Abstract
Genomic analyses of primary liver cancer samples reveal a complex mutational landscape with vast intertumor and intratumor heterogeneity. Different primary liver tumors and subclones within each tumor display striking molecular and biological variations. Consequently, tumor molecular heterogeneity contributes to drug resistance and tumor relapse following therapy, which poses a substantial obstruction to improving outcomes of patients with liver cancer. There is an urgent need to the compositional and functional understanding of tumor heterogeneity. In this review, we summarize genomic and non-genomic diversities, which include stemness and microenvironmental causes of the functional heterogeneity of the primary liver cancer ecosystem. We discuss the importance and intricacy of intratumor heterogeneity in the context of cancer cell evolution. We also discuss methodologies applicable to determine intratumor heterogeneity and highlight the best-fit patient-derived in vivo and in vitro models to recapture the functional heterogeneity of primary liver cancer with the aim to improve future therapeutic strategies.
Introduction
Approximately one million cases of primary liver cancer (PLC) occurs annually and ranks the second most lethal tumor type in the world.1 It is predicted that the incidence of PLC will increase by more than 50% by 2030 in the United States.2, 3, 4 Notably, in the past 20 years, PLC has been the only cancer with the fastest rising incidence and mortality and with a 5-year survival rate of less than 15% in the United States.5, 6, 7, 8 Tumor heterogeneity is the major contributing factor for the refractory nature of PLC to treatment.
Tumor heterogeneity consists of intertumor (tumor by tumor) and intratumor (within a tumor) heterogeneity. Intertumor heterogeneity refers to PLCs from different patients whose altered genotype and phenotype are induced by diverse etiological and environmental factors.9, 10 In contrast, intratumor heterogeneity refers to genomic and biological variations within a tumor lesion gained by tumor cell evolution under diverse microenvironments linked to different etiologies. According to the Darwinian evolution selection, a PLC lesion evolves from a single malignant cell into a functionally heterogenous tumor mass with a hierarchically organized tumor cell community, promoting its survival and fitness in response to the various microenvironments.11
The most current understanding of PLC heterogeneity is limited to intertumor heterogeneity, mostly focused on molecular subclassification based on genomic profiling. Accordingly, molecular subclassification has identified different patient subtypes according to their genomic profiles with targeted therapy choice. Notably, subclass-related oncogene addiction loops have been identified based on genomic profiling of various tumor subtypes.12, 13, 14, 15, 16, 17, 18 This approach has been successfully applied to the management of chronic myeloid leukemia (CML) and breast cancer.19, 20, 21 For example, most of CMLs are driven by a fused BCR-ABL oncoprotein, a constitutively activated tyrosine kinase. Ninety-eight percent of BCR-ABL CML responds favorably to the initial treatment of tyrosine kinase inhibitors (TKIs). While BCR-ABL-based subclassification and associated target therapy remain the standard care for CML patients, there is growing evidence showing that a minority of mutational subclones within CML are highly resistant to tyrosine kinase inhibitors,22, 23, 24 suggesting that subclassification based on intertumor heterogeneity may not capture the full tumor spectrum. Thus, we need to integrate molecular features of both intertumor and intratumor heterogeneity with functional heterogeneity to improve patient subclassification and response to therapy.
PLC consists of two major histological types,10 hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (ICC), each of which is treated differently, according to their clinical guidelines. Currently, no biomarker-guided targeted therapy is available for HCC or ICC. However, emerging studies aim to HCC or ICC molecular subclassification associated with subtype-related target therapy.15, 25, 26, 27, 28, 29 Studies such as whole-genome sequencing (WGS) of HCC reveal frequent mutations in various tumor suppressor genes and oncogenes, including telomerase reverse transcriptase promoter mutations (54–60%), catenin beta 1 (CTNNB1) mutations (11–37%), tumor protein P53 (TP53) mutations (12–48%) and AT-rich interactive domain-containing protein 1A (ARID1A) mutations (4–17%).30, 31, 32, 33 However, mutation-specific subtypes are not evident. In contrast, transcriptome-based studies have revealed stable molecular subtypes of HCC and ICC. For example, HCC can be divided into several molecular subtypes based on stemness gene expression patterns.12, 13, 14, 15, 34, 35 Similarly, ICC have been categorized to two major subclasses linked to patient outcomes, that is, proliferation subtype and inflammation subtype.36, 37 While subclassification based on intertumor heterogeneity is stable among different data sets, improved PLC treatment remains to be seen. Furthermore, much less is known about intratumor molecular heterogeneity of HCC or ICC. It is unclear whether the degree of intratumor heterogeneity is associated with specific tumor types and patient outcomes. Thus, understanding the link between intertumor and intratumor heterogeneity may help improve PLC subclassification and treatment stratification.
