Abstract
We reported modified endpoint PCR results analyzed by universal and human-, swine-, and cattle-specific Bacteroidales gene markers with human sewage and animal fecal samples (i.e., swine, cattle, chicken, goat, sheep, buffalo, and duck) from Tha Chin and Chao Phraya watersheds. Annealing locations of PCR primers were illustrated by maps of 16s rRNA Bacteroidales genes. We also summarized previously published work on the performance of the PCR assays. For further discussion of the data presented here, please refer to Somnark et al., Performance evaluation of Bacteroidales genetic markers for human and animal microbial source tracking in tropical agricultural watersheds, Environ. Pollut. 236 (2018) 100–110.
Keywords: Endpoint PCR, Fecal pollution, Microbial source tracking, Bacteroidales, Sensitivity, Specificity, Water quality
Specifications Table
| Subject area | Biology |
| More specific subject area | Applied microbiology |
| Type of data | Tables and figures |
| How data were acquired | PCR instrument (Mastercycler Pro thermocycler, Eppendorf), and literature review |
| Data format | Analyzed |
| Experimental factors | Composite fecal and sewage samples were collected, and DNA extraction was performed |
| Experimental features | PCR primers originally designed as endpoint and quantitative PCR were used in the modified endpoint PCR assays. |
| Data source location | Samples were collected from Tha Chin (Chai Nat, Suphan Buri, Nakhon Pathom, and Samut Sakhon provinces) and Chao Phraya (Phra Nakhon Si Ayutthaya, Pathum Thani, and Bangkok provinces) watersheds, located in the central part of Thailand. |
| Data accessibility | Data are with this article |
Value of the data
-
–
PCR results of Bacteroidales-modified endpoint PCR markers could be compared with microbial source tracking (MST) studies in other geographic areas for further development of region-specific MST methods.
-
–
Bacteroidales PCR primer maps could offer an insight into annealing regions of primers for further design of new primers or evaluating currently available primers with their performance.
-
–
A summary of PCR assays that are originally designed and adopted to other regions could serve as a database for comparing the MST method performance in different geographical areas.
1. Data
We performed endpoint PCR assays modified from published methods originally in PCR and qPCR platforms. PCR results of ten good-performing modified endpoint PCR assays against human sewage and animal fecal samples from Tha Chin and Chao Phraya watersheds are shown (Table 1). There were six modified endpoint PCR assays that demonstrated potentially low sensitivity or specificity during the process of testing against a limited number of samples and therefore were not further tested with total samples (Table 2). We also compiled sensitivity and specificity data of previously published Bacteroidales genetic markers from both studies that originally designed the assays and studies that adopted the designed assays to be used in another geographic location (Table 3). To provide further insight into PCR performance, we mapped PCR primers to 16 s rRNA gene of human-, swine-, and cattle-associated Bacteroidales (Fig. 1, Fig. 2). Amplified PCR products with universal and human-, swine-, and cattle-specific Bacteroidales PCR assays were presented (Fig. 3).
Table 1.
Positive PCR results of modified endpoint PCR markers showing good performance with samples from Tha Chin and Chao Phraya watersheds.
| Host | Assay name | Tha Chin watershed |
Chao Phraya watershed |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human (19) | Swine (20) | Cattle (20) | Chicken (19) | Goat (7) | Sheep (5) | Buffalo (5) | Duck (5) | Human (9) | Swine (8) | Cattle (5) | Chicken (2) | Goat (3) | Buffalo (1) | ||
| Universal | BacUni EP | 19 | 20 | 20 | 20 | 7 | 5 | 5 | 5 | 9 | 8 | 5 | 1 | 3 | 1 |
| GenBac3 EP | 19 | 20 | 20 | 20 | 7 | 5 | 5 | 5 | 9 | 8 | 5 | 2 | 3 | 1 | |
| Bac32F/Bac708R | 15 | 20 | 20 | 20 | 7 | 5 | 5 | 5 | 9 | 8 | 5 | 2 | 3 | 1 | |
| Human | BacHum EP | 18 | 17 | 1 | 9 | 5 | 2 | 1 | 2 | 9 | 7 | 0 | 0 | 2 | 0 |
| HF183F/BFDrev EP | 16 | 4 | 1 | 9 | 2 | 1 | 0 | 2 | 9 | 0 | 0 | 0 | 0 | 0 | |
| Modified HF183F/Bac708R | 18 | 4 | 4 | 12 | 1 | 1 | 0 | 2 | 9 | 5 | 0 | 0 | 1 | 0 | |
| Swine | Pig-2-Bac EP | 0 | 20 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 |
| Cattle | Bac2 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 |
| Bac3 | 0 | 0 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | |
| Cow-Bac2 EP | 0 | 1 | 18 | 18 | 7 | 5 | 5 | 4 | 0 | 0 | 5 | 1 | 3 | 1 | |
Table 2.
