Abstract
FOXM1 (Forkhead box M1) is a key regulator of tumorigenesis. Previous studies demonstrated that FOXM1 overexpression was strongly correlated with poor prognosis in various cancers, including hepatocellular carcinoma (HCC). In this study, we examined an association between the gene expression signature of FOXM1 and HCC patient outcome. The co-expressed gene set of FOXM1, which is significantly associated with the prognosis of HCC patients, was identified by analyzing the gene expression profiles of 100 patients with HCC, and this gene set was validated in two independent HCC patient cohorts (n=573). In multivariate analysis, the co-expressed gene set of FOXM1 was the most significant prognostic factor for overall survival in patients with HCC (hazard ratio=1.706, 95% confidence intervals=1.176–2.475, P=0.005). We also analyzed different types of cancer, including pancreatic adenocarcinoma, lung adenocarcinoma, breast carcinoma and bladder urothelial carcinoma, to verify the association between the co-expressed gene set of FOXM1 and patient prognosis, and we found a consistent prognostic significance, regardless of tumor type. Finally, we identified a putative signaling pathway in which miR-34a acts as an upstream regulator of the FOXM1-MYC signaling network; this pathway may be ultimately responsible for the poor prognosis of HCC patients. The prognostic subgroups defined by the gene expression signature of FOXM1 could help predict high-risk patients and may guide selection of the best treatment strategy.
Introduction
Hepatocellular carcinoma (HCC) is a major public health problem; it is the primary malignancy of the liver, one of the most common tumors worldwide, and one of the fastest growing causes of cancer-related death. In 2012, 782,500 new cases of liver cancer were diagnosed, and 745,500 deaths occurred worldwide.1 Because of the clinical heterogeneity of this malignant disease, precisely predicting the prognosis of HCC patients remains challenging.
With the advent of new technologies over the past decade, many genome-wide expression profiling studies have revealed the existence of biologically distinct subtypes of HCC, with prognostic implications.2, 3, 4 Furthermore, genome-wide gene expression profiling studies have discovered several genomic predictors for the recurrence or survival of HCC.5, 6 However, it is currently unknown whether this signature is applicable to the development of new treatments. Recently, a number of putative prognostic markers for HCC, including FOXM1, have been proposed in various investigations. Although several studies have shown that FOXM1 overexpression was associated with poor outcomes in many types of cancer,7, 8, 9, 10 its clinical relevance as a prognostic and predictive biomarker is not yet fully understood. Furthermore, no studies have examined the prognostic impact of the co-expressed gene set of FOXM1 and whether it can provide additional insights on the potential signaling pathway that is responsible for the poor prognosis of HCC patients.
Here we examined an association between the gene expression signature of FOXM1 and HCC patient outcome using gene expression data sets. We also analyzed four different cancer types to verify the association between the gene expression signature of FOXM1 and patient outcome. To further validate the utility of the signature, we tested whether the gene expression signature of FOXM1 could identify the distinct subgroups of patients with different types of cancer in The Cancer Genome Atlas (TCGA) pan-cancer gene expression data set. Through gene-to-gene network and upstream regulator analyses, we also identified a putative signaling pathway composed of FOXM1, MYC and miR-34a that potentially mediates HCC patient outcome.
Materials and methods
Patients and gene expression data
Gene expression data sets from the Korean and Fudan HCC cohorts, as reported in previous studies, were acquired from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database.6, 11 The mRNA-seq and miRNA-seq data sets of HCC patients from the TCGA project were obtained from Cancer Browser (https://genome-cancer.uscs.edu). The Korean cohort (GSE16757, n=100) was used as the exploration data set. The Fudan (GSE14520, n=242) and TCGA (n=331) cohorts were used for validation. Supplementary Table 1 shows the pathologic and clinical characteristics of the patients in all three HCC cohorts.