Sources of intratumor heterogeneity
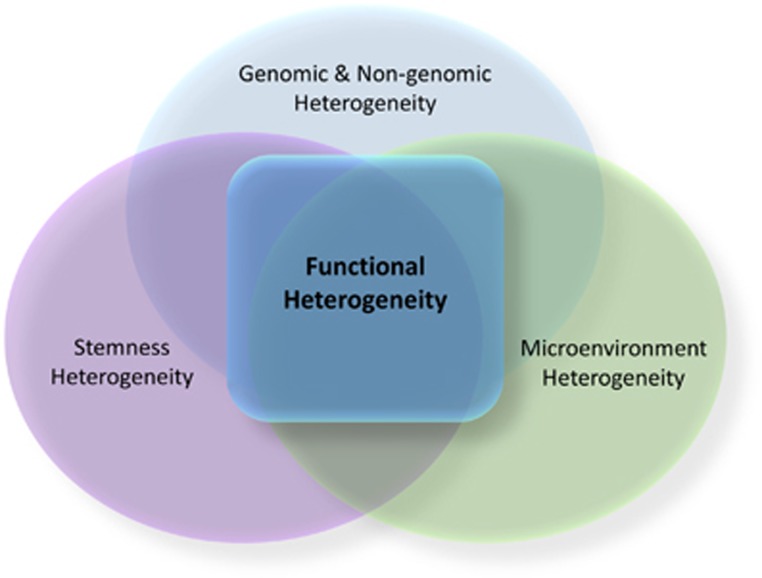
Intratumor heterogeneity has been observed in PLCs in many studies, including histology, ploidy patterns analyses, DNA fingerprinting and WGS.38, 39, 40, 41, 42 Here we propose a concept of intratumor ‘functional’ heterogeneity that describes the abilities of cancer cells to undergo cellular proliferation, adaption and drug resistance within a defined microenvironment linked to a specific etiology. We argue that the compositional heterogeneity helps to understand functional heterogeneity, which is key to understanding PLC heterogeneity (Figure 1).
Figure 1.
Sources of intratumor heterogeneity of liver cancer. Sources of intratumor heterogeneity of liver cancer include genomic, non-genomic, stemness and microenvironment heterogeneity. Functional heterogeneity refers to abilities of cancer cell populations to undergo cellular proliferation, adaption and drug resistance within a defined microenvironment linked to a specific etiology. These abilities of functional heterogeneity are linked to different sources, including genomic and non-genomic, stemness and microenvironment heterogeneity.
Intratumor genomic heterogeneity
One important concept about intratumor genomic heterogeneity is the genome-axis evolution model, which describes that gene evolution can increase the survival adaptive function of a cell known as ‘genomic heterogeneity’.43 This model suggests that genomic heterogeneity is composed of oncogenes and tumor suppressors that have multiple alterations, including mutation and copy number alterations. Together, these alterations maximize tumor proliferation and survival.44, 45 One intriguing example of intratumor genomic heterogeneity and functional heterogeneity is demonstrated in glioblastoma. Accordingly, glioblastoma intratumor with varied epidermal growth factor receptor copy number gain has been measured by Shannon diversity index, demonstrating that functional heterogeneity is linked to increased cell proliferation, resistance to epidermal growth factor receptor inhibitor treatment and aggressive tumor relapse.46, 47 Similarly, it has been demonstrated that larger PLC cell populations exhibit greater genomic heterogeneity.48, 49 Furthermore, genomic sequencing analyses of HCC specimens have revealed identical mutational profiles of multiregional HCC and ICC and distinct CNVs from single-cell analysis.50, 51 However, only a small percentage of mutations were found differently among tumor cell subclones.42 While much is known about the heterogeneity for PLCs, the functional consequence of genomic heterogeneity remains unclear.
Intratumor non-genomic heterogeneity
The dysregulation of the epigenome is one mechanism that contributes to intratumor subclonal variations. Non-genomic intratumor heterogeneity, including histone modifications, DNA hypo- or hyper- methylation, non-coding RNAs, and transcriptional regulators, contribute intratumor heterogeneity by regulating the spatial chromatin organization and altering the transcriptome. Interestingly, aberrant DNA methylation patterns in PLCs are associated with chronic viral hepatitis infections.52 Moreover, other studies show that CpG methylation status within the E-cadherin gene promoters is strongly correlated with the heterogeneous expression of E-cadherin in HCC.53 Furthermore, promoter hypermethylation of Hexokinase 2 (HK2), an aerobic glycolysis process enzyme gene, has been demonstrated to be associated with poor survival, compared with the hypomethylation of HK2 promoter.54 As these studies demonstrate the importance of intertumor heterogeneity in PLCs, more functional studies are required to further understand the association between intratumor non-genomic heterogeneity and its related functions.
Intratumor heterogeneity and cancer stem cells
Cancer stem cells (CSCs) have been demonstrated to contribute to tumor heterogeneity.55 CSCs acquire stem-like characteristics (or stemness) similar to normal stem/progenitor cells, which have been demonstrated to promote liver cancer progression, relapse and intratumor heterogeneity.35, 56, 57 Accordingly, Fan’s group recently showed that spatial heterogeneity of CD13+ CSCs is associated with long non-coding RNAs (lncRNAs), including lnc-β-Catm, lncTCF7 and lncBRM, which have been demonstrated to promote the PLC organoid formation.58, 59, 60 Although the spatial CSCs heterogeneity has been demonstrated, the functional diverse distribution of CSCs remains unclear.
