Abstract
Recurrence and metastasis are major challenges in the management of hepatocellular carcinoma (HCC) patients after resection. To identify a metastasis-associated gene signature, we performed comparative gene expression analysis with recurrent HCC tissues from HCC patients who underwent partial or total hepatectomy and from non-metastatic primary HCC tissues. From this, we were able to identify genes associated with HCC recurrence. TCIRG1 (T-Cell Immune Regulator 1) was one of the aberrantly overexpressed genes in patients with recurrent HCC who had undergone total hepatectomy. The significant overexpression of TCIRG1 was confirmed using the Liver Hepatocellular Carcinoma dataset from The Cancer Genome Atlas. High expression of TCIRG1 was significantly associated with poor 5-year disease-free and recurrence-free survival of HCC patients. TCIRG1 knockdown suppressed tumor cell growth and proliferation in HCC cell lines; caused a significant increase in the proportion of cells in the G1/S phase of cell cycle; induced cell death; suppressed the metastatic potential of HCC cells by selectively regulating the epithelial–mesenchymal transition (EMT) regulatory proteins E-cadherin, N-cadherin, Fibronectin, Snail and Slug; and significantly attenuated the metastatic potential of ras-transformed NIH-3T3 cells in vitro and in vivo. These findings suggest that TCIRG1 functions as a metastatic enhancer by modulating growth, death and EMT in HCC cells. TCIRG1 could be a therapeutic target for the treatment of liver malignancy and metastasis.
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide and the second most common cause of cancer-related death.1 The primary curative treatment for HCC is surgical resection in a minority of patients diagnosed at early stages, when curative treatments are feasible. However, resection and percutaneous ablation are hampered by a recurrence rate of up to 70% at 5 years, which is closely linked to survival.2 Effective palliative treatment is hindered in HCC, which is frequently resistant to conventional chemotherapy and radiotherapy, because of toxicity in the presence of underlying liver disease.3 Only a minority of patients are eligible for curative treatment. Thus, there is an urgent need for new medical treatments. Randomized trials conducted with sorafenib have shown survival benefits in advanced HCC, which suggests that molecular-targeted therapies could be effective, and investigations are actively being conducted on the molecular pathogenesis and treatment of HCC.4
Classically, two types of recurrence are identified: early and late. Early recurrence, within 2 years after local treatment, is related to metastatic spread. Late recurrence, which occurs after 2 years, is related to the emergence of de novo HCC.5 Vascular invasion (both macroscopic and microscopic) is the strongest predictor of recurrence. Other associated variables are baseline serum alpha-fetoprotein (AFP) level, multinodularity, tumor size, poor degree of differentiation, presence of satellite nodules and presence of microvascular invasion.6 Unfortunately, microvascular invasion and satellites can be assessed only with the full pathologic specimen, which reduces the odds of an accurate preoperative prediction of HCC recurrence.
In addition to this diagnostic problem, the molecular mechanisms of liver metastasis are far from clear. A molecular understanding of the metastatic behavior of HCC is an important step toward the identification of additional biomarkers and new therapeutic targets with increased specificity for HCC metastasis. Transcriptomic analysis has proven useful in investigating the carcinogenesis of several malignant diseases. Many studies have addressed recurrence prediction in primary HCC using comparative transcriptome profiling of tissues obtained from primary HCC and metastatic HCC. However, several of these identified signatures were frequently ill-defined, being generated in patients at different stages of disease and with distinct etiologies for their underlying liver damage. Hence, the concordance of these signatures on a patient-by-patient basis remains unknown. The prognostic performance of these signatures in patients with early HCC is important, as they may be eligible for liver resection.
The present study aimed to identify potential markers in predicting the metastatic potential of HCC. For this, comprehensive transcriptomic analyses were conducted on primary HCC samples from patients who underwent total or partial hepatectomy but appeared to have early or late recurrence. The molecular signature of the recurrence group was compared to that of the non-recurrence group. Many genes associated with recurrence were identified; these were assumed to be metastasis-enhancing genes. T-cell immune regulator 1 (TCIRG1) was aberrantly upregulated in patients with recurrence of HCC who had undergone total hepatectomy. Overexpression of TCIRG1 in overt HCC was confirmed using data from large cohorts of HCC patients available from The Cancer Genome Atlas (TCGA) dataset and the Gene Expression Omnibus (GEO) database from the National Center for Biotechnology Information (NCBI). Further clinical interrogations suggested that the high expression of TCIRG1 was significantly associated with poor prognosis of HCC patients. Targeted disruption of TCIRG1 gene caused suppression of the neoplastic and metastatic properties of HCC cells. TCIRG1 inactivation selectively regulated epithelial–mesenchymal transition (EMT) regulatory proteins in HCC cells. These results suggest that TCIRG1 functions as a metastasis-enhancing gene by modulating cellular growth and EMT in HCC progression. TCIRG1 may have prognostic and therapeutic value in the treatment of liver malignancy.
Materials and methods
Tissue samples
HCC tissues were collected from HCC patients who had undergone surgical resection at Seoul National University Hospital from January 1995 to May 2006. The tissues were immediately snap-frozen and stored in liquid nitrogen. Informed consent was obtained from patients enrolled in this study. Written informed consent was obtained from each subject according to the Declaration of Helsinki, and the study was approved by the Institutional Review of Board of the College of Medicine (Songeui Campus) of the Catholic University of Korea (IRB approval number: MC12SNMI0184).
Molecular pathway mining and gene set enrichment analysis
To investigate gene signatures that were enriched from known molecular databases, we downloaded gene sets from MSigDB (http://software.broadinstitute.org/gsea/msigdb) at the Broad Institute Gene Set Enrichment Analysis (GSEA) (http://www.broadinstitute.org/gsea). GSEA was performed to access the relevance of the LIAO_METASTASIS gene set and overt metastatic gene signatures.7 Given a data set in which genes are rank-ordered by the correlation of their expression levels with the phenotype of interest, a basic GSEA provides a score that quantifies the degree of enrichment of a given gene set at the top (positive correlation) or bottom (negative correlation) of the rank-ordered data set. The proximities of gene sets were measured using Kolmogorov–Smirnoff (KS) scores (higher scores correspond to greater proximity). The observed KS scores were compared with the distribution of 1000 permuted KS scores for all gene sets to assess significance. FDR q-values of <0.05 were regarded as statistically significant.
Tissue microarray for TCIRG1
Hematoxylin and eosin-stained slides were reviewed, and one representative formalin-fixed, paraffin-embedded archival block was selected for each case. Core tissue biopsies (2 mm in diameter) were taken from individual formalin-fixed, paraffin-embedded blocks (donor blocks) and arranged in recipient paraffin blocks (tissue array blocks) using a trephine. Eight array blocks containing tissues from 302 HCC cases were prepared (Superbiochips Laboratories, Seoul, Korea).