The four gene expression data sets, which consist of pancreatic adenocarcinoma (PAAD, GSE21501, n=132), lung adenocarcinoma (LUAD, GSE31210, n=246), breast invasive carcinoma (BRCA, GSE20486, n=97), and bladder urothelial carcinoma (BLCA, GSE13507, n=165) samples, were obtained from the NCBI GEO database. Of the 97 patients with BRCA, we only used the 81 patients who had estrogen-receptor-positive tumors.
The TCGA pan-cancer gene expression data set (n=9,755) was obtained from Cancer Browser. This data set is composed of 32 different cancer types. Among these, we focused on five cancer types: HCC (the TCGA cohort mentioned above, n=331), PAAD (n=161), LUAD (n=468), BRCA (n=1,055), and BLCA (n=327). Of the 1,055 patients with BRCA, only 779 patients who had estrogen-receptor-positive tumors were selected. Therefore, a total of 2,066 patient samples were examined in this study (the Pan-cancer cohort, n=2,066).
Clinical data were obtained from the Supplementary Information of the corresponding literature or was requested from the authors. The patients with available survival time data were subjected to statistical analysis.
Statistical analysis
The non-tumor and replicated samples were removed based on the sample annotations attached to each data set. Gene expression profile data, regardless of platform, was separately log2 transformed and quantile normalized. For the exploration data set, genes were filtered out if they did not have at least a two-fold change relative to the median in more than 5% of the samples. For gene expression profiling from the RNA-seq data set, genes with a value of zero were set to the missing value after log2 transformation, and genes were filtered out if they had missing data in >30% of the samples.
To identify the co-expressed gene set of FOXM1, we analyzed the gene expression profile data by employing the same procedure described in a previous study.12 Briefly, we selected genes that had a significant correlation with the FOXM1 gene (P<0.001 by Pearson correlation coefficient; r<−0.4 or r>0.4). Then, hierarchical clustering analysis was performed using a gene expression data matrix that consisted of FOXM1 and its correlated genes. We estimated patient prognosis using Kaplan–Meier plots and the log-rank test. The prognostic association between the signature and risk factors was assessed using multivariate Cox proportional hazard regression models. All statistical analyses were performed in the R language environment.
Using the Ingenuity Pathway Analysis (IPA) tool (www.ingenuity.com), gene set enrichment and upstream regulator analyses were performed as described in a previous study.13
Results
Baseline characteristics
The baseline characteristics of all eight data sets analyzed in this study are summarized in Table 1. In each data set, overall survival was defined as the number of months from the initial pathological diagnosis of the tumor until the patient was deceased or lost to follow-up. In the Fudan and TCGA cohorts, recurrence-free survival was defined as the time from surgery to the first confirmed recurrence.
Table 1. Characteristics of data sets and patients included in this study.
| Data set | Data source | Platform description | GEO platform ID | No. of patients |
|---|---|---|---|---|
| Korean cohort | GSE16757 | Illumina Human-6 v2.0 | GPL6102 | 100 |
| Fudan cohort | GSE14520 | Affymetrix Human Genome U133A Array | GPL571/GPL3921 | 242 |
| TCGA cohort | a | Illumina RNA-seq | NA | 331 |
| PAAD | GSE21501 | Agilent-014850 Whole Human Genome Microarray | GPL4133 | 102 |
| LUAD | GSE31210 | Affymetrix Human Genome U133 Plus 2.0 Array | GPL570 | 226 |
| BRCA | GSE20486 | Illumina HumanHT-12 V3.0 | GPL6947 | 81 |
| BLCA | GSE13507 | Illumina Human-6 v2.0 | GPL6102 | 165 |
| Pan-cancer cohort | a | Illumina RNA-seq | NA | 2,066 |
Abbreviations: BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; GEO, National Center for Biotechnology Information’s Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/); LUAD, lung adenocarcinoma; PAAD, pancreatic adenocarcinoma.