Intratumor heterogeneity contributed by tumor microenvironment
Genomic and non-genomic heterogeneity and the presence of stemness from CSCs have provided some degrees of understanding of PLC. Importantly, microenvironment heterogeneity, which can directly interact with tumor cells and influence therapeutic response, has been considered as a novel hallmark of cancer.61, 62 Notably, the peritumor microenvironment, from the seed and soil concept, can foster tumor development by reprograming the stromal with the aid of cancer-associated fibroblasts.63, 64 In vivo studies have demonstrated that tumor cells benefit from the microenvironment, which contributes to promoting cellular diversity by supporting the tumor vasculature and reprogramming immune cell.62 Interestingly, tumor-infiltrating lymphocytes can be recruited by HCC cells without secreting the tumor-toxic cytokine interferon gamma (IFN-γ).65, 66 These studies suggest that PLC cells could recruit non-toxic infiltrating lymphocytes to promote intratumor heterogeneity and evade from the immune system. HCC studies show that diverse microenvironments are composed of immuno-regulating cytokines, growth factors, immunosuppressors and heterogenous and naturalized stromal cell types. These diverse microenvironments functionally suppress the immune system, by diminishing natural killer cells (NK cells) or enhancing the immunosuppressive regulatory T cells (Treg cells).67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79
Tumor cell clonal architecture
The process of tumor initiation has not been fully understood, as not much is known about the cellular origin of a tumor mass. In 1976, Nowell11 proposed that most tumors arise from a single cell which gains genetic instability within the original clone with sequential selection of branching subclones evolving over time. Notably, recent studies from WGS of different lesions of a tumor mass have extended this model, further suggesting that a tumor cell community is hierarchically organized through a branching or paralleling evolution.80, 81 It has been shown that PLC has the propensity to develop multiple subclones, resulting in extended intratumor heterogeneity.48, 49 A systematic analysis of PLC intratumor heterogeneity came from a study by Xue et al.,42 in which the authors performed somatic mutations and copy number variations (CNVs) using a low-depth whole genome and exome sequencing in 43 tumor lesions derived from 10 HBV-positive HCC cases and found evidence of branched evolution linked to HCC. This comprehensive study provides evidence of branched evolution linked to HCC and demonstrate that while tumors from each patient have different clonal hierarchies, satellite nodules share more than 90% of somatic mutations with their primary tumors. Intriguingly, in a patient with multicentric tumors, six lesions show parallel evolution patterns that can be divided into two major subclones, each with its own unique genomic characteristics. In contrast, PLCs with histologically mixed HCC and ICC, satellite nodules shared more than 100 somatic mutations, indicating that subsequent diverse subclones are evident through the genetic evolution of tumor nodules within each PLC. In another study, Ling et al.41 have analyzed 286 tumor regions from one HCC sample using whole-exome sequencing and genotyping and discovered the evidence of high genetic diversity within a single tumor, indicative of non-Darwinian cell evolution prevalence.41 These results suggest that the extent of intratumor heterogeneity varies considerably among patients with HCC.
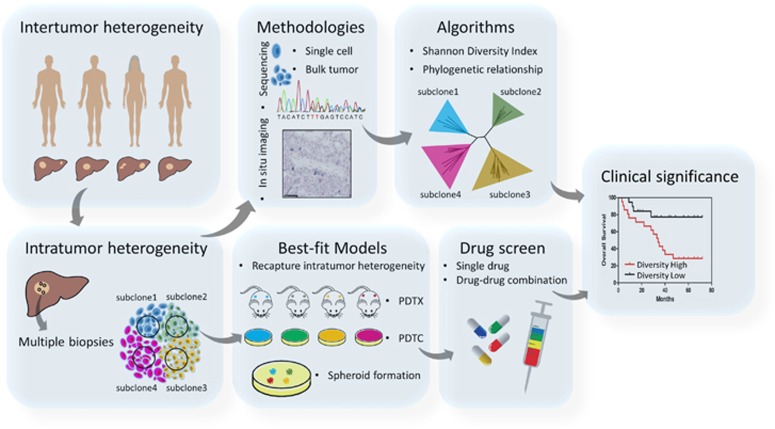
To address the extent of PLC heterogeneity, multiple biopsies from a single tumor is required. This can be accomplished by collecting multiple biopsies from different geographical regions within a tumor or patient. Such analyses will reveal tumor cell hierarchies at the genomic and non-genomic level, including the presence of CSCs and the diversities within the tumor microenvironment at a given time (Figure 2).80 In addition, longitudinal sampling from each patient including prior and post treatment can be implemented.44 This approach improves our understanding of the PLC evolution during tumor progression. Notably, some has suggested that the use of imaging techniques is sufficient for PLC diagnosis. Given the extent of genomic and biological heterogeneity associated with PLC, such a view is shortsighted.82 Therefore, a comprehensive and systematic analysis of multiple biopsies is recommended to uncover the PLC functional heterogeneity that drives tumor evolution.
Figure 2.