Cell culture and transfection
Hep3B, HepG2, Huh7, PLC/PRF/5, SK-Hep1, SNU354, SNU368, SNU387, SNU398, SNU423, SNU449 and SNU475 HCC cell lines were acquired from the Korean Cell Line Bank (KCLB, Seoul, South Korea). The THLE3 normal liver cell line was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA), and the immortalized human hepatocyte line MIHA was kindly provided by Dr Roy-Chowdhury (Albert Einstein College of Medicine, Bronx, NY, USA). All of the cell lines were maintained in RPMI 1640, DMEM (GenDEPOT, Barker, TX, USA) or EMEM (ATCC) with 10% fetal bovine serum and 100 U ml−1 of penicillin–streptomycin (GenDEPOT). All cells were cultured at 37 °C in a humidified incubator with 5% CO2. For transfection, small interfering RNA molecules (siRNA) and negative-control RNA duplexes were synthesized by Bioneer (Dajeon, Korea). The RNA duplexes were transfected at a concentration of 100 nM. All transfection procedures were conducted with Lipofectamine RNAiMAX reagents (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
TCGA and GEO data analyses
To determine the expression level of TCIRG1 mRNA in HCC, the data were obtained from The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA_LIHC) project and NCBI GEO databases (Accession Nos GSE14520,8, 9 GSE16757,10 GSE22058,11 GSE25097,12 GSE36376,13 GSE36411, GSE45436, GSE7731414 and GSE89337). cBioPortal for Cancer Genomics website (http://www.cbioportal.org/public-portal/) was used to access TCGA mRNA expression data. The provisional RNASeq V2 RSEM z-score for each gene was used. The z-score represents a normalized relative expression level, which was used as a surrogate for the mRNA level.
RNA isolation and quantitative real-time polymerase chain reaction
Total RNA was isolated from frozen tissues and cell lines using TRIzol reagent (Invitrogen) according to the manufacturer’s instruction. For synthesis of cDNAs specific to mRNAs, a Tetro cDNA synthesis kit (Bioline, London, UK) was used. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed with a SensiFAST SYBR NoROX Kit (Bioline). To normalize differences in the amount of total cDNA added to each reaction; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the endogenous control.
Western blot analysis
Proteins from tissues and whole-cell lysates were prepared with radio immunoprecipitation (RIPA) buffer containing phenylmethane-sulfonylfluoride (PMSF; Sigma-Aldrich, St Louis, MO, USA) and 1 × complete protease inhibitor cocktail tablets (Roche, Indianapolis, IN, USA). Total proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories, Hercules, CA, USA). The membrane was blocked with blocking buffer (5% skimmed milk in Tris-buffered saline, 0.1% Tween-20) and incubated with the following antibodies: anti-TCIRG1 (ab139812), anti-Snail (ab78105) (both from Abcam, Cambridge, MA, USA), anti-GAPDH (sc-32233), anti-fibronectin (sc-29011) (both from Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-E-cadherin (#610181), anti-N-cadherin (#610920) (both from BD Transduction, San Jose, CA, USA), anti-Vimentin (#5741) and anti-Slug (#9585) (both from Cell Signaling Technology, Danvers, MA, USA). Chemiluminescence signals were detected with Immobilon horseradish peroxidase substrate (Millipore, Billerica, MA, USA) and visualized using an LAS-4000 image analyzer (Fuji Photo Film, Tokyo, Japan).
Clonogenic assay
Cells were transfected with TCIRG1-targeting siRNA in 60 mm2 cell culture plates. After transfection for 24 h, 1 × 103 and 2 × 103 cells were reseeded in 6-well plates and incubated for 2 weeks. Next, the cells were washed with phosphate-buffered saline (PBS) and fixed with 1% paraformaldehyde for 30 min at room temperature. The fixed cells were stained with 0.5% crystal violet for 1 h at room temperature. Colonies were counted using a clono-counter program as previously described.15
Cell viability
Cells were seeded in wells of a six-well plate and transfected with negative-control siRNA or TCIRG1-targeting siRNA. After transfection, cells were incubated for 72 h, collected by trypsinization and stained using trypan blue solution (Sigma-Aldrich). After staining, the cells were counted using a hemocytometer (Marienfeld Superior, LaudaKönigshofen, Germany)
Cell proliferation
Cells were seeded in a 12-well plate for transfection. After transfection, cells were incubated with 5 mg ml−1 of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] solution (Sigma-Aldrich) for 1 h. The dark blue formazan products formed by viable cells were dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich), and absorbance was measured using a VICTOR3 Multilabel plate reader (PerkinElmer, Boston, MA, USA).
Fluorescence-activated cell sorting
To determine cell cycle distribution, cells seeded in 60 mm2 plates were transfected with negative control and TCIRG1-targeting siRNA. After incubation for 72 h, the cells were collected by trypsinization. The collected cells were fixed using 70% ethanol, washed in PBS and resuspended with 200 μl PBS containing 1 mg ml−1 RNase (Sigma-Aldrich), 0.05% Triton X-100 and 50 μg ml−1 propidium iodide (BD Biosciences, San Jose, CA, USA). The suspended cells were incubated in a 37 °C incubator without CO2 for 1 h, and fluorescence-activated cell sorting (FACS) analysis was performed using a FACSCalibur flow cytometer (BD Biosciences) with FlowJo software (Tree Star, Ashland, OR, USA).
The Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit I (BD Biosciences) was used to measure the level of programmed cell death. After transfection for 72 h, cells were trypsinized, rinsed in PBS, and resuspended using 1 × binding buffer (1 × 106 cells ml−1), and 100 μl containing 1 × 105 cells was transferred to a 5-ml culture tube. Then, 5 μl of Annexin V-FITC and 5 μl of propidium iodide (PI) solution were added. The cells were incubated at room temperature for 15 min in the dark. After incubation, 400 μl of 1 × binding buffer was added to each tube, and apoptotic fractions were detected using a FACSCanto flow cytometer (BD Biosciences).
Migration and invasion assay
For in vitro cell migration and invasion assay, cell motility was measured using a modified Boyden chamber assay (BD Biosciences). For the invasion assay, Matrigel (BD Biosciences) was diluted to a concentration of 0.3 mg ml−1 with coating buffer. One hundred microliter aliquots of Matrigel were used to coat the upper surface of the Transwell cell culture inserts. After incubation for 2 h at 37 °C, the inserts were ready to be seeded with cells. After preparation, cells were plated on the top surfaces of Transwell inserts, and the inserts were placed in a 24-well plate. The lower wells contained 2% FBS as a chemoattractant. The plate was incubated overnight and stained using a Diff-Quik staining kit (Sysmex, Chuo-ku, Kobe, Japan). The cell images were captured using an Axiovert 200 inverted microscope (Zeiss, Oberkochen, Germany) at × 200 magnification, and the number of cells was counted in three random image fields.