UCSC Cancer Genome Browser (https://genome-cancer.ucsc.edu).
Prognostic utility of FOXM1 and its associated genes on HCC patients in the exploration data set
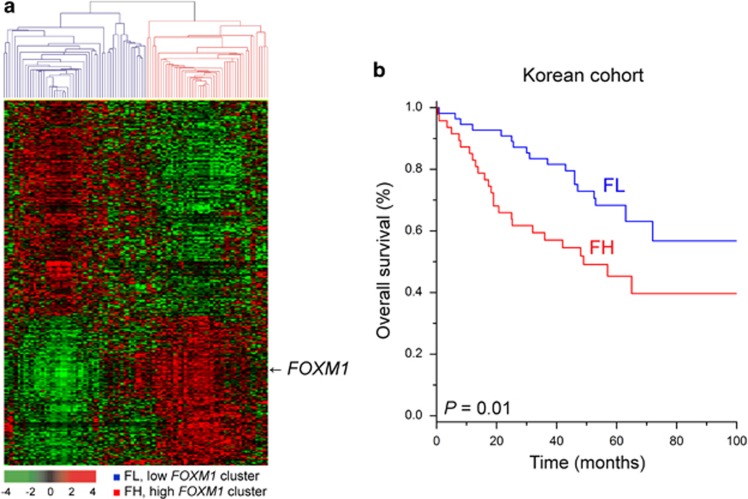
We first tried to identify a gene set that was highly correlated with FOXM1 expression, then used that gene set to predict the prognosis of HCC patients. We identified 1,185 genes that were highly correlated with FOXM1 expression in the exploration data set. Based on hierarchical clustering analysis of the expression patterns of these genes, patients with HCC were divided into two subgroups: FOXM1-high (FH) and FOXM1-low (FL) (Figure 1a). The survival rate of the FH subgroup was significantly worse than that of the FL subgroup (P=0.01 by log-rank test; Figure 1b).
Figure 1.
Gene expression patterns of the co-expressed gene set of FOXM1 and the overall survival of HCC patients in the exploration data set (n=100). (a) Gene expression patterns of FOXM1 and its associated genes. A total of 1,185 genes with expression patterns that highly correlated with FOXM1 were selected for cluster analysis. The patients were divided into two subgroups: FOXM1-high (FH) and FOXM1-low (FL). (b) Kaplan–Meier plot of the overall survival of HCC patients, stratified by the co-expressed gene set of FOXM1. The survival rate of the FH subgroup was significantly worse than that of the FL subgroup (P=0.01 by log-rank test).
Validation of the gene expression signature of FOXM1 on HCC patient survival using the independent cohorts
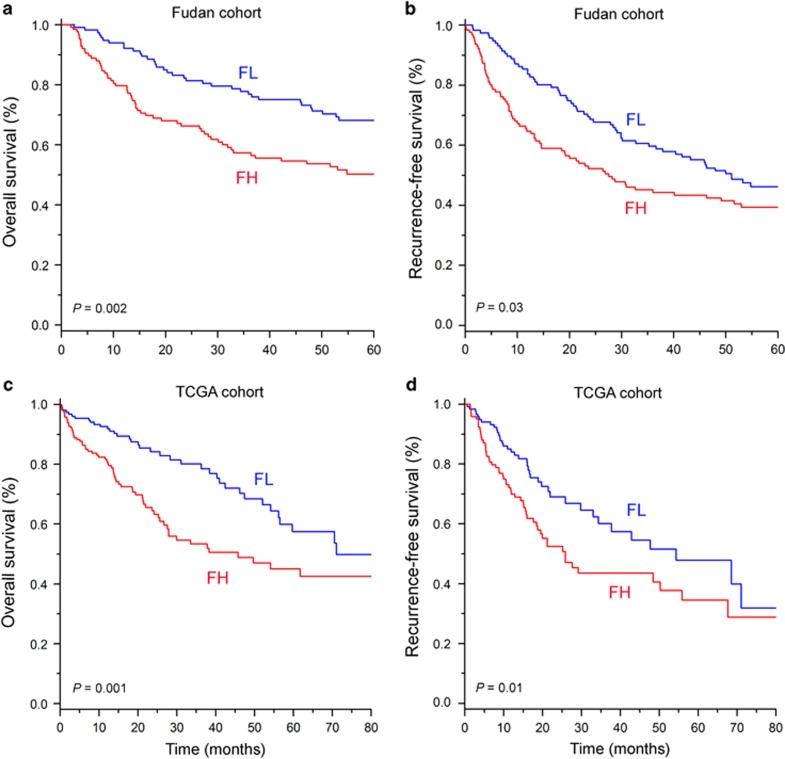
To validate our findings, we used gene expression profile data from an independent cohort of HCC patients. The gene expression profile data of the Fudan cohort (Affymetrix microarray platform) does not contain all of the gene symbols in the exploration data set (Illumina microarray platform). For this reason, we selected the 826 genes that overlapped with the 1,185 genes derived from the exploration data set. Applying the same procedure employed in the previous result, the