Positive PCR results of modified endpoint PCR markers showing relatively poor performance with limited numbers of samples from the Tha Chin watershed.
| Fecal origin | Assay name | No. of positive samples/no. of samples testeda |
Sensitivity | Specificity | Accuracy | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | Swine | Cattle | Chicken | Goat | Sheep | Buffalo | Duck | |||||
| Swine | PF163F/Bac708R | 0/0 | 20/20 | 6/20 | 3/19 | 0/7 | 1/5 | 4/5 | 0/5 | 1.00 | 0.77 | 0.83 |
| Cattle | CowM2 EP | 0/0 | 3/3 | 6/7 | 1/1 | 2/2 | 0/0 | 0/0 | 0/0 | 0.86 | 0.00 | 0.47 |
| BacCow EP | 0/0 | 3/3 | 7/7 | 1/1 | 2/2 | 0/0 | 0/0 | 0/0 | 1.00 | 0.00 | 0.54 | |
| CF193F/Bac708R | 0/0 | 0/3 | 0/7 | 0/1 | 0/2 | 0/0 | 0/0 | 0/0 | 0.00 | 1.00 | 0.46 | |
| CF128F/Bac708R | 0/0 | 3/3 | 7/7 | 1/1 | 2/2 | 0/0 | 0/0 | 0/0 | 1.00 | 0.00 | 0.54 | |
| BoBac EP | 0/0 | 18/19 | 20/20 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 1.00 | 0.05 | 0.54 | |
Limited number of animal samples tested for certain assays due to potentially low sensitivity or specificity.
Table 3.
Sensitivity and specificity of Bacteroidales markers in studied in which the assays were originally designed and adopted to other geographic regions.
| Host source | Assay name | Platform | Geographical region | Original/ Adopted | Sensitivity (n)a | Specificity (n) | Non-target hosts | Reference |
|---|---|---|---|---|---|---|---|---|
| Universal | BacUni | qPCR | California, USA | Original | 1.00 (n=73) | NAb | Humanc, cow, horse, dog, cat, seagull, WWTPd (primary influent) | [1] |
| PCR | Tha Chin watershed, Thailand | Adopted | 1.00 (n=100, composite) | NA | Swine, cattle, chicken, goat, sheep, buffalo, duck, sewagee | [2] | ||
| PCR | Chao Phraya watershed, Thailand | Adopted | 0.96 (n=28, composite) | NA | Swine, cattle, chicken, goat, buffalo, sewage | [2] | ||
| Universal | GenBac3 | qPCR | Louisiana, Michigan, Mississippi, USA | Original | NA | NA | Surface water sample | [3] |
| PCR | Tha Chin watershed, Thailand | Adopted | 1.00 (n=100, composite) | NA | Swine, cattle, chicken, goat, sheep, buffalo, duck, sewage | [2] | ||
| Chao Phraya watershed, Thailand | Adopted | 1.00 (n=28, composite) | NA | Swine, cattle, chicken, goat, buffalo, sewage | [2] | |||
| Universal | Bac32F/Bac708R | PCR | Oregon, USA | Original | 1.00 (n=30) | NA | Human, cow | [4] |
| PCR | Southeast Queensland, Australia | Adopted | 1.00 (n=186) | NA | Cattle, pig, sheep, goat, horse, chicken, dog, duck, pelican, kangaroo, WWTP | [5] (one base pair mismatch for Bac32F primer) | ||
| PCR | Wisconsin, USA | Adopted | 1.00 (n=89) | NA | Cow, WWTP | [6] | ||
| PCR | Missouri, USA | Adopted | 0.89 (n=286) | NA | Human, sewage, dog, beef cattle, dairy cattle, chicken, turkey, horse, swine, goose | [7] | ||
| PCR | Britanny and Normandy, France | Adopted | 0.96 (n=136) | NA | Pig, cow, sheep, chicken, wild bird | [8] | ||
| PCR | Saskatchewan, Canada | Adopted | 1.00 (n=273) | NA | Human, WWTP, cow, pig, chicken, goose, moose, deer, caribou, bison, goat | [9] | ||
| PCR | Illinois, Nebraska, Ohio, Texas, Delaware, and West Virginia, USA | Adopted | 0.78 (n=222) | NA | Cattle, human, chicken, raccoon, horse, pig, pig manure pit, pig waste lagoon | [10] | ||
| PCR | Puerto Rico, USA | Adopted | 0.89 (n=356) | NA | Cow, goat, horse, swine, monkey, fish, pigeon, chicken, guinea fowl, duck, turkey, swan, WWTP | [11] | ||
| PCR | Tha Chin watershed, Thailand | Adopted | 0.96 (n=100, composite) | NA | Swine, cattle, chicken, goat, sheep, buffalo, duck, sewage | [2] | ||