A schematic diagram of understanding, recapturing intratumor heterogeneity of PLC on best-fit models and their applications in drug screen and prognosis. PLC heterogeneity includes intertumor and intratumor heterogeneity. The latter can be dissected by multiple biopsies, which can catch the compositional subclones within each tumor. In situ imaging, single cell and bulk tumor sequencing can be performed on the multiple biopsies to help determine the compositional subclones. Intratumor heterogeneity can be quantified by Shannon diversity index and compositional subclones can be categorized by using phylogenetic relationship. In vivo PDTX, in vitro PDTC and spheroid formation are the preclinically relevant best-fit models, which mostly recapture and preserve the compositional heterogeneity within a tumor and can be used for drug screen. PLC, primary liver cancer; PDTC, patient-derived tumor cell; PDTX, patient-derived tumor xenograft.
Approaches to study intratumor heterogeneity
Several studies in recent years have begun to explore methodologies to study intratumor heterogeneity. Here we outline some of the current methodologies that have been successfully used to characterize intratumor heterogeneity (Figure 2).
Intratumor heterogeneity from genome-wide studies
Morphological intratumor heterogeneity has long been observed by pathologists. However, intratumor molecular heterogeneity has only been appreciated in recent years due to WGS technologies. The first study to address intratumor molecular heterogeneity was based on multiregional sequencing of renal carcinomas and associated metastatic sites.80 Mutational intratumor heterogeneity was observed for multiple tumor suppressor genes and further analysis of multiple regions of PLCs provide evidence of the trunk and branch mutational profile.41, 42 Moreover, Zhang’s group studied HCC heterogeneity using allelic frequency profiles of frequently mutated genes across genomes and exomes based on deep sequencing of two separate biopsies from each tumor.83 Among 42 HCC cases analyzed, they found evidence of diverse modes of genomic alteration in HCC. These studies suggest that given the extent of intratumor mutational heterogeneity, multiple sampling of only a few regions to determine driver mutations may not necessarily be adequate to capture the true mutational landscape of a tumor type.
Intratumor heterogeneity at single-cell level
The recent development of single-cell genome sequencing technologies has generated many new insights into complex biological systems including human cancer.84 Single-cell analysis may provide the level of sensitivity and specificity to study tumor cell diversity in a tumor cell ecosystem.85, 86, 87 One latest technological advance includes the development of scTrio-seq, a single cell triple omics method developed by Hou et al.51 to simultaneously analyze genome (CNVs), DNA methylome and transcriptome in a single cell. While single-cell technology is useful to study tumor cell diversity in each tumor cell ecosystem, it lacks information about topological space within a tumor cell community. New approaches such as single-cell gene expression profiling with multiplexed error-robust fluorescence in situ hybridization may help resolving this issue.88, 89 In addition, while single-cell RNA transcriptome has provided sufficient resolution to distinguish single cells, whole genome or exosome sequencing technologies are not sufficient to provide a comprehensive view of global genomic landscape of a tumor at the single-cell level. Therefore, identifying cancer driver genes linked to single tumor cells is a challenge.
Circulating tumor cells
Circulating tumor cells (CTCs), found in blood and originated from aggressive subclones of the primary tumor, are responsible for metastasis and tumor relapse. A meta-analysis of HCC CTC studies revealed that CTCs are positively correlated with poor prognosis.90 However, not all CTCs are tumorigenic, suggesting that these cells are heterogeneous.91, 92, 93 Molecular analysis of CTCs may provide information on tumor cell heterogeneity. A deep understanding of CTC heterogeneity may facilitate our ability to identify key molecular targets to improve cancer therapy. Interestingly, Miyamoto et al.94 demonstrated considerable heterogeneity among CTCs by analyzing 77 intact CTCs from 13 prostate cancer patients by single-cell RNA sequencing. As CTCs are rare, therefore methods that can accurately detect and isolate them for downstream analysis are crucial. In a study by Lohr et al.,95 the authors developed a microscope-based technology to detect and isolate CTCs from multiple myeloma (MM) patients for single-cell RNA-sequencing analysis. They found that CTCs provide the same genetic information as bone marrow multiple myeloma cells, providing a more clear overview of the tumor itself than bone marrow biopsies thus enabling the classification of MM to improve precision medicine.95 In addition, Kalinich et al.96 developed a high-throughput microfluidic CTC-iChip, which can be used to enrich HCC CTC while excluding hematopoietic cells from the blood. It will be interesting in determining if these methods allow assessment of CTC heterogeneity. Moreover, whether these methods can be incorporated into clinical practice, especially PLC management, remains to be determined.
Spatial organization of a tumor cell community
In situ methods are desirable techniques for understanding PLC heterogeneity because it can address the spatial organization of a tumor cell community. This method has been used to detect DNA and RNA using formalin-fixed paraffin-embedded tissue microarrays in a high-throughput manner.97, 98 With the use of high-resolution microscopy, CNVs, genetic mutation profile and RNA quantification can be profiled at the single-cell level. This is evident in a study by Kimura et al.99 who demonstrated the presence of molecular heterogeneity by in situ hybridization with a chromosome 17-specific DNA probe. The authors found that distinct subclones in 2/25 HCC samples with chromosome 17 copy number alterations and ploidy patterns.99 Others have also determined topological features of intratumor subclones by in situ approaches.44 While the current in situ hybridization method still has a poor resolution as only a few molecular features can be assessed at one time. More sensitive methods with higher densities of DNA/RNA probes are being developed to help further our understanding of spatial organization of a tumor subclone.89 Additional mathematical models such as the Shannon diversity index method are being explored to precisely measure the degree of intratumor heterogeneity in a tumor cell ecosystem. New models to incorporate spatial organization with Shannon diversity index should be exploited to measure the true dynamic of a tumor cell community.