Lung metastasis mouse model
For in vivo lung metastasis assay, 3 × 105 transfected ras-transformed NIH-3T3 cells (NC (Negative Control) or siTcirg1) were mixed with 0.1 ml PBS, and the cell suspension was injected into 5-week-old athymic male nude mice through tail veins. After injection, the mice were examined daily. After 12 days of inoculation, the mice were anesthetized for micro-computed tomography (micro-CT) imaging. The mice were placed on the scanner bed, and micro-CT images were acquired using a Triumph II PET/CT System (Trifoil Imaging, Chatsworth, CA, USA). Images were reconstructed, and the total lung tumor volume per mouse was quantified using MicroView analysis software (GE Healthcare, Pittsburgh, PA, USA). After CT imaging, the mice were euthanized to obtain lung images, and the number of lung surface nodules were counted.
Statistical analysis
All experiments were performed at least three times, and all samples were analyzed in triplicate. The results are presented as the mean±s.d. Statistical differences between groups were evaluated via an unpaired two-tailed Student’s t-test using GraphPad 5.0 (GraphPad software, San Diego, CA, USA). P-values <0.05 were considered statistically significant. Receiver operating characteristic curve (ROC) analysis was performed with MedCalc version 12.1.4.0 (MedCalc Software, Mariakerke, Belgium). Continuous variables are reported as the mean±s.e. and interquartile range. Categorical variables are reported as numbers and percentages. Univariable and multivariable logistic regression analyses were performed to identify preoperative independent predictors for HCC patient prognosis. All preoperative predictor variables that were identified as significant by a two-tailed nominal probability value of <0.05 in univariable regression analyses were then used in a multivariable logistic regression analysis model.
Results
Identification of genes associated with recurrence of HCC in patients
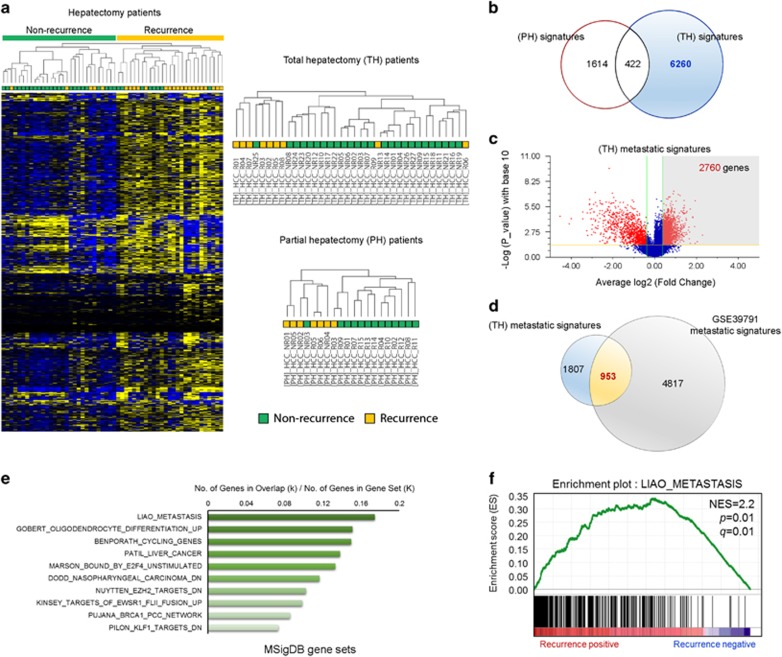
Liver transplantation is theoretically the ideal treatment for HCC. However, recurrence and metastasis are common lethal outcomes after liver transplantation.16 Thus, it is important to identify molecular markers specific to HCC metastasis and/or recurrence that are possibly detected in the primary tumor lesions. To this end, we collected 56 HCC tissues from patients who underwent partial or total hepatectomy and subjected these to transcriptomic analysis to identify gene expression signatures associated with recurrence after liver resection. The 56 HCC tissues included 9 recurrent specimens from 36 total hepatectomy patients and 15 recurrent specimens from 20 partial hepatectomy patients (Supplementary Table 1). Hierarchical clustering of 7550 genes obtained from the statistical analysis between the non-recurrence and recurrence groups (one-way ANOVA, P<0.05) resulted in two distinct subclusters within the dendrogram: the non-recurrence group (Figure 1a; right, green) and the recurrence group (Figure 1a; right, yellow). Reanalysis with both hepatectomy groups also showed distinct clusters in dendrograms indicating a recurrence-related molecular signature within both groups (Figure 1a, upper right and lower right). The analysis identified differentially expressed gene (DEG) signatures in the total hepatectomy (6682 genes) and partial hepatectomy (2036) groups by Welch’s t-test. Venn diagram analysis with the two molecular signatures of total hepatectomy and partial hepatectomy groups revealed 422 genes as part of a common molecular signature for both hepatectomy groups (Figure 1b). These 422 genes were discarded from the 6682 genes in the total hepatectomy signature, as recurrence after partial hepatectomy may occur from the remaining cancer cells of degenerating nodules in premalignant HCC lesions. Volcano plot analysis (P<0.05 and 1.3-fold upregulated) of the total hepatectomy signature revealed 2760 genes that were overexpressed in total hepatectomy patients with HCC recurrence compared with non-recurred total hepatectomy patients (Figure 1c). Lastly, to identify genes specific to recurrence of HCC, we performed comparative analysis with the GSE39791 data set available from the NCBI GEO database, which was previously used to study genomic predictors for recurrence patterns of HCC.17 From this, we were able to identify 953 genes as part of the recurrence-associated molecular signature (Figure 1d). Next, to investigate the biological relevance of the recurrence-associated molecular signature, we performed GSEA with the 953 genes. The analysis identified intracellular signaling pathways enriched in the recurrence-associated molecular signature (953 genes) of recurred HCC, and LIAO_METASTASIS was identified as a highly significantly enriched gene set (Figure 1e and f).
Figure 1.
Identification and selection of genes associated with the recurrence of hepatocellular carcinoma (HCC). (a) Two-dimensional diagram of the differential expression of 7550 genes; the data were organized according to non-recurrence or recurrence HCC tissues from patients who underwent liver resection. Dendrograms were derived from clustering the data for patients with total hepatectomy (TH) or partial hepatectomy (PH). Blue and yellow colors indicate downregulation and upregulation, respectively. (b) Venn diagram of genes altered between the TH and PH groups. (c) Volcano plot of differentially upregulated genes between non-recurrence and recurrence groups in patient with total hepatectomy. (d) The 953 common upregulated genes between the metastatic signature from patients with TH and that from the GSE39791 data set. (e) Bar chart of the top 10 genes among the 953 genes identified in the overt metastatic molecular signature using MSigDB. (f) Gene Set Enrichment Analysis (GSEA) was performed with the recurrence-associated molecular signature and LIAO_METASTASIS gene set. The barcode indicates gene positions. The y axis indicates the extent of enrichment.