| Chao Phraya watershed, Thailand | Adopted | 1.00 (n=28, composite) | NA | Swine, cattle, chicken, goat, buffalo, sewage | [2] | |||
| Human | BacHum | qPCR | California, USA | Original | 0.67 (n=18); | 0.98 (n=41) | Cow, horse, dog, cat, seagull | [1] |
| 1.00 (n=14, sewage) | ||||||||
| PCR | Southeast Queensland, Australia | Adopted | 1.00 (n=50, WWTP) | 0.96 (n=136) | Cattle, pig, sheep, goat, horse, chicken, dog, duck, pelican, kangaroo | [5] | ||
| PCR | Tha Chin watershed, Thailand | Adopted | 0.95 (n=19, sewage) | 0.54 (n=81, composite) | Swine, cattle, chicken, goat, sheep, buffalo, duck | [2] | ||
| Chao Phraya watershed, Thailand | Adopted | 1.00 (n=9, sewage) | 0.53 (n=19, composite) | Swine, cattle, chicken, goat, buffalo | [2] | |||
| Human | HF183/BFDrev | qPCR | Michigan, Minnesota, Colorado, South Dakota, Wyoming, Hawaii, Virginia, Ohio, Florida, North Carolina, and New York, USA | Original | 1.00 (n=14, WWTP) | 0.60 (n=5, composite) | Cow, pig, chicken, dog, cat | [12] |
| PCR | Tha Chin watershed, Thailand | Adopted | 0.84 (n=19, sewage) | 0.77 (n=81, composite) | Swine, cattle, chicken, goat, sheep, buffalo, duck | [2] | ||
| Chao Phraya watershed, Thailand | Adopted | 1.00 (n=9, sewage) | 1.00 (n=19, composite) | Swine, cattle, chicken, goat, buffalo | [2] | |||
| Human | HF183/Bac708R | PCR | Oregon, USA | original | 0.85 (n=13); 1.00 (n=3, WWTP) | 1.00 (n=46) | Cow, deer, elk, cat, dog, duck, pig, gull, goat, llama, sheep | [13] |
| PCR | Southeast Queensland, Australia | Adopted | 1.00 (n=52, WWTP) | 1.00 (n=155) | Duck, kangaroos, cattle, horse, dog, chicken, pig, pelican, goat, deer, wild birds, sheep | [14] | ||
| PCR | Spain | Adopted | 0.50 (n=40, WWTP) | 0.71 (n=73) | Poultry, pig, cow | [15] | ||
| PCR | Southeast Queensland, Australia | Adopted | 1.00 (n=59, WWTP); 0.80 (n=20) | 0.95 (n=214) | Bird, camel, cattle, chicken, dog, duck, horse, kangaroo, pig, possom | [16] | ||
| PCR | Britanny and Normandy, France | Adopted | 0.98 (n=44) | 0.99 (n=86) | Pig, cow, sheep, chicken, wild bird | [8] | ||
| PCR | Puerto Rico, USA | Adopted | 0.75 (n=16, sewage WWTP) | 1.00 (n=340) | Cow, goat, horse, swine, monkey, fish, pigeon, chicken, guinea fowl, duck, turkey, swan | [11] | ||
| PCR | Wisconsin, USA | Adopted | 1.00 (n=14, WWTP) | 1.00 (n=75) | Cow | [6] | ||
| PCR | Saskatchewan, Canada | Adopted | 1.00 (n=8, WWTP); | 1.00 (n=211) | Cow, pig, chicken, goose, moose, deer, caribou, bison, goat | [9] | ||
| 0.94 (n=54) | ||||||||
| PCR | Tha Chin watershed, Thailand | Adopted | 0.95 (n=19, sewage) | 0.70 (n=81, composite) | Swine, cattle, chicken, goat, sheep, buffalo, duck | [2] | ||
| Chao Phraya watershed, Thailand | Adopted | 1.00 (n=9, sewage) | 0.68 (n=19, composite) | Swine, cattle, chicken, goat, buffalo | [2] | |||
| Swine | PF163F/Bac708R | PCR | Cincinnati, Ohio | Original | 1.00 (n=19) | NA | NA | [17] |
| PCR | Saskatchewan, Canada | Adopted | 1.00 (n=50) | 1.00 (n=223) | Human, WWTP, cow, chicken, goose, moose, deer, caribou, bison, goat | [9] | ||
| PCR | Illinois, Nebraska, Ohio, Texas, Delaware, and West Virginia, USA | Adopted | 0.87 (n=97); 1.00 (n=6, slurry) | 0.77 (n=119) | Cattle, cattle lagoon, human, chicken, raccoon, horse | [10] | ||
| PCR | Puerto Rico, USA | Adopted | 1.00 (n=30) | 0.75 (n=261) | Cow, goat, horse, monkey, fish, pigeon, chicken, guinea fowl, duck, turkey, swan, WWTP | [11] | ||