patients in the Fudan cohort were divided into two subgroups (FH and FL). The FH subgroup in the Fudan cohort also had significantly worse survival than the FL subgroup (P=0.002 by log-rank test, Figure 2a). Furthermore, we also estimated the recurrence-free survival of each subgroup to assess whether the co-expressed genes could identify patients at high risk for recurrence. The recurrence rate was significantly higher in the FH subgroup than the FL subgroup (P=0.03 by log-rank test; Figure 2b). The survival analysis was further validated with the TCGA RNA-seq data set (P=0.001 for the survival rate and P=0.01 for the recurrence rate by log-rank test; Figure 2c and d). Taken together, these results demonstrate that the gene expression signature of FOXM1 is a robust prognostic indicator of HCC patient outcome.
Figure 2.
The validation of the gene expression signature of FOXM1 in independent cohorts. (a) Overall survival and (b) recurrence-free survival in HCC patients, stratified by the co-expressed gene set of FOXM1, were estimated in the Fudan cohort (n=242). (c) Overall survival and (d) recurrence-free survival were also estimated in the TCGA cohort (n=331). FH, FOXM1-high; FL, FOXM1-low.
We next combined the clinical data from the Korean and Fudan cohorts (total n=321) and assessed the prognostic association between the co-expressed gene set of FOXM1 and other known clinical and pathologic factors for HCC using univariate Cox regression analyses. In addition to tumor size, Tumor-Node-Metastasis (TNM) stage, and Barcelona Clinical Liver Cancer (BCLC) stage, all of which are already well-known risk factors, the co-expressed gene set of FOXM1 was a significant indicator for overall survival (Supplementary Table 2). We also applied multivariate Cox regression analyses to verify the independent utility of the gene set. Importantly, the co-expressed gene set of FOXM1 was the most important independent prognostic factor for overall survival (HR=1.706, 95% CI=1.176–2.475, P=0.005; Supplementary Table 2).