TCIRG1 is overexpressed in HCC and is associated with recurrence of HCC after liver resection
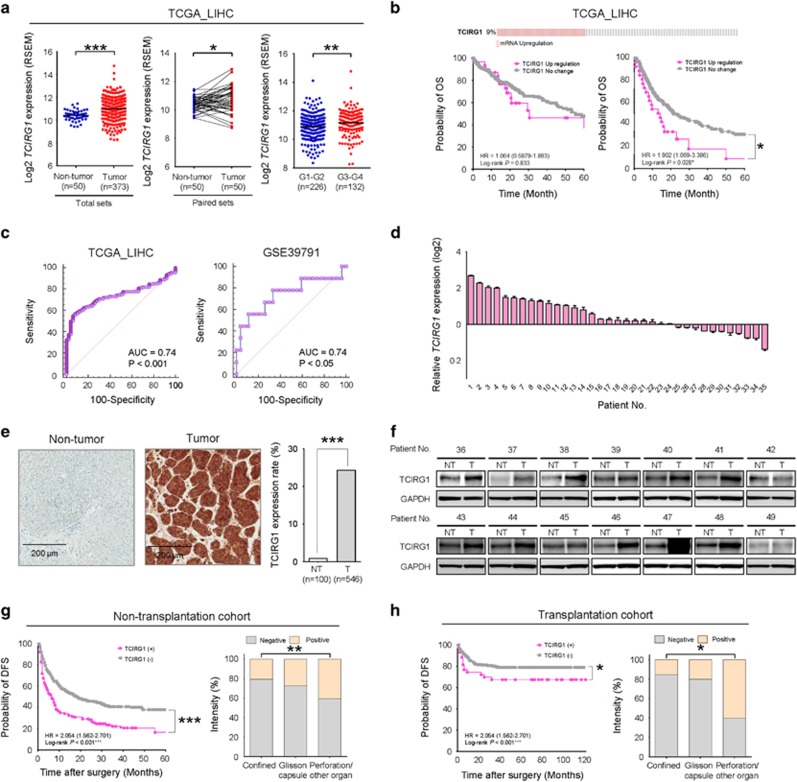
Among 953 recurrence-associated genes, the TCIRG1 gene was found to be significantly overexpressed in the TCGA_LIHC data set. TCGA database includes gene expression (mRNA) data sets from 50 non-tumor liver tissues and 373 HCC tissues including 50 matched pairs (50 HCCs with corresponding non-tumor tissues). The matched pairs also showed significantly differential expression between the non-cancer and HCC tissues. Interestingly, in a subset of HCC defined by Edmondson grade I–II (G1–G2, n=225) and grade III–VI (G3–G4, n=132), TCIRG1 gene expression was significantly higher in G3–G4 than that in G1–G2 (Figure 2a). To generalize TCIRG1 overexpression in HCC, we validated the differential TCIRG1 gene expression in various GEO data sets (Accession Nos GSE14520,8, 9 GSE16757,10 GSE22058,11 GSE25097,12 GSE36376,13 GSE36411, GSE45436, GSE7731414 and GSE89337); the data are presented as scatter plots. As expected, TCIRG1 was significantly overexpressed in these eight different HCC cohorts (Supplementary Figure 1a). Because TCIRG1 expression was upregulated in TCGA_LIHC data set, we next assessed the prognostic association of TCIRG1 expression using the data for HCC patients included in TCGA_LIHC data set available on cBioPortal (www.cbioportal.org). Nine percent of HCC patients exhibited overexpression of TCIRG1. Kaplan–Meier survival curves of HCC patients indicated that the 5-year disease-free survival (DFS) rates of patients with upregulated TCIRG1 expression was significantly lower than that of HCC patients with no change in TCIRG1 expression (Figure 2b). Upregulated TCIRG1 expression had no effect on the 5-year overall survival (OS) rate. To validate TCIRG1 as a biomarker of HCC recurrence after liver resection, comparison analysis of ROC curves was performed (Figure 2c). The area under the curve (AUC) was 0.74 in the TCGA_LIHC cohort and 0.74 in the GSE39791 cohort, implicating TCIRG1 as a potential valuable biomarker for the diagnosis of HCC.18 In addition, when TCIRG1 expression was analyzed in two large HCC patient cohorts using gene expression of matched paired samples (GSE39791 and GSE77314), TCIRG1 was found to be significantly overexpressed in the HCC tissues compared with non-tumor tissues (Supplementary Figure 1b). The TCIRG1 gene was significantly overexpressed in higher grade HCC in patients (G3–G4; Figure 2a). Consistently, TCIRG1 was significantly overexpressed in overt HCC samples (ovHCC, n=26) compared to that in normal liver tissues (normal, n=13) or early HCC samples (eHCC, n=25) (Supplementary Figure 1c, left). Differential gene expression analysis according to non-recurrence or recurrence in the same patients (GSE89337) revealed significant overexpression of TCIRG1 in HCC patients whose cancer recurred following total hepatectomy (Supplementary Figure 1c, right). To verify the overexpression of TCIRG1 in HCC patients, the expression of TCIRG1 was investigated by qRT-PCR in 35 randomly selected HCC tissues paired with adjacent non-cancerous liver tissues. Twenty-four of the 35 tested HCC tissues (68.6%) showed TCIRG1 overexpression (Figure 2d). Next, to validate TCIRG1 protein overexpression in HCC tissues, immunostaining with TCIRG1-specific antibody was performed. The positive detection rate of TCIRG1 in HCC tissues was 24.4% (133/546), whereas the positive detection rate of TCIRG1 in the non-tumor tissues was 1.0% (1/100) (Figure 2e). Increased expression of TCIRG1 protein was confirmed by western blotting in 14 randomly selected human HCC tissues with corresponding non-cancerous liver tissues (Figure 2f).
Figure 2.
Validation of TCIRG1 overexpression in hepatocellular carcinoma (HCC) and its clinical relevance. (a) Analysis of TCGA_LIHC data. TCGA_LIHC data showed that TCIRG1 was significantly overexpressed in HCC tissues compared with that in non-tumor tissues in all the analyzed samples (n=423, left) and paired samples (n=100, middle), and TCIRG1 was overexpressed in higher tumor grade (right) (mean±s.d.; *P<0.05; ***P<0.001). (b) Kaplan–Meier survival analysis of TCIRG1 mRNA expression in TCGA_LIHC for overall survival (left) and disease-free survival (right). The P-value was obtained with the log-rank test. (c) Receiver operating characteristic curve (ROC) analysis of TCIRG1 in TCGA_LIHC (left) and GSE39791 (right). Statistically significant difference of the area under the curve (AUC) compared with reference line (*P<0.05; ***P<0.001). (d) The mRNA expression of TCIRG1 was determined by qRT-PCR in a subset of HCC tissues. (e) Representative photomicrographs of immunohistochemical analysis of non-tumor or tumor tissues (left) and extent of TCIRG1 expression (right) (***P<0.001). (f) Western blot analysis of TCIRG1 in a subset of HCC tissues (NT: adjacent non-tumor tissues; T: tumor tissues). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the loading control. (g) Kaplan–Meier survival curves (left) for patients with positive or negative expression of TCIRG1, and the extent of TCIRG1 expression (right) in the tumors of the non-transplanted cohort. (h) Kaplan–Meier survival curves (left) for patients with positive or negative expression of TCIRG1, and the extent of TCIRG1 expression (right) in the tumors of the transplantation cohort.