| PCR | Britanny and Normandy, France | Adopted | 1.00 (n=25) | 0.98 (n=105) | Human, cow, sheep, chicken, wild bird | [8] | ||
| PCR | Tha Chin watershed, Thailand | Adopted | 1.00 (n=20, composite) | 0.77 (n=61, composite) | Cattle, chicken, goat, sheep, buffalo, duck | [2] | ||
| Swine | Pig-2-Bac | qPCR | Brittany, France | Original | 1.00 (n=25); | 1.00 (n=54) | Human, bovine, horse, sheep | [18] |
| 1.00 (n=23, slurry) | ||||||||
| PCR | Tha Chin watershed, Thailand | Adopted | 1.00 (n=20, composite) | 0.98 (n=80, composite) | Cattle, chicken, goat, sheep, buffalo, duck, sewage | [2] | ||
| Chao Phraya watershed, Thailand | Adopted | 1.00 (n=8, composite) | 1.00 (n=20, composite) | Cattle, chicken, goat, buffalo, sewage | [2] | |||
| Cattle | CowM2 | qPCR | West Virginia, Georgia, Wyoming, Delaware, Florida, and Ohio, USA | Original | 1.00 (n=60) | 1.00 (n=139); 1.00 (n=5, WWTP (primary effluent) | Alpaca, goat, mule deer, sheep, Canadian goose, cat, chicken, dog, duck, horse, human, pelican, pig, sea gull, turkey | [19] |
| PCR | Tha Chin watershed, Thailand | Adopted | 0.86 (n=7, composite) | 0.00 (n=6, composite) | Swine, chicken, goat | [2] | ||
| Cattle | BacCow | qPCR | California, USA | Original | 1.00 (n=8) | 0.95 (n=65) | Human, horse, dog, cat, seagull, WWTP (primary effluent) | [1] |
| PCR | Tha Chin watershed, Thailand | Adopted | 1.00 (n=7, composite) | 0.00 (n=6, composite) | Swine, chicken, goat | [2] | ||
| Cattle | CF193/Bac708R | PCR | Oregon, USA | Original | 1.00 (n=19) | 0.72 (n=43) | Human, WWTP, deer, elk, cat, dog, duck, pig, gull, goat, llama, sheep | [13] |
| PCR | Wisconsin, USA | Adopted | 0.85 (n=75) | NA | NA | [6] | ||
| PCR | Saskatchewan, Canada | Adopted | 0.16 (n=32) | NA | NA | [9] | ||
| PCR | Spain, UK, Cyprus, France, and Sweden | Adopted | 0.00 (n=19, ruminant) | 0.99 (n=94) | WWTP, poultry, pig | [15] | ||
| PCR | USA | Adopted | 0.68 (n=247) | 1.00 (n=175) | Alpaca, pronghorn, elk, gazelle, giraffe, goat, mule deer, okapi, sheep, takin, tufted deer, moose, white-tailed deer, Canadian goose, cat, chicken, dog, duck, horse, human, pelican, pig, raccoons, sea gull, turkey | [20] | ||
| PCR | Tha Chin watershed, Thailand | Adopted | 0.00 (n=7, composite) | 1.00 (n=6, composite) | Swine, chicken, goat | [2] | ||
| Cattle | CF128F/Bac708R | PCR | Oregon, USA | Original | 1.00 (n=19) | 0.77 (n=43) | Human, WWTP, deer, elk, cat, dog, duck, pig, gull, goat, llama, sheep | [13] |
| PCR | Wisconsin, USA | Adopted | 1.00 (n=75) | 0.93 (n=14) | WWTP | [6] | ||
| PCR | Britanny and Normandy, France | Adopted | 1.00 (n=32) | 0.60 (n=98) | Human, pig, chicken, sheep, wild bird | [8] | ||
| PCR | Saskatchewan, Canada | Adopted | 0.96 (n=51, cow); | 0.62 (n=222, cow); | Human, WWTP, pig, chicken, goose | [9] | ||
| 0.98 (n=121, ruminant=cow, deer, caribou, bison, moose, goat) | 0.93 (n=152, ruminant=cow, deer, caribou, bison, moose, goat) | |||||||
| PCR | Spain | Adopted | 0.26 (n=19, ruminant) | 1.00 (n=95) | WWTP, poultry, pig | [15] | ||
| PCR | USA | Adopted | 0.85 (n=247) | 0.76 (n=175) | Alpaca, pronghorn, elk, gazelle, giraffe, goat, mule deer, okapi, sheep, takin, tufted deer, moose, white-tailed deer, Canadian goose, cat, chicken, dog, duck, horse, human, pelican, pig, raccoons, sea gull, turkey | [20] | ||
| PCR | Puerto Rico, USA | Adopted | 0.64 (n=66) | 0.90 (n=290) | Goat, horse, swine, monkey, fish, pigeon, chicken, guinea fowl, duck, turkey, swan, WWTP | [11] | ||