Prognostic utility of the gene expression signature of FOXM1 in different types of cancer
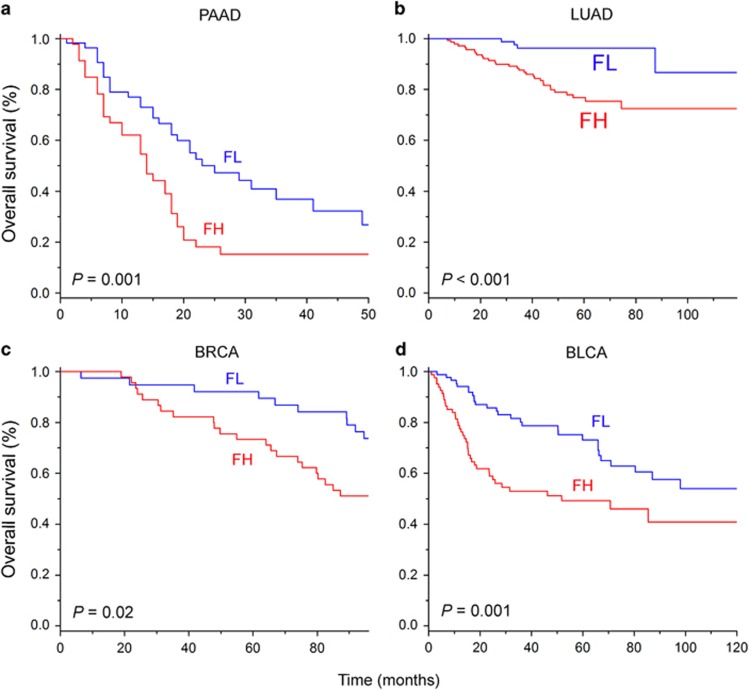
Because the dysregulated expression and function of FOXM1 are not confined to certain types of cancer,14 we next asked whether the co-expressed gene set of FOXM1 was associated with the prognosis of patients with different types of cancer. From the NCBI GEO database, we selected the gene expression data sets of four cancers and identified 2,689, 4,211, 760 and 1,349 co-expressed genes of FOXM1 in pancreatic adenocarcinoma (PAAD), lung adenocarcinoma (LUAD), breast invasive carcinoma (BRCA) and bladder urothelial carcinoma (BLCA), respectively. We did not include other cancer types in this study because we could not identify a sufficient co-expressed gene set of FOXM1 in their data sets (due to a limited sample size or an inadequate follow-up time). Consistent with our previous result, survival analysis revealed that the co-expressed gene set of FOXM1 was a significant predictor of overall survival in patients with different types of cancer (each P<0.05 by log-rank test, Figure 3). This finding strongly demonstrates that the gene expression signature of FOXM1 has prognostic significance, regardless of tumor type.
Figure 3.
Kaplan–Meier plots showing differences in overall survival for patients diagnosed with different types of cancer based on the gene expression signature of FOXM1. Data from (a) PAAD, (b) LUAD, (c) BRCA and (d) BLCA are shown. FH, FOXM1-high, FL, FOXM1-low.
Two distinct subgroups of patients with different types of cancer in the Pan-cancer cohort
Having identified two subgroups that show considerable differences in clinical outcomes in the five cancer types, we next examined whether we could identify two subgroups of patients with different cancer types in a large, pooled data set. Therefore, we used a gene expression data set that was generated by merging multiple TCGA RNA-seq data sets, including HCC, PAAD, LUAD, BRCA and BLCA. The co-expressed gene set of FOXM1 was identified by selecting the genes that highly correlated with FOXM1 expression in the Pan-cancer cohort. We applied hierarchical cluster analysis to the gene expression data matrix, which consisted of FOXM1 and its correlated genes, and the results showed that the cancer patients were divided into two distinct subgroups (FH and FL) based on the co-expressed gene set of FOXM1, not on the histological cancer type (Supplementary Figure 1). The results based on the gene expression profile data suggest that FOXM1 may be a master regulator of expression changes in genes associated with patient survival across cancer types.
Biological insight into the gene expression signature of FOXM1 that is associated with the prognosis of HCC patients
Gene set enrichment analysis was performed to identity the biological characteristics of the co-expressed gene set of FOXM1 using the IPA tool. When applying the 1,185 genes obtained from the exploration data set to the IPA tool, genes involved in organismal injury and abnormalities, gastrointestinal disease and hepatic system disease were significantly enriched. Furthermore, genes involved in DNA replication, recombination and repair, such as BRCA1, EXO1, POLQ, POLD1, POLE, FANCD2, PRKDC and PARP1, were also significantly enriched. These results indicate that the biological processes associated with the DNA repair system might be closely linked to the poor prognosis of HCC patients (Supplementary Figure 2).