Preclinically relevant best-fit models to study intratumor heterogeneity
Experimental models are very useful tools to explore molecular mechanisms that drive tumorigenesis, and ideally used to study intratumor heterogeneity. Appropriate models that mimic human diseases are necessary to provide the clinical relevance.100, 101 Among various available models, patient-derived tumor xenografts (PDTXs) and matched patient-derived tumor cells (PDTCs) have been the preferable choices. Accordingly, Bruna et al.102 have generated a living biobank of human breast cancers in order to adequately capture the inter- and intratumor heterogeneity in pre-clinical models. Remarkably, the intratumor genomic clonal architecture of the originating tumors is largely preserved in PDTXs and matched PDTCs.102 Using this resource coupled with a high-throughput response and resistance screens, they have successfully assessed drug responses in these models.102 In addition, in vivo PDTX can recapture up to 82.5% of ex vivo PDTCs in terms of the drug response. These results suggest that both PDTC and PDTX represent a powerful resource for pre-clinical pharmacogenomic studies. Most recently, Gao et al.103 also observed considerable intratumor heterogeneity and branched evolution in HCC samples that correspond to existing pharmacologic agents using PDTCs. These studies indicate that PDTX and PDTC coupled with the use of their genomic information are useful platforms for pharmacologic assessments linked to intratumor heterogeneity. Thus, improving assessment of the complexity of PLC intratumor heterogeneity will provide a better understanding of functional heterogeneity, which may be linked to therapeutic responses.
Future perspective
Our understanding of PLC heterogeneity has been improved in recent years, but many challenges are still ahead such as a precision measurement to recapture and preserve the diversity of a tumor cell community linked to patient outcomes including treatment response. The PLC ecosystem is dynamically changing during therapy as resistant clones may be selected and expanded throughout tumor evolution. To improve HCC therapies, understanding the impact of tumor heterogeneity must be appreciated. First, we need to understand the diversity of a given tumor cell community and to learn their collective behaviors. By measuring the phylogenetic relationship among different cells and using the Shannon diversity index through subclone bulk or single-cell sequencing (Figure 2), we may understand the complexity of a tumor cell community and determine the degree of diversity among individual patients. Second, to further understand the pattern of intratumor molecular heterogeneity of a given tumor and its relationship to other tumor types, we need to better understand the dynamics of the molecular subtypes of PLC linked to ethnicity, etiological factors and patient outcomes. A well-defined patient data ecosystem including diverse patient populations is vital step to achieve this goal.82, 104 Third, given the importance of the molecular and phenotypical intratumor heterogeneity of PLCs, the access to a living biobank of PLCs will help build the much-needed database to further understand features of intratumor heterogeneity. Furthermore, a drug response database from high throughput single drug and drug–drug combination screen using in vivo PDTX and in vitro PDTC models will prove effective for developing targeted therapies (Figure 2). Together, these efforts will improve PLC preclinical trial design, consequently, improving our discovery of new and effective therapeutics for PLC. Finally, a better understanding of the microenvironment diversity, and the dynamic interplay between the microenvironment and tumor ecosystem, will help improve our understanding of a patient’s response to immune therapy or molecularly targeted therapies.105 With the development of better molecular technologies, we may be able to ultimately find solutions to improve outcomes of patients who suffer from this dreadful disease once for all.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
We thank the NIH Fellows Editorial Board for editorial assistance. This work was supported by a grant (Z01 BC 01031) from the Intramural Research Program of the Center for Cancer Research of the National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- Weir HK, Thompson TD, Soman A, Moller B, Leadbetter S. The past, present, and future of cancer incidence in the United States: 1975 through 2020. Cancer 2015; 121: 1827–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BD, Smith GL, Hurria A, Hortobagyi GN, Buchholz TA. Future of cancer incidence in the United States: burdens upon an aging, changing nation. J Clin Oncol 2009; 27: 2758–2765. [DOI] [PubMed] [Google Scholar]
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014; 74: 2913–2921. [DOI] [PubMed] [Google Scholar]
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012; 380: 2095–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–E386. [DOI] [PubMed] [Google Scholar]
- Committee ACPRW, Sawyers CL, Abate-Shen C, Anderson KC, Barker A, Baselga J et al. AACR Cancer Progress Report 2013. Clin Cancer Res 2013; 19: S4–98. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics. 2015CA Cancer J Clin 2015; 65: 5–29. [DOI] [PubMed] [Google Scholar]