| PCR | Tha Chin watershed, Thailand | Adopted | 1.00 (n=7, composite) | 0.00 (n=6, composite) | Swine, chicken, goat | [2] | ||
| Cattle | Bac2 | PCR | USA | Adopted | 0.54 (n=148) | 1.00 (n=279) | Bird, human, domestic, wildlife, pets, water by cattle | [21] |
| PCR | USA | Adopted | 0.54 (n=247) | 1.00 (n=175) | Alpaca, pronghorn, elk, gazelle, giraffe, goat, mule deer, okapi, sheep, takin, tufted deer, moose, white-tailed deer, Canadian goose, cat, chicken, dog, duck, horse, human, pelican, pig, raccoons, sea gull, turkey | [20] | ||
| PCR | Tha Chin watershed, Thailand | Adopted | 0.70 (n=20, composite) | 1.00 (n=80, composite) | Swine chicken, goat, sheep, buffalo, duck, sewage | [2] | ||
| Chao Phraya watershed, Thailand | Adopted | 1.00 (n=5, composite) | 1.00 (n=23, composite) | Swine chicken, goat, buffalo, sewage | [2] | |||
| Cattle | Bac3 | PCR | USA | Original | 0.91 (n=148) | 0.99 (n=245) | Human, sewage, bovine, chicken, black vulture, Canadian goose, peacock, pigeon, dog, cat, guinea pig, domestic goat, pig, sheep, horse, alpaca, llama, armadillo, bobcat, coyote, gray squirrel, rabbit, opossum, raccoon, whitetail deer, wild turkey, hedgehog, prairie dog | [21] |
| PCR | USA | Adopted | 0.69 (n=247, ind) | 0.99 (n=175, ind) | Alpaca, pronghorn, elk, gazelle, giraffe, goat, mule deer, okapi, sheep, takin, tufted deer, moose, white-tailed deer, canadian goose, cat, chicken, dog, duck, horse, human, pelican, pig, raccoons, sea gull, turkey | [20] | ||
| PCR | Tha Chin watershed, Thailand | Adopted | 0.85 (n=20, composite) | 1.00 (n=80, composite) | Swine chicken, goat, sheep, buffalo, duck, sewage | [2] | ||
| Chao Phraya watershed, Thailand | Adopted | 1.00 (n=5, composite) | 1.00 (n=23, composite) | Swine chicken, goat, buffalo, sewage | [2] | |||
| Cattle | Cow-Bac2 | qPCR | Sapporo and Ebetsu Cities, Japan | Original | 1.00 (n=7) | 1.00 (n=9) | Human, pig | [22] |
| PCR | Tha Chin watershed, Thailand | Adopted | 0.90 (n=20, composite) | 0.50 (n=80, composite) | Swine chicken, goat, sheep, buffalo, duck, sewage | [2] | ||
| Chao Phraya watershed, Thailand | Adopted | 1.00 (n=5, composite) | 0.78 (n=23, composite) | Swine chicken, goat, buffalo, sewage | [2] | |||
| Cattle | BoBac | qPCR | Tennessee, Pennsylvania, and Texas, USA | Adopted | 1.00 (n=11) | 0.87 (n=15) | Human, swine, canine, equine | [23] |
| PCR | Tha Chin watershed, Thailand | Adopted | 1.00 (n=20, composite) | 0.05 (n=19, composite) | Swine | [2] |
Total number of samples being tested.
Not applicable.
Human individual fecal sample.
Influent of municipal wastewater treatment plant, unless stated otherwise.
Influent of wastewater treatment system in buildings or septic tanks.
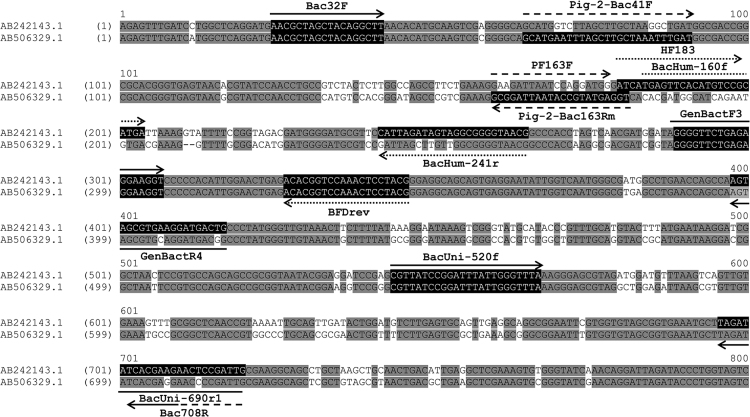
Fig. 1.
Primer map targeting the 16 S rRNA gene of human- and swine-associated Bacteroidales. All primers were BLASTed against the NCBI database. The representative sequences from human feces (Accession no. AB242143.1 [24]) and swine feces (AB506329.1 [25]) were selected to align with specific primers. Human-specific, swine-specific and universal Bacteroidales primers are indicated in dotted, dashed and solid arrows, respectively.
Fig. 2.
Primer map targeting the 16 S rRNA gene of cattle-associated Bacteroidales. All primers were BLASTed against the NCBI database. The representative sequences (Accession nos. GQ921871.1 [26], KR514419.1, LC028711.1, and LC028829.1) were selected to align with specific primers.
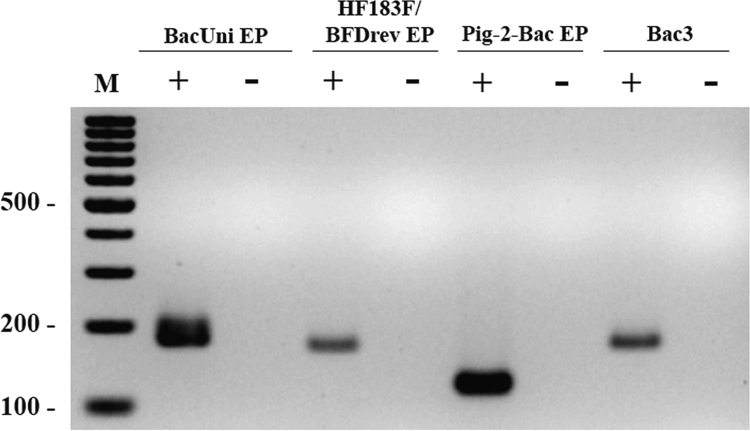
Fig. 3.