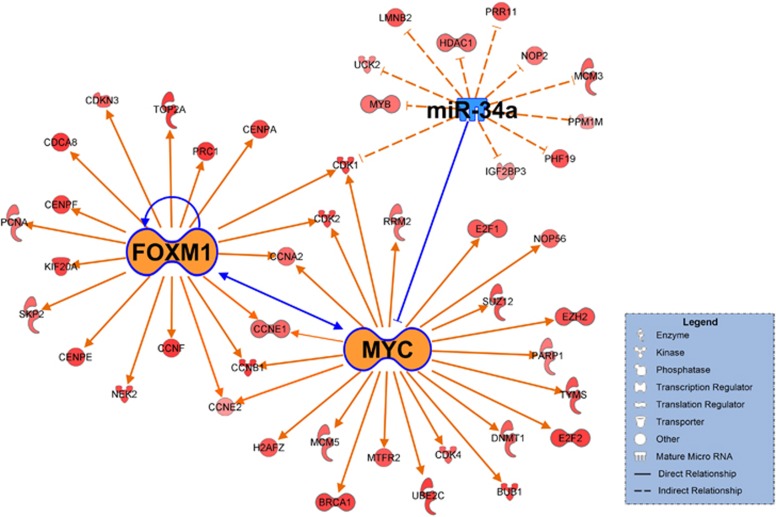
We next conducted upstream regulator analysis of the 1,185 genes to uncover potential upstream regulators of the molecular signature based on a co-expressed gene set of FOXM1. An examination of the enriched genes revealed the involvement of several important activated regulators; the strongest overrepresentation of these regulators was the predominant activation of MYC (Figure 4). The downstream effectors of MYC, such as E2F1, SUZ12, EZH2 and PARP1, were previously reported to be associated with tumor aggressiveness and metastasis,15, 16, 17 indicating that MYC is a key factor associated with the gene signature of FOXM1. Although the expression of MYC downstream genes is higher in the FH subgroup than in the FL subgroup, the expression of MYC is not altered in either the FH or FL subgroups, suggesting that MYC activation might not be driven by changes in gene expression levels, but by other mechanisms, such as amplifications or translocations.18 FOXM1 was also a predominant regulator, and it was interconnected with another gene network hub that included MYC (Figure 4). All of the satellite genes in the gene network participate in the cell cycle, cellular growth and proliferation, and the DNA repair system, which corresponds to the well-known activities of FOXM1 and MYC.18, 19 These results indicate that activation of the FOXM1-MYC signaling network may be a key event associated with a poor prognosis in HCC.
Figure 4.
The putative signaling pathway composed of FOXM1, MYC and miR-34a. Upregulated genes in the FOXM1-high (FH) subgroup are indicated in red. The intensity of color is indicative of the degree of overexpression. Orange- or blue-colored elements indicate predicted activation or inhibition, respectively. Each line and arrow represents the functional and physical interactions between the genes and the direction of regulation reported in the literature. The networks were generated using the Ingenuity Pathways Analysis tool (www.ingenuity.com).
To identify an upstream regulator of the FOXM1-MYC signaling network, we used the path-exploring function of the IPA tool and observed the functional connectivity between MYC and miR-34a; MYC is a downstream effector of miR-34a (Figure 4). miR-34a is reported as a direct transcriptional target of p53, and it is frequently downregulated in various types of cancer,20, 21, 22 including HCC.21 In the TCGA cohort, the expression level of miR-34a was significantly lower in the FH subgroup than in the FL subgroup (P<0.001 by two-sample t-test; Supplementary Figure 3).
