.M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itoh T., Takeda S., Akao Y. MicroRNA-208 modulates BMP-2-stimulated mouse preosteoblast differentiation by directly targeting V-ets erythroblastosis virus E26 oncogene homolog 1. J. Biol. Chem. 2010;285:27745–27752. doi: 10.1074/jbc.M110.105080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J., Zhao L., Xing L., Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordeladze J.O., Reseland J.E., Duroux-Richard I., Apparailly F., Jorgensen C. From stem cells to bone: phenotype acquisition, stabilization, and tissue engineering in animal models. ILAR J. 2009;51:42–61. doi: 10.1093/ilar.51.1.42. [DOI] [PubMed] [Google Scholar]
- 26.Eskildsen T., Taipaleenmäki H., Stenvang J., Abdallah B.M., Ditzel N., Nossent A.Y., Bak M., Kauppinen S., Kassem M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl. Acad. Sci. USA. 2011;108:6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macías-Silva M., Hoodless P.A., Tang S.J., Buchwald M., Wrana J.L. Specific activation of Smad1 signaling pathways by the BMP7 type I receptor, ALK2. J. Biol. Chem. 1998;273:25628–25636. doi: 10.1074/jbc.273.40.25628. [DOI] [PubMed] [Google Scholar]
- 28.Shore E.M., Gannon F.H., Kaplan F.S. Fibrodysplasia ossificans progressiva why do some people have two skeletons? J. Clin. Rheumatol. 1997;3(2, Suppl):84–89. doi: 10.1097/00124743-199704001-00019. [DOI] [PubMed] [Google Scholar]
- 29.Shore E.M., Xu M., Feldman G.J., Fenstermacher D.A., Cho T.J., Choi I.H., Connor J.M., Delai P., Glaser D.L., LeMerrer M. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat. Genet. 2006;38:525–527. doi: 10.1038/ng1783. [DOI] [PubMed] [Google Scholar]
- 30.Yu P.B., Deng D.Y., Lai C.S., Hong C.C., Cuny G.D., Bouxsein M.L., Hong D.W., McManus P.M., Katagiri T., Sachidanandan C. BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nat. Med. 2008;14:1363–1369. doi: 10.1038/nm.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karbiener M., Neuhold C., Opriessnig P., Prokesch A., Bogner-Strauss J.G., Scheideler M. MicroRNA-30c promotes human adipocyte differentiation and co-represses PAI-1 and ALK2. RNA Biol. 2011;8:850–860. doi: 10.4161/rna.8.5.16153. [DOI] [PubMed] [Google Scholar]
- 32.Zumbrennen-Bullough K.B., Wu Q., Core A.B., Canali S., Chen W., Theurl I., Meynard D., Babitt J.L. MicroRNA-130a is up-regulated in mouse liver by iron deficiency and targets the bone morphogenetic protein (BMP) receptor ALK2 to attenuate BMP signaling and hepcidin transcription. J. Biol. Chem. 2014;289:23796–23808. doi: 10.1074/jbc.M114.577387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamiya N., Kaartinen V.M., Mishina Y. Loss-of-function of ACVR1 in osteoblasts increases bone mass and activates canonical Wnt signaling through suppression of Wnt inhibitors SOST and DKK1. Biochem. Biophys. Res. Commun. 2011;414:326–330. doi: 10.1016/j.bbrc.2011.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baron R., Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat. Med. 2013;19:179–192. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
- 35.Morse A., McDonald M.M., Kelly N.H., Melville K.M., Schindeler A., Kramer I., Kneissel M., van der Meulen M.C., Little D.G. Mechanical load increases in bone formation via a sclerostin-independent pathway. J. Bone Miner. Res. 2014;29:2456–2467. doi: 10.1002/jbmr.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X., Guo B., Li Q., Peng J., Yang Z., Wang A., Li D., Hou Z., Lv K., Kan G. miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 2013;19:93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- 37.Kureel J., Dixit M., Tyagi A.M., Mansoori M.N., Srivastava K., Raghuvanshi A., Maurya R., Trivedi R., Goel A., Singh D. miR-542-3p suppresses osteoblast cell proliferation and differentiation, targets BMP-7 signaling and inhibits bone formation. Cell Death Dis. 2014;5:e1050. doi: 10.1038/cddis.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;12:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Betel D., Koppal A., Agius P., Sander C., Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao F., Zuo Z., Cai G., Kang S., Gao X., Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohberger B., Kaltenegger H., Stuendl N., Payer M., Rinner B., Leithner A. Effect of cyclic mechanical stimulation on the expression of osteogenesis genes in human intraoral mesenchymal stromal and progenitor cells. BioMed Res. Int. 2014;2014:189516. doi: 10.1155/2014/189516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z., Hassan M.Q., Volinia S., van Wijnen A.J., Stein J.L., Croce C.M., Lian J.B., Stein G.S. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc. Natl. Acad. Sci. USA. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salingcarnboriboon R., Tsuji K., Komori T., Nakashima K., Ezura Y., Noda M. Runx2 is a target of mechanical unloading to alter osteoblastic activity and bone formation in vivo. Endocrinology. 2006;147:2296–2305. doi: 10.1210/en.2005-1020. [DOI] [PubMed] [Google Scholar]