- Davidson NE, Armstrong SA, Coussens LM, Cruz-Correa MR, DeBerardinis RJ, Doroshow JH et al. AACR Cancer Progress Report 2016. Clin Cancer Res 2016 22: S1–S137. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016; 2: 16018. [DOI] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science 1976; 194: 23–28. [DOI] [PubMed] [Google Scholar]
- Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med 2003; 9: 416–423. [DOI] [PubMed] [Google Scholar]
- Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology 2004; 40: 667–676. [DOI] [PubMed] [Google Scholar]
- Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med 2006; 12: 410–416. [DOI] [PubMed] [Google Scholar]
- Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 2009; 69: 7385–7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucman-Rossi J. Molecular classification of hepatocellular carcinoma. Dig Liver Dis 2010; 42(Suppl 3): S235–S241. [DOI] [PubMed] [Google Scholar]
- Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014; 46: 1267–1273. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M et al. Genomic spectra of biliary tract cancer. Nat Genet 2015; 47: 1003–1010. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Guilhot F, O’Brien SG, Gathmann I, Kantarjian H, Gattermann N et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med 2006; 355: 2408–2417. [DOI] [PubMed] [Google Scholar]
- Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci USA 2012; 109: 2724–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard HM, Lewis GD, Sarup JC, Fendly BM, Maneval D, Mordenti J et al. Monoclonal antibody therapy of human cancer: taking the HER2 protooncogene to the clinic. J Clin Immunol 1991; 11: 117–127. [DOI] [PubMed] [Google Scholar]
- Soverini S, Branford S, Nicolini FE, Talpaz M, Deininger MW, Martinelli G et al. Implications of BCR-ABL1 kinase domain-mediated resistance in chronic myeloid leukemia. Leuk Res 2014; 38: 10–20. [DOI] [PubMed] [Google Scholar]
- Roche-Lestienne C, Lai JL, Darre S, Facon T, Preudhomme C. A mutation conferring resistance to imatinib at the time of diagnosis of chronic myelogenous leukemia. N Engl J Med 2003; 348: 2265–2266. [DOI] [PubMed] [Google Scholar]
- Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell 2002; 2: 117–125. [DOI] [PubMed] [Google Scholar]
- Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012; 44: 694–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res 2008; 68: 1451–1461. [DOI] [PubMed] [Google Scholar]
- Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology 2011; 140: 1501–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachenmayer A, Alsinet C, Savic R, Cabellos L, Toffanin S, Hoshida Y et al. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin Cancer Res 2012; 18: 4997–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology 2017; 152: 745–761. [DOI] [PubMed] [Google Scholar]
- Nault JC, Mallet M, Pilati C, Calderaro J, Bioulac-Sage P, Laurent C et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun 2013; 4: 2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Coste A, Romagnolo B, Billuart P, Renard CA, Buendia MA, Soubrane O et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci USA 1998; 95: 8847–8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature 1991; 350: 429–431. [DOI] [PubMed] [Google Scholar]
- Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015; 47: 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res 2008; 68: 1451–1461. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 2009; 136: 1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia D, Hoshida Y, Villanueva A, Roayaie S, Ferrer J, Tabak B et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology 2013; 144: 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JB, Spee B, Blechacz BR, Avital I, Komuta M, Barbour A et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology 2012; 142: 1021–1031.e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Toh Y, Adachi E, Matsumata T, Mori R, Sugimachi K. Tumor progression in hepatocellular carcinoma may be mediated by p53 mutation. Cancer Res 1993; 53: 2884–2887. [PubMed] [Google Scholar]
- Hsu HC, Chiou TJ, Chen JY, Lee CS, Lee PH, Peng SY. Clonality and clonal evolution of hepatocellular carcinoma with multiple nodules. Hepatology 1991; 13: 923–928. [PubMed] [Google Scholar]
- Carotenuto P, Fassan M, Pandolfo R, Lampis A, Vicentini C, Cascione L et al. Wnt signalling modulates transcribed-ultraconserved regions in hepatobiliary cancers. Gut 2017; 66: 1268–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S, Hu Z, Yang Z, Yang F, Li Y, Lin P et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc Natl Acad Sci USA 2015; 112: E6496–E6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue R, Li R, Guo H, Guo L, Su Z, Ni X et al. Variable intra-tumor genomic heterogeneity of multiple lesions in patients with hepatocellular carcinoma. Gastroenterology 2016; 150: 998–1008. [DOI] [PubMed] [Google Scholar]
- Wolman SR, Heppner GH. Genetic heterogeneity in breast cancer. J Natl Cancer Inst 1992; 84: 469–470. [DOI] [PubMed] [Google Scholar]
- Janiszewska M, Liu L, Almendro V, Kuang Y, Paweletz C, Sakr RA et al. in situ single-cell analysis identifies heterogeneity for PIK3CA mutation and HER2 amplification in HER2-positive breast cancer. Nat Genet 2015; 47: 1212–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017; 168: 613–628. [DOI] [PubMed] [Google Scholar]
- Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 2017; 543: 122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 2014; 343: 72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenmochi K, Sugihara S, Kojiro M. Relationship of histologic grade of hepatocellular carcinoma (HCC) to tumor size, and demonstration of tumor cells of multiple different grades in single small HCC. Liver 1987; 7: 18–26. [DOI] [PubMed] [Google Scholar]
- Sirivatanauksorn Y, Sirivatanauksorn V, Bhattacharya S, Davidson BR, Dhillon AP, Kakkar AK et al. Evolution of genetic abnormalities in hepatocellular carcinomas demonstrated by DNA fingerprinting. J Pathol 1999; 189: 344–350. [DOI] [PubMed] [Google Scholar]
- Shi JY, Xing Q, Duan M, Wang ZC, Yang LX, Zhao YJ et al. Inferring the progression of multifocal liver cancer from spatial and temporal genomic heterogeneity. Oncotarget 2016; 7: 2867–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res 2016; 26: 304–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis—A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology 2000; 32: 970–979. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Ushijima S, Hui AM, Ochiai A, Tsuda H, Sakamoto M et al. The E-cadherin gene is silenced by CpG methylation in human hepatocellular carcinomas. Int J Cancer 1997; 71: 355–359. [DOI] [PubMed] [Google Scholar]
- Lee HG, Kim H, Son T, Jeong Y, Kim SU, Dong SM et al. Regulation of HK2 expression through alterations in CpG methylation of the HK2 promoter during progression of hepatocellular carcinoma. Oncotarget 2016; 7: 41798–41810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell 2014; 14: 275–291. [DOI] [PubMed] [Google Scholar]
- Oishi N, Kumar MR, Roessler S, Ji J, Forgues M, Budhu A et al. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology 2012; 56: 1792–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Shen J, Liu L, Xia F, Xu C, Duan G et al. Nanog regulates self-renewal of cancer stem cells through the insulin-like growth factor pathway in human hepatocellular carcinoma. Hepatology 2012; 56: 1004–1014. [DOI] [PubMed] [Google Scholar]
- Zhu P, Wang Y, Huang G, Ye B, Liu B, Wu J et al. lnc-beta-Catm elicits EZH2-dependent beta-catenin stabilization and sustains liver CSC self-renewal. Nat Struct Mol Biol 2016; 23: 631–639. [DOI] [PubMed] [Google Scholar]
- Wang Y, He L, Du Y, Zhu P, Huang G, Luo J et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 2015; 16: 413–425. [DOI] [PubMed] [Google Scholar]
- Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun 2016; 7: 13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013; 501: 346–354. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 2012; 21: 309–322. [DOI] [PubMed] [Google Scholar]
- Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science 2013; 339: 286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med 2014; 211: 1503–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoong KF, McNab G, Hubscher SG, Adams DH. Vascular adhesion protein-1 and ICAM-1 support the adhesion of tumor-infiltrating lymphocytes to tumor endothelium in human hepatocellular carcinoma. J Immunol 1998; 160: 3978–3988. [PubMed] [Google Scholar]
- Flecken T, Schmidt N, Hild S, Gostick E, Drognitz O, Zeiser R et al. Immunodominance and functional alterations of tumor-associated antigen-specific CD8+ T-cell responses in hepatocellular carcinoma. Hepatology 2014; 59: 1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hato T, Goyal L, Greten TF, Duda DG, Zhu AX. Immune checkpoint blockade in hepatocellular carcinoma: current progress and future directions. Hepatology 2014; 60: 1776–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura H, Yamagiwa S, Tsuchiya A, Takamura M, Matsuda Y, Ohkoshi S et al. Reduced NKG2D ligand expression in hepatocellular carcinoma correlates with early recurrence. J Hepatol 2012; 56: 381–388. [DOI] [PubMed] [Google Scholar]
- Yan W, Liu X, Ma H, Zhang H, Song X, Gao L et al. Tim-3 fosters HCC development by enhancing TGF-beta-mediated alternative activation of macrophages. Gut 2015; 64: 1593–1604. [DOI] [PubMed] [Google Scholar]
- Yan W, Han P, Zhou Z, Tu W, Liao J, Li P et al. Netrin-1 induces epithelial-mesenchymal transition and promotes hepatocellular carcinoma invasiveness. Dig Dis Sci 2014; 59: 1213–1221. [DOI] [PubMed] [Google Scholar]
- Pan K, Wang H, Chen MS, Zhang HK, Weng DS, Zhou J et al. Expression and prognosis role of indoleamine 2,3-dioxygenase in hepatocellular carcinoma. J Cancer Res Clin Oncol 2008; 134: 1247–1253. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhang Y, Sun XX, Ma X, Chen ZN. microRNA-146a inhibits cancer metastasis by downregulating VEGF through dual pathways in hepatocellular carcinoma. Mol Cancer 2015; 14: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Lin N, Chen Z, Xu R. Hypoxia-induced secretion of platelet-derived growth factor-BB by hepatocellular carcinoma cells increases activated hepatic stellate cell proliferation, migration and expression of vascular endothelial growth factor-A. Mol Med Rep 2015; 11: 691–697. [DOI] [PubMed] [Google Scholar]
- Hoechst B, Ormandy LA, Ballmaier M, Lehner F, Kruger C, Manns MP et al. A new population of myeloid-derived suppressor cells in hepatocellular carcinoma patients induces CD4(+)CD25(+)Foxp3(+) T cells. Gastroenterology 2008; 135: 234–243. [DOI] [PubMed] [Google Scholar]