PCR results showing amplification products for universal and human-, swine-, and cattle-specific Bacteroidales markers (see [2] for related information).
2. Experimental design, materials and methods
2.1. Sample collection and DNA extraction
Raw human sewage and non-human fecal samples were collected from Tha Chin and Chao Phraya watersheds. One composite fecal sample was prepared by mixing fresh feces of at least 20 individuals. Samples were transported on ice to the laboratory. DNA extraction of composite fecal samples and 0.22-µm-pore-size mixed cellulose ester membrane (Merck Millipore, Billerica, MA, USA) after 50–100 mL human sewage filtration was performed with a ZR Fecal DNA MiniPrep kit (Zymo Research, Irvine, CA, USA). DNA concentrations were measured using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
2.2. PCR method modification and performance criteria
PCR primers targeting universal and human-, swine-, and cattle-specific fecal markers were selected from both endpoint and quantitative PCR platforms (Table 4). A 10-μL PCR is composed of 0.5 μL each of 10 μM forward primers and 10 μM reverse primers, 1-μL of DNA template (corresponding to 0.2, 2.0 or 20 ng total DNA), 5 μL of DreamTaq PCR Master Mix (2×; Thermo Fisher Scientific, Waltham, MA, USA), and sterile water. The reaction was processed in a Mastercycler Pro thermocycler (Eppendorf, Hamburg, Germany). PCR cycling conditions were modified as follows: initial denaturation at 95 °C for 3 min; 30 cycles of a denaturation step at 95 °C for 30 s, an annealing step at varying temperature and time (Table 4), and an elongation step at 72 °C for 30 s; and a final extension at 72 °C for 10 min. PCR products were visualized with a Gel Doc XR system (BIO-RAD, Hercules, CA, USA). PCRs were run in duplicate. No-template controls and extraction blanks were included for quality control. Performance criteria including sensitivity, specificity, and accuracy were calculated as TP/(TP+FN), TN/(TN+FP), and (TP+TN)/(TP+FP+TN+FN), respectively, where TP, FN, TN, and FP, are true positive, false negative, true negative, and false positive, respectively.
Table 4.
Primer sequences and PCR cycling conditions.
| Host | Assay name | Primer name | Primer sequence (5′ - 3′) | Annealing temperature (°C) | Annealing time (s) | Original platform | Reference |
|---|---|---|---|---|---|---|---|
| Universal | BacUni EP | BacUni-520f | CGT-TAT-CCG-GAT-TTA-TTG-GGT-TTA | 60.0 | 30 | qPCR | [1] |
| BacUni-690r1 | CAA-TCG-GAG-TTC-TTC-GTG-ATA-TCT-A | ||||||
| GenBac3 EP | GenBac3F | GGG-GTT-CTG-AGA-GGA-AGG-T | 60.0 | 30 | qPCR | [3] | |
| GenBac3R | CCG-TCA-TCC-TTC-ACG-CTA-CT | ||||||
| Bac32F/Bac708R | Bac32F | AAC-GCT-AGC-TAC-AGG-CTT | 53.7 | 60 | PCR | [4], [27] | |
| Bac708R | CAA-TCG-GAG-TTC-TTC-GTG | ||||||
| Human sewage | BacHum EP | BacHum-160f | TGA-GTT-CAC-ATG-TCC-GCA-TGA | 60.0 | 30 | qPCR | [1] |
| BacHum-241r | CGT-TAC-CCC-GCC-TAC-TAT-CTA-ATG | ||||||
| HF183/BFDrev EP | HF183 | ATC-ATG-AGT-TCA-CAT-GTC-CG | 60.0 | 30 | qPCR | [12] | |
| BFDrev | CGT-AGG-AGT-TTG-GAC-CGT-GT | ||||||
| Modified HF183F/Bac708R | HF183F | ATC-ATG-AGT-TCA-CAT-GTC-CG | 55.3 | 60 | PCR | [13], [27] | |
| Bac708R | CAA-TCG-GAG-TTC-TTC-GTG | ||||||
| Swine | PF163F/Bac708R | PF163F | GCG-GAT-TAA-TAC-CGT-ATG-A | 52.4 | 60 | PCR | [17], [27] |
| Bac708R | CAA-TCG-GAG-TTC-TTC-GTG | ||||||
| Pig-2-Bac EP | Pig-2-Bac41F | GCA-TGA-ATT-TAG-CTT-GCT-AAA-TTT-GAT | 60.0 | 30 | qPCR | [18] | |
| Pig-2-Bac163Rm | ACC-TCA-TAC-GGT-ATT-AAT-CCG-C | ||||||
| Cattle | CowM2 EP | CowM2F | CGG-CCA-AAT-ACT-CCT-GAT-CGT | 60.0 | 30 | qPCR | [19] |
| CowM2R | GCT-TGT-TGC-GTT-CCT-TGA-GAT-AAT | ||||||
| BacCow EP | CF128F | CCA-ACY-TTC-CCG-WTA-CTC | 60.0 | 30 | qPCR | [1] | |
| BacCow 305r | GGA-CCG-TGT-CTC-AGT-TCC-AGT-G | ||||||
| CF193F/Bac708R | CF193 | TAT-GAA-AGC-TCC-GGC-C | 55.0 | 30 | PCR | [13] | |
| Bac708R | CAA-TCG-GAG-TTC-TTC-GTG | ||||||
| Modified CF128F/Bac708R | CF128F | CCA-ACY-TTC-CCG-WTA-CTC | 62.0 | 60 | PCR | [13], [28] | |
| Bac708R | CAA-TCG-GAG-TTC-TTC-GTG | ||||||
| Bac2 | Bac2F | GCT-TGT-TGC-GTT-CCT-TGAGAT-AAT | 62.0 | 30 | PCR | [21] | |
| Bac2R | ACA-AGC-CAG-GTG-ATA-CAG-AAA-G | ||||||
| Bac3 | Bac3F | CTA-ATG-GAA-AAT-GGA-TGG-TAT-CT | 60.0 | 30 | PCR | [21] | |
| Bac3R | GCC-GCC-CAG-CTC-AAA-TAG | ||||||
| Cow-Bac2 EP | qCS621F | AAC-CAC-AGC-CCG-CGA-TT | 62.0 | 30 | SYBR qPCR | [22] | |