In addition, the close examination of gene networks revealed that CDK1 was a common downstream effector controlled by FOXM1, MYC and miR-34a (Figure 4). CDK1 has previously been reported to play an important role in cell cycle regulation.23 Thus, CDK1 activation might account for the poorer prognosis of the FH subgroup than the FL subgroup. Among the other satellite genes in the gene network, CCNB1 and CCNE1, which have been shown to play critical roles in cancer progression,12, 13 were identified as common downstream effectors of FOXM1 and MYC. Taken together, these results suggest that the complicated molecular interactions of multiple gene networks are associated with the poor prognosis of HCC patients.
Discussion
FOXM1 (Forkhead box M1) is one of the most commonly overexpressed genes in various cancer types, based on genome-wide gene expression profile analysis. In addition, it has been shown that the overexpression of FOXM1 is associated with aggressive tumor features and with the eventual prognosis of cancer patients, including those with HCC.7 Importantly, the prognosis of patients may be affected not only by the overexpression of FOXM1 but also by other genes associated with its expression. Here we identified two distinct subgroups of cancer patients based on the gene expression signature of FOXM1. Survival analysis revealed a significant survival difference between the cancer patient subgroups. Finally, we identified a putative signaling pathway that might be responsible for the poor prognosis of HCC patients.
FOXM1, a major oncogenic transcription factor, plays an essential role in the regulation of a wide spectrum of biological processes, suggesting that it has a fundamental role in tumor development and progression.24 Although an association between the overexpression of FOXM1 and patient prognosis has been observed in several studies,25 the prognostic relevance of a co-expressed gene set of FOXM1 has not been demonstrated. Through an estimation of prognosis using a co-expressed gene set of FOXM1, we discovered that an expression signature of FOXM1 and its associated genes has prognostic significance in various cancer types. By applying hierarchical cluster analysis to the gene expression profile data from a pool of five different cancer types from the TCGA RNA-seq data set, we showed the presence of two subgroups based on a co-expressed gene set of FOXM1, regardless of tumor type. These data suggested that the gene expression signature of FOXM1 could be used as a potential prognostic and predictive indicator in various cancers.
Gene network analysis of the gene expression signature of FOXM1 identified a putative signaling pathway composed of FOXM1, MYC and miR-34a; this pathway is associated with the poor prognosis of HCC. Among several upstream candidates, FOXM1 and MYC were revealed as the two predominant regulators of the overexpressed genes in the FH subgroup, which is consistent with previous studies that show their regulatory role in HCC patients with poor prognosis.7, 26 miR-34a, an upstream regulator of MYC, was reported as a tumor suppressor gene by targeting many oncogenes related to proliferation, apoptosis and invasion. A recent study suggested that miR-34a directly targets FOXM1 and MYC via binding the 3′ untranslated regions in liver cancer cells,27 which indicates that expression changes in miR-34a might be a critical factor for the diagnosis of HCC patients.
By applying the co-expressed gene set of FOXM1, we were able to divide individuals with cancer into two subgroups that showed a significant difference in their clinical outcomes. These findings support the notion that FOXM1 is a key regulator of genes associated with patient survival in various types of cancer. Furthermore, the good or poor prognostic subgroups defined by the gene expression signature of FOXM1 could be exploited for use as a diagnostic tool to predict high-risk patients and to improve treatment responses.
Acknowledgments
This research was supported by a grant from KRIBB Research Initiative Program.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
The authors declare no conflict of interest.