- 45.Blaber E.A., Dvorochkin N., Torres M.L., Yousuf R., Burns B.P., Globus R.K., Almeida E.A. Mechanical unloading of bone in microgravity reduces mesenchymal and hematopoietic stem cell-mediated tissue regeneration. Stem Cell Res. (Amst.) 2014;13:181–201. doi: 10.1016/j.scr.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Spatz J.M., Wein M.N., Gooi J.H., Qu Y., Garr J.L., Liu S., Barry K.J., Uda Y., Lai F., Dedic C. The Wnt inhibitor sclerostin is up-regulated by mechanical unloading in osteocytes in vitro. J. Biol. Chem. 2015;290:16744–16758. doi: 10.1074/jbc.M114.628313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Z., Cao X., Zhang Z., Hu Z., Zhang L., Wang H., Zhou H., Li D., Zhang S., Xie M. Simulated microgravity inhibits L-type calcium channel currents partially by the up-regulation of miR-103 in MC3T3-E1 osteoblasts. Sci. Rep. 2015;5:8077. doi: 10.1038/srep08077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Z., Cao X., Hu Z., Zhang L., Wang H., Zhou H., Li D., Zhang S., Xie M. MiR-103 inhibits osteoblast proliferation mainly through suppressing Cav1.2 expression in simulated microgravity. Bone. 2015;76:121–128. doi: 10.1016/j.bone.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 49.Sayed D., Abdellatif M. MicroRNAs in development and disease. Physiol. Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 50.Song H., Wang Q., Wen J., Liu S., Gao X., Cheng J., Zhang D. ACVR1, a therapeutic target of fibrodysplasia ossificans progressiva, is negatively regulated by miR-148a. Int. J. Mol. Sci. 2012;13:2063–2077. doi: 10.3390/ijms13022063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Culbert A.L., Chakkalakal S.A., Theosmy E.G., Brennan T.A., Kaplan F.S., Shore E.M. Alk2 regulates early chondrogenic fate in fibrodysplasia ossificans progressiva heterotopic endochondral ossification. Stem Cells. 2014;32:1289–1300. doi: 10.1002/stem.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu P., Wu D., You Y., Sun J., Lu L., Tan J., Bie P. miR-208-3p promotes hepatocellular carcinoma cell proliferation and invasion through regulating ARID2 expression. Exp. Cell Res. 2015;336:232–241. doi: 10.1016/j.yexcr.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Montgomery R.L., Hullinger T.G., Semus H.M., Dickinson B.A., Seto A.G., Lynch J.M., Stack C., Latimer P.A., Olson E.N., van Rooij E. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H., Zheng D., Zhang B., Liu L., Ou J., Chen W., Xiong S., Gu Y., Yang J. Mir-208 promotes cell proliferation by repressing SOX6 expression in human esophageal squamous cell carcinoma. J. Transl. Med. 2014;12:196. doi: 10.1186/1479-5876-12-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yehya N., Yerrapureddy A., Tobias J., Margulies S.S. MicroRNA modulate alveolar epithelial response to cyclic stretch. BMC Genomics. 2012;13:154. doi: 10.1186/1471-2164-13-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mohamed J.S., Lopez M.A., Boriek A.M. Mechanical stretch up-regulates microRNA-26a and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3β. J. Biol. Chem. 2010;285:29336–29347. doi: 10.1074/jbc.M110.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shyu K.G., Wang B.W., Wu G.J., Lin C.M., Chang H. Mechanical stretch via transforming growth factor-β1 activates microRNA208a to regulate endoglin expression in cultured rat cardiac myoblasts. Eur. J. Heart Fail. 2013;15:36–45. doi: 10.1093/eurjhf/hfs143. [DOI] [PubMed] [Google Scholar]
- 58.Li H., Xie H., Liu W., Hu R., Huang B., Tan Y.F., Xu K., Sheng Z.F., Zhou H.D., Wu X.P., Luo X.H. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J. Clin. Invest. 2009;119:3666–3677. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 60.Morey-Holton E.R., Globus R.K. Hindlimb unloading rodent model: technical aspects. J. Appl. Physiol. 2002;92(1985):1367–1377. doi: 10.1152/japplphysiol.00969.2001. [DOI] [PubMed] [Google Scholar]
- 61.Jepsen K.J., Silva M.J., Vashishth D., Guo X.E., van der Meulen M.C. Establishing biomechanical mechanisms in mouse models: practical guidelines for systematically evaluating phenotypic changes in the diaphyses of long bones. J. Bone Miner. Res. 2015;30:951–966. doi: 10.1002/jbmr.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao X.Y., Li W., Lv Z., Liu L., Tong M., Hai T., Hao J., Guo C.L., Ma Q.W., Wang L. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 63.Chen M., Wang X., Ye Z., Zhang Y., Zhou Y., Tan W.S. A modular approach to the engineering of a centimeter-sized bone tissue construct with human amniotic mesenchymal stem cells-laden microcarriers. Biomaterials. 2011;32:7532–7542. doi: 10.1016/j.biomaterials.2011.06.054. [DOI] [PubMed] [Google Scholar]
- 64.Hu L.F., Qian A.R., Wang Y., Di S.M., Shang P. Inhibitory effect of simulated microgravity on differentiating preosteoblasts. Adv. Space Res. 2013;51:107–114. [Google Scholar]
- 65.Hu L.F., Li J.B., Qian A.R., Wang F., Shang P. Mineralization initiation of MC3T3-E1 preosteoblast is suppressed under simulated microgravity condition. Cell Biol. Int. 2015;39:364–372. doi: 10.1002/cbin.10391. [DOI] [PubMed] [Google Scholar]
- 66.van Loon J.J.W.A. Some history and use of the random positioning machine, RPM, in gravity related research. Adv. Space Res. 2007;39:1161–1165. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.