| qBac725R | CAA-TCG-GAG-TTC-TTC-GTG-ATA-TCT-A | ||||||
| BoBac EP | BoBac367f | GAA-GAC-TGA-ACC-AGC-CAA-GTA | 57.0 | 30 | qPCR | [23] | |
| BoBac467r | GCT-TAT-TCA-TAC-GGT-ACA-TAC-AAG |
Acknowledgements
This research was financially supported by the Thailand Research Fund (Contract no. SRI5930305) and the Kurita Water and Environmental Foundation (KWEF) – Asian Institute of Technology (AIT) research grant.
Author's statement
The authors declare that they have no competing interests.
Footnotes
Transparency data associated with this article can be found in the online version at doi:10.1016/j.dib.2018.04.129.
Transparency document. Supplementary material
Supplementary material
References
- 1.Kildare B.J., Leutenegger C.M., McSwain B.S., Bambic D.G., Rajal V.B., Wuertz S. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 2007;41:3701–3715. doi: 10.1016/j.watres.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 2.Somnark P., Chyerochana N., Mongkolsuk S., Sirikanchana K. Performance evaluation of Bacteroidales genetic markers for human and animal microbial source tracking in tropical agricultural watersheds. Environ. Pollut. 2018;236:100–110. doi: 10.1016/j.envpol.2018.01.052. [DOI] [PubMed] [Google Scholar]
- 3.Siefring S., Varma M., Atikovic E., Wymer L., Haugland R.A. Improved real-time PCR assays for the detection of fecal indicator bacteria in surface waters with different instrument and reagent systems. J. Water Health. 2008;6:225–237. doi: 10.2166/wh.2008.022. [DOI] [PubMed] [Google Scholar]
- 4.Bernhard A.E., Field K.G. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 2000;66:1587–1594. doi: 10.1128/aem.66.4.1587-1594.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed W., Goonetilleke A., Powell D., Gardner T. Evaluation of multiple sewage-associated Bacteroides PCR markers for sewage pollution tracking. Water Res. 2009;43:4872–4877. doi: 10.1016/j.watres.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 6.Bower P., Scopel C.O., Jensen E.T., Depas M.M., Mclellan S.L. Detection of genetic markers of fecal indicator bacteria in Lake Michigan and determination of their relationship to Escherichia coli densities using standard microbiological methods. Appl. Environ. Microbiol. 2005;71:8305–8313. doi: 10.1128/AEM.71.12.8305-8313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carson C.A., Christiansen J.M., Benson V.W., Baffaut C., Jerri V., Broz R.R., Kurtz W.B., Rogers W.M., Fales W.H., Yampara-iquise H., Davis J.V. Specificity of a Bacteroides the taiotaomicron marker for human feces. Appl. Environ. Microbiol. 2005;71:4945–4949. doi: 10.1128/AEM.71.8.4945-4949.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gourmelon M., Caprais M.P., Segura R., Le Mennec C., Lozach S., Piriou J.Y., Rince A. Evaluation of two library-independent microbial source tracking methods to identify sources of fecal contamination in French estuaries. Appl. Environ. Microbiol. 2007;73:4857–4866. doi: 10.1128/AEM.03003-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fremaux B., Gritzfeld J., Boa T., Yost C.K. Evaluation of host-specific Bacteroidales 16S rRNA gene markers as a complementary tool for detecting fecal pollution in a prairie watershed. Water Res. 2009;43:4838–4849. doi: 10.1016/j.watres.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 10.Lamendella R., Santo Domingo J.W., Yannarell A.C., Ghosh S., Di Giovanni G., Mackie R.I., Oerther D.B. Evaluation of swine-specific PCR assays used for fecal source tracking and analysis of molecular diversity of swine-specific “Bacteroidales” populations. Appl. Environ. Microbiol. 2009;75:5787–5796. doi: 10.1128/AEM.00448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toledo-Hernandez C., Ryu H., Gonzalez-Nieves J., Huertas E., Toranzos G., Domingo J.W. Santo. Tracking the primary sources of fecal pollution in a tropical watershed in a one-year study. Appl. Environ. Microbiol. 2013;79:1689–1696. doi: 10.1128/AEM.03070-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haugland R.A., Varma M., Sivaganesan M., Kelty C., Peed L., Shanks O.C. Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by qPCR. Syst. Appl. Microbiol. 2010;33:348–357. doi: 10.1016/j.syapm.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Bernhard A.E., Field K.G., PCR A. assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 2000;66:4571–4574. doi: 10.1128/aem.66.10.4571-4574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed W., Stewart J., Powell D., Gardner T. Evaluation of Bacteroides markers for the detection of human faecal pollution. Lett. Appl. Microbiol. 2008;46:237–242. doi: 10.1111/j.1472-765X.2007.02287.x. [DOI] [PubMed] [Google Scholar]