Multivariable logistic regression analysis for various clinical parameters associated with liver metastasis including TCIRG1 showed that TCIRG1 overexpression had the strongest association (odds ratio: 1.768, P<0.001) among the six different metastatic characteristics (Table 1). Consistently, the DFS rate of TCIRG1-positive patients was significantly lower than that of TCIRG1-negative patients in cohorts of 546 non-transplantation HCC patients and 285 transplantation HCC patients (Figure 2g, h and Supplementary Table 2). Tumor progression was significantly higher in TCIRG1-positive patients in both cohorts (Supplementary Table 2). These results indicate TCIRG1 as an informative marker of HCC that has a high potential of recurrence after total hepatectomy.
Table 1. Logistic regression analysis for identifying TCIRG1 as the metastatic biomarkers.
| Characteristics | OR (95% CI) | P-value |
|---|---|---|
| Multiplicity (single vs multiple) | 1.712 (1.352–2.167) | <0.001 |
| Angioinvasion (absent vs present) | 1.223 (0.972–1.539) | 0.086 |
| ES nuclear grade (grade 1–2 vs 3–4) | 1.194 (1.006–1.418) | 0.042 |
| Size of tumor (⩽5 cm vs >5 cm) | 1.037 (1.005–1.071) | 0.023 |
| Extent of tumor | 1.412 (1.179–1.690) | <0.001 |
| TCIRG1 (negative vs positive) | 1.768 (1.397–2.237) | <0.001 |
Abbreviations: ES nuclear grade, Edmonson–Steiner nuclear grade; OR (95% CI), odds ratio (95% confidence interval).
Targeted disruption of TCIRG1 elicits a tumor suppressive effect on HCC cells
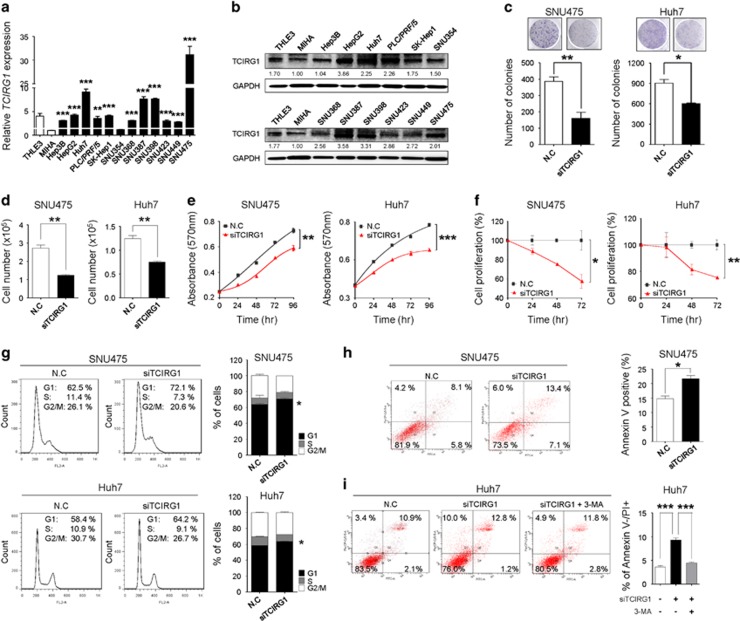
To better understand the molecular functions of TCIRG1 in liver tumorigenesis, TCIRG1 was knocked down using RNA-interference. To select HCC cell lines overexpressing TCIRG1, the endogenous level of TCIRG1 was measured by qRT-PCR and western blotting in 14 different liver cell lines, including MIHA and THLE3 immortalized normal hepatic cell lines. Human HCC cells (Hep3B, HepG2, Huh7, PLC/PRF/5, SK-Hep1, SNU354, SNU368, SNU387, SNU398, SNU423, SNU449 and SNU475) exhibited relatively high levels of TCIRG1 compared with MIHA cells by both methods (Figure 3a and b). TCIRG1 knockdown reduced growth and proliferation of SNU475 and Huh7 HCC cells (Figure 3c–f). The growth-suppressive effect could be partially explained by the disruption of regulation of cell growth due to cell cycle arrest, cellular senescence or apoptosis, upon TCIRG1-silencing. FACS analysis showed that TCIRG1 knockdown led to a significant increase in the number of cells in the G1 phase with a concomitant decrease in the number of cells in the S phase and G2/M phase in both SNU475 and Huh7 HCC cell lines (Figure 3g). Cells were stained with Annexin V-FITC and PI after transfection with TCIRG1-targeting siRNA (siTCIRG1) for cell death analysis. FACS analysis revealed that transfection with TCIRG1-targeting siRNA significantly induced apoptosis compared with negative-control siRNA (NC) in SNU475 cells (Figure 3h). TCIRG1-targeting siRNA transfection also caused a significant induction of autophagic cell death (upper left quadrant of the scatter plot) in Huh7 cells. Specific inhibition of autophagy by treatment with 3-methyladenine blocked autophagic cell death in the same cells (Figure 3i). These results indicate that the overexpression of TCIRG1 may exert an anti-apoptotic and/or anti-autophagic effect on liver tumorigenesis.
Figure 3.
TCIRG1 knockdown has anti-tumorigenic effects on hepatocellular carcinoma (HCC) cells. (a) qRT-PCR analysis of TCIRG1 in 14 hepatic cell lines, including two normal (THLE3 and MIHA) and 12 hepatoma cell lines (Hep3B, HepG2, Huh7, PLC/PRF/5, SK-Hep1, SNU354, SNU368, SNU387, SNU398, SNU423, SNU449 and SNU475). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for normalization (mean±s.d.; n=3, **P<0.01; ***P<0.001). (b) Endogenous protein expression of TCIRG1 in hepatic cell lines was analyzed by western blotting. GAPDH was used as a loading control, and the numbers under the blot indicate the relative expression level of each protein. (c) Clonogenic assays were performed in SNU475 and Huh7 cell lines. Upper panel: representative images of colonies. Lower panel: the data from three randomly selected images shown in the form of a graph (mean±s.d.; n=3, *P<0.05, **P<0.01). (d) SNU475 and Huh7 cell lines were transfected with negative-control siRNA or TCIRG1-specific siRNA. Cell number was determined using trypan blue-based cell counting (mean±s.d., n=3, **P<0.01). (e) SNU475 and Huh7 cell lines were transfected with negative-control siRNA or TCIRG1-specific siRNA. Cell growth was measured by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. (f) Bromodeoxyuridine (BrdU) incorporation assays were carried out using SNU475 and Huh7 cell lines (mean±s.d.; n=3, *P<0.05, **P<0.01). (g) Fluorescence-activated cell sorting (FACS) analysis was conducted after negative-control siRNA or TCIRG1-specific siRNA transfection. When TCIRG1 was knocked down, G1/S arrest was observed in SNU475 and Huh7 cell lines. The percentage indicates the distribution of cells in the different phases of cell cycle (mean±s.d., n=3, *P<0.05). (h) Negative-control siRNA (NC) or TCIRG1-specific siRNA (siTCIRG1) were transfected into cells. FACS analysis was performed using PI and Annexin V staining. The bar graphs indicate the percentage of annexin V-positive cells (mean±s.d., n=3, *P<0.05). (i) Following transfection, cells were treated with 3-MA (5 mM), and FACS analysis was performed. The bar graph indicates the percentage of Annexin V negative and PI-positive cells (mean±s.d., n=3, ***P<0.001).