- Neaud V, Faouzi S, Guirouilh J, Le Bail B, Balabaud C, Bioulac-Sage P et al. Human hepatic myofibroblasts increase invasiveness of hepatocellular carcinoma cells: evidence for a role of hepatocyte growth factor. Hepatology 1997; 26: 1458–1466. [DOI] [PubMed] [Google Scholar]
- Hochst B, Schildberg FA, Sauerborn P, Gabel YA, Gevensleben H, Goltz D et al. Activated human hepatic stellate cells induce myeloid derived suppressor cells from peripheral blood monocytes in a CD44-dependent fashion. J Hepatol 2013; 59: 528–535. [DOI] [PubMed] [Google Scholar]
- Dunham RM, Thapa M, Velazquez VM, Elrod EJ, Denning TL, Pulendran B et al. Hepatic stellate cells preferentially induce Foxp3+ regulatory T cells by production of retinoic acid. J Immunol 2013; 190: 2009–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhu A, Forgues M, Ye QH, Jia HL, He P, Zanetti KA et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell 2006; 10: 99–111. [DOI] [PubMed] [Google Scholar]
- Prieto J, Melero I, Sangro B. Immunological landscape and immunotherapy of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2015; 12: 681–700. [DOI] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med 2012; 4: 127ps110. [DOI] [PubMed] [Google Scholar]
- Wang XW, Thorrgeirsson SS. The biological and clinical challenge of liver cancer heterogeneity. Hepatic Oncol 2014; 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S, Jiang Z, Stawiski EW, Gnad F, Liu J, Mayba O et al. Diverse modes of genomic alteration in hepatocellular carcinoma. Genome Biol 2014; 15: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet 2016; 17: 175–188. [DOI] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet 2013; 14: 618–630. [DOI] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J et al. Tumour evolution inferred by single-cell sequencing. Nature 2011; 472: 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 2009; 461: 809–813. [DOI] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015; 348: aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Hao J, Wang G, Chen KH, Babcock HP, Zhuang X. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci USA 2016; 113: 11046–11051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JL, Yang YF, Yuan CH, Chen H, Wang FB. Circulating tumor cells for predicting the prognostic of patients with hepatocellular carcinoma: a meta analysis. Cell Physiol Biochem 2015; 37: 629–640. [DOI] [PubMed] [Google Scholar]
- Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res 2013; 73: 2965–2975. [DOI] [PubMed] [Google Scholar]
- Mostert B, Jiang Y, Sieuwerts AM, Wang H, Bolt-de Vries J, Biermann K et al. KRAS and BRAF mutation status in circulating colorectal tumor cells and their correlation with primary and metastatic tumor tissue. Int J Cancer 2013; 133: 130–141. [DOI] [PubMed] [Google Scholar]
- Mohamed Suhaimi NA, Foong YM, Lee DY, Phyo WM, Cima I, Lee EX et al. Non-invasive sensitive detection of KRAS and BRAF mutation in circulating tumor cells of colorectal cancer patients. Mol Oncol 2015; 9: 850–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 2015; 349: 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Kim S, Gould J, Knoechel B, Drier Y, Cotton MJ et al. Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. Sci Transl Med 2016; 8: 363ra147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinich M, Bhan I, Kwan TT, Miyamoto DT, Javaid S, LiCausi JA et al. An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proc Natl Acad Sci USA 2017; 114: 1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubendorf L, Nocito A, Moch H, Sauter G. Tissue microarray (TMA) technology: miniaturized pathology archives for high-throughput in situ studies. J Pathol 2001; 195: 72–79. [DOI] [PubMed] [Google Scholar]
- Anderson CM, Zhang B, Miller M, Butko E, Wu X, Laver T et al. Fully automated RNAscope in situ hybridization assays for formalin-fixed paraffin-embedded cells and tissues. J Cell Biochem 2016; 117: 2201–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Kagawa K, Deguchi T, Nakajima T, Kakusui M, Ohkawara T et al. Cytogenetic analyses of hepatocellular carcinoma by in situ hybridization with a chromosome-specific DNA probe. Cancer 1996; 77: 271–277. [DOI] [PubMed] [Google Scholar]
- Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet 2004; 36: 1306–1311. [DOI] [PubMed] [Google Scholar]
- Xue W, Wang XW. The search for precision models clinically relevant to human liver cancer. Hepatic Oncol 2015; 2: 315–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruna A, Rueda OM, Greenwood W, Batra AS, Callari M, Batra RN et al. A Biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell 2016; 167: 260–274 e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Wang ZC, Duan M, Lin YH, Zhou XY, Worthley DL et al. Cell culture system for analysis of genetic heterogeneity within hepatocellular carcinomas and response to pharmacologic agents. Gastroenterology 2017; 152: 232–242 e234. [DOI] [PubMed] [Google Scholar]
- Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015; 149: 1226–1239 e1224. [DOI] [PubMed] [Google Scholar]
- Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 2015; 161: 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]