Supplementary Material
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med 2008; 359: 1995–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 2007; 45: 42–52. [DOI] [PubMed] [Google Scholar]
- Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med 2006; 12: 410–416. [DOI] [PubMed] [Google Scholar]
- Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology 2004; 40: 667–676. [DOI] [PubMed] [Google Scholar]
- Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res 2010; 70: 10202–10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HC, Li M, Lu JL, Yan DW, Zhou CZ, Fan JW et al. Overexpression of Forkhead box M1 protein associates with aggressive tumor features and poor prognosis of hepatocellular carcinoma. Oncol Rep 2011; 25: 1533–1539. [DOI] [PubMed] [Google Scholar]
- Bektas N, Haaf AT, Veeck J, Wild PJ, Lüscher-Firzlaff J, Hartmann A et al. Tight correlation between expression of the Forkhead transcription factor FOXM1 and HER2 in human breast cancer. BMC Cancer 2008; 8: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Fujiwara Y, Takahashi T, Nakamura Y, Takiguchi S, Nakajima K et al. Overexpression of forkhead box M1 transcription factor (FOXM1) is a potential prognostic marker and enhances chemoresistance for docetaxel in gastric cancer. Ann Surg Oncol 2013; 20: 1035–1043. [DOI] [PubMed] [Google Scholar]
- Xu N, Jia D, Chen W, Wang H, Liu F, Ge H et al. FoxM1 is associated with poor prognosis of non-small cell lung cancer patients through promoting tumor metastasis. PLoS ONE 2013; 8: e59412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Leem SH, Chu IS, Park YY, Kim SC, Kim SB et al. Sixty-five gene-based risk score classifier predicts overall survival in hepatocellular carcinoma. Hepatology 2012; 55: 1443–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song BN, Kim SK, Chu IS. Bioinformatic identification of prognostic signature defined by copy number alteration and expression of CCNE1 in non-muscle invasive bladder cancer. Exp Mol Med 2017; 49: e282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Roh YG, Park K, Kang TH, Kim WJ, Lee JS et al. Expression signature defined by FOXM1-CCNB1 activation predicts disease recurrence in non-muscle-invasive bladder cancer. Clin Cancer Res 2014; 20: 3233–3243. [DOI] [PubMed] [Google Scholar]
- Kalin TV, Ustiyan V, Kalinichenko VV. Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle 2011; 10: 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Leem SH, Lee SY, Kim SC, Park ES, Kim SB et al. Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. J Clin Oncol 2010; 28: 2660–2667. [DOI] [PubMed] [Google Scholar]
- Lee SR, Roh YG, Kim SK, Lee JS, Seol SY, Lee HH et al. Activation of EZH2 and SUZ12 regulated by E2F1 predicts the disease progression and aggressive characteristics of bladder cancer. Clin Cancer Res 2015; 21: 5391–5403. [DOI] [PubMed] [Google Scholar]
- Choi EB, Yang AY, Kim SC, Lee J, Choi JK, Choi C et al. PARP1 enhances lung adenocarcinoma metastasis by novel mechanisms independent of DNA repair. Oncogene 2016; 35: 4569–4579. [DOI] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer 2008; 8: 976–990. [DOI] [PubMed] [Google Scholar]
- Lam EWF, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer 2013; 13: 482–495. [DOI] [PubMed] [Google Scholar]
- Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA 2007; 104: 15472–15477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y et al. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett 2008; 275: 44–53. [DOI] [PubMed] [Google Scholar]
- Gallardo E, Navarro A, Vinolas N, Marrades RM, Diaz T, Gel B et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis 2009; 30: 1903–1909. [DOI] [PubMed] [Google Scholar]
- Liu P, Kao TP, Huang H. CDK1 promotes cell proliferation and survival via phosphorylation and inhibition of FOXO1 transcription factor. Oncogene 2008; 27: 4733–4744. [DOI] [PubMed] [Google Scholar]
- Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer 2007; 7: 847–859. [DOI] [PubMed] [Google Scholar]
- Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015; 21: 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang KY, Noh SJ, Lehwald N, Tao GZ, Bellovin DI, Park HS et al. SIRT1 and c-Myc promote liver tumor cell survival and predict poor survival of human hepatocellular carcinomas. PLoS ONE 2012; 7: e45119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen W, Miao R, Zhou Y, Wang Z, Zhang L et al. miR-34a induces cellular senescence via modulation of telomerase activity in human hepatocellular carcinoma by targeting FoxM1/c-Myc pathway. Oncotarget 2015; 6: 3988–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.