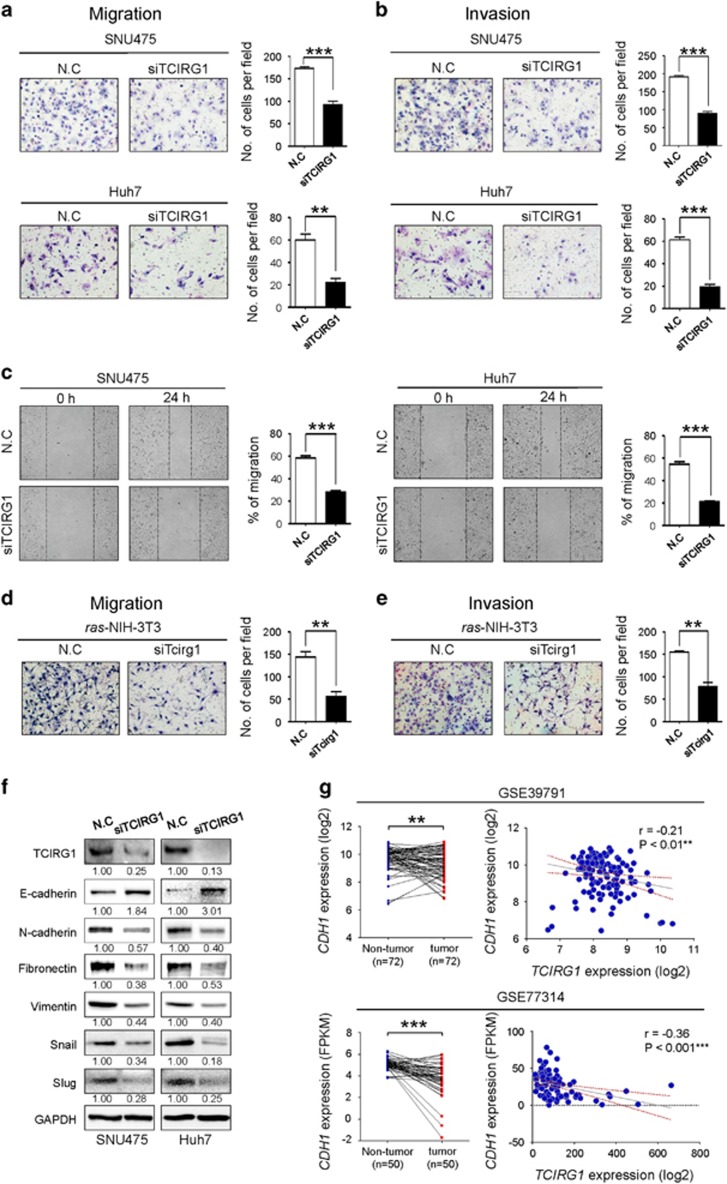
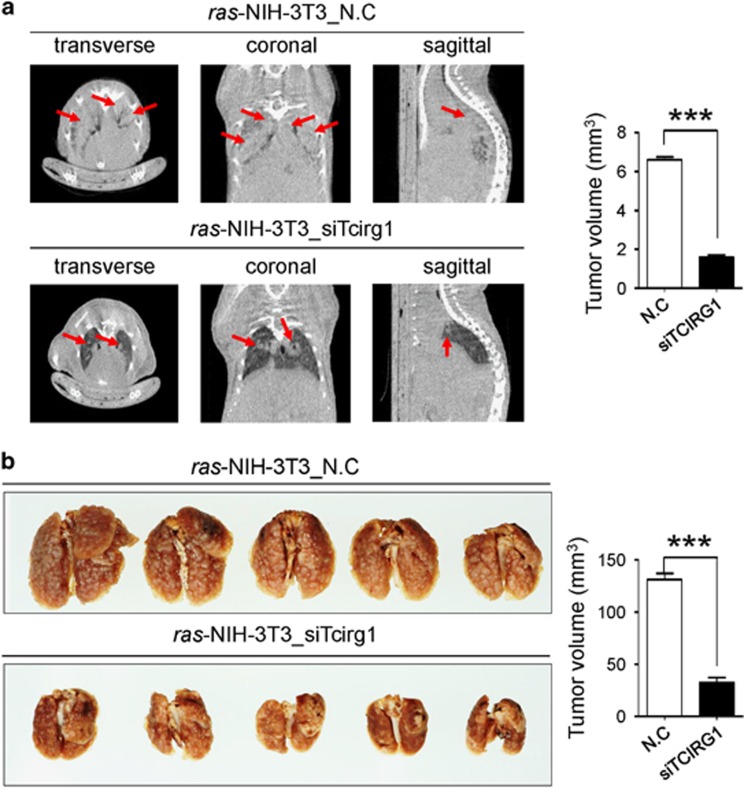
GSEA with the recurrence-associated molecular signature showed that the LIAO_METASTASIS gene set was significantly enriched in the recurrence signature (Figure 1f). Thus, to elucidate the role of TCIRG1 in the malignant behavior of HCC cells, we performed in vitro migration and invasion assays. Modified Boyden chamber assay revealed that TCIRG1 knockdown significantly suppressed chemoattractant (2% FBS)-stimulated migratory and invasive responses of both SNU475 and Huh7 cells (Figure 4a and b). Similarly, wound healing assay also showed that TCIRG1 knockdown reduced chemoattractant-stimulated wound healing efficiency of the HCC cell lines (Figure 4c). To further validate the metastatic potential of TCIRG1, the Boyden chamber assay was also performed with ras-transformed NIH-3T3 cells. TCIRG1 knockdown significantly suppressed chemoattractant-stimulated migratory and invasive responses of ras-transformed NIH-3T3 cells (Figure 4d and e). To clarify the regulatory effect of TCIRG1 on EMT, western blot analysis was performed to detect EMT regulatory proteins in HCC cells. E-cadherin was dramatically increased in TCIRG1 knockdown SNU475 and Huh7 cells, whereas N-cadherin, Fibronectin, Vimentin, Snail and Slug were decreased in the same cells (Figure 4f). TCIRG1 expression analysis in two large HCC patient cohorts with gene expression data of matched paired samples (GSE39791 and GSE77314) showed that E-cadherin (CDH1) expression was significantly decreased in HCC compared with non-tumor tissues, and an inverse correlation of CDH1 with TCIRG1 was observed in the data sets (Figure 4g). Lastly, in vivo metastasis analysis also showed that TCIRG1 knockdown significantly attenuated the metastatic potential of ras-transformed NIH-3T3 cells observed by both micro-CT image analysis and lung nodule quantification (Figure 5a and b).