- 15.Ballesté E., Bonjoch X., Belanche L.A., Blanch A.R. Molecular indicators used in the development of predictive models for microbial source tracking. Appl. Environ. Microbiol. 2010;76:1789–1795. doi: 10.1128/AEM.02350-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed W., Masters N., Toze S. Consistency in the host specificity and host sensitivity of the Bacteroides HF183 marker for sewage pollution tracking. Lett. Appl. Microbiol. 2012;55:283–289. doi: 10.1111/j.1472-765X.2012.03291.x. [DOI] [PubMed] [Google Scholar]
- 17.Dick L.K., Bernhard A.E., Brodeur T.J., Santo Domingo J.W., Simpson J.M., Walters S.P., Field K.G. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 2005;71:3184–3191. doi: 10.1128/AEM.71.6.3184-3191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mieszkin S., Furet J.P., Corthier G., Gourmelon M. Estimation of pig fecal contamination in a river catchment by real-time PCR using two pig-specific Bacteroidales 16S rRNA genetic markers. Appl. Environ. Microbiol. 2009;75:3045–3054. doi: 10.1128/AEM.02343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shanks O.C., Atikovic E., Blackwood A.D., Lu J., Noble R.T., Domingo J.S., Seifring S., Sivaganesan M., Haugland R.A. Quantitative PCR for detection and enumeration of genetic markers of bovine fecal pollution. Appl. Environ. Microbiol. 2008;74:745–752. doi: 10.1128/AEM.01843-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shanks O.C., White K., Kelty C.A., Hayes S., Sivaganesan M., Jenkins M., Varma M., Haugland R.A. Performance assessment PCR-based assays targeting Bacteroidales genetic markers of bovine fecal pollution. Appl. Environ. Microbiol. 2010;76:1359–1366. doi: 10.1128/AEM.02033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shanks O.C., Santo Domingo J.W., Lamendella R., Kelty C.A., Graham J.E. Competitive metagenomic DNA hybridization identifies host-specific microbial genetic markers in cow fecal samples. Appl. Environ. Microbiol. 2006;72:4054–4060. doi: 10.1128/AEM.00023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okabe S., Okayama N., Savichtcheva O., Ito T. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl. Microbiol. Biotechnol. 2007;74:890–901. doi: 10.1007/s00253-006-0714-x. [DOI] [PubMed] [Google Scholar]
- 23.Layton A., McKay L., Williams D., Garrett V., Gentry R., Sayler G. Development of Bacteroides 16S rRNA gene taqman-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 2006;72:4214–4224. doi: 10.1128/AEM.01036-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bakir M.A., Sakamoto M., Kitahara M., Matsumoto M., Benno Y. Bacteroides dorei sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2006;56:1639–1643. doi: 10.1099/ijs.0.64257-0. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi Y., Itoh A., Miyawaki K., Koike S., Iwabuchi O., Iimura Y., Kobashi Y., Kawashima T., Wakamatsu J., Hattori A., Murakami H., Morimatsu F., Nakaebisu T., Hishinuma T. Effect of liquid whey feeding on fecal microbiota of mature and growing pigs. Anim. Sci. J. 2011;82:607–615. doi: 10.1111/j.1740-0929.2011.00876.x. [DOI] [PubMed] [Google Scholar]
- 26.Jeong J.Y., Park H.D., Lee K.H., Hwang J.H., Ka J.O. Quantitative analysis of human- and cow-specific 16S rRNA gene markers for assessment of fecal pollution in river waters by real-time PCR. J. Microbiol. Biotechnol. 2010;20:245–253. [PubMed] [Google Scholar]
- 27.Hussein K.R., Waines P.L., Nisr R.B., Glegg G., Bradley G. Development and use of Bacteroides 16S rRNA polymerase chain reaction assay for source tracking dog faecal pollution in bathing waters. Hydrol.: Curr. Res. 2014;5:1–8. [Google Scholar]
- 28.Lamendella R., Domingo J.W.S., Oerther D.B., Vogel J.R., Stoeckel D.M. Assessment of fecal pollution sources in a small northern-plains watershed using PCR and phylogenetic analyses of Bacteroidetes 16S rRNA gene. FEMS Microbiol. Ecol. 2007;59:651–660. doi: 10.1111/j.1574-6941.2006.00211.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material