Figure 4.
TCIRG1 knockdown attenuates the metastatic potential of hepatocellular carcinoma (HCC) cells. (a, b) TCIRG1 knockdown inhibited migration (a) and invasion (b) of SNU475 and Huh7 cell lines in vitro. The number of migrated and invaded cells was determined. Three randomly selected fields were captured, and the results were graphically presented (mean±s.d., n=3, **P<0.01, ***P<0.001). Representative images are shown. (c) Wound healing assay. The bar graphs show the ratios of the recovered area (mean±s.d., n=3, ***P<0.001). (d, e) Effect of TCIRG1 knockdown on cell migration and invasion in vitro. TCIRG1 knockdown inhibited migration (d) and invasion (e) of ras-transformed NIH-3T3 mouse fibroblasts. The number of migrated and invaded cells was determined. Three randomly selected fields were captured, and the results were presented in the form of a graph (mean±s.d., n=3, **P<0.01). Representative images. (f) Western blot analysis of epithelial–mesenchymal transition (EMT) markers. The protein level of TCIRG1, E-cadherin, N-cadherin, Fibronectin, Vimentin, Snail and Slug was detected with their specific antibodies. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the loading control. The numbers under the blot indicate the relative expression level of each protein. (g) Analysis of the NCBI GEO database. GEO data sets with the accession numbers GSE39791 and GSE77314 were used for expression analysis of TCIRG1 and CDH1. Left panel, CDH1 expression in GSE39791 and GSE77314. Right panel, negative correlation of TCIRG1 and CDH1 expression in GSE39791 (Pearson’s correlation coefficient, r=−0.21, **P<0.01) and in GSE77314 (Pearson’s correlation coefficient, r=−0.36, ***P<0.001).
Figure 5.
Effect on Tcirg1 knockdown in an experimental model of metastasis. (a) Representative CT image of ras-transformed NIH-3T3 cells after transient suppression of Tcirg1 (left). The volume of each tumor was calculated, and the total tumor volume was determined (right). The arrows indicate tumor mass. (b) In vivo mages of ras-transformed NIH-3T3 cells using a lung metastasis mouse model after transfection with negative-control siRNA (n=5) and Tcirg1-targeting siRNA (n=5) (left). The number of nodules in mouse lungs is shown by the bar graph (right) (mean±s.d., n=5, ***P<0.001).
Discussion
Predictive factors of recurrence after treatment of early-stage HCC are needed to improve therapeutic strategies. Liver hepatectomy is a treatment modality for HCC. However, even after liver transplantation, recurrence and metastasis remain high; nearly 70% of patients relapse within 5 years after surgery.5, 19 Predicting recurrence in patients undergoing resection is clinically challenging. Genomic profiling has an enormous potential as a diagnostic and prognostic tool in molecular medicine. Gene signatures have allowed the accurate prediction of prognosis and response to therapy in oncology, clearing the path for personalized cancer medicine.20 Likewise, the prognostic prediction in HCC has been studied using gene expression profiling using tumor tissues or non-tumoral adjacent cirrhotic tissues.21 However, these signatures can frequently be ill-defined, being generated in patients with different disease stages and with distinct etiologies for their underlying liver damage. Hence, the concordance of these signatures on a patient-by-patient basis is lacking. In addition, the prognostic performance of these signature in patients with early HCC is unclear; these patients are important to study, as they could be candidates for liver resection.
In this study, we obtained transcriptome data from HCC tissues of 56 patients who underwent partial or total hepatectomy. The DEGs between recurrence and non-recurrence groups were probed to retrieve a comprehensive gene signature associated with HCC recurrence. These genes were comparatively analyzed with a previously published data set to obtain a stringent molecular signature for HCC recurrence (Figure 1). Various metastasis-related genes were highly enriched in the HCC recurrence-associated molecular signature similar to the LIAO_METASTASIS gene set. These results indicate TCIRG1 as a potential deciding factor in the recurrence of primary HCC (Figure 2 and Supplementary Figure 1).
TCIRG1 is one of the V-ATPase subunits that has a peripheral domain (V1) to hydrolyze ATP and an integral domain (V0) to transport protons.22 The V0 domain contains five subunits (a, d, c, c″ and e) in mammals. Among these subunits, subunit a is an integral membrane protein containing a 50 kDa hydrophilic N-terminal domain and 50 kDa C-terminal domain with multiple membrane-spanning regions,23 and it has four isoforms (a1–a4).24, 25, 26, 27 TCIRG1 has been reported in breast cancer and melanoma cells using in vivo mouse models.28, 29 TCIRG1 was found to be overexpressed in the highly invasive MDA-MB-231 breast cancer cell line but not in the poorly invasive MCF7 cell line. Furthermore, knockdown of TCIRG1 using siRNA inhibited the in vitro invasion of MDA-MB-231 cells. In contrast, TCIRG1 overexpression in non-invasive MCF7 cells enhanced invasiveness.28 In addition, in the highly metastatic B16-F10 mouse melanoma cell line, expression of Tcirg1 was upregulated relative to that in the poorly metastatic B16 mouse melanoma cell line, and knockdown of Tcirg1 in B16-F10 cells inhibited metastasis in an in vivo mouse model.29 Consistent with these reports, our results indicated that TCIRG1 was significantly overexpressed in HCC tissues compared with that in corresponding adjacent non-cancerous liver tissues, and high expression of TCIRG1 was significantly correlated with reduced survival in patients with HCC (Figure 2). Using in vitro cell growth and flow cytometry assays, inhibition of TCIRG1 expression was found to reduce growth and proliferation of HCC cells and induce G1/S cell cycle arrest. In a previous report, Tcirg1 knockout was found to increase autophagic sequestration and impair autophagic degradation, as evidenced by the accumulation of autophagy markers including ATG12-ATG5 conjugates, LC3-II and p62.30 Our results also showed that TCIRG1 knockdown activated caspase-dependent and/or caspase-independent cell death (Figure 3). Intriguingly, targeted disruption of TCIRG1 also attenuated the metastatic potential of SNU475 and Huh7 HCC cells by modulating the EMT regulatory proteins E-cadherin, N-cadherin, Fibronectin, Vimentin, Snail and Slug. In vivo in a lung metastasis mouse model, Tcirg1 knockdown elicited suppression of distant metastasis of ras-transformed NIH-3T3 cells (Figures 4 and 5). These results strongly support the pivotal role of TCIRG1 in early metastasis and its value as a decision-making biomarker for the recurrence of HCC after surgical resection.
In conclusion, TCIRG1 is a deciding factor for early recurrence of HCC after surgical resection of the primary lesion. TCIRG1 is upregulated in HCC, and its targeted inactivation inhibits liver tumorigenesis in vitro and in vivo by selectively modulating EMT-related proteins in HCC cells. This finding indicates that the development of specific modulators of TCIRG1 may be crucial to control tumor progression or even the recurrence of HCC after resection. Altogether, these findings define TCIRG1 as a novel predictive marker of early recurrence of HCC and suggest that TCIRG1 has potential therapeutic value for the treatment of HCC.
Acknowledgments
This study was supported by grants from the National Research Foundation (NRF) of Korea (2012M3A9D1054476, 2012R1A5A2047939, 2016R1D1A1B03932532 and 2017R1A2B3002989) and by grants of the Korea Health Technology R & D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, South of Korea (grant number: HI14C1920).
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
The authors declare no conflict of interest.
Supplementary Material
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology 2011; 140: 1501–1512 e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin 2012; 62: 394–399. [DOI] [PubMed] [Google Scholar]
- Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012; 379: 1245–1255. [DOI] [PubMed] [Google Scholar]
- Imamura H, Matsuyama Y, Tanaka E, Ohkubo T, Hasegawa K, Miyagawa S et al. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol 2003; 38: 200–207. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Schwartz M, Mazzaferro V. Resection and liver transplantation for hepatocellular carcinoma. Semin Liver Dis 2005; 25: 181–200. [DOI] [PubMed] [Google Scholar]
- Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene 2008; 27: 5578–5589. [DOI] [PubMed] [Google Scholar]
- Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res 2010; 70: 10202–10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology 2012; 142: 957–966 e912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YY, Sohn BH, Johnson RL, Kang MH, Kim SB, Shim JJ et al. Yes-associated protein 1 and transcriptional coactivator with PDZ-binding motif activate the mammalian target of rapamycin complex 1 pathway by regulating amino acid transporters in hepatocellular carcinoma. Hepatology 2016; 63: 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, Fan ST et al. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open 2012; 2: e000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 2012; 44: 765–769. [DOI] [PubMed] [Google Scholar]
- Lim HY, Sohn I, Deng S, Lee J, Jung SH, Mao M et al. Prediction of disease-free survival in hepatocellular carcinoma by gene expression profiling. Ann Surg Oncol 2013; 20: 3747–3753. [DOI] [PubMed] [Google Scholar]
- Liu G, Hou G, Li L, Li Y, Zhou W, Liu L. Potential diagnostic and prognostic marker dimethylglycine dehydrogenase (DMGDH) suppresses hepatocellular carcinoma metastasis in vitro and in vivo. Oncotarget 2016; 7: 32607–32616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyazi M, Niyazi I, Belka C. Counting colonies of clonogenic assays by using densitometric software. Radiat Oncol 2007; 2: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SG, Moon DB, Hwang S, Ahn CS, Kim KH, Song GW et al. Liver transplantation in Korea: past, present, and future. Transplant Proc 2015; 47: 705–708. [DOI] [PubMed] [Google Scholar]
- Kim JH, Sohn BH, Lee HS, Kim SB, Yoo JE, Park YY et al. Genomic predictors for recurrence patterns of hepatocellular carcinoma: model derivation and validation. PLoS Med 2014; 11: e1001770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol 2010; 5: 1315–1316. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Fuster J, Bruix J. Intention-to-treat analysis of surgical treatment for early hepatocellular carcinoma: resection versus transplantation. Hepatology 1999; 30: 1434–1440. [DOI] [PubMed] [Google Scholar]
- van't Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature 2008; 452: 564–570. [DOI] [PubMed] [Google Scholar]
- Villanueva A, Minguez B, Forner A, Reig M, Llovet JM. Hepatocellular carcinoma: novel molecular approaches for diagnosis, prognosis, and therapy. Annu Rev Med 2010; 61: 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol 2007; 8: 917–929. [DOI] [PubMed] [Google Scholar]
- Wang Y, Toei M, Forgac M. Analysis of the membrane topology of transmembrane segments in the C-terminal hydrophobic domain of the yeast vacuolar ATPase subunit a (Vph1p) by chemical modification. J Biol Chem 2008; 283: 20696–20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Murata Y, Namba M, Yoshimizu T, Toyomura T, Yamamoto A et al. a4, a unique kidney-specific isoform of mouse vacuolar H+-ATPase subunit a. J Biol Chem 2001; 276: 40050–40054. [DOI] [PubMed] [Google Scholar]
- Smith AN, Finberg KE, Wagner CA, Lifton RP, Devonald MA, Su Y et al. Molecular cloning and characterization of Atp6n1b: a novel fourth murine vacuolar H+-ATPase a-subunit gene. J Biol Chem 2001; 276: 42382–42388. [DOI] [PubMed] [Google Scholar]
- Sun-Wada GH, Toyomura T, Murata Y, Yamamoto A, Futai M, Wada Y. The a3 isoform of V-ATPase regulates insulin secretion from pancreatic beta-cells. J Cell Sci 2006; 119: 4531–4540. [DOI] [PubMed] [Google Scholar]
- Toyomura T, Murata Y, Yamamoto A, Oka T, Sun-Wada GH, Wada Y et al. From lysosomes to the plasma membrane: localization of vacuolar-type H+-ATPase with the a3 isoform during osteoclast differentiation. J Biol Chem 2003; 278: 22023–22030. [DOI] [PubMed] [Google Scholar]
- Hinton A, Sennoune SR, Bond S, Fang M, Reuveni M, Sahagian GG et al. Function of a subunit isoforms of the V-ATPase in pH homeostasis and in vitro invasion of MDA-MB231 human breast cancer cells. J Biol Chem 2009; 284: 16400–16408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishisho T, Hata K, Nakanishi M, Morita Y, Sun-Wada GH, Wada Y et al. The a3 isoform vacuolar type H(+)-ATPase promotes distant metastasis in the mouse B16 melanoma cells. Mol Cancer Res 2011; 9: 845–855. [DOI] [PubMed] [Google Scholar]
- Belaid A, Cerezo M, Chargui A, Corcelle-Termeau E, Pedeutour F, Giuliano S et al. Autophagy plays a critical role in the degradation of active RHOA, the control of cell cytokinesis, and genomic stability. Cancer Res 2013; 73: 